Abstract
A direct and quantitative analysis of the internal structure and dynamics of a polyunsaturated lipid bilayer composed of 1-stearoyl-2-docosahexaenoyl-sn-glycero-3-phosphocholine (18:0-22:6n3-PC) containing 29 mol% cholesterol was carried out by neutron diffraction, 2H-NMR and 13C-MAS NMR. Scattering length distribution functions of cholesterol segments as well as of the sn-1 and sn-2 hydrocarbon chains of 18:0-22:6n3-PC were obtained by conducting experiments with specifically deuterated cholesterol and lipids. Cholesterol orients parallel to the phospholipids, with the A-ring near the lipid glycerol and the terminal methyl groups 3 Å away from the bilayer center. Previously, we reported that the density of polyunsaturated docosahexaenoic acid (DHA, 22:6n3) chains was higher near the lipid–water interface. Addition of cholesterol partially redistributes DHA density from near the lipid–water interface to the center of the hydrocarbon region. Cholesterol raises chain-order parameters of both stearic acid and DHA chains. The fractional order increase for stearic acid methylene carbons C8–C18 is larger, reflecting the redistribution of DHA chain density toward the bilayer center. The correlation times of DHA chain isomerization are short and mostly unperturbed by the presence of cholesterol. The uneven distribution of saturated and polyunsaturated chain densities and the cholesterol-induced balancing of chain distributions may have important implications for the function and integrity of membrane receptors, such as rhodopsin.
Keywords: Docosahexaenoic acid, Polyunsaturation, Phosphatidylcholine, Membrane, Cholesterol, Neutron diffraction, NMR
Introduction
The lipid bilayer of cell membranes is important for both the structural integrity of cells as well as the function of integral and peripheral membrane proteins. The complexity of structural and functional phenomena in cell membranes requires a proper balance of lipid composition. Almost all membranes contain a fraction of lipids with polyunsaturated hydrocarbon chains. Membranes with the highest content of polyunsaturated fatty acids are found in the brain, retina and sperm. The dominant fatty acid in those membranes is docosahexaenoic acid (DHA), a linear hydrocarbon chain with 22 carbons and six cis-locked double bonds (22:6n3).
DHA belongs to the class of ω-3 fatty acids. It is taken up directly from food sources such as fish, or it is synthesized from ω-3 fatty acid precursors such as α-linolenic acid (18:2n3, ALA). The lack of DHA or its ω-3 fatty acid precursors in the diet, in particular during periods of rapid brain development, has been associated with a variety of neurological disorders including impaired vision and mental retardation (Salem et al. 2001; Yamamoto et al. 1998; Umezawa et al. 1995; Enslen et al. 1991). Synaptosomal membranes as well as the membranes of rod outer segment (ROS) disks of the retina typically contain DHA at concentrations of 30–50 mol% of all fatty acids in phospholipids (Miljanich et al. 1979; Anderson et al. 1976). The synaptosomes and ROS disks contain not only high concentrations of polyunsaturated lipids but also significant amounts of cholesterol. For ROS it was observed that cholesterol has a concentration gradient over the ROS disks (Boesze-Battaglia and Albert 1990). Newly formed basal disks as well as the plasma membrane are high in cholesterol. With apical replacement of disks, cholesterol is rapidly depleted and the disk phospholipids are further modified to favor an unsaturated disk membrane. Those changes are known to favor formation of meta-rhodopsin-II, the photointermediate that activates G protein (Mitchell et al. 1992).
The molecular mechanisms of lipid–protein interaction and of membrane protein function and the role of cholesterol as a modulator of lateral organization in membranes are research areas of high current interest. Recently, we have shown that lipids with DHA chains are special in their influence on rhodopsin function because of both their lipid monolayer elastic properties and their ability to engage in direct interactions with the G protein–coupled receptors (GPCRs) that favor formation of the meta-rhodopsin-II state, the photoisomer that activates G protein (Soubias et al. 2006). The influence of cholesterol in 1-stearoyl-2-docosahexaenoyl-sn-glycero-3-phosphocholine (18:0-22:6n3-PC) membranes is currently investigated at our lab at the NIH. It was reported that cholesterol-containing membranes increase the stability of rhodopsin to thermal denaturation (Bennett and Mitchell 2008). Cholesteryl hemisuccinate (CHS) is frequently added to detergents for stabilization of GPCRs during solubilization, purification and reconstitution (Grisshammer 2009). The mechanism responsible for this cholesterol-induced stabilization of GPCR is unknown.
In mixtures of polyunsaturated lipids with a composition similar to ROS disk membranes and synaptosomal membranes, cholesterol interacts preferentially with the DHA-containing phosphatidylcholines (PCs) (Huster et al. 1998). Evidence for the formation of microscopic lipid clusters enriched in PC and cholesterol was obtained. Several studies presented evidence for a preferential interaction of cholesterol with the saturated lipid chains of mixed chain lipids or with the lipid species with saturated hydrocarbon chains in mixtures of saturated and polyunsaturated lipids (Huster et al. 1998; Brzustowicz et al. 2002; Pitman et al. 2004; Polozova and Litman 2000). While the typical orientation of cholesterol molecules in bilayers is parallel to the bilayer normal, in bilayers formed from lipids with two polyunsaturated hydrocarbon chains a different orientation was found. The laboratories of Katsaras and Wassall reported recently that cholesterol in such bilayers is located flat in the middle of bilayers (Harroun et al. 2008).
