Abstract
Virus-induced gene silencing (VIGS) is a recently developed technique for characterizing the function of plant genes by gene transcript suppression and is increasingly used to generate transient loss-of-function assays. Here we report that the 2mDNA1, a geminivirus satellite vector, can induce efficient gene silencing in Nicotiana tabacum with Tobacco curly shoot virus. We have successfully silenced the β-glucuronidase (GUS) gene in GUS transgenic N. tabacum plants and the sulphur desaturase (Su) gene in five different N. tabacum cultivars. These pronounced and severe silencing phenotypes are persistent and ubiquitous. Once initiated in seedlings, the silencing phenotype lasted for the entire life span of the plants and silencing could be induced in a variety of tissues and organs including leaf, shoot, stem, root, and flower, and achieved at any growth stage. This system works well between 18–32 °C. We also silenced the NtEDS1 gene and demonstrated that NtEDS1 is essential for N gene mediated resistance against Tobacco mosaic virus in N. tabacum. The above results indicate that this system has great potential as a versatile VIGS system for routine functional analysis of genes in N. tabacum.
Keywords: Virus induced gene silencing, Geminivirus, Alphasatellite, Nicotiana tabacum
1. Introduction
Post-transcriptional gene silencing (PTGS), also known as quelling in fungi and RNA interference in animals (Cogoni and Macino, 1997; Fire et al., 1998), was initially described as a unique artifact of transgenic expression in petunia, and is now known as a widespread phenomenon in many organisms to serve as a natural defense response (Napoli et al., 1990; Cogoni and Macino, 2000; Voinnet, 2001). PTGS functions via a sequence-specific RNA degradation mechanism that is triggered by double stranded RNA (dsRNA). dsRNA is cleaved into 21–25 nucleotides (nt) small RNA (sRNA) molecules by dicer-like enzymes. sRNAs act post-transcriptionally to direct the cleavage or translational repression of target RNAs (Xie et al., 2004; Ramachandran and Chen, 2008). In plants, PTGS can be accomplished by creating stable transgene expression hairpin RNA molecules or dsRNA. In the recent decade, virus-induced gene silencing (VIGS), a transient reverse genetics tool for characterizing gene functions, has been widely used in many plants (Kumagai et al., 1995; Burch-Smith et al., 2004; Ding et al., 2006; Becker and Lange, 2010). When a virus infects a plant tissue and spreads systemically throughout the tissue, the endogenous gene transcripts, which are homologous to the fragment inserted into viral vector, are degraded by PTGS. Compared to other techniques such as antisense and hairpin RNA expression techniques, VIGS has many advantages, including time saving and avoidance of laborious plant transformation work. It can also knockdown either a single copy gene or a gene family, and can be used for studying some genes whose traditional knockout is embryo-lethal or sterile (Burch-Smith et al., 2004; Becker and Lange, 2010). Many viruses or virus satellites have been modified into gene silencing vectors. Most of them are applicable to Solanaceous species, particularly in Nicotiana plants such as Nicotiana benthamiana, N. glutinosa, and N. attenuate, but few of them can induce efficient gene silencing into N. tabacum (Purkayastha and Dasgupta, 2009).
Begomoviruses (family Geminiviridae) are plant viruses with circular single-stranded DNA genome encapsidated in unique twinned particles. Many begomoviruses contain two components referred to as DNA A and DNA B that are essential for virus proliferation, but some have only a single genomic component, equivalent to the DNA A of their bipartite virus counterparts (Mansoor et al., 2003; Mansoor et al., 2006; Fauquet et al., 2008). Some monopartite begomoviruses are associated with satellite molecules and two types of satellites, namely alphasatellites (formerly called DNA1) and betasatellites (formerly called DNAβ), have been identified. These satellites are approximately half the size of the genomes of their helper begomoviruses, on which they depend for replication and movement in host plants, as well as for insect transmission between plants (Briddon and Stanley, 2006; Fauquet et al., 2008). Alphasatellites make no significant contribution to the pathogenicity of begomovirus, but most betasatellites are essential for begomovirus to induce typical disease symptoms in the host from which they were isolated (Cui et al., 2004; Wu and Zhou, 2005; Briddon and Stanley, 2006). Begomoviruses and their associated satellite molecules have been successfully modified into VIGS vectors which induce efficient gene silencing in diverse plant species such as Solanum lycopersicum, Nicotiana benthamiana, Arabidopsis thaliana, Manihot esculenta, and Gossypium hirsutum (Carrillo-Tripp et al., 2006; Tuttle et al., 2008).
