Abstract
A group A Streptococcus(GAS) isolate,serotypeM12,recovered from a patient with streptococcal toxic shock syndrome was analyzed for superantigen-carrying prophages, revealing  149, which encodes superantigen SSA. Sequence analysis of the att-L proximal region of
149, which encodes superantigen SSA. Sequence analysis of the att-L proximal region of  149 showed that the phage had a mosaic nature. Remarkably, we successfully obtained lysogenic conversion of GAS clinical isolates of various M serotypes (M1, M3, M5, M12, M19, M28, and M94), as well as of group C Streptococcus equisimilis (GCSE) clinical isolates, via transfer of a recombinant phage
149 showed that the phage had a mosaic nature. Remarkably, we successfully obtained lysogenic conversion of GAS clinical isolates of various M serotypes (M1, M3, M5, M12, M19, M28, and M94), as well as of group C Streptococcus equisimilis (GCSE) clinical isolates, via transfer of a recombinant phage  149::Kmr. Phage
149::Kmr. Phage 149::Kmr from selected lysogenized GAS and GCSE strains could be transferred back to M12 GAS strains. Our data indicate that horizontal transfer of lysogenic phages among GAS can occur across the M-type barrier; these data also provide further support for the hypothesis that toxigenic conversion can occur via lysogeny between species. Streptococci might employ this mechanism specifically to allow more efficient adaptation to changing host challenges, potentially leading to fitter and more virulent clones.
149::Kmr from selected lysogenized GAS and GCSE strains could be transferred back to M12 GAS strains. Our data indicate that horizontal transfer of lysogenic phages among GAS can occur across the M-type barrier; these data also provide further support for the hypothesis that toxigenic conversion can occur via lysogeny between species. Streptococci might employ this mechanism specifically to allow more efficient adaptation to changing host challenges, potentially leading to fitter and more virulent clones.
Streptococcus pyogenes, or β-hemolytic group A Streptococcus (GAS), is a notorious human pathogen, responsible for a number of different infections that range from mild skin diseases to fulminant, severe, invasive syndromes. Since the mid-1980s, a remarkable increase in the incidence of the severe forms of GAS infections (in particular, necrotizing fasciitis and streptococcal toxic shock syndrome [STSS]) has been observed worldwide [1, 2]. Many hypotheses have emerged to explain this phenomenon; some focus on impaired host defense against specific strains or specific virulence determinants (e.g., immunogenetic background or predisposing factors), whereas others focus on changes in bacterial virulence itself [3–5].
Fluctuations in the severity and character of streptococcal virulence have also been linked to the plasticity of the streptococcal genome. Mobile genetic elements, including lysogenic bacteriophages, that have integrated into the GAS genome over time were found to be almost exclusively responsible for the genetic differences among strains [6–11]. Recent genomic analyses indicate that GAS is a polylysogenic organism in which prophage sequences constitute ~10% of the total genome [1, 3, 7, 12]. In GAS, prophages encode 1 or more putative or established virulence factors, including phospholipases, streptodornases, and superantigens, in addition to their essential viral proteins [1, 12]. It is now widely acknowledged that virulence gene transfer may occur among GAS when favorable conditions are encountered in the human host [13–18]. By means of lysogenic conversion, the toxin-encoding bacteriophages can convert their bacterial host from a nonpathogenic strain to a virulent strain or a strain with increased virulence [12, 13, 19]. Along these lines, prophages have been shown to play a key role in subclone diversification and to be largely responsible for the uniqueness of clinical isolates [3]. Consequently, GAS prophages play a critical role in determining the distinct disease pathologies associated with otherwise similar strains [1].
The critical role of superantigens in severe GAS infection underlines the clinical relevance of prophages [2, 20, 21]. In GAS, the majority of superantigen genes are located on prophage genomes. Experimental ex vivo toxigenic conversion by lysogeny has been demonstrated only for speA and speC and almost exclusively among strains belonging to the same M serotype. The suggested bacteriophage-mediated transfer of other superantigens is based only on indirect evidence: for example, their association with prophage sequences, their distribution pattern throughout the streptococcal population, or the inducible character of prophages carrying the respective genes [13, 15, 22–25]. Further, a number of studies indicate that M proteins could function, directly or indirectly, as barriers to horizontal gene exchange. A phenotypic correlation between resistance to bacteriophage infection and M-protein surface expression has been described [26, 27]. Nonrandom associations between exotoxin alleles and emm patterns were also observed, thus suggesting some direct or indirect biological interactions between M-protein surface structures and bacteriophage-associated properties [28–30]. In this study, we focus on the mobility of superantigen SSA, which has been associated with GAS isolated from patients with STSS and which has highly potent superantigenic activity that has been proven experimentally [31–33]. Presence of the ssa gene has been detected across the GAS population, as well as in clinical isolates of group C (GCS) and group G (GGS) streptococci (Streptococcus dysgalactiae subspecies equisimilis GCSE and GGSE, respectively), thus suggesting the possibility of interspecies ssa gene transfer [14, 29, 30, 34–36].
We report here the characterization of an inducible chimeric prophage,  149, carrying the superantigen-encoding ssa-3 allele from a GAS clinical isolate of serotype M12, RDN149. We describe the host range of this phage by investigating its transfer-ability to GAS strains of various M types, as well as to GCSE and GGSE. This study experimentally confirms previous indications that lysogenic toxin conversion can occur across the M-type barrier in GAS and between GAS and GCSE.
149, carrying the superantigen-encoding ssa-3 allele from a GAS clinical isolate of serotype M12, RDN149. We describe the host range of this phage by investigating its transfer-ability to GAS strains of various M types, as well as to GCSE and GGSE. This study experimentally confirms previous indications that lysogenic toxin conversion can occur across the M-type barrier in GAS and between GAS and GCSE.
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions
Streptococcal strains (table 1; [29, 37, 38]) were grown at 37°C with 5% CO2 without agitation in Todd-Hewitt broth either with or without 0.2% yeast extract supplementation and on tryptic soy agar supplemented with 3% sheep blood or THY agar. Transformation of Escherichia coli and S. pyogenes with plasmid DNA was performed as described elsewhere [39, 40]. Whenever required, antibiotics were added to the media: erythromycin at 3 μg/mL for S. pyogenes; and kanamycin at 25 μg/mL for E. coli and at 300 μg/mL for streptococcal strains.
Table 1.
Bacterial strains and plasmids used in this study.