In a previous study we showed that the density distributions of the stearoyl and DHA chains in lipid bilayers formed of 18:0-22:6n3-PC differ dramatically. The segments of DHA chains bend and coil such that their density is higher near the lipid–water interface (Mihailescu and Gawrisch 2006). In contrast, the stearoyl chains prefer an extended conformation that may even reach into the apposing monolayer, filling up the voids in the bilayer center caused by the preferential location of DHA chains near the lipid–water interface. This results in a higher stearic acid chain density in the bilayer center. Here, we show by neutron diffraction and NMR spectroscopy that addition of cholesterol to membranes of 18:0-22:6n3-PC increases DHA and stearyl acid chain order parameters and partially redistributes DHA chain density. The DHA chains maintain a high level of motional flexibility in the presence of cholesterol. The location and orientation of cholesterol in the bilayer were determined using specifically A-ring and methyl terminal group-labeled cholesterols. A comprehensive view of the structure and dynamics of a cholesterol-containing, mixed-chain 18:0-22:6n3-PC bilayer is provided.
Materials and Methods
Materials
The lipid 18:0-22:6n3-PC, with protonated hydrocarbon chains, or specifically deuterated stearic acid (18:0-d35), or DHA (22:6n3-d31), was synthesized by Avanti Polar Lipids (Alabaster, AL). Deuterated DHA was produced in collaboration with Martek Biosciences (Columbia, MD). The DHA-producing dinoflagellate Crypthecodinium cohnii was grown in partially deuterated medium. It yielded a DHA that was deuterated at random to 38%, except for the terminal methyl group that was deuterated to 30% as determined by 2H-NMR. After fractional distillation and preparative HPLC separation, the perdeuterated DHA had a purity of better than 98% as determined by NMR and gas chromatography. Cholesterol in the natural protonated form was procured from Sigma-Aldrich (St. Louis, MO). Two forms of deuterium-labeled cholesterol were used: A-ring deuterated (2,2,3,4,4,6-d6) from Cambridge Isotope Laboratories (Andover, MA) and methyl-tail deuterated (25,25,25,26,26,26-d6) from C/D/N Isotopes (Pointe-Claire, Canada). For preparation of mixed samples, cholesterol and 18:0-22:6n3-PC were codissolved in chloroform. The composition was adjusted to 29 ± 1 mol% cholesterol in the 18:0-22:6n3-PC/cholesterol mixture based on precise 1H-NMR measurements of the concentration ratio. Oriented stacks of bilayers were prepared by deposition from solvent of 1–2 mg of 18:0-22:6n3-PC/cholesterol mixture to glass coverslips. The solvent was removed in a stream of N2 gas. Samples contained the antioxidant BHT at a lipid/BHT molar ratio of 100:1 and were prepared and investigated in an oxygen-free environment.
Methods
Neutron diffraction experiments were conducted on the advanced neutron diffractometer/reflectometer operating at the NIST Center for Neutron Research (Gaithersburg, MD) (Dura et al. 2006). Samples for both neutron diffraction and solid-state 2H-NMR at reduced water content consisted of oriented stacks of ~1,000–2,000 lipid bilayers on glass coverslips. For neutron diffraction, each sample was mounted vertically in the neutron beam and enclosed in a sealed chamber filled with argon gas. A relative humidity of 86% was maintained during the experiments by placing beakers with a saturated KCl solution into the chamber. Diffraction spectra were recorded in a θ (sample angle) to 2θ (detection angle) configuration relative to the incident beam. In this configuration, the momentum transfer (Q) is oriented parallel to the normal of the sample surface, which allows for probing the bilayer density profile along the normal to the bilayer plane (z axis). The hydration and equilibration of the sample were followed by periodically recording diffraction patterns. Only data acquired after complete stabilization of diffraction patterns were retained for analysis. A Fourier synthesis of the diffraction signal sampled at the diffraction peaks yields the scattering length density (SLD) profile of the bilayer projected on the z axis. Specific deuteration of different parts of the lipid bilayer (hydrocarbon chains, cholesterol and water of hydration) was used for direct determination of the position and distribution of those deuterated components relative to the bilayer center. Their individual profiles were determined by subtracting the SLD profile of the fully protonated (reference) sample from the SLD profile of a selectively deuterated sample (Eq. 3 in Appendix).
NMR measurements were carried out on AV800 and DMX500 NMR spectrometers (Bruker Biospin, Billerica, MA) at NIAAA, NIH (Bethesda, MD). NMR experiments on unoriented samples containing 50 wt% deuterium-depleted water as well as on oriented samples hydrated at 86% relative humidity were conducted at a magnetic field strength of 18.8 T with a cylindrical or flat r.f. coil, respectively. Hydration of the oriented sample was controlled by a saturated KCl solution in the sample container. Spectra were acquired with a quadrupolar echo sequence with a π/2 pulse length of 2.5 μs for unoriented samples and 4.5 μs for oriented samples, a delay time between the π/2 pulses of 50 μs and a pulse sequence repetition delay of 250 ms. The water content of the oriented sample, hydrated at 86% relative humidity, was determined by 1H-magic angle spinning (MAS) NMR on bilayers oriented at the inner surface of a MAS rotor and exposed for several hours to the water vapor pressure of a saturated KCl solution before data acquisition. The number of water molecules per lipid was determined by comparison of integral signal intensities of the water and the lipid choline resonance.
