Abstract
Host populations with high genetic diversity are predicted to have lower levels of infection prevalence. This theory assumes that host genetic diversity results in variation in susceptibility and that parasites exhibit variation in infectivity. Empirical studies on the effects of host heterogeneity typically neglect the role of parasite diversity. We conducted three laboratory experiments designed to test if genetic variation in Daphnia magna populations and genetic variation in its parasites together influence the course of parasite spread after introduction. We found that a natural D. magna population exhibited variation in susceptibility to infection by three parasite species and had strong host clone–parasite species interactions. There was no effect of host heterogeneity in experimental host populations (polycultures and monocultures) separately exposed to single strains of three parasite species. When we manipulated the genetic diversity of a single parasite species and exposed them to host monocultures and polycultures, we found that parasite prevalence increased with the number of parasite strains. Host monocultures exposed to several parasite strains had higher mean parasite prevalence and higher variance than polycultures. These results indicate that effect of host genetic diversity on the spread of infection depends on the level of genetic diversity in the parasite population.
Keywords: Daphnia magna, genetic diversity, Glugoides intestinalis, heterogeneity, host-parasite, infection, microsporidia, monoculture, Ordospora colligata, prevalence
Introduction
Host populations with low genetic diversity are predicted to have to higher infection prevalence and intensity of disease outbreaks than populations with greater diversity in host susceptibility (Elton 1958, Leonard 1969, Hamilton 1987, Sherman et al. 1988, Schmid-Hempel 1998). The effects of host genetic diversity on disease invasion was first described in agriculture by Elton (1958), where crops grown in genetically homogeneous monocultures are typically more susceptible to severe disease outbreaks than mixtures (e.g., Browning and Frey 1969, Leonard 1969, Wolfe 1985, McDonald et al. 1988, Mahmood et al. 1991, Garrett and Mundt 1999, Zhu et al. 2000, Mundt 2002, Pilet et al. 2006). Support for the effects of host genetic heterogeneity on the spread of disease has also been found in social insects (e.g., ants [Hughes and Boomsma 2004, Reber et al. 2008], bumble bees [Liersch and Schmid-Hempel 1998, Baer and Schmid-Hempel 1999], and honeybees [Tarpy 2003, Tarpy and Seeley 2006, Seeley and Tarpy 2007]) and in other hosts, including gypsy moths (Dwyer et al. 1997), water fleas (Altermatt and Ebert 2008), water buffalos (Capparelli et al. 2009) and humans (Lloyd-Smith et al. 2005). However, the effect of host heterogeneity on infection is often small and negative results are less likely to be published (e.g., Ratnieks 1989). Van Baalen and Beekman (2006) showed that the effects of host heterogeneity depend on a balance between reduced damage as a result of infection and increased frequency of infection. While genetic heterogeneity in host populations may slow the spread of infection, more diverse host populations are more susceptible to a greater range of parasites (Van Baalen and Beekman 2006). Moreover, the advantage of host heterogeneity depends on variation in parasite strains, which is not addressed explicitly in most empirical studies. Host populations composed entirely of resistant individuals will have the lowest infection levels if there is no variation in parasite infectivity (Boomsma and Ratnieks 1996).
We performed three experimental studies designed to test whether genetic variation in populations of the crustacean, Daphna magna influences the invasion success of microsporidian gut parasites and explicitly investigate the role of parasite diversity. We use microcosms to test whether monoclonal host populations exhibit variation in susceptibility to novel (allopatric) pathogen genotypes, how host genetic diversity affects the outcome of parasite introductions, and whether parasite diversity influences infection prevalence. This system is ideal for studying the effects of host genetic heterogeneity on infection dynamics because we can conduct population-level experiments in well-mixed populations with minimal spatial effects. Furthermore, we can assemble monoclonal host populations because Daphnia reproduce by cyclical parthenogenesis and can be raised clonally under laboratory conditions.