| Strain or plasmid | Relevant characteristic(s) | Source or reference |
|---|---|---|
| Strain | ||
| E. coli | Host strain for cloning | Lab collection |
| DH5-α | ||
| TOP10 | Host strain for cloning | Lab collection |
| S. pyogenes | ||
| RDN149 | M12,  149 donor, ssa+, speH+ 149 donor, ssa+, speH+
|
[29] |
| EC516 | RDN149 ( 149::Kmr) 149::Kmr) |
This study |
| RDN151 | M12, indicator strain for  149, ssa−, speH+ 149, ssa−, speH+ |
[29] |
| EC627 | RDN151 (pMSP3535) | This study |
| EC496 | RDN151 lysogenized with  149 149 |
This study |
| EC497 | RDN151 lysogenized with  149 149 |
This study |
| CS112 | M12,  CS112 donor, speC+ CS112 donor, speC+ |
[38] |
| CS24 | M12, indicator strain for  CS112 and CS112 and  149, speC−, 149, speC−,ssa−, speH− |
[38] |
| RDN313 | CS24 lysogenized with  CS112 CS112 |
This study |
| RDN312 | CS24 lysogenized with  149 149 |
This study |
| EC500 | CS24 lysogenized with  149 149 |
This study |
| RDN1 | M1, clinical isolate | Lab collection |
| RDN2 | M19, clinical isolate, recipient for  149 149 |
Lab collection |
| RDN3 | M1, clinical isolate | Lab collection |
| RDN4 | M49, clinical isolate | Lab collection |
| RDN5 | M3, clinical isolate, recipient for  149 149 |
Lab collection |
| RDN6 | M3, clinical isolate, recipient for  149 149 |
Lab collection |
| RDN8 | M1, clinical isolate, recipient for  149 149 |
Lab collection |
| RDN27 | M12, clinical isolate, recipient for  149 149 |
Lab collection |
| RDN29 | M1, clinical isolate Lab | collection |
| RDN57 | M49, clinical isolate | Lab collection |
| RDN60 | M5, clinical isolate, recipient for  149 149 |
Lab collection |
| RDN74 | M89, clinical isolate | Lab collection |
| RDN78 | M3, clinical isolate Lab | collection |
| RDN83 | M89, clinical isolate | Lab collection |
| RDN86 | M1, clinical isolate | Lab collection |
| RDN89 | M3, clinical isolate | Lab collection |
| RDN100 | M83, clinical isolate | Lab collection |
| RDN104 | M81, clinical isolate | Lab collection |
| RDN106 | M94, clinical isolate, recipient for  149 149 |
Lab collection |
| RDN107 | M1, clinical isolate | Lab collection |
| RDN114 | M3, clinical isolate | Lab collection |
| RDN116 | M94, clinical isolate, recipient for  149 149 |
Lab collection |
| RDN119 | M1, clinical isolate | Lab collection |
| RDN138 | M12, clinical isolate, recipient for  149 149 |
Lab collection |
| RDN139 | M28, clinical isolate | Lab collection |
| RDN144 | M28, clinical isolate, recipient for  149 149 |
Lab collection |
| RDN145 | M12, clinical isolate, recipient for  149 149 |
Lab collection |
| RDN146 | M12, clinical isolate, recipient for  149 149 |
Lab collection |
| RDN147 | M12, clinical isolate, recipient for  149 149 |
Lab collection |
| RDN148 | M12, clinical isolate, recipient for  149 149 |
Lab collection |
| RDN152 | M22, clinical isolate | Lab collection |
| RDN156 | M28, clinical isolate, recipient for  149 149 |
Lab collection |
RDN148 lysogenized with  149::Kmr 149::Kmr
|
This study | |
| EC1092 | RDN148 lysogenized with  149::Kmr 149::Kmr
|
This study |
| EC1093 | RDN148 lysogenized with  149::Kmr 149::Kmr
|
This study |
| EC749 | RDN156 lysogenized with  149::Kmr 149::Kmr
|
This study |
| EC917 | RDN156 lysogenized with  149::Kmr 149::Kmr
|
This study |
| EC727 | RDN116 lysogenized with  149::Kmr 149::Kmr
|
This study |
| EC1083 | RDN116 lysogenized with  149::Kmr 149::Kmr
|
This study |
|
S. dysgalactiae
subsp. equisimilis |
||
| EC783 | Clinical isolate | Lab collection |
| EC788 | Clinical isolate | Lab collection |
| Plasmid | ||
| pUC19 | ColE1ori, Ampr, lacZ | Lab collection |
| pMSP3535 | ermB, repDEG-pAMβ-1, nisRK, ColE1ori, PnisA | [37] |
| pEC61 | pUC19ΩaphIII | This study |
| pEC96 | pEC61Ωssa-up | This study |
| pEC97 | pEC96Ωssa-down | This study |
NOTE. Lab, laboratory.
General DNA manipulation and analysis
Isolation of streptococcal DNA (genomic and phage) and general DNA manipulations were performed as described elsewhere [16, 39], with minor modifications. VBC-Biotech Services supplied the primers used in this study (table 2) and performed sequencing reactions. General sequence analysis was performed by use of the DNA* (DNAStar) software package and the Basic Local Alignment Search Tool (BLAST) algorithm.
Table 2.
Oligonucleotides used in this study.
| Oligonucleotide | Direction | Sequence (5′ to 3′ direction) | Specific gene target |
|---|---|---|---|
| oliRN70 | F | GCTATTTTTTGACTTACTGGG | aphIII |
| oliRN69 | R | TCCGTATCTTTTACGCAGCGG | aphIII |
| oliRN110 | F | CGCTGCGTAAAAGATACG | aphIII |
| oliRN111 | R | AATGGAGTGTCTTCTTCC | aphIII |
| oliRN133 | F | TATTCGCTTAGAAAATTAA | emm |
| oliRN134 | R | GCAAGTTCTTCAGCTTGTTT | emm |
| oliRN39 | F | AAAAATGACATTGCTTGGGC | hki |
| oliRN40 | R | TTTAGTAGATATACTTTATTGGC | hki |
| oliRN233 | F | AGATTGGATATCACAGG | speH |
| oliRN234 | R | CTATTCTCTCGTTATTGG | speH |
| oliRN237 | F | GTGTAGAATTGAGGTAATTG | ssa |
| oliRN238 | R | TAATATAGCCTGTCTCGTAC | ssa |
| OLEC141 | F | AACAAGTCACTGTTCAAG | ssa |
| OLEC142 | R | GCTACAACGAGTATTCTT | ssa |
| OLEC166 | F | GTACGAGACAGGCTATAT | ssa |
| OLEC211 | F | TGCACAATTATTATCGATTAG | ssa |
| OLEC153 | F | TAACTTGGATCCGTTCAATAGTAAAGATTTAGCTGC | Insertion of aphIII into ss |
| OLEC154 | R | TTCTTACTGCAGGCTGACCTGTGGATCTTACATTAG | Insertion of aphIII into ss |
| OLEC155 | F | AAGTAAGCATGCACCCAATATTTTTCGTTAGTGAAG | Insertion of aphIII into ss |
| OLEC159 | R | TATGAAGGTACCTTAACTAGTATCATGGATTTGATAC | Insertion of aphIII into ss |
| OLEC179 | F | TTGCTCATGCGCAATTCG | Spy1278 |
| OLEC497 | R | AATATTGGGTGTTTTAGAAGCC | Spy1278 |
| OLEC178 | R | ATCGCTTGCTTAAACTCG | prx |
| OLEC395 | F | ATGGAGAAGTTGTGACTAAGG | prx |
| OLEC396 | F | TGACAATGGATATATCGCAGG | prx |
| OLEC156 | R | TTAACTAGTATCATGGAT | Intergenic ssa-prx |
| OLEC167 | R | CAATTACCTCAATTCTAC | Intergenic ssa-Spy1278 |
| OLEC267 | R | AAGAACCTTGTCGCTATC | Intergenic attR-Spy1275 |
| OLEC325 | R | GATGACTGACCATTATAG | Intergenic attR-Spy1275 |
| OLEC324 | R | GCTACAAAATAACGAAAC | Intergenic attR-prx |
| OLEC500 | F | AGAAGCAACTGAATTGA | Spy1329 |
NOTE. The underlined sequences indicate restriction sites. Spy1275, Spy1278, and Spy1329 refer to annotated open reading frames in strain MGAS10750F. F, forward; prx, paratox gene; R, reverse.
Construction of  149::Kmr
149::Kmr
A kanamycin-resistance (Kmr) cassette, aphIII, was inserted into the ssa gene of phage  149 in strain RDN149, thus generating strain EC516, which carried recombinant phage
149 in strain RDN149, thus generating strain EC516, which carried recombinant phage 149::Kmr. For this purpose, an 856-bp fragment upstream of ssa and a 569-bp fragment downstream of ssa were amplified by use of RDN149 genomic DNA and primers (containing flanking restriction sites) OLEC154/OLEC155 and OLEC153/OLEC159, respectively. After digestion with appropriate restriction enzymes, fragments were ligated and cloned into pEC61, a suicide plasmid for GAS. The resulting plasmid, pEC97, was digested with ScaI (which interrupts the ampicillin-resistance cassette) and used to transform electrocompetent RDN149. Kmr clones were then selected and analyzed by polymerase chain reaction (PCR) and Southern blot analyses. This ensured that the recombination event by double crossover was correct and did not affect the DNA regions located upstream and downstream of ssa
149::Kmr. For this purpose, an 856-bp fragment upstream of ssa and a 569-bp fragment downstream of ssa were amplified by use of RDN149 genomic DNA and primers (containing flanking restriction sites) OLEC154/OLEC155 and OLEC153/OLEC159, respectively. After digestion with appropriate restriction enzymes, fragments were ligated and cloned into pEC61, a suicide plasmid for GAS. The resulting plasmid, pEC97, was digested with ScaI (which interrupts the ampicillin-resistance cassette) and used to transform electrocompetent RDN149. Kmr clones were then selected and analyzed by polymerase chain reaction (PCR) and Southern blot analyses. This ensured that the recombination event by double crossover was correct and did not affect the DNA regions located upstream and downstream of ssa
Phage induction by mitomycin C treatment
Mitomycin C (Sigma) was added (0.2 μg/mL) to cultures grown to early–mid-logarithmic phase [24, 41]. After incubation for 3 hat 37°C, cells were removed by centrifugation and supernatants were sterile-filtered through 0.45-μm pores (Iwaki Glass). Possible bacterial contamination of filtrates was excluded by inoculating an aliquot of the filtrates in liquid and on solid media.