Correlation times of DHA chain isomerization as well as chain-order parameter profiles were determined as reported previously (Soubias and Gawrisch 2007; Eldho et al. 2003). In brief, the well-resolved resonances of the 13C MAS NMR spectra of DHA were acquired at a temperature of 30°C and a MAS frequency of 10 kHz. The H-C bond order parameters of DHA chains defined on a time scale of 10−5 s were derived from 2H-NMR experiments on 18:0-22:6n3-d31-PC using signal assignments published in Eldho et al. (2003). For determination of the correlation times of DHA chain isomerization, spin-lattice (T1) and spin–spin relaxation times (T2) were measured under proton decoupling at two values of magnetic field strength, 11.8 and 18.8 T. The T1 values were measured by the inversion-recovery pulse sequence and the T2 values, by a Hahn echo pulse sequence. Correlation times of DHA chain isomerization were calculated from the relaxation data as described in Soubias and Gawrisch (2007). The relaxation data yielded also an order parameter profile defined on a time scale of 10−8 s (Soubias and Gawrisch 2007).
Results and Discussion
Neutron Diffraction
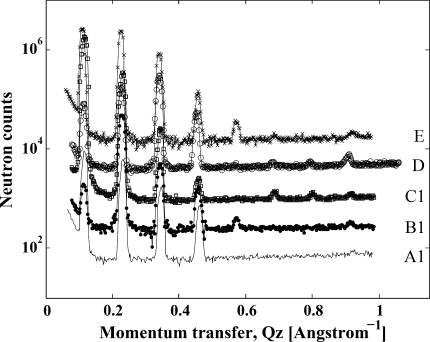
A set of five different samples of equivalent composition containing 29 mol% cholesterol were investigated by neutron diffraction. The set consisted of a fully hydrogenous sample, which constituted the reference, and four other samples specifically deuterated in either of the two lipid chains (steroyl or DHA) or the cholesterol molecule (A-ring or methyl-tail). Samples hydrated at 86% relative humidity formed a single lamellar phase. In contrast, attempts to conduct measurements at 66% relative humidity resulted in samples with coexistence of two phases. This was confirmed in NMR experiments. Therefore, only experiments at 86% relative humidity were conducted. Several H2O/D2O ratios were used for sample hydration, to enable determination of the phase of diffraction peaks and to scale the data (see Appendix). Samples were kept in an argon atmosphere throughout the measurements. This prevented oxidation of the lipid for about 30 h. Acquisition of a complete data set per sample took about 10 h. For every sample, several scans over the whole angular range were acquired. This enabled us to follow water uptake by the samples until an equilibrium was reached. It also allowed detection of sample degradation from oxidation toward the end of the useful lifetime of samples. Oxidation was readily detected as broadening or splitting of diffraction peaks. This was confirmed in high-resolution NMR experiments on the lipids from the oriented samples dissolved in methanol. Any results that were possibly tainted by oxidation were discarded and the experiment was repeated. Figure 1 shows typical diffraction data for selected samples.
Fig. 1.
Diffraction data of 18:0-22:6n3-PC bilayers containing 29 mol% cholesterol and hydrated at 86% relative humidity but with different deuterium labeling: black, all protonated components; blue, DHA chain deuterated (11 deuterium atoms evenly distributed along the chain); red, stearoyl chain perdeuterated (35 deuterium atoms); green, cholesterol deuterated at the A-ring (carbon positions 2, 2, 3, 4, 4, 6); ochre, cholesterol deuterated at the methyl end (carbon positions 25, 25, 25, 26, 26, 26)
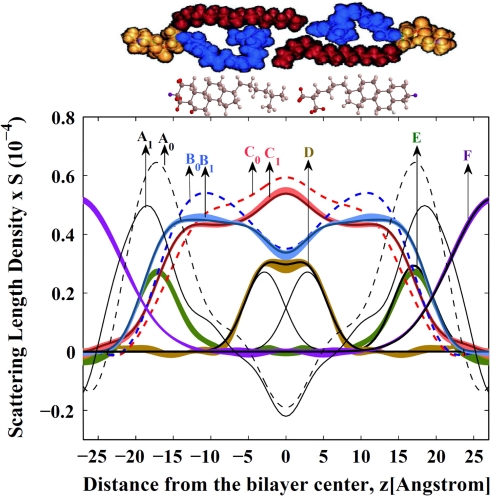
The large differences in SLD between protonated and deuterated hydrocarbon chains and cholesterol permitted calculation of the SLD distribution of saturated stearoyl and polyunsaturated DHA chains, as well as location of the A-ring and the terminal methyl end of cholesterol. Results are presented in Fig. 2. For comparison, the stearoyl and DHA chain distributions in 18:0-22:6n3-PC bilayers without cholesterol, reported by us previously (Mihailescu and Gawrisch 2006), are also shown. The uncertainty bands associated with the density profile were determined from the standard deviations in the experimental structure factors, including uncertainties in the scale factors, determined by a Monte-Carlo sampling procedure (Wiener and White 1992). The data show that addition of cholesterol triggers a redistribution of hydrocarbon chain densities as well as an increase in bilayer repeat spacing from 52.3 ± 0.1 Å without cholesterol to 55.3 ± 0.1 Å with 29 mol% cholesterol. The DHA chain density significantly redistributes from the interface region to the center of the bilayer with addition of cholesterol. The stearoyl chains slightly extend with cholesterol addition, while maintaining a maximum of density in the middle of the bilayer.
Fig. 2.