Any hypothesis related to the spread of infectious agents in populations with different genetic diversity requires some form of genetic variation among hosts with respect to disease related traits. Therefore, we first tested for genotypic interactions in an experiment that measured parasite spread in different monoclonal populations of the host, D. magna. Although, genetic variation for host-parasite interactions are known in the Daphnia–microparasite system (Ebert 1994, Little and Ebert 2001, Decaestecker et al. 2003), variation in the ability to spread within populations has not been tested before. In a second experiment, we manipulated the genetic structure of the host population (Appendix A). We predicted that monoclonal (monocultures) and polyclonal (polycultures) populations of hosts would not differ in mean parasite prevalence but would differ in variance (Appendix B). We expected to find higher variance among monocultures because monocultures may be composed entirely of resistant or susceptible host genotypes, in which case parasite prevalence will be low or high, respectively. Due to this variation in susceptibility among hosts, we expected to find medium mean prevalence and high variance across populations of monocultures. In contrast, polycultures are mixtures of host genotypes and each host population will have a greater probability of containing susceptible genotypes. Here, we predicted that polycultures would have low to medium mean prevalence with lower variance among populations (Appendix B). In a third experiment, we manipulated genetic variation in the parasite populations as well as host populations. In this case, we predicted that host populations (monocultures and polycultures) would differ in mean parasite prevalence when exposed to higher parasite diversity (Appendix B). We expected that a host monoculture would be more likely to be susceptible to one of several parasite strains, which would result in higher mean parasite prevalence compared with host polycultures.
Methods
We studied D. magna and several microsporidian parasites. Microsporidians are the most abundant parasites in natural D. magna populations (Stirnadel and Ebert 1997, Bengtsson and Ebert 1998, Decaestecker et al. 2005). Clones were hatched from resting eggs collected from a large pond (about 4 ha) near Munich, Germany (Ismaninger Speichersee area). Each clone originated from a different resting egg. Eggs are produced by sexual reproduction and are genetically distinct. We reared more than 200 clones in isolation (500-mL jars) and randomly selected clones for each experimental treatment. No host clone was used more than once and polycultures were composed sets of ten different clones (Appendix A). We used 10 rearing jars per clone to reduce possible jar effects causing differences between clones (experiment 2 used 165 clones and 1650 jars). Experimental populations were kept in 1.2 L of artificial culture medium (Ebert et al. 1998) in 1.5-L jars. We randomized the distribution of jars in a climate-controlled room (20° ± 1°C and a 16:8 hours light: dark cycle). We fed all populations by adding 1×108 cells of the unicellular green alga, Scenedesmus obliquus three times weekly. The culture medium was augmented weekly to maintain the starting volume.
After 30 days (experiments one and two) and 24 days (experiment three), we removed 25% of the population (and culture medium) from each 1.5-L jar by splitting the contents of the jar twice with a Folsom’s plankton sample divider. The Daphnia culture medium was replaced with fresh media after sampling. We selected the sampling date to coincide approximately with the parasite reaching mid-level prevalences in the host populations. We also wanted to measure prevalence before intraspecific competition reduced clonal diversity. Individual hosts were examined for signs of infection using a phase contrast microscope at 600× magnification. Host population size was counted for each replicate population and ranged from 36 to 144 individuals.
Experiment one
In the first experiment, we tested for differences in susceptibility of different clones in monoculture to three different microsporidian gut parasites (Glugoides intestinalis, Ordospora colligata, and an undescribed species, Microsporidia sp.). Each parasite species was isolated from a single D. magna clone in a host population in northern Germany (G. intestinalis and O. colligata) or a host clone collected from a population in England (Microsporidia sp.). Our experimental design included 15 clones, four replicates per clone, and three parasite treatments, for a total of 180 experimental populations. Hosts were exposed to gastrointestinal parasites that were defecated by ten infected hosts that were encaged in nets hanging in the 1.5-L jars and removed ten days after the start of the experiment.
Experiment two
In the second experiment, we exposed two host population treatments (monoculture and polyculture) to four different parasite treatments (single strains of G. intestinalis, O. colligata, and Microsporidia sp., and parasite absent) with 15 replicates, for a total of 120 experimental populations. Every jar contained different host clones; we did not replicate on a clonal level in order to maximize power to see treatment effects. As in the first experiment, host populations were exposed to parasites that were defecated by ten infected hosts that were held in nets at the top of the microcosm and removed ten days after the start of the experiment (Appendix A).