Detection of phage release in mitomycin C–treated bacterial cultures
Two approaches were used. (1) A phage indicator strain culture grown to mid-logarithmic phase was mixed with sterile water, poured over THY agar plates, and allowed to sediment. Superfluous bacterial suspension was carefully removed and plates were left to dry. Supernatants of mitomycin C–treated phage donor cultures were then spotted on the indicator lawn. (2) Supernatants of mitomycin C–treated phage donor cultures were mixed with cultures of phage indicator strain and added to molten top agar. The mixture was then poured over THY agar plates and allowed to dry. In both approaches, the final step was to incubate the plates at 37°C until plaques were observed.
Lysogenization of recipient strains with phage  149
149
By use of the first approach described above, secondary growth colonies were recovered from the areas of lysis and analyzed for sensitivity to phage infection. Lysogenized strains for which no lysis was observed were then selected and analyzed by PCR and Southern blot analysis for the presence of the phage.
Determination of the host range of  149::Kmr
149::Kmr
Supernatants of mitomycin C–treated EC516 cultures were mixed with recipient cultures grown to early–mid-logarithmic phase and further incubated for 3 hours at 37°C. Aliquots of the mixture were then plated on tryptic soy agar that contained kanamycin. Selected Kmr-positive colonies were analyzed by PCR and Southern blot analysis to determine whether the recombinant phage was present. Lysogenization was confirmed for 2–3 independent lysogens per infection experiment. Lysogenization experiments were performed independently 3–4 times.
RESULTS
Phage release after mitomycin C treatment of an M12 clinical isolate
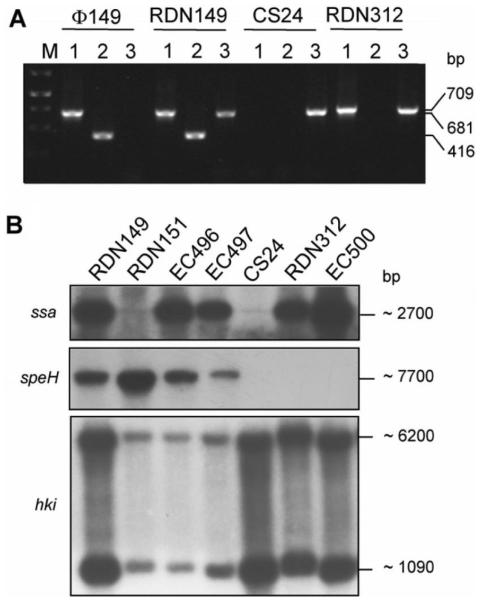
The analysis of a type M12 clinical GAS isolate, RDN149, recovered from a patient with STSS, revealed the superantigen genes ssa and speH (table 1). Subsequent treatment of a bacterial culture of RDN149 with mitomycin C resulted in the release of phage particles in the culture supernatant, which could be demonstrated by plaque detection with the phage indicator strains CS24 and RDN151 (M12 types). The plaques obtained with both indicator strains were turbid, ~2 mm in diameter, and the bacteriophage titer was estimated at 7 × 104 pfu/mL (data not shown). The presence of phage particles in the supernatant of mitomycin C–induced RDN149 cultures was further confirmed by isolation of phage DNA. In contrast, no phage DNA could be detected in the supernatant of cultures that were not subjected to mitomycin C treatment (data not shown). Further characterization of the released prophage(s) by PCR analysis indicated the possible presence of 2 phages in the supernatant (figure 1A).
Figure 1.
Gel electrophoresis patterns demonstrating ex vivo lysogenic transfer of ssa-3– carrying phage  149 to group A Streptococcus (GAS) clinical isolates of type M12. A, Results of polymerase chain reaction analysis of isolated phage DNA from strain RDN149 and genomic DNA from phage donor strain RDN149, phage indicator strain CS24, and lysogenized strain RDN312 with primers specific to the ssa gene (lane 1), the speH gene (lane 2) and the hki gene (lane 3). hki encoding a putative histidine kinase served as positive control for GAS genomic DNA. M, 1-kb DNA ladder (Fermentas Life Sciences). The 709-, 681-, and 416-bp DNA fragments correspond to the ssa-, hki-, and speH-specific amplified products, respectively. B, Results of Southern blot analysis of HindIII-digested genomic DNA of phage donor strain RDN149; phage indicator strains RDN151 and CS24; and lysogenized strains EC496, EC497, RDN312, and EC500, with α-32P labeled DNA probes specific to the ssa, speH, and hki genes. The approximate sizes of the hybridizing fragments are indicated in bp.
149 to group A Streptococcus (GAS) clinical isolates of type M12. A, Results of polymerase chain reaction analysis of isolated phage DNA from strain RDN149 and genomic DNA from phage donor strain RDN149, phage indicator strain CS24, and lysogenized strain RDN312 with primers specific to the ssa gene (lane 1), the speH gene (lane 2) and the hki gene (lane 3). hki encoding a putative histidine kinase served as positive control for GAS genomic DNA. M, 1-kb DNA ladder (Fermentas Life Sciences). The 709-, 681-, and 416-bp DNA fragments correspond to the ssa-, hki-, and speH-specific amplified products, respectively. B, Results of Southern blot analysis of HindIII-digested genomic DNA of phage donor strain RDN149; phage indicator strains RDN151 and CS24; and lysogenized strains EC496, EC497, RDN312, and EC500, with α-32P labeled DNA probes specific to the ssa, speH, and hki genes. The approximate sizes of the hybridizing fragments are indicated in bp.
Ex vivo lysogeny of phage  149 associated with ssa-toxigenic conversion
149 associated with ssa-toxigenic conversion
To investigate whether phage(s) released from strain RDN149 could convert speH-negative and ssa-negative strains into speH-positive and/or ssa-positive strains, lysogenization experiments involving the phage indicator strain CS24 with mitomycin C–induced supernatants of RDN149 were conducted. Each lysogenization experiment performed was positively controlled with  CS112 lysogenization of recipient strain CS24 with strain CS112 used as phage donor [24]. Lysogenic clones were obtained successfully and further analyzed for transfer of the superantigen genes. PCR analysis of genomic DNA from the phage donor strain (RDN149), phage indicator strain (CS24), and lysogenized CS24 strains (RDN312 and EC500), as well as purified phage DNA from RDN149, showed that only ssa, not speH, was transferred to the phage indicator strain (figure 1A).
CS112 lysogenization of recipient strain CS24 with strain CS112 used as phage donor [24]. Lysogenic clones were obtained successfully and further analyzed for transfer of the superantigen genes. PCR analysis of genomic DNA from the phage donor strain (RDN149), phage indicator strain (CS24), and lysogenized CS24 strains (RDN312 and EC500), as well as purified phage DNA from RDN149, showed that only ssa, not speH, was transferred to the phage indicator strain (figure 1A).