SLD profiles and distribution of deuterated segments in a 18:0-22:6n3-PC bilayer containing 29 mol% cholesterol, measured at 86% relative humidity and 22°C. Profiles are presented on an absolute (per lipid) scale, up to an unknown multiplicative factor, S, accounting for the area per lipid. A0 and A1, SLD profiles for a protonated lipid bilayer without cholesterol (dashed curve, determined previously [Mihailescu and Gawrisch 2006]) and with cholesterol (solid curve); B0 and B1, DHA chain density distribution in the absence of cholesterol (Mihailescu and Gawrisch 2006) and in the presence of cholesterol, respectively; C0 and C1, stearoyl chain density distribution in the absence of cholesterol (determined previously for 66% relative humidity [Mihailescu and Gawrisch 2006]) and in the presence of cholesterol, respectively. The scale for the stearoyl chain distribution was adjusted by a factor 0.3 to account for the difference in the degree of deuteration between both chains. D, Cholesterol’s methyl-tail distribution. Note that the distribution is best described by two gaussians located at z = ±2.9 Å from the bilayer center, shown as solid black lines. E, Cholesterol’s A-ring distribution. The slightly asymmetrical shape is likely due the uneven distribution of deuterium labels over the A-ring. Solid black line shows the result of a fit to a gaussian. F, Water distribution. The uncertainty bands associated with the profiles were calculated from the standard deviation of the bilayer structure factors. Upper image is a cartoon of the 18:0-22:6n3-PC lipid conformation in the bilayer, according to molecular dynamics simulations (Eldho et al. 2003). Cholesterol molecules are shown in ball-and-stick representation with the deuterated positions highlighted in red
The distributions of the A-ring and methyl-tail of cholesterol in 18:0-22:6n3-PC bilayers are shown in Fig. 2 as well. Due to thermal disorder, the lipid molecules and cholesterol have relatively wide distribution functions. Nevertheless, the mean position of a specific label can be determined with a resolution to a fraction of 1 Å. The data were modeled using gaussian functions, with the height, midpoint and width as fit parameters (see Table 1, Appendix). The cholesterol A-ring is positioned with highest probability at a distance of 1.4 Å inward from the position of the lipid carbonyl groups. The carbonyl location (z = 18.4 Å) is easily identified as the point with the highest SLD in bilayers. The preferred location of the A-ring confirms earlier predictions from molecular simulations that found a preferred location 1.5 Å inward from carbonyls (Pitman et al. 2004). The density profile of methyl groups was best described by two gaussian functions positioned, symmetrically, at z = ±2.9 (±0.2) Å from the bilayer center. It clearly indicates that the cholesterol tails reside in the individual monolayers. The projected distance between the A-ring and the methyl-tail, on the bilayer normal, is 14.1 Å, which is somewhat smaller than the distance of 14.6 Å calculated from the atomic coordinates of a cholesterol model. It suggests that, on average, the cholesterol molecules in lipid bilayers are slightly tilted with respect to the bilayer normal, most likely as the result of thermal motions.
Table 1.
Selected parameters of 18:0-22:6n3-PC bilayers with and without 29 mol% cholesterol
| d (Å) | Nwa | Water | A-ring | Methyl-tail | ||||
|---|---|---|---|---|---|---|---|---|
| Z (Å)b | A (Å) | Z (Å) | A (Å) | Z (Å) | A (Å) | |||
| Pure SDPC | 52.3 ± 0.1 | 9.4 ± 0.1 | d/2 | 7.8 ± 0.3 | – | – | – | – |
| 29%Chol/SDPC | 55.3 ± 0.1 | 8.9 ± 0.1 | d/2 | 8.0 ± 0.2 | 17.0 ± 0.3 | 3.8 ± 0.1 | 2.9 ± 0.2 | 3.8 ± 0.2 |
d Repeat distance, Z mean position, A 1/e half-width, A and Z were determined by gaussian model fitting over the structure factor difference (ΔF(n)) in the reciprocal space, using Eq. 4
aNumber of waters per lipid determined by 1H-MAS NMR
bWater mean position was fixed at d/2 in the fit; thus, A corresponds to the envelope of the water hydrating two apposing bilayers
The uneven distribution of the chains across the bilayer for 18:0-22:6n3-PC lipid is not observed for a monounsaturated lipid such as 16:0-18:1n9-PC. In order to demonstrate this, we performed a series of additional experiments to determine the palmitoyl chain density distributions in 16:0-18:1n9-PC bilayers hydrated at 86% relative humidity, with and without addition of 33 mol% cholesterol (Fig. 3). Unlike the stearoyl chain in 18:0-22:6n3-PC bilayers, the palmitoyl chain is fully extended and its density evenly distributed in both the presence and the absence of cholesterol. The repeat spacing is found to increase from 52.8 (±0.1) Å for the pure lipid to 55.5 (±0.1) Å when 33 mol% cholesterol is present in this lipid.
Fig. 3.
SLD profiles for a 16:0-18:1n9-PC (POPC) bilayer measured at 86% relative humidity: black solid line, bilayer profile with 33 mol% cholesterol; red dashed line, palmitoyl chain distribution in the absence of cholesterol; red solid line, palmitoyl chain distribution in the presence of cholesterol
The distribution function of the water of hydration does not change significantly in the presence of cholesterol (Table 1). However, cholesterol addition slightly decreases the total amount of water taken up by the bilayers at 86% relative humidity. The increase in the repeat spacing of 3 Å upon incorporation of 29 mol% cholesterol is primarily the result of a cholesterol-induced area condensation. A similar influence of cholesterol on the bilayer repeat spacing and water distribution was observed for 16:0-18:1n9-PC and 18:1n9-18:1n9-PC bilayers (M. M., unpublished results).