Experiment three
Host populations were exposed to 16 different parasite treatments: all combinations of four strains of O. colligata and a control. Parasite strains were isolated from individual D. magna clones collected from populations in Belgium, Germany, England, and Scotland. The experimental design included four replicates of two host diversity treatments (monoculture or polyculture) and 16 parasite treatments (one negative control, four single infections, six double infections, four triple infections and one quadruple infection), for a total of 128 experimental populations. As before, we did not replicate on a clonal level. Host populations were exposed to parasites by the placement of infected hosts in nets at the top of the jars for the first ten days of the experiment. To minimize dosage effects, we placed 20 infected hosts above single (20 hosts infected by one strain), double (ten hosts of each of two strains) and quadruple infections (five hosts of each of four strains) and 21 infected hosts (seven individuals of each of three strains) were placed above triple infections.
Results
Experiment one
Monoclonal host populations differed strongly in parasite prevalence after 30 days or about two host generations (logistic regression; host clone: df = 14, χ2 = 145.6, P < 0.0001; Fig. 1). There was also a strong parasite species effect on prevalence (logistic regression; parasite, df = 2, χ2 = 565.4, P < 0.0001). Higher prevalence levels were reached in host populations exposed to O. colligata and G. intestinalis, while lower levels were found in populations infected by Microsporidium sp. (Tukey’s hsd; α = 0.05, Q = 2.37). There were significant host clone by parasite species interactions (logistic regression; host clone × parasite: df = 28, χ2 = 135.6, P < 0.0001; Fig. 1). No significant treatment effects on host population size were detected (ANOVA; host clone, F14, 134 = 1.11, P = 0.36; parasite species, F2, 134 = 2.87, P = 0.06; host clone × parasite species, F28, 134 = 1.04, P = 0.42).
Fig. 1.
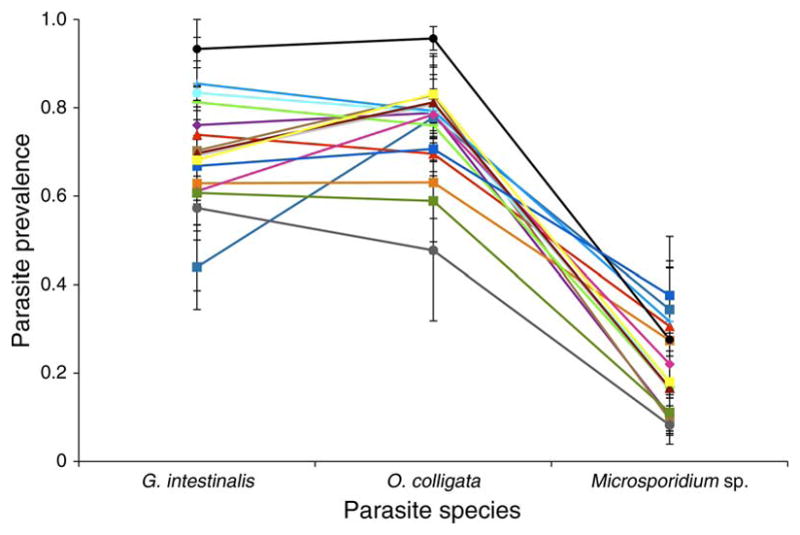
Mean parasite prevalence (±SE) in monoclonal host populations 30 days after they were exposed to three gut parasite species (Glugoides intestinalis, Ordospora colligata, and Microsporidium sp.). Fifteen different host clones are indicated by color.
Experiment two
Prevalences differed among parasite species (logistic regression; parasite, df = 2, χ2 = 98.1, P < 0.0001; Fig. 2). As found in the first experiment, all host populations were infected at higher prevalence levels by O. colligata and G. intestinalis than Microsporidium sp. (Tukey’s hsd; α = 0.05, Q = 2.39). Polycultures tended to have slightly higher, but not significantly different, parasite prevalence levels than monocultures (logistic regression; host treatment, df = 1, χ2 = 1.76, P = 0.18). There was no significant interaction between host treatment and parasite species (logistic regression; df = 2, χ2 = 0.70, P = 0.71). Contrary to expectations, monocultures and polycultures did not differ in variance of parasite prevalence (Levene test; F1,87 = 0.27, P = 0.60). Host population size was not affected by parasite species (ANOVA; F2,84 = 1.6, P = 0.21), host treatment (monoculture or polyculture, ANOVA; F1,84 = 0.87, P = 0.35), or their interaction (ANOVA; F2,84 = 1.09, P = 0.34).