These results, confirmed by Southern blot analysis, suggested that in RDN149, speH and ssa are encoded on 2 distinct prophages (figure 1B). The transferable, lysogenic, ssa-carrying phage was designated  149. To further verify that lysogeny was responsible for the observed ssa-toxigenic conversion, we repeated the experiments with the second phage indicator strain, RDN151. Toxigenic conversion of RDN151 with
149. To further verify that lysogeny was responsible for the observed ssa-toxigenic conversion, we repeated the experiments with the second phage indicator strain, RDN151. Toxigenic conversion of RDN151 with  149 was also observed (figure 1). Thus, we have identified ssa-carrying phage
149 was also observed (figure 1). Thus, we have identified ssa-carrying phage  149 in strain RDN149, which can convert ssa-negative clinical strains of type M12 to ssa-positive strains via lysogenization.
149 in strain RDN149, which can convert ssa-negative clinical strains of type M12 to ssa-positive strains via lysogenization.
Characterization of the ssa-carrying phage  149
149
The superantigen SSA shares considerable homology with the S. aureus enterotoxins SEB and SEC [33, 42]. Three alleles of ssa have already been identified in natural GAS populations [35]. The alleles ssa-1 and ssa-3 differ by a single synonymous substitution in codon 94, and both encode SSA-1. The ssa-2 allele is identical to ssa-3 at codon 94, but has a nonsynonymous substitution at codon 28, which changes the second amino acid of the mature protein from serine to arginine [35]. PCR amplification of the ssa coding sequence in RDN149 with primers oliRN237 and OLEC156, followed by sequencing, revealed the ssa-3 allele. To determine the attachment site of  149 and its location on the chromosome of strain RDN149, we took advantage of the fact that, in all GAS prophages, the genes encoding virulence factors are located at one extremity of the prophage, between the phage lysis cassette and the phage attachment site [12]. At the time of the study, 2 GAS prophages,
149 and its location on the chromosome of strain RDN149, we took advantage of the fact that, in all GAS prophages, the genes encoding virulence factors are located at one extremity of the prophage, between the phage lysis cassette and the phage attachment site [12]. At the time of the study, 2 GAS prophages,  315.2 (strain MGAS315 [M3 type]) and
315.2 (strain MGAS315 [M3 type]) and  SPsP6 (strain SSI-1 [M3 type]), had already been described that carried the ssa gene (ssa-1 allele) [7, 10].
SPsP6 (strain SSI-1 [M3 type]), had already been described that carried the ssa gene (ssa-1 allele) [7, 10].
Southern blot analysis of HindIII-digested genomic DNA of strain RDN149 revealed an ssa-hybridizing DNA fragment, the size of which (~2700 bp) was different from that expected in strains MGAS315 and SSI-1 (5090 bp) (figure 1). There are several possible explanations for this result: (1)  149 is identical to
149 is identical to  315.2 or
315.2 or  SPsP6 but is integrated at a different attachment site; (2)
SPsP6 but is integrated at a different attachment site; (2)  149 is identical to
149 is identical to  315.2 or
315.2 or  SPsP6 and is integrated at the same attachment site, but the chromosomal region surrounding the attachment sites differs among strains; or (3)
SPsP6 and is integrated at the same attachment site, but the chromosomal region surrounding the attachment sites differs among strains; or (3)  149 is a novel GAS prophage that has not yet been characterized.
149 is a novel GAS prophage that has not yet been characterized.
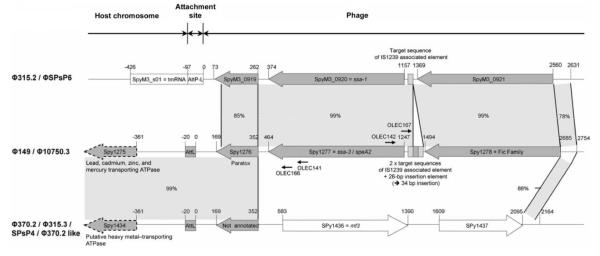
We performed inverse PCR using religated, HindIII-digested genomic fragments of strain RDN149 as DNA templates and ssa-specific outward primers OLEC141 and OLEC142 (table 2; figure 2). The PCR fragment obtained was purified and used as a DNA template in PCR reactions with embedded primers OLEC166 and OLEC167 (table 2; figure 2). Sequence analysis of the resulting PCR fragment indicated that  149 consisted of a chimeric GAS prophage. The downstream region of the ssa coding sequence, which includes a hypothetical phage protein referred to as paratox, the left
149 consisted of a chimeric GAS prophage. The downstream region of the ssa coding sequence, which includes a hypothetical phage protein referred to as paratox, the left  149 attachment site, and the adjacent sequence on the bacterial chromosome showed identity to the DNA region upstream of the streptodornase gene (mf3)in4 GAS prophages that have already been described:
149 attachment site, and the adjacent sequence on the bacterial chromosome showed identity to the DNA region upstream of the streptodornase gene (mf3)in4 GAS prophages that have already been described:  370.2 (SF370 [M1]),
370.2 (SF370 [M1]),  SPsP4 (SSI-1 [M3]),
SPsP4 (SSI-1 [M3]),  315.3 (MGAS315 [M3]), and
315.3 (MGAS315 [M3]), and  370.2-like prophage (MGAS8232 [M18]) (figure 2) [7, 8, 10, 11].
370.2-like prophage (MGAS8232 [M18]) (figure 2) [7, 8, 10, 11].
Figure 2.
Comparison of the att-L proximal region of ssa-3–carrying phage  149 with the att-L proximal regions of ssa-1 –carrying phages
149 with the att-L proximal regions of ssa-1 –carrying phages  315.2 (MGAS315; M3) and SPsP6 (SSI-1, M3) and mf3-carrying phages
315.2 (MGAS315; M3) and SPsP6 (SSI-1, M3) and mf3-carrying phages  315.3 (MGAS315; M3), SPsP4 (SSI-1; M3),
315.3 (MGAS315; M3), SPsP4 (SSI-1; M3),  370.2 (SF370; M1), and
370.2 (SF370; M1), and  370.2-like (MGAS8232; M18). The shaded areas indicate the regions of similarity with the percentage of nucleotide identity given. The 34 bp IS1239-related sequence located 103 bp upstream of the ssa coding sequence is shown. Annotated open reading frames refer to the genomes of MGAS315 strain (
370.2-like (MGAS8232; M18). The shaded areas indicate the regions of similarity with the percentage of nucleotide identity given. The 34 bp IS1239-related sequence located 103 bp upstream of the ssa coding sequence is shown. Annotated open reading frames refer to the genomes of MGAS315 strain ( 315.2) and SF370 (
315.2) and SF370 ( 370.2). Small, black arrows, positions of primers.
370.2). Small, black arrows, positions of primers.
All 4 prophages, and therefore also  149, are integrated into the GAS bacterial genome at the same attachment site, which is located 341 bp upstream of a putative heavy metal–transporting, ATPase-encoding gene. Nucleotide sequence analysis of the upstream region of ssa-3 in
149, are integrated into the GAS bacterial genome at the same attachment site, which is located 341 bp upstream of a putative heavy metal–transporting, ATPase-encoding gene. Nucleotide sequence analysis of the upstream region of ssa-3 in  149 revealed 99% identity with the upstream region of ssa-1 in
149 revealed 99% identity with the upstream region of ssa-1 in  315.2 (MGAS315 [M3]) and
315.2 (MGAS315 [M3]) and  SPsP6 (SSI-1 [M3]). The difference consists of a 34-bp insertion located 103 bp upstream of the ssa-3 start codon, which is not present upstream of ssa-1. This 34-bp insertion comprises a 26-bp insertion element (5′- CTCTTTTAAAATTAAAACATTGATTT-3′), which doubles the sequence 5′-AATTTTAT-3′. The 26-bp insertion element was previously identified upstream of the insertion sequence IS1239 and, on this basis, it was suggested that it may function as a target sequence [42]. It was shown to be more frequently associated with ssa-3 than with ssa-1 or ssa-2 [35]. In addition, the presence of IS1239 associated with the 26-bp insertion element was identified upstream of ssa-3 in several serotypes—M4, M15, M23, M33 and M41—leading to the suggestion that IS1239 might have contributed to the horizontal transfer of ssa among GAS [35]. Taken together, these observations suggest that recombination events may have taken place, leading to the mosaic nature of
SPsP6 (SSI-1 [M3]). The difference consists of a 34-bp insertion located 103 bp upstream of the ssa-3 start codon, which is not present upstream of ssa-1. This 34-bp insertion comprises a 26-bp insertion element (5′- CTCTTTTAAAATTAAAACATTGATTT-3′), which doubles the sequence 5′-AATTTTAT-3′. The 26-bp insertion element was previously identified upstream of the insertion sequence IS1239 and, on this basis, it was suggested that it may function as a target sequence [42]. It was shown to be more frequently associated with ssa-3 than with ssa-1 or ssa-2 [35]. In addition, the presence of IS1239 associated with the 26-bp insertion element was identified upstream of ssa-3 in several serotypes—M4, M15, M23, M33 and M41—leading to the suggestion that IS1239 might have contributed to the horizontal transfer of ssa among GAS [35]. Taken together, these observations suggest that recombination events may have taken place, leading to the mosaic nature of  149.