NMR
Figure 4a shows the 2H-NMR spectra of 18:0-d35-22:6n3-PC with and without 29 mol% cholesterol and hydrated with 50 wt% deuterium-depleted water. It is clearly visible that addition of cholesterol increases all quadrupolar splittings of stearoyl chains by a significant amount. The corresponding smoothed order parameter profiles are shown in Fig. 4b. The data points presented as filled squares are from a sample without cholesterol and those presented as open squares are with cholesterol. Addition of cholesterol not only results in an overall increase of chain order parameters but also alters the shape of the order parameter profile. This is demonstrated by the profile with open circles in Fig. 4a. These data points were calculated from the order profile with addition of cholesterol but attenuated by a factor to match order parameter values of the so-called order parameter plateau (carbons C2–C7). The comparison shows a preferential increase of chain order for the second half of the hydrocarbon chain, C8–C18. A similar behavior was observed in experiments conducted on oriented bilayers at 86% relative humidity (Fig. 4c). We interpret the preferential increase of order parameters C8–C18 as the result of redistribution of DHA chain density that was observed in the neutron diffraction studies. With increasing DHA chain density in the bilayer center, the stearoyl chain segments C8–C18 have less freedom for movement, which raises their order parameters.
Fig. 4.
a 2H spectra of 18:0d35-22:6n3-PC bilayers recorded at a temperature of 30°C, (top) without cholesterol and (bottom) with 29 mol% cholesterol. b Order parameter profile of the sn-1 stearoyl chain with 29 mol% cholesterol (open squares) and without cholesterol (filled squares). Open circles are an order parameter profile in the presence of cholesterol with adjustment by a constant factor to equalize order in the presence and absence of cholesterol. c Order parameter profile of the sn-1 stearoyl chains with 29 mol% cholesterol (open squares) and without cholesterol (filled squares) measured on a sample of oriented bilayers with the bilayer normal oriented parallel to the magnetic field. The sample was hydrated at a relative humidity of 86%, and spectra were recorded at 22°C to mimic the conditions of the neutron diffraction experiments. Open circles are an order parameter profile in the presence of cholesterol with adjustment by a constant factor to equalize order in the presence and absence of cholesterol
While the neutron diffraction data were obtained at 86% relative humidity for better resolution, the resolution of NMR data does not depend on water content and the results were obtained on both fully hydrated bilayers and slightly dehydrated bilayers. Dehydration increases chain order somewhat, but it does not alter the overall trend of a larger increase in stearoyl chain order for chain segments C8–C18 with addition of cholesterol. It indicates that the redistribution of DHA chain density with cholesterol addition is a general phenomenon that is not limited to slightly dehydrated bilayers.
The correlation times of DHA chain isomerization and DHA order parameters were determined by 13C MAS NMR as described in Materials and Methods. Figure 5a shows the correlation times with (open triangles) and without (filled squares) cholesterol. There is a trend toward slightly shorter correlation times of chain isomerization for segments near the DHA chain carbonyl group after addition of cholesterol, but the differences are small. The DHA chains isomerize quite rapidly in both the presence and the absence of cholesterol. We showed earlier that rapid isomerization is the result of low energetic barriers to rotation about vinyl-methylene bonds in DHA (Feller et al. 2003).
Fig. 5.
a Dependence of correlation times of DHA isomerization on the chain carbon position measured on 18:0-22:6n3-PC vesicles with (open triangles) and without (filled squares) 29 mol% cholesterol measured at a temperature of 30°C. b DHA order parameter profile in 18:0-22:6n3-PC bilayers extracted from the relaxation analysis (open squares) and from 2H-NMR order parameter measurements (filled squares). Data for bilayers with 29 mol% cholesterol (b1) and without cholesterol (b2)
Figure 5 shows DAH chain order parameter profiles measured with (Fig. 5b1) and without (Fig. 5b2) addition of cholesterol. The open squares are from relaxation measurements that yield order parameters on a time scale of 10−8 s. The filled squares are derived from 2H-NMR order parameter experiments with an intrinsic time scale of 10−5 s. Addition of cholesterol increases order parameters of DHA chains for chain segments C6–C22. This is more visible for the order parameters determined on the shorter time scale of 10−8 s. These order parameters are larger because collective modes of lipid motions like tumbling of individual lipids about the bilayer normal and motions related to bilayer undulation with correlation times from 10−8 to 10−5 s have not contributed yet to the lipid order. Order profiles on both time scales look similar, suggesting that DHA chains explore their conformational space reasonably well even on the much shorter time scale. Order parameters of DHA chains in membranes are much lower than those of stearoyl chain segments. The differences are caused in part by the different geometry of double bonds. However, the low order parameters of methylene groups of DHA clearly point at the importance of rapid chain isomerization for low DHA order (Eldho et al. 2003). The increase in DHA chain order with cholesterol addition correlates nicely with the redistribution of DHA chain density toward the bilayer center observed in the neutron diffraction studies.