Fig. 2.
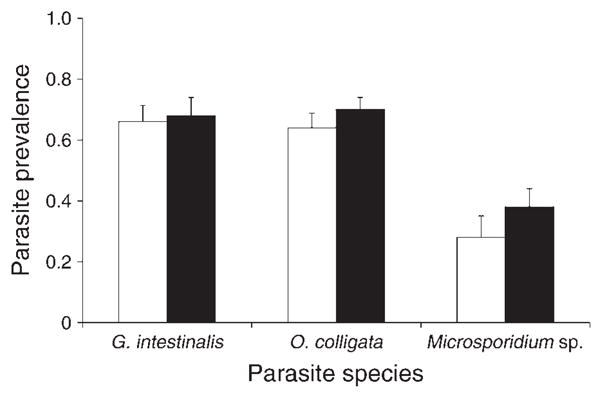
Mean prevalence (and SE) in monoclonal (white bars) and polyclonal (black bars) populations of D. magna 30 days after exposure to three different parasite species (G. intestinalis, O. colligata, and Microsporidium sp.).
Experiment three
In both host population treatment groups, parasite prevalence increased strongly with number of strains of the gut parasite, O. colligata (Kruskal-Wallace test; χ2 = 10.94, df = 3, P = 0.012; Fig. 3). Overall, parasite prevalence was greater in host monocultures than host polycultures (logistic regression; host population treatment, df = 1, χ2 = 6.24, P = 0.01). Host population sizes were not affected by parasite treatment (ANOVA; F15, 110 = 0.35, P = 0.56). Although monocultures and polycultures differed in mean prevalence, variances did not differ as predicted; host polycultures had significantly lower variance than monocultures when exposed to three parasite strains (Levene test; F1,30 = 4.5, P = 0.04) but not when exposed to one strain (Levene test; F1,30 = 0.53, P = 0.47), two strains (Levene test; F1,44 = 0.21, P = 0.65), or four strains (Levene test; F1,6 = 0.88, P = 0.38).
Fig. 3.
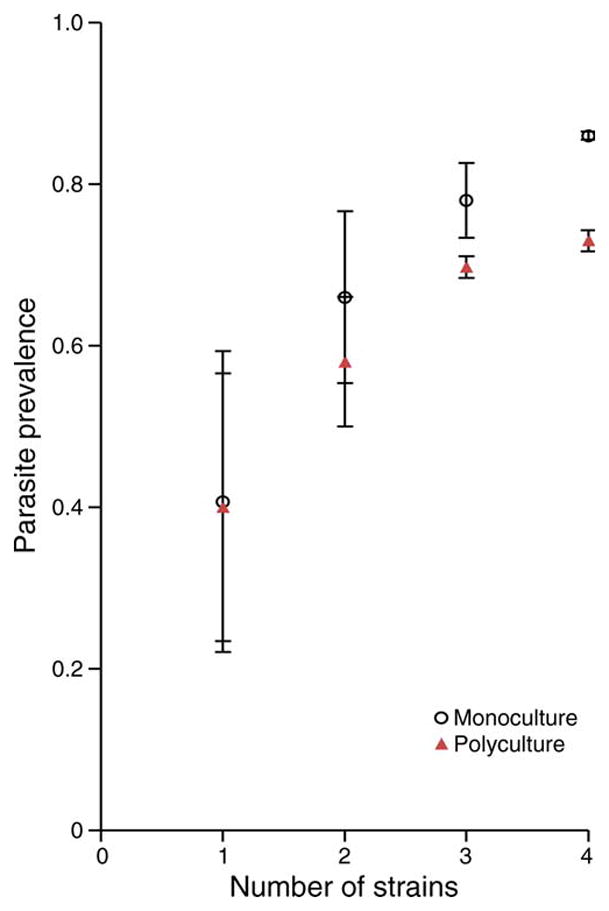
Mean parasite prevalence (± variance) in monoclonal (black circles) and polyclonal (red triangles) populations of D. magna 24 days after exposure to as many as four different strains of a single parasite species (O. colligata).