149.
Determination of host range of  149 among clinical isolates of GAS
149 among clinical isolates of GAS
We were interested in the possibility that  149 could be transferred ex vivo to clinical isolates of GAS. As described above, we showed transfer of
149 could be transferred ex vivo to clinical isolates of GAS. As described above, we showed transfer of  149 from the donor strain RDN149 (type M12) to the phage indicator strains CS24 and RDN151 (type M12). To facilitate the detection of toxin-converted strains, a selective marker (the kanamycin-resistance cassette, apiII) was inserted into the ssa-3 coding sequence of
149 from the donor strain RDN149 (type M12) to the phage indicator strains CS24 and RDN151 (type M12). To facilitate the detection of toxin-converted strains, a selective marker (the kanamycin-resistance cassette, apiII) was inserted into the ssa-3 coding sequence of  149, thus creating strain EC516. Strain EC516 was then used as a donor strain of
149, thus creating strain EC516. Strain EC516 was then used as a donor strain of  149::Kmr in infection experiments involving other GAS clinical isolates of various M serotypes. Lysogenic strains were selected on the basis of their Kmr phenotype, which was verified by PCR scoring for the presence of the Kmr cassette flanked by the interrupted ssa-3 sequence and further verified by subsequent Southern blot analysis. In this study, 34 clinical isolates, which represented 12 M types, were analyzed for lysogenic conversion. A total of 17 strains that comprised 7 different M types and were classified as pattern A-C/SOF- strains (M1, M3, M5, M12, and M19) and pattern E/SOF[H11001] strains (M28 and M94) [43, 44] were successfully lysogenized (table 3; figure 3). Further, the transfer of
149::Kmr in infection experiments involving other GAS clinical isolates of various M serotypes. Lysogenic strains were selected on the basis of their Kmr phenotype, which was verified by PCR scoring for the presence of the Kmr cassette flanked by the interrupted ssa-3 sequence and further verified by subsequent Southern blot analysis. In this study, 34 clinical isolates, which represented 12 M types, were analyzed for lysogenic conversion. A total of 17 strains that comprised 7 different M types and were classified as pattern A-C/SOF- strains (M1, M3, M5, M12, and M19) and pattern E/SOF[H11001] strains (M28 and M94) [43, 44] were successfully lysogenized (table 3; figure 3). Further, the transfer of  149::Kmr from selected M1, M12, and M94 lysogens back to clinical isolates of type M12 (RDN151 and RDN138) was obtained. This indicated that the recombinant phage was still functional once inserted in the chromosome of the lysogenic strains. In addition, infection experiments with UV-induced and hydrogen peroxide (H2O2)–induced EC516 culture supernatants led successfully to lysogenic clones among M12 strains (data not shown).
149::Kmr from selected M1, M12, and M94 lysogens back to clinical isolates of type M12 (RDN151 and RDN138) was obtained. This indicated that the recombinant phage was still functional once inserted in the chromosome of the lysogenic strains. In addition, infection experiments with UV-induced and hydrogen peroxide (H2O2)–induced EC516 culture supernatants led successfully to lysogenic clones among M12 strains (data not shown).
Table 3.
Ex vivo lysogenic transfer of  149::Kmr between streptococcal species.
149::Kmr between streptococcal species.
| Group, M type | Clinical isolates, no. |
Serum opacity factor |
Lysogenizationa with  149::Kmr 149::Kmr |
|
|---|---|---|---|---|
| Positive | Negative | |||
| Group A Streptococcus | ||||
| M1 | 7 | − | 1 | 6 |
| M3 | 5 | − | 2 | 3 |
| M5 | 1 | − | 1 | 0 |
| M12 | 8 | + | 8 | 0 |
| M19 | 1 | − | 1 | 0 |
| M22 | 1 | + | 0 | 1 |
| M28 | 3 | + | 2 | 1 |
| M49 | 2 | + | 0 | 2 |
| M81 | 1 | + | 0 | 1 |
| M83 | 1 | − | 0 | 1 |
| M89 | 2 | + | 0 | 2 |
| M94 | 2 | + | 2 | 0 |
| Group C Streptococcus | 10 | … | 2 | 8 |
| Group G Streptococcus | 10 | … | 0 | 10 |
Each lysogenization experiment was performed at least 3 times independently. Lysogens were analyzed by polymerase chain reaction and Southern blot analysis for the presence of the recombinant phage.
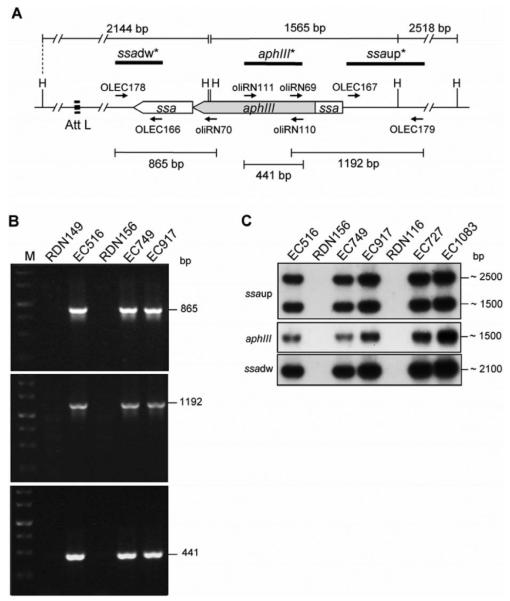
Figure 3.
Analysis of the broad host range of recombinant phage  149::Kmr. A, Representative restriction map of the att-L proximal region of
149::Kmr. A, Representative restriction map of the att-L proximal region of  149::Kmr. Genes are drawn to scale. Small, black arrows, positions of primers. Sizes of PCR and hybridizing restriction fragments are indicated in bp. B, Results of polymerase chain reaction analysis of genomic DNA from
149::Kmr. Genes are drawn to scale. Small, black arrows, positions of primers. Sizes of PCR and hybridizing restriction fragments are indicated in bp. B, Results of polymerase chain reaction analysis of genomic DNA from  149 donor strain RDN149,
149 donor strain RDN149,  149::Kmr donor strain EC516, recipient strain RDN156 (M28), and lysogenized strains EC749 and EC917, with primers OLEC178 and oliRN70 (top), oliRN69 and OLEC179 (middle), and oliRN111 and oliRN110 (bottom). M, 1-kb DNA ladder (Fermentas Life Sciences). C, Results of Southern blot analysis of HindIII-digested genomic DNA from
149::Kmr donor strain EC516, recipient strain RDN156 (M28), and lysogenized strains EC749 and EC917, with primers OLEC178 and oliRN70 (top), oliRN69 and OLEC179 (middle), and oliRN111 and oliRN110 (bottom). M, 1-kb DNA ladder (Fermentas Life Sciences). C, Results of Southern blot analysis of HindIII-digested genomic DNA from  149::Kmr donor strain EC516, phage recipient strain RDN156 (M28) and corresponding lysogenized strains EC749 and EC917, phage recipient strain RDN116 (M94) and corresponding lysogenized strains EC727 and EC1083 with α-32P labelled DNA probes specific to the ssa upstream fragment (ssaup), aphIII gene, and ssa downstream fragment (ssadw).