Conclusions
The present results provide a compelling view of the internal organization of mixed-chain polyunsaturated 18:0-22:6n3-PC bilayers and the role that cholesterol plays for in-plane and in-depth organization of bilayers. The DHA chains are somewhat polar because of their six cis-locked double bonds, and they are more flexible due to low barriers for rotation about vinyl-methylene bonds. Both properties are responsible for the higher DHA density near the lipid–water interface compared to stearoyl chains. The data in this report show that addition of cholesterol to bilayers pushes DHA chains away from the interface toward the bilayer center. This is accompanied by an increase of order parameters for both stearoyl and DHA chains. Part of that increase is caused by a cholesterol-induced condensation of the lipid lateral area. The condensation effect is not as large as was observed for saturated and monounsaturated bilayers, but it is still significant. The somewhat larger fractional increase of lipid order for chain segments toward the bilayer center with cholesterol addition is the direct result of higher DHA chain density in the bilayer center. The methylene segments C8–C18 of stearoyl chains have less freedom for movement because of higher DHA chain densities in the center of bilayers. DHA chain order increases because of the somewhat higher preference for extended DHA conformations in the presence of cholesterol. The motional correlation times of DHA chain isomerization remain short in the presence of cholesterol.
The mean position of cholesterol molecules in the bilayers was pinpointed by selective labeling. The A-ring of cholesterol is located slightly deeper in the lipid–water interface than lipid carbonyl groups. Its broad distribution superimposes not only with the density distribution of carbonyls but also with lipid phosphate groups and water. This points to the importance of the cholesterol hydroxyl group for cholesterol–phospholipid and cholesterol–water interactions via hydrogen bonds (Soubias et al. 2004). Although the distribution functions of cholesterol A-rings and terminal methyl groups are wide, the data clearly show that cholesterol molecules in 18:0-22:6n3-PC bilayers remain centered in monolayers with a low probability of passing into the apposing monolayer.
Both neutron diffraction and NMR results have shown that cholesterol is an important modulator of the structure of polyunsaturated lipid bilayers. The redistribution of chain densities from cholesterol addition is likely to change the microenvironment for function of membrane proteins like GPCRs. Cholesterol also modulates membrane elasticity and transport of substances across cell membranes, which is of functional importance as well.
Acknowledgements
This work was supported in part by U.S. National Institutes of Health grants GM74737 (to S. H. W.) from NIGMS and by the Intramural Research Program of NIAAA (to O. S. and K. G.). We are grateful for the support of the National Institute of Standards and Technology, U.S. Department of Commerce, in providing the neutron research facilities used for neutron diffraction experiments. The identification of any commercial product or trade name does not imply endorsement or recommendation by the U.S. National Institute of Standards and Technology.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Appendix
The diffraction signal from oriented lipid multilayers is sampled, according to the Bragg law, in intense diffraction peaks which occur at Q z = n * π/d, where n denotes the diffraction order and d is the repeat distance of the structure (i.e., the bilayer thickness plus the thickness of the water layer). In kinematical approximation, the coherently diffracted intensity at each diffraction order (n) is related to the structure factor of a bilayer (F) and the number of bilayers (N):
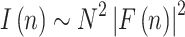 |
1 |
Since the natural diffraction peak width is broadened due to the instrumental resolution and the effective mosaic spread of the sample, the coherent reflected intensity at each Bragg order was determined by integrating the counts under the peak, I(n). The structure factors F(n) are then determined—up to a phase factor—as the square root of the peak integrals (in variable θ), after background extraction, absorption correction (A) and Lorentz factor correction, sin(2θn), where θn represents the angle of incidence corresponding to the n-th order of diffraction. The absorption correction was calculated as A(n) = sin(θn)/(2μt) * [1 − exp(−2 μt/sin(θn)], where t is the sample thickness and μ, the linear absorption coefficient (Worcester and Franks 1976).
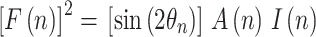 |
2 |
The scattering length density (SLD) of the bilayer was determined from a Fourier synthesis of the bilayer structure factors as
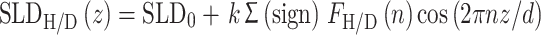 |
3 |
For a centrosymmetric unit cell, such as a lipid bilayer, there are only two possible signs (+1 or −1) of the structure factors, corresponding to cos(0o) and cos(180o). Methods to determine those signs have been discussed in detail elsewhere (Franks and Lieb 1979; Worcester and Franks 1976; Blasie et al. 1975). We determined phases from measurements using 1H2O–2H2O contrast of at least three different mole fractions of 2H2O. The Fourier transform of the bilayer density is a linear function of the isotopic composition of water. Assuming a gaussian distribution of water near the lipid head groups (Wiener et al. 1991), the difference structure factors corresponding to a lipid bilayer hydrated with 2H2O (FD) and 1H2O (F H), respectively, are
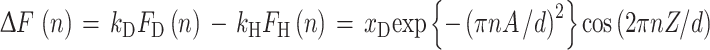 |
4 |
where A and Z denote the 1/e half-width and the mean position of the gaussian water distribution and x D is the fraction of 2H2O. The cosine factor in Eq. 4 determines the slope of the linear dependence, ΔF(n) = f(x D), and thus the appropriate signs for the structure factors in Eq. 3.