Discussion
We tested the hypothesis that genetic variation in both host and parasite populations affects infection dynamics in host populations exposed to novel parasite strains. Our results indicate that levels of parasite prevalence depended on the genetic variation in populations of D. magna and its parasites. While infection prevalence increased with parasite strain diversity in all host populations, prevalence was significantly lower in genetically diverse host populations when they were exposed to more than one parasite strain. Thus, genetically diverse host populations were more resilient to introductions of novel parasite genotypes than monocultures when there was variation in the parasite population. Invasions from different sources, as mimicked in our experiment by the addition of isolates from different geographic areas, can augment genetic diversity and contribute to the success of the invader (reviewed in Dlugosch and Parker 2008).
In order to test for effects of diversity on the success of introductions of novel parasite strains in experimental host populations, we selected host clones randomly, relying upon the naturally occurring variation in host susceptibility arising after sexual reproduction in one large D. magna population. We showed in the first experiment that this population exhibited variation in host susceptibility to infection by three microsporidian parasite species. In addition, we found a strong interaction between host clone and parasite species such that the level of parasite prevalence attained by one species in a specific host clone is a poor predictor of prevalence by another parasite species in the same host clone. Such interactions in parasite performance traits were found in another D. magna population (Decaestecker et al. 2003) and may be the rule in this system (Ebert 2005). These results indicate that there is genetic variation in host susceptibility in the receiving population and that host resistance is specific in its interactions with different parasite species. Further, no host clone was superior in its resistance to infection.
When we manipulated the diversity of host populations and exposed them to single parasite strains, we predicted that host monocultures and polycultures would not differ in mean infection prevalence but would differ in their variances (Appendix B). In agreement with our prediction, polycultures had similar infection prevalences as monocultures when exposed to a single strain of each of three parasite species. However, contrary to our prediction, there was no difference in variance of parasite prevalence. This finding was explained by the results of our third experiment where we also manipulated parasite diversity and exposed host populations to up to four parasite strains. Here, we found that monocultures and polycultures differed in both mean prevalence overall and variance at intermediate levels of strain number, as predicted. Overall parasite prevalence increased as expected with the number of parasite strains in both monocultures and polycultures (Fig. 3). Prevalence may increase with number of strains due to an increased likelihood of matching between host susceptibility and parasite infectivity as strain number increases. Such a sampling effect would also affect the chance that one of the host clones in a polyculture would be susceptible to a parasite strain, which might account for the higher than expected variance detected in polycultures exposed to low parasite diversity.
In the present study, the effect of host heterogeneity in D. magna populations exposed to multiple strains of a parasite depended on parasite diversity. In a field experiment with the same host, Altermatt and Ebert (2008) showed that a different parasite (Octosporea bayeri) spread faster in D. magna populations of low diversity compared with host populations of high diversity but they detected no effect of parasite diversity. The differences in outcomes may reflect our experimental design. We were more likely to detect an effect with our factorial design (all combinations of four parasite strains or sixteen parasite treatments) than their unreplicated parasite treatment (low and high). Altermatt and Ebert (2008) further suggest that their treatment of parasite diversity may have differed less than expected if there was unidentified diversity within the parasite isolate used for the low treatment. For many infectious diseases, individual hosts are commonly infected by multiple genotypes of the same pathogen (reviewed in Read and Taylor 2001). Finally, we may have obtained different results because our experiments model the invasion of highly diverse parasite strains collected from different countries (the closest parasite population was more than 850 km away from the host population), while Altermatt and Ebert (2008) studied parasite stains that were collected locally (but not from the same rock pools).