149::Kmr donor strain EC516, phage recipient strain RDN156 (M28) and corresponding lysogenized strains EC749 and EC917, phage recipient strain RDN116 (M94) and corresponding lysogenized strains EC727 and EC1083 with α-32P labelled DNA probes specific to the ssa upstream fragment (ssaup), aphIII gene, and ssa downstream fragment (ssadw).
Ex vivo lysogenic transfer  149 to other streptococcal species
149 to other streptococcal species
Several studies report the presence of ssa-3 among the emerging human pathogens GCSE and GGSE, thus suggesting a temporal dynamic for ssa-3 exchange across streptococcal species [14]. Here, we asked whether  149 could be lysogenically transferred ex vivo from GAS to other streptococcal species. Infection experiments were conducted that used the
149 could be lysogenically transferred ex vivo from GAS to other streptococcal species. Infection experiments were conducted that used the  149::Kmr phage donor strain EC516 and 10 GCSE clinical isolates and 10 GGSE clinical isolates as recipient strains, by use of the same approach described above. Two GCSE strains were successfully lysogenized (table 3).
149::Kmr phage donor strain EC516 and 10 GCSE clinical isolates and 10 GGSE clinical isolates as recipient strains, by use of the same approach described above. Two GCSE strains were successfully lysogenized (table 3).  149::Kmr was also transferred from selected GCSE lysogens back to clinical isolates of type M12 (RDN151 and RDN138). Taken together, the ability of
149::Kmr was also transferred from selected GCSE lysogens back to clinical isolates of type M12 (RDN151 and RDN138). Taken together, the ability of  149:: Kmr to lysogenize GAS strains of different clonal lineages and different M types as well as GCSE isolates indicate that phage transfer most probably played a critical role in mediating the spread of ssa across the streptococcal population.
149:: Kmr to lysogenize GAS strains of different clonal lineages and different M types as well as GCSE isolates indicate that phage transfer most probably played a critical role in mediating the spread of ssa across the streptococcal population.
DISCUSSION
The recent reemergence of severe, invasive GAS diseases observed during the mid-1980s has encouraged many researchers to investigate this phenomenon. Some studies questioned whether enhanced virulence could be associated with a particular M type or with certain virulence factors. Reports show the existence of particular virulent and invasive clones that have successfully spread worldwide [3, 4, 7, 45]. However, despite the dominance of certain M types, such as M1, M3, M12, and M28, among endemic and invasive GAS isolates, no exclusive link between M type and specific diseases could be demonstrated. Only certain M types seem to exhibit a correlation with invasive diseases (e.g., M1 and M3) or postinfectious sequelae from superficial GAS infection (e.g., M18) [11, 29, 30, 45]. Furthermore, invasive GAS isolates do not seem to be characterized by a common toxin-gene profile. Although reports show the dominance of 1 or 2 toxin-gene profiles in all M types, strains that did not share the predominant profile still showed nonrandom distribution of key toxin genes that were characteristic of the M type [29].
Among bacterial factors that can determine enhanced virulence are superantigen-carrying bacteriophages that can, via lysogenic transfer, convert a nontoxinogenic strain to a toxin-producing strain [19]. In this study, the analysis of a clinical GAS isolate, type M12, to determine its superantigen-encoding bacteriophage profile uncovered prophage  149, which carried the allele ssa-3 that encodes the streptococcal superantigen SSA. Sequence analysis of the att-L proximal region of
149, which carried the allele ssa-3 that encodes the streptococcal superantigen SSA. Sequence analysis of the att-L proximal region of  149 indicated that the phage was a mosaic prophage that shared characteristics of 2 different types of prophages:
149 indicated that the phage was a mosaic prophage that shared characteristics of 2 different types of prophages:  370.2,
370.2,  SPsP4,
SPsP4,  315.3, and
315.3, and  370.2–like phages (already identified in M1, M3, and M18 strains) and
370.2–like phages (already identified in M1, M3, and M18 strains) and  315.2 and SPsP6 phages (M3 strains). This indicates that phage genomic rearrangements occurred, leading to genetic mosaicism in
315.2 and SPsP6 phages (M3 strains). This indicates that phage genomic rearrangements occurred, leading to genetic mosaicism in  149. Recently, the complete genome sequence of an M4-type strain, MGAS10750, revealed that the strain harbors an ssa-3– carrying prophage,
149. Recently, the complete genome sequence of an M4-type strain, MGAS10750, revealed that the strain harbors an ssa-3– carrying prophage,  10750.3 [46], whose att-L proximal region is 100% identical to that of
10750.3 [46], whose att-L proximal region is 100% identical to that of  149 (figure 2). The contribution that the genetic mosaicism of GAS prophages makes to the emergence and diversification of the globally disseminated clonal M1T1 strain has already been demonstrated, indicating the exchange of genetic material among GAS prophages [1, 3, 12].
149 (figure 2). The contribution that the genetic mosaicism of GAS prophages makes to the emergence and diversification of the globally disseminated clonal M1T1 strain has already been demonstrated, indicating the exchange of genetic material among GAS prophages [1, 3, 12].
Superantigen SSA produced by GAS shares greater homology with staphylococcal enterotoxins SEB and SEC than it does with streptococcal superantigens. Several studies during the past decade have further documented the presence of the ssa gene among GAS isolates of different M types, as well as in emerging clinical isolates of GCSE and GGSE [14, 29, 30, 32–35]. In our study, we performed infection experiments on GAS clinical isolates of various M types, as well as experiments on GCSE and GGSE clinical isolates using mitomycin C–induced cell-free culture supernatants of strain EC516 harboring the selective recombinant phage  149::Kmr. Our results show the ability of
149::Kmr. Our results show the ability of  149:Kmr to lysogenize GAS clinical isolates of 7 different M types (M1, M3, M5, M12, M19, M28, and M94), as well as the GCSE clinical isolates. To our knowledge, this is the first direct experimental evidence for toxigenic conversion via lysogeny across a diverse range of strains of distinct M types and between GAS and GCSE. However, no lysogenic conversion of GGSE clinical isolates could be observed. We hypothesize that such events could take place, but the experimental conditions in our study might have limited their detection.
149:Kmr to lysogenize GAS clinical isolates of 7 different M types (M1, M3, M5, M12, M19, M28, and M94), as well as the GCSE clinical isolates. To our knowledge, this is the first direct experimental evidence for toxigenic conversion via lysogeny across a diverse range of strains of distinct M types and between GAS and GCSE. However, no lysogenic conversion of GGSE clinical isolates could be observed. We hypothesize that such events could take place, but the experimental conditions in our study might have limited their detection.
Although some studies indicate that M proteins could function as barriers to horizontal gene exchange, recent genomic sequence analysis strongly suggests that extensive lateral gene transfer occurred among GAS isolates of different M types [1]. Along these lines, lysogenic conversion of 1 T25-type strain with SpeC toxin originating from an M12-type isolate was described; however, the lysogenic transfer event was not confirmed by DNA analysis [31]. Reports on intergroup streptococcal bacteriophage transfer remain limited. Intergroup transduction of markers for streptomycin and bacitracin resistance between GCS and GAS using A-25 lytic phage and propagation of a temperate phage isolated from GGS on either GAS or GCS have been reported [47, 48]. GCSE and GGSE, although generally considered to be commensal organisms, have also been isolated from infections in humans that had clinical manifestations similar to those caused by GAS. In particular, reports of GCSE and GGSE isolates recovered from patients with severe forms of infection and of the presence of superantigen genes in these isolates have increased in recent years [49]. Our data strongly support the hypothesis that the emergence of superantigen genes in GCSE and GGSE may occur by means of horizontal transfer from GAS via temperate phage. Accordingly, it has been suggested that housekeeping genes are exchanged among GCSE, GGSE, and GAS via lateral transfer with a strong net directionality of gene movement from GAS donors to GCS and GGS recipients, but only on the basis of phylogenetic studies [50]. Our experimental findings raise questions about the extent to which M type and streptococcal species are barriers to the horizontal transfer of lysogenic phages, as well as questions about the assumption of phage specificity within a given streptococcal group.