Gaussian model fitting based on Eq. 4 was then used to determine the parameters describing the deuterium label distribution (e.g., water of hydration): mean position (Z) and 1/e half-width (A) but also the scale factors (k). This procedure is applied for determining the distribution of all the other deuterated components, which could be approximated by a gaussian—i.e., the cholesterol A-ring and methyl-tail. A series of H2O–2H2O water contrast measurements was run for each sample for the purpose of phase assignment and absolute scaling (Wiener et al. 1991), as above. Then, the stearoyl and DHA chain distributions were determined by direct subtraction of the real space profile (Eq. 3) of the protonated lipid from the profile of the chain-deuterated lipid, measured under the same hydration conditions. Table 2 presents a list of structure factors that were used to produce the SLD profiles in Figs. 1–3. Each set of eight observable structure factors corresponds to a given sample or deuteration scheme. Equation 3 can be used to calculate the SLD profile for each set. Direct subtraction of the all-protonated profile (SLDH) from the specifically deuterated profiles (SLDD) yields the individual distribution for the chains, cholesterol and water. The number of water/lipid can be determined from the neutron data, but in this case was significantly less accurate than the NMR determination (Table 1) due to the oxidation-limited measuring times.
Table 2.
Structure factors
| Diffraction order (n) |
F(n) protonated lipid H2O |
F(n) protonated lipid 20%D2O |
F(n) DHA-deuterated H2O |
F(n) SA-deuterated H2O |
F(n) cholesterol A-ring deuterated H2O |
F(n) cholesterol CH3-tail deuterated H2O |
|---|---|---|---|---|---|---|
| 1 | −4.52 ± 0.01 | −9.13 ± 0.01 | 1.05 ± 0.03 | 18.37 ± 0.08 | −5.91 ± 0.01 | −1.35 ± 0.02 |
| 2 | −5.35 ± 0.01 | −3.36 ± 0.01 | −9.19 ± 0.04 | −10.86 ± 0.08 | −8.27 ± 0.02 | −3.11 ± 0.02 |
| 3 | 3.57 ± 0.01 | 3.02 ± 0.02 | 3.53 ± 0.04 | 3.51 ± 0.01 | 6.08 ± 0.02 | 4.92 ± 0.03 |
| 4 | −2.79 ± 0.01 | −2.79 ± 0.01 | −2.29 ± 0.05 | 1.05 ± 0.01 | −2.69 ± 0.03 | −2.44 ± 0.03 |
| 5 | 0 ± 0.06 | 0 ± 0.10 | −0.87 ± 0.05 | 0.31 ± 0.07 | −1.32 ± 0.04 | 0 ± 0.15 |
| 6 | 0 ± 0.13 | 0 ± 0.14 | 0 ± 0.10 | −0.67 ± 0.04 | 0.60 ± 0.08 | −0.48 ± 0.06 |
| 7 | 0 ± 0.07 | 0 ± 0.11 | 0 ± 0.17 | 0.65 ± 0.05 | 0 ± 0.14 | −0.38 ± 0.07 |
| 8 | −0.58 ± 0.06 | −0.58 ± 0.09 | −0.72 ± 0.08 | −0.53 ± 0.07 | −0.89 ± 0.10 | −0.86 ± 0.04 |
References
- Anderson RE, Landis DJ, Dudley PA. Essential fatty acid deficiency and renewal of rod outer segments in the albino rat. Inv Ophtal. 1976;15:232–236. [PubMed] [Google Scholar]
- Bennett MP, Mitchell DC. Regulation of membrane proteins by dietary lipids: effects of cholesterol and docosahexaenoic acid acyl chain-containing phospholipids on rhodopsin stability and function. Biophys J. 2008;95:1206–1216. doi: 10.1529/biophysj.107.122788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasie JK, Schoenborn BP, Zaccai G. Direct methods for the analysis of lamellar neutron diffraction from oriented multilayers: a difference Patterson deconvolution approach. Brookhaven Symp Biol. 1975;27:III58–III67. [PubMed] [Google Scholar]
- Boesze-Battaglia K, Albert AD. Cholesterol modulation of photoreceptor function in bovine retinal rod outer segments. J Biol Chem. 1990;265:20727–20730. [PubMed] [Google Scholar]
- Brzustowicz MR, Cherezov V, Caffrey M, Stillwell W, Wassall SR. Molecular organization of cholesterol in polyunsaturated membranes: microdomain formation. Biophys J. 2002;82:285–298. doi: 10.1016/S0006-3495(02)75394-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dura JA, Pierce DJ, Majkrzak CF, Maliszewskyj NC, McGillivray DJ, Losche M, O’Donovan KV, Mihailescu M, Perez-Salas U, Worcester DL, White SH. AND/R: advanced neutron diffractometer/reflectometer for investigation of thin films and multilayers for the life sciences. Rev Sci Instrum. 2006;77:074301. doi: 10.1063/1.2219744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eldho NV, Feller SE, Tristram-Nagle S, Polozov IV, Gawrisch K. Polyunsaturated docosahexaenoic vs docosapentaenoic acid-differences in lipid matrix properties from the loss of one double bond. J Am Chem Soc. 2003;125:6409–6421. doi: 10.1021/ja029029o. [DOI] [PubMed] [Google Scholar]
- Enslen M, Milon H, Malnoe A. Effect of low intake of n-3 fatty acids during development on brain phospholipid fatty acid composition and exploratory behavior in rats. Lipids. 1991;26:203–208. doi: 10.1007/BF02543972. [DOI] [PubMed] [Google Scholar]
- Feller ES, Gawrisch K, Woolf TB. Rhodopsin exhibits a preference for solvation by polyunsaturated docosahexaenoic acid. J Am Chem Soc. 2003;125:4434–4435. doi: 10.1021/ja0345874. [DOI] [PubMed] [Google Scholar]