In summary, the ability of three different parasite species to spread in D. magna populations depended on host clone with strong host clone by parasite species interactions. In agreement with theoretical predictions, there was no effect of host genetic diversity in D. magna populations exposed to single strains of three parasite species. When parasite diversity is low, heterogeneous host populations may experience no benefit of diversity because they are more likely to contain a susceptible host (Van Baalen and Beekman 2006) and homogeneous host populations are predicted have the lowest infection levels if they contain a resistant host genotype (Boomsma and Ratnieks 1996). When we manipulated parasite diversity in experimental D. magna populations, we observed that mean parasite prevalence increased and variance in parasite prevalence decreased as the number of strains of O. colligata increased. Host monocultures exposed to several parasite strains had higher mean parasite prevalence than host polycultures. Thus, the benefit of host heterogeneity in susceptibility arose when parasite diversity was higher, which supports the prediction that genetic diversity in parasite populations increases the probability that a homogeneous host population is susceptible to one of the parasite strains (Van Baalen and Beekman 2006). The effect of host genetic diversity on disease risk is conceptually similar to the dilution effect (e.g., Van Buskirk and Ostfeld 1995), which predicts that higher species diversity lowers infection prevalence. Many studies of the dilution effect have found that disease risk decreases with higher host diversity but species diversity has also been shown to increase disease prevalence (e.g., Bowers and Begon 1991, Begon et al. 1992, Power and Mitchell 2004). The effect of an additional species on infection prevalence in a population depends on its ability to serve as a pathogen reservoir (Power and Mitchell 2004, Keesing et al. 2006). The results presented here suggest that future studies on the effects of host diversity on the spread of infectious disease should also explicitly consider the role of parasite diversity.
Supplementary Material
A figure showing how 10 jars of host clones were combined to create each experimental polyculture and monoculture (Ecological Archives E091-088-A1).
A figure showing predicted mean parasite prevalence and its variance across replicates in experimental host populations in relation to host and parasite diversity (Ecological Archives E091-088-A2).
Acknowledgments
We are grateful to J. Hottinger, S. Lass, D. Refardt, M. Zbinden, F. Parpan, L. Sygnarski, A. Eggel, and N. Basieux for their help in the lab. In addition, conversations with S. Lass, D. Refardt, and M. Zbinden improved the experimental design. T. Fabro lent us the 2000 jars needed to set up the second experiment. We thank P. Kamath, S. Lass, S. Lawler, C. Lively, W. Turner, G. Wittemeyer, and an anonymous reviewer for helpful comments on the manuscript. This research was supported by a U.S. NSF International Research Fellowship Program Grant INT-0301934 to H. H. Ganz and a Swiss National Fonds Grant to D. Ebert. While writing the manuscript, H. H. Ganz was funded by U.S. NIH Ecology of Infectious Diseases grant R01 GM083863-01 to Wayne M. Getz.
Literature Cited
- Altermatt F, Ebert D. Genetic diversity of Daphnia magna populations enhances resistance to parasites. Ecology Letters. 2008;11:918–928. doi: 10.1111/j.1461-0248.2008.01203.x. [DOI] [PubMed] [Google Scholar]
- Baer B, Schmid-Hempel P. Experimental variation in polyandry affects parasite load and fitness in a bumblebee. Nature. 1999;397:151–154. [Google Scholar]
- Begon M, Bowers RG, Kadianakis N, Hodgkinson DE. Disease and community structure the importance of host self-regulation in a host–host–pathogen model. American Naturalist. 1992;139:1131–1150. [Google Scholar]
- Bengtsson J, Ebert D. Distributions and impacts of microparasites on Daphnia in a rockpool metapopulation. Oecologia. 1998;115:213–221. doi: 10.1007/s004420050510. [DOI] [PubMed] [Google Scholar]