In our previous study, an alphasatellite- and a betasatellite-based VIGS vectors were developed and they induced efficient gene silencing in Nicotiana spp., tomato, and petunia when co-inoculated with the helper virus Tomato yellow leaf curl China virus (TYLCCNV) (Tao and Zhou, 2004; Cai et al., 2007; Huang et al., 2009). However, the low inoculation efficiencies of TYLCCNV and the satellite vectors in N. tabacum limit the use of VIGS in N. tabacum. In this paper, we describe an alphasatellite vector (2mDNA1) that can trigger high silencing efficiency into N. tabacum with the helper virus Tobacco curly shoot virus (TbCSV). By using a silencing system based on 2mDNA1 and TbCSV, we ubiquitously silenced the β-glucuronidase (GUS) gene in transgenic plants and the sulphur desaturase (Su) gene in different N. tabacum cultivars. This system can induce persistent silencing in a wide range of temperatures. By using this system, we also confirmed that NtEDS1 is essential for N gene-mediated resistance against Tobacco mosaic virus (TMV) in N. tabacum.
2. Materials and methods
2.1. Plasmid construction
2mDNA1 vector (Fig. 1a, GenBank accession No. FM212564.1) and TbCSV infectious clone were described in our previous study (Li et al., 2005; Huang et al., 2009). To generate 2mDNA1-GUS, 2mDNA1-NtSu, and 2mDNA1-NtEDS1, a 320-base pair (bp) GUS gene DNA fragment from pINT121 plasmid (Ding et al., 2009), a 351-bp Su gene DNA fragment and a 400-bp NtEDS1 DNA fragment from N. tabacum cv. NC89 plant complementary DNA (cDNA) synthesized from total RNA using the oligo(dT) primer and reverse transcriptase (TaKaRa, Dalian, China) were polymerase chain reaction (PCR) amplified using primer pairs GUSF (5′-TCTAGATAATGTTCTGCGACGCTCAC-3′, XbaI site was introduced)/GUSR (5′-GGATCCGGCGAAATTCCATACCTGTTC-3′, BamHI site was introduced), SuF (5′-GGATCCTCTAGACAGGGCAGAGTCAAGGGAGG-3′, BamHI and XbaI sites were introduced)/Su351R (5′-GGATCCTGGATCTGAATTGAACGGATC-3′, BamHI site was introduced), and NtEDS1F (5′-TCTAGAGAATTGAAGAGGGCAGAGAAG-3′, XbaI site was introduced)/NtEDS1R (5′-GGATCCGTTCCAGACACCATAGGGCT-3′, BamHI site was introduced), respectively. The resulting PCR products were separately cloned into XbaI-BamHI-cut 2mDNA1 in sense orientation. All PCR amplifications were performed using Taq DNA polymerase (TaKaRa).
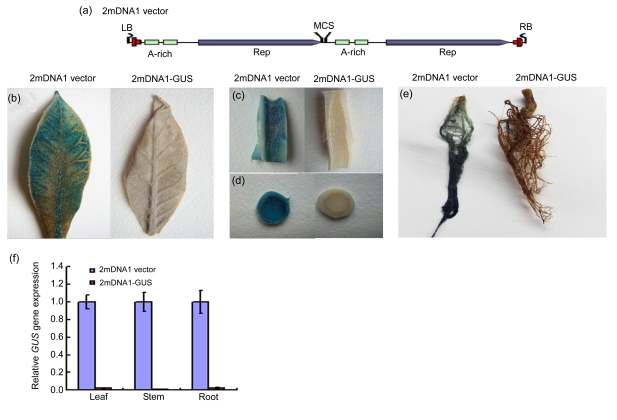
Fig. 1.