Given the critical role of temperate bacteriophages in toxi-genic conversion and in the mediation of strain diversity and evolution in GAS, it will be important to obtain additional knowledge of the molecular mechanisms underlying these events [19, 23]. Further investigation is also needed to unravel the complex interplay between strain genotype, in vivo toxin-gene transfer, regulation of toxin expression, and host factors (i.e., genetics and underlying conditions) that is responsible for the development of severe streptococcal disease.
Acknowledgments
We thank Dr. Schlievert for his gift of the  CS112 phage donor strain CS112 and indicator strain CS24; Dr. Garnier and Dr. Ploy for their gift of group B, C, and G streptococcal clinical isolates; and Dr. Dunny for his gift of vector pMSP3535. We also thank Dr. Eckert for her help with sequence analyses; Dr. Bläsi, Dr. Novak, and Dr. Witte for helpful discussions; and Dr. Normark for critical reading of the manuscript.
CS112 phage donor strain CS112 and indicator strain CS24; Dr. Garnier and Dr. Ploy for their gift of group B, C, and G streptococcal clinical isolates; and Dr. Dunny for his gift of vector pMSP3535. We also thank Dr. Eckert for her help with sequence analyses; Dr. Bläsi, Dr. Novak, and Dr. Witte for helpful discussions; and Dr. Normark for critical reading of the manuscript.
Financial support: Austrian Science Fund Project (grant P17238-B09 to E.C.); Austrian National Bank (grant 10802 to E.C.); Higher Education Commission of Pakistan (scholarship to Z.A.P.); Austrian Exchange Service (scholarship to R.P.J.).
Footnotes
Potential conflicts of interest: none reported.
Presented in part: 7th American Society for Microbiology Conference on Streptococcal Genetics, Saint-Malo, France, 18 –21 June 2006 (abstract A126).
References
- 1.Banks DJ, Beres SB, Musser JM. The fundamental contribution of phages to GAS evolution, genome diversification and strain emergence. Trends Microbiol. 2002;10:515–21. doi: 10.1016/s0966-842x(02)02461-7. [DOI] [PubMed] [Google Scholar]
- 2.Cunningham MW. Pathogenesis of group A streptococcal infections. Clin Microbiol Rev. 2000;13:470–511. doi: 10.1128/cmr.13.3.470-511.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aziz RK, Edwards RA, Taylor WW, Low DE, McGeer A, Kotb M. Mosaic prophages with horizontally acquired genes account for the emergence and diversification of the globally disseminated M1T1 clone of Streptococcus pyogenes. J Bacteriol. 2005;187:3311–8. doi: 10.1128/JB.187.10.3311-3318.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aziz RK, Ismail SA, Park HW, Kotb M. Post-proteomic identification of a novel phage-encoded streptodornase, Sda1, in invasive M1T1 Streptococcus pyogenes. Mol Microbiol. 2004;54:184–97. doi: 10.1111/j.1365-2958.2004.04255.x. [DOI] [PubMed] [Google Scholar]
- 5.Kotb M, Norrby-Teglund A, McGeer A. An immunogenetic and molecular basis for differences in outcomes of invasive group A streptococcal infections. Nat Med. 2002;8:1398–404. doi: 10.1038/nm1202-800. [DOI] [PubMed] [Google Scholar]
- 6.Banks DJ, Porcella SF, Barbian KD, et al. Progress toward characterization of the group A Streptococcus metagenome: complete genome sequence of a macrolide-resistant serotype M6 strain. J Infect Dis. 2004;190:727–38. doi: 10.1086/422697. [DOI] [PubMed] [Google Scholar]
- 7.Beres SB, Sylva GL, Barbian KD, et al. Genome sequence of a serotype M3 strain of group A Streptococcus: phage-encoded toxins, the high-virulence phenotype, and clone emergence. Proc Natl Acad Sci USA. 2002;99:10078–83. doi: 10.1073/pnas.152298499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ferretti JJ, McShan WM, Ajdic D, et al. Complete genome sequence of an M1 strain of Streptococcus pyogenes. Proc Natl Acad Sci USA. 2001;98:4658–63. doi: 10.1073/pnas.071559398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Green NM, Zhang S, Porcella SF, et al. Genome sequence of a serotype M28 strain of group A Streptococcus: potential new insights into puerperal sepsis and bacterial disease specificity. J Infect Dis. 2005;192:760–70. doi: 10.1086/430618. [DOI] [PubMed] [Google Scholar]
- 10.Nakagawa I, Kurokawa K, Yamashita A, et al. Genome sequence of an M3 strain of Streptococcus pyogenes reveals a large-scale genomic rearrangement in invasive strains and new insights into phage evolution. Genome Res. 2003;13:1042–55. doi: 10.1101/gr.1096703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smoot JC, Barbian KD, Van Gompel JJ, et al. Genome sequence and comparative microarray analysis of serotype M18 group A Streptococcus strains associated with acute rheumatic fever outbreaks. Proc Natl Acad Sci USA. 2002;99:4668–73. doi: 10.1073/pnas.062526099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brussow H, Canchaya C, Hardt WD. Phages and the evolution of bacterial pathogens: from genomic rearrangements to lysogenic conversion. Microbiol Mol Biol Rev. 2004;68:560–602. doi: 10.1128/MMBR.68.3.560-602.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Broudy TB, Fischetti VA. In vivo lysogenic conversion of Tox(−) Streptococcus pyogenes to Tox(+) with lysogenic streptococci or free phage. Infect Immun. 2003;71:3782–6. doi: 10.1128/IAI.71.7.3782-3786.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Igwe EI, Shewmaker PL, Facklam RR, Farley MM, van Beneden C, Beall B. Identification of superantigen genes speM, ssa, and smeZ in invasive strains of beta-hemolytic group C and G streptococci recovered from humans. FEMS Microbiol Lett. 2003;229:259–64. doi: 10.1016/S0378-1097(03)00842-5. [DOI] [PubMed] [Google Scholar]
- 15.Kapur V, Nelson K, Schlievert PM, Selander RK, Musser JM. Molecular population genetic evidence of horizontal spread of two alleles of the pyrogenic exotoxin C gene (speC) among pathogenic clones of Streptococcus pyogenes. Infect Immun. 1992;60:3513–7. doi: 10.1128/iai.60.9.3513-3517.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ikebe T, Wada A, Inagaki Y, et al. Dissemination of the phage-associated novel superantigen gene speL in recent invasive and noninvasive Streptococcus pyogenes M3/T3 isolates in Japan. Infect Immun. 2002;70:3227–33. doi: 10.1128/IAI.70.6.3227-3233.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Musser JM, Hauser AR, Kim MH, Schlievert PM, Nelson K, Selander RK. Streptococcus pyogenes causing toxic-shock-like syndrome and other invasive diseases: clonal diversity and pyrogenic exotoxin expression. Proc Natl Acad Sci USA. 1991;88:2668–72. doi: 10.1073/pnas.88.7.2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Voyich JM, Sturdevant DE, Braughton KR, et al. Genome-wide protective response used by group A Streptococcus to evade destruction by human polymorphonuclear leukocytes. Proc Natl Acad Sci USA. 2003;100:1996–2001. doi: 10.1073/pnas.0337370100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zabriskie JB. The role of temperate bacteriophage in the production of erythrogenic toxin by group A streptococci. J Exp Med. 1964;119:761–80. doi: 10.1084/jem.119.5.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Proft T, Sriskandan S, Yang L, Fraser JD. Superantigens and streptococcal toxic shock syndrome. Emerg Infect Dis. 2003;9:1211–8. doi: 10.3201/eid0910.030042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schlievert PM. Role of superantigens in human disease. J Infect Dis. 1993;167:997–1002. doi: 10.1093/infdis/167.5.997. [DOI] [PubMed] [Google Scholar]