- Boomsma JJ, Ratnieks FLW. Paternity in eusocial Hymenoptera. Philosophical Transactions of the Royal Society B. 1996;351:947–975. [Google Scholar]
- Bowers RG, Begon M. A host–host–pathogen model with free-living infective stages applicable to microbial pest control. Journal of Theoretical Biology. 1991;148:305–330. doi: 10.1016/s0022-5193(05)80240-1. [DOI] [PubMed] [Google Scholar]
- Browning JA, Frey KJ. Multiline cultivars as a means of disease control. Annual Reviews of Phytopathology. 1969;7:355–382. [Google Scholar]
- Capparelli R, Parlato M, Iannaccone M, Roperto S, Marabelli R, Roperto F, Iannelli D. Heterogeneous shedding of Brucella abortus in milk and its effect on the control of animal brucellosis. Journal of Applied Microbiology. 2009;106:2041–2047. doi: 10.1111/j.1365-2672.2009.04177.x. [DOI] [PubMed] [Google Scholar]
- Decaestecker E, Declerck S, De Meester L, Ebert D. Ecological implications of parasites in natural Daphnia populations. Oecologia. 2005;144:382–390. doi: 10.1007/s00442-005-0083-7. [DOI] [PubMed] [Google Scholar]
- Decaestecker E, Vergote A, Ebert D, De Meester L. Evidence for strong host clone–parasite species interactions in the Daphnia microparasite system. Evolution. 2003;57:784–792. doi: 10.1111/j.0014-3820.2003.tb00290.x. [DOI] [PubMed] [Google Scholar]
- Dlugosch KM, I, Parker M. Founding events in species invasions: genetic variation, adaptive evolution, and the role of multiple introductions. Molecular Ecology. 2008;17:431–449. doi: 10.1111/j.1365-294X.2007.03538.x. [DOI] [PubMed] [Google Scholar]
- Dwyer G, Elkinton JS, Buonaccorsi JP. Host heterogeneity in susceptibility and disease dynamics: tests of a mathematical model. American Naturalist. 1997;150:685–707. doi: 10.1086/286089. [DOI] [PubMed] [Google Scholar]
- Ebert D. Virulence and local adaptation of a horizontally transmitted parasite. Science. 1994;265:1084–1086. doi: 10.1126/science.265.5175.1084. [DOI] [PubMed] [Google Scholar]
- Ebert D. Daphnia. National Library of Medicine, National Center for Biotechnology Information; Bethesda, Maryland, USA: 2005. Ecology, epidemiology and evolution of parasitism. [Google Scholar]
- Ebert D, Zschokke-Rohringer CD, Carius HJ. Within- and between-population variation for resistance of Daphnia magna to the bacterial endoparasite Pasteuria ramosa. Proceedings of the Royal Society B. 1998;265:2127–2134. [Google Scholar]
- Elton CS. The ecology of invasions by animals and plants. John Wiley; New York, New York, USA: 1958. [Google Scholar]
- Garrett KA, Mundt CC. Epidemiology in mixed host populations. Phytopathology. 1999;89:984–990. doi: 10.1094/PHYTO.1999.89.11.984. [DOI] [PubMed] [Google Scholar]
- Hamilton WD. Kinship, recognition, disease, and intelligence: constraints of social evolution. In: Ito Y, Brown JL, Kikkawa J, editors. Animal societies: theory and facts. Japanese Scientific Society Press; Tokyo, Japan: 1987. pp. 81–102. [Google Scholar]
- Hughes WOH, Boomsma JJ. Genetic diversity and disease resistance in leaf-cutting ant societies. Evolution. 2004;58:1251–1260. doi: 10.1554/03-546. [DOI] [PubMed] [Google Scholar]
- Keesing F, Holt RD, Ostfeld RS. Effects of species diversity on disease risk. Ecology Letters. 2006;9:485–498. doi: 10.1111/j.1461-0248.2006.00885.x. [DOI] [PubMed] [Google Scholar]
- Leonard KJ. Factors affecting rates of stem rust increase in mixed plantings of susceptible and resistant oat varieties. Phytopathology. 1969;59:1845–1850. [Google Scholar]
- Liersch S, Schmid-Hempel P. Genetic variation within social insect colonies reduces parasite load. Proceedings of the Royal Society B. 1998;265:221–225. [Google Scholar]
- Little TJ, Ebert D. Temporal patterns of genetic variation for resistance and infectivity in a Daphnia–microparasite system. Evolution. 2001;55:1146–1152. doi: 10.1111/j.0014-3820.2001.tb00634.x. [DOI] [PubMed] [Google Scholar]