Alphasatellite-mediated VIGS of the β-glucuronidase (GUS) gene in the N. tabacum T19 line after co-agroinoculation with TbCSV
(a) Diagram of the 2mDNA1 structure. 2mDNA1 contains two direct repeats of the TbCSV DNA1 and a multiple cloning site (MCS) downstream of Rep for subsequent insertion of a DNA fragment targeted for silencing (Huang et al., 2009); (b–e) VIGS of the GUS gene in N. tabacum T19 line leaf (b), stem (c, d), and root (e) co-infected with TbCSV and either 2mDNA1 vector or 2mDNA1-GUS; (f) Silencing efficiency in GUS-silenced or non-silenced (control) N. tabacum T19 plants as revealed by real-time RT-PCR at 30 dpi. Three samples were tested and error bars represent the standard deviation of the mean
2.2. Plant growth and inoculation
N. tabacum plants were grown in pots at 25 °C in an insect-free chamber under a 16/8-h photoperiod with 60% humidity. The temperature comparison experiments were carried out in six temperature-controlled chambers with either 35, 32, 30, 20, 18, or 15 °C. Four-leaf seedlings were used for VIGS. For the VIGS assay, TbCSV and 2mDNA1 or their derivatives were introduced into the Agrobacterium tumefaciens strain EHA105 by electroporation as described (Cui et al., 2004). A. tumefaciens cells were cultured in yeast extract peptone (YEP) medium supplemented with kanamycin (50 mg/L) and rifampicin (50 mg/L) and shaken at 250 r/min (28 °C) overnight. Then the bacterial cells were harvested by centrifugation, resuspended in infiltration media (10 mmol/L MgCl2, 10 mmol/L MES, and 200 μmol/L acetosyringone), adjusted to an optical density at 600 nm (OD600) of 1.5, and left at room temperature for 3 to 4 h. Agroinoculation was done as described (Cui et al., 2004).
2.3. GUS staining
Leaves were assayed for GUS activity as described (Jefferson et al., 1987) with minor modifications. For staining of GUS, leaves were carefully and uniformly abraded on the lower side with carborundum, fixed for 20 min in 90% acetone, vacuum-infiltrated with a buffer containing 50 mmol/L sodium phosphate (pH 7.2), 0.5 mmol/L K3Fe(CN)6, 0.5 mmol/L K4Fe(CN)6, and 1 mmol/L 5-bromo-4-chloro-3-indolyl β-D-glucuronide, and then incubated for 12–24 h at 37 °C. Leaf pieces were subsequently treated with 95% ethanol to remove chlorophyll.
2.4. DNA extraction and PCR detection
Total DNA was extracted from tissues of infected symptomatic plants as described (Zhou et al., 2001). Viral infection was detected by PCR-mediated amplification with TbCSV specific primers TbCSVF (5′-CGTAGGCCTGTGGATAAACCTCAAGAT-3′) and Y6R2 (5′-GGAAGCCAGTTCAAATTAAAGG-3′) and alphasatellite universal primers UN101 (5′-AAGCTTGCGACTATTGTATGAAAGAGG-3′) and UN102 (5′-AAGCTTCGTCTGTCTTACGAGCTCGCTG-3′) as described (Li et al., 2005; Wu and Zhou, 2005).