- 22.Banks DJ, Lei B, Musser JM. Prophage induction and expression of prophage-encoded virulence factors in group A Streptococcus serotype M3 strain MGAS315. Infect Immun. 2003;71:7079–86. doi: 10.1128/IAI.71.12.7079-7086.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Broudy TB, Pancholi V, Fischetti VA. The in vitro interaction of Streptococcus pyogenes with human pharyngeal cells induces a phage-encoded extracellular DNase. Infect Immun. 2002;70:2805–11. doi: 10.1128/IAI.70.6.2805-2811.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson LP, Schlievert PM, Watson DW. Transfer of group A streptococcal pyrogenic exotoxin production to nontoxigenic strains of lysogenic conversion. Infect Immun. 1980;28:254–7. doi: 10.1128/iai.28.1.254-257.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Musser JM, Kapur V, Kanjilal S, et al. Geograpic and temporal distribution and molecular characterization of two highly pathogenic clones of Streptococcus pyogenes expressing allelic variants of pyrogenic exotoxin A (scarlet fever toxin) J Infect Dis. 1993;167:337–46. doi: 10.1093/infdis/167.2.337. [DOI] [PubMed] [Google Scholar]
- 26.Cleary PP, Johnson Z. Possible dual function of M protein: resistance to bacteriophage A25 and resistance to phagocytosis by human leukocytes. Infect Immun. 1977;16:280–92. doi: 10.1128/iai.16.1.280-292.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Spanier JG, Cleary PP. Bacteriophage control of antiphagocytic determinants in group A streptococci. J Exp Med. 1980;152:1393–1406. doi: 10.1084/jem.152.5.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bessen DE, Izzo MW, Fiorentino TR, Caringal RM, Hollingshead SK, Beall B. Genetic linkage of exotoxin alleles and emm gene markers for tissue tropism in group A streptococci. J Infect Dis. 1999;179:627–36. doi: 10.1086/314631. [DOI] [PubMed] [Google Scholar]
- 29.Schmitz FJ, Beyer A, Charpentier E, et al. Toxin-gene profile heterogeneity among endemic invasive European group A streptococcal isolates. J Infect Dis. 2003;188:1578–86. doi: 10.1086/379230. [DOI] [PubMed] [Google Scholar]
- 30.Vlaminckx BJ, Mascini EM, Schellekens J, et al. Site-specific manifestations of invasive group a streptococcal disease: type distribution and corresponding patterns of virulence determinants. J Clin Microbiol. 2003;41:4941–9. doi: 10.1128/JCM.41.11.4941-4949.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.De Marzi MC, Fernandez MM, Sundberg EJ, et al. Cloning, expression and interaction of human T-cell receptors with the bacterial superantigen SSA. Eur J Biochem. 2004;271:4075–83. doi: 10.1111/j.1432-1033.2004.04345.x. [DOI] [PubMed] [Google Scholar]
- 32.Mollick JA, Miller GG, Musser JM, Cook RG, Rich RR. Isolation and characterization of a novel streptococcal superantigen. Trans Assoc Am Physicians. 1992;105:110–22. [PubMed] [Google Scholar]
- 33.Mollick JA, Miller GG, Musser JM, Cook RG, Grossman D, Rich RR. A novel superantigen isolated from pathogenic strains of Streptococcus pyogenes with aminoterminal homology to staphylococcal enterotoxins B and C. J Clin Invest. 1993;92:710–9. doi: 10.1172/JCI116641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reda KB, Kapur V, Mollick JA, Lamphear JG, Musser JM, Rich RR. Molecular characterization and phylogenetic distribution of the streptococcal superantigen gene (ssa) from Streptococcus pyogenes. Infect Immun. 1994;62:1867–74. doi: 10.1128/iai.62.5.1867-1874.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reda KB, Kapur V, Goela D, Lamphear JG, Musser JM, Rich RR. Phylogenetic distribution of streptococcal superantigen SSA allelic variants provides evidence for horizontal transfer of ssa within Streptococcus pyogenes. Infect Immun. 1996;64:1161–5. doi: 10.1128/iai.64.4.1161-1165.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Davies MR, Tran TN, McMillan DJ, Gardiner DL, Currie BJ, Sriprakash KS. Inter-species genetic movement may blur the epidemiology of streptococcal diseases in endemic regions. Microbes Infect. 2005;7:1128–38. doi: 10.1016/j.micinf.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 37.Bryan EM, Bae T, Kleerebezem M, Dunny GM. Improved vectors for nisin-controlled expression in gram-positive bacteria. Plasmid. 2000;44:183–90. doi: 10.1006/plas.2000.1484. [DOI] [PubMed] [Google Scholar]
- 38.Goshorn SC, Schlievert PM. Bacteriophage association of streptococcal pyrogenic exotoxin type C. J Bacteriol. 1989;171:3068–73. doi: 10.1128/jb.171.6.3068-3073.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Caparon MG, Scott JR. Genetic manipulation of pathogenic streptococci. Methods Enzymol. 1991;204:556–86. doi: 10.1016/0076-6879(91)04028-m. [DOI] [PubMed] [Google Scholar]
- 40.Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed Cold Spring Harbor Laboratory Press; Cold Spring Harbor: 1989. [Google Scholar]
- 41.Weeks CR, Ferretti JJ. The gene for type A streptococcal exotoxin (erythrogenic toxin) is located in bacteriophage T12. Infect Immun. 1984;46:531–6. doi: 10.1128/iai.46.2.531-536.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kapur V, Reda KB, Li LL, Ho LJ, Rich RR, Musser JM. Characterization and distribution of insertion sequence IS1239 in Streptococcus pyogenes. Gene. 1994;150:135–40. doi: 10.1016/0378-1119(94)90872-9. [DOI] [PubMed] [Google Scholar]
- 43.Johnson DR, Kaplan EL, VanGheem A, Facklam RR, Beall B. Characterization of group A streptococci (Streptococcus pyogenes): correlation of M-protein and emm-gene type with T-protein agglutination pattern and serum opacity factor. J Med Microbiol. 2006;55:157–64. doi: 10.1099/jmm.0.46224-0. [DOI] [PubMed] [Google Scholar]
- 44.McGregor KF, Spratt BG, Kalia A, et al. Multilocus sequence typing of Streptococcus pyogenes representing most known emm types and distinctions among subpopulation genetic structures. J Bacteriol. 2004;186:4285–594. doi: 10.1128/JB.186.13.4285-4294.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schwartz B, Facklam RR, Breiman RF. Changing epidemiology of group A streptococcal infection in the USA. Lancet. 1990;336:1167–671. doi: 10.1016/0140-6736(90)92777-f. [DOI] [PubMed] [Google Scholar]
- 46.Beres SB, Richter EW, Nagiec MJ, et al. Molecular genetic anatomy of inter- and intraserotype variation in the human bacterial pathogen group A Streptococcus. Proc Natl Acad Sci USA. 2006;103:7059–64. doi: 10.1073/pnas.0510279103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wannamaker LW, Almquist S, Skjold S. Intergroup phage reactions and transduction between group C and group A streptococci. J Exp Med. 1973;137:1338–53. doi: 10.1084/jem.137.6.1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Colon AE, Cole RM, Leonard CG. Lysis and lysogenization of groups A, C, and G streptococci by a transducing bacteriophage induced from a group G streptococcus. J Virol. 1971;8:103–10. doi: 10.1128/jvi.8.1.103-110.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sachse S, Seidel P, Gerlach D, et al. Superantigen-like gene(s) in human pathogenic Streptococcus dysgalactiae, subsp equisimilis: genomic localisation of the gene encoding streptococcal pyrogenic exotoxin G (speG-(dys)) FEMS Immunol Med Microbiol. 2002;34:159–67. doi: 10.1111/j.1574-695X.2002.tb00618.x. [DOI] [PubMed] [Google Scholar]
- 50.Kalia A, Enright MC, Spratt BG, Bessen DE. Directional gene movement from human-pathogenic to commensal-like streptococci. Infect Immun. 2001;69:4858–69. doi: 10.1128/IAI.69.8.4858-4869.2001. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]