- Lloyd-Smith JO, Schreiber SJ, Kopp PE, Getz WM. Superspreading and the effect of individual variation on disease emergence. Nature. 2005;438:355–359. doi: 10.1038/nature04153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmood T, Marshall D, McDaniel ME. Effect of winter wheat cultivar mixtures on leaf rust severity and grain yield. Phytopathology. 1991;81:470–474. [Google Scholar]
- McDonald BA, Allard RW, Webster RK. Responses of two-, three-, and four-component barley mixtures to a variable pathogen population. Crop Science. 1988;28:447–452. [Google Scholar]
- Mundt CC. Use of multiline cultivars and cultivar mixtures for disease management. Annual Reviews of Phytopathology. 2002;40:381–410. doi: 10.1146/annurev.phyto.40.011402.113723. [DOI] [PubMed] [Google Scholar]
- Pilet F, Chacon G, Forbes GA, Andrivon D. Protection of susceptible potato cultivars against late blight in mixtures increases with decreasing disease pressure. Phytopathology. 2006;96:777–783. doi: 10.1094/PHYTO-96-0777. [DOI] [PubMed] [Google Scholar]
- Power AG, Mitchell CE. Pathogen spillover in disease epidemics. American Naturalist. 2004;164:S79–S89. doi: 10.1086/424610. [DOI] [PubMed] [Google Scholar]
- Ratnieks FLW. Dissertation. Cornell University; Ithaca, New York, USA: 1989. Conflict and cooperation in insect societies. [Google Scholar]
- Read AF, Taylor LR. The ecology of genetically diverse infections. Science. 2001;292:1099–1102. doi: 10.1126/science.1059410. [DOI] [PubMed] [Google Scholar]
- Reber A, Castella G, Christe P, Chapuisat M. Experimentally increased group diversity improves disease resistance in an ant species. Ecology Letters. 2008;11:682–689. doi: 10.1111/j.1461-0248.2008.01177.x. [DOI] [PubMed] [Google Scholar]
- Schmid-Hempel P. Parasites in social insects. Princeton University Press; Princeton, New Jersey, USA: 1998. [Google Scholar]
- Seeley TD, Tarpy DR. Queen promiscuity lowers disease resistance within honeybee colonies. Proceedings of the Royal Society B. 2007;274:67–72. doi: 10.1098/rspb.2006.3702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman PW, Seeley TD, Reeve HK. Parasites, pathogens, and polyandry in social Hymenoptera. American Naturalist. 1988;131:602–610. doi: 10.1086/286127. [DOI] [PubMed] [Google Scholar]
- Stirnadel HA, Ebert D. Prevalence, host specificity and impact on host fecundity of microparasites and epibionts in three sympatric Daphnia species. Journal of Animal Ecology. 1997;66:212–222. [Google Scholar]
- Tarpy DR. Genetic diversity within honeybee colonies prevents severe infections and promotes colony growth. Proceedings of the Royal Society B. 2003;270:99–10. doi: 10.1098/rspb.2002.2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarpy DR, Seeley TD. Lower disease infections in honeybee (Apis mellifera) colonies headed by polyandrous vs monandrous queens. Naturwissenschaften. 2006;93:195–199. doi: 10.1007/s00114-006-0091-4. [DOI] [PubMed] [Google Scholar]
- Van Baalen M, Beekman M. The costs and benefits of genetic heterogeneity in resistance against parasites in social insects. American Naturalist. 2006;167:568–577. doi: 10.1086/501169. [DOI] [PubMed] [Google Scholar]
- Van Buskirk J, Ostfeld RS. Controlling Lyme disease by modifying the density and species composition of tick hosts. Ecological Applications. 1995;5:1133–1140. [Google Scholar]
- Wolfe MS. The current status and prospects of multiline cultivars and variety mixtures for disease resistance. Annual Reviews of Phytopathology. 1985;23:251–273. [Google Scholar]
- Zhu YY, et al. Genetic diversity and disease control in rice. Nature. 2000;406:718–722. doi: 10.1038/35021046. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A figure showing how 10 jars of host clones were combined to create each experimental polyculture and monoculture (Ecological Archives E091-088-A1).
A figure showing predicted mean parasite prevalence and its variance across replicates in experimental host populations in relation to host and parasite diversity (Ecological Archives E091-088-A2).