- Franks NP, Lieb WR. The structure of lipid bilayers and the effects of general anesthetics. An X-ray and neutron diffraction study. J Mol Biol. 1979;133:469–500. doi: 10.1016/0022-2836(79)90403-0. [DOI] [PubMed] [Google Scholar]
- Grisshammer R. Purification of recombinant G-protein coupled receptors. Methods Enzymol. 2009;463:631–645. doi: 10.1016/S0076-6879(09)63036-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harroun TA, Katsaras J, Wassall SR. Cholesterol is found to reside in the center of a polyunsaturated lipid membrane. Biochemistry. 2008;47:7090–7096. doi: 10.1021/bi800123b. [DOI] [PubMed] [Google Scholar]
- Huster D, Arnold K, Gawrisch K. Influence of docosahexaenoic acid and cholesterol on lateral lipid organization in phospholipid mixtures. Biochemistry. 1998;37:17299–17308. doi: 10.1021/bi980078g. [DOI] [PubMed] [Google Scholar]
- Mihailescu M, Gawrisch K. The structure of polyunsaturated lipid bilayers important for rhodopsin function: a neutron diffraction study. Biophys J. 2006;90:L4–L6. doi: 10.1529/biophysj.105.071712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miljanich GP, Sklar LA, White DL, Dratz EA. Disaturated and dipolyunsaturated phospholipids in the bovine retinal rod outer segment disk membrane. Biochim Biophys Acta. 1979;552:294–306. doi: 10.1016/0005-2736(79)90284-0. [DOI] [PubMed] [Google Scholar]
- Mitchell DC, Straume M, Litman BJ. Role of sn-1-saturated, sn-2-polyunsaturated phospholipids in control of membrane receptor conformational equilibrium: effects of cholesterol and acyl chain unsaturation on the metarhodopsin I in equilibrium with metarhodopsin II equilibrium. Biochemistry. 1992;31:662–670. doi: 10.1021/bi00118a005. [DOI] [PubMed] [Google Scholar]
- Pitman MC, Suits F, Mackerrel AD, Jr, Feller SE. Molecular-level organization of saturated and polyunsaturated fatty acids in a phosphatidylcholine bilayer containing cholesterol. Biochemistry. 2004;43:15318–15328. doi: 10.1021/bi048231w. [DOI] [PubMed] [Google Scholar]
- Polozova A, Litman BJ. Cholesterol dependent recruitment of di22:6-PC by a G protein–coupled receptor into lateral domains. Biophys J. 2000;79:2632–2643. doi: 10.1016/S0006-3495(00)76502-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salem N, Jr, Litman B, Kim H-Y, Gawrisch K. Mechanisms of action of docosahexaenoic acid in the nervous system. Lipids. 2001;36:945–959. doi: 10.1007/s11745-001-0805-6. [DOI] [PubMed] [Google Scholar]
- Soubias O, Gawrisch K. Docosahexaenoyl chains isomerize on the sub-nanosecond time scale. J Am Chem Soc. 2007;129:6678–6679. doi: 10.1021/ja068856c. [DOI] [PubMed] [Google Scholar]
- Soubias O, Jolibois F, Reat V, Milon A. Understanding sterol–membrane interactions, part II: complete 1H and 13C assignments by solid-state NMR spectroscopy and determination of the hydrogen-bonding partners of cholesterol in a lipid bilayer. Chemistry. 2004;10:6005–6014. doi: 10.1002/chem.200400246. [DOI] [PubMed] [Google Scholar]
- Soubias O, Teague WE, Gawrisch K. Evidence for specificity in lipid-rhodopsin interactions. J Biol Chem. 2006;281:33233–33241. doi: 10.1074/jbc.M603059200. [DOI] [PubMed] [Google Scholar]
- Umezawa M, Ohta A, Tojo H, Yagi H, Hosokawa M, Takeda T. Dietary alpha-linolenate/linoleate balance influences learning and memory in the senescence-accelerated mouse (SAM) Brain Res. 1995;669:225–233. doi: 10.1016/0006-8993(94)01250-L. [DOI] [PubMed] [Google Scholar]
- Wiener MC, White SH. Structure of a fluid dioleoylphosphatidylcholine bilayer determined by joint refinement of X-ray and neutron diffraction data. III. Complete structure. Biophys J. 1992;61:434–447. doi: 10.1016/S0006-3495(92)81849-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiener MC, King GI, White SH. Structure of a fluid dioleoylphosphatidylcholine bilayer determined by joint refinement of X-ray and neutron diffraction data. I. Scaling of neutron data and the distributions of double bonds and water. Biophys J. 1991;60:568–576. doi: 10.1016/S0006-3495(91)82086-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worcester DL, Franks NP. Structural analysis of hydrated egg lecithin and cholesterol bilayers. II. Neutron diffraction. J Mol Biol. 1976;100:359–378. doi: 10.1016/S0022-2836(76)80068-X. [DOI] [PubMed] [Google Scholar]
- Yamamoto N, Hashimoto A, Takemoto Y, Okuyama H, Nomura M, Kitajima R, Togashi T, Tamai Y. Effect of the dietary alpha-linolenate/linoleate balance on lipid compositions and learning ability of rats. II. Discrimination process, extinction process, and glycolipid compositions. J Lipid Res. 1988;29:1013–1021. [PubMed] [Google Scholar]