2.5. Quantitive real-time PCR and reverse transcriptase (RT)-PCR analyses
Total RNA was isolated as described (Huang et al., 2009). The first strand cDNA was synthesized from total RNA using the oligo(dT) primer and reverse transcriptase (TaKaRa). SYBR® Green real-time RT-PCR analysis was carried out with a real-time PCR detection system (MJ Research, Waltham, Mass, USA) using primers 5′-TGCTGTCGGCTTTAACCTCTCT-3′ and 5′-TGAGCGTCGCAGAACATTACAT-3′ for GUS, 5′-GCTTCTACACCCTTGTCTTCTCG-3′ and 5′-CCCCTATCACCCATTATCATCAC-3′ for Su, 5′-TGTTGGCACAGATGAGGTAGC-3′ and 5′-GCAAGAGGAGATCCAAAGGTTA-3′ for NtEDS1. These primers were designed to exclude the region of the cDNA cloned into the 2mDNA1 vector to ensure that only the endogenous mRNA was amplified. The glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene was used as an internal constitutively expressed positive control. The primers 5′-GCAGTGAACGACCCATTTATCTC-3′ and 5′-AACCTTCTTGGCACCACCCT-3′ were designed to amplify GAPDH DNA fragment. All PCR products were sequenced and confirmed to be the correct clones for use as templates to generate a calibration curves. All gene expression analyses were repeated three times with different sets of silenced plants, and the data presented are means of triplicates for each condition in one representative experiment. All the studies were performed in accordance with minimum information for publication of quantitative real-time PCR experiments (MIQE) guidelines (Bustin et al., 2009). RT-PCR assay was performed as described by Tao and Zhou (2004). The intensities of PCR-generated fragments were analyzed and quantified using Gel Doc 2000 and Quantity One Version 4.3 (Bio-Rad, Hercules, California, USA).
2.6. Serological analysis of TMV
Triple antibody sandwich-enzyme-linked immunosorbent assay (TAS-ELISA) (Zhou et al., 1997) was used for TMV detection. TMV monoclonal antibody was prepared in the authors′ lab and the goat-anti-mouse immunoglobulin G (IgG)-alkaline phosphatase conjugate was purchased from Sigma (St. Louis, MO, USA). Absorbance readings (OD405) were made with a microplate reader (Bio-Rad). The positive threshold was fixed at twice the average of the OD obtained with the healthy plant controls. All tests were duplicated.
2.7. Northern blot analysis for TMV RNA
Total RNA was extracted from leaf tissues with TRIzol reagent following the manufacturer′s protocol (Invitrogen, USA). Equal amounts of total RNA (5 μg) were subjected to 12 g/L agarose gel electrophoresis under denaturing conditions and subsequently transferred to Hybond N+ membranes. Membranes were hybridized with [32P]ATP-labeled probes specific for the TMV movement protein (MP) gene which was synthesized by the random-priming method using a Prime-a-Gene labeling system kit (Promega, Madison, WI, USA). Hybridization signals were detected by phosphorimaging using a Typhoon 9200 imager (Amersham Pharmacia Biotech, Uppsala, USA).
3. Results
3.1. Suppression of GUS gene expression in transgenic N. tabacum plants expressing GUS by 2mDNA1 and TbCSV
To determine whether 2mDNA1 and TbCSV can be used to silence gene expression in N. tabacum, we initially targeted the transgenic N. tabacum line T19 carrying a GUS transgene (English et al., 1996). A 320-bp fragment of the GUS coding sequence was cloned and inserted into the 2mDNA1 (Fig. 1a) in sense orientation to produce construct 2mDNA1-GUS. Approximately two weeks after agroinoculation of T19 plants with 2mDNA1-GUS and TbCSV, loss of blue color was observed in newly grown systemic leaf (Fig. 1b). The GUS silencing phenotype was also observed in stem and root (Figs. 1c–1e). By contrast, the control plants agroinoculated with 2mDNA1 and TbCSV gave a strong blue signal due to GUS expression. The silencing phenotype was confirmed by analysis of GUS transcript levels by SYBR real-time RT-PCR of RNA derived from leaf, stem and root tissues of inoculated plants. The silenced plants showed more than 95% reduction in GUS transcript levels as compared with the control plants (Fig. 1f). The results indicate that 2mDNA1 and TbCSV can be used to silence transgene in N. tabacum efficiently.
3.2. Induction of efficient and persistent endogenous gene silencing in N. tabacum by 2mDNA1 and TbCSV
We further tested whether the 2mDNA1 vector can induce endogenous gene silencing in N. tabacum. We examined the ability of 2mDNA1 vector to suppress expression of the endogenous Su gene, which encodes a component of the magnesium chelatase complex and is essential for chlorophyll II biosynthesis (Koncz et al., 1990). A mixture of Agrobacterium cultures containing 2mDNA1-NtSu and TbCSV was injected into the stem and petiole of 3-week-old N. tabacum plants. At 10–12 d post inoculation (dpi), the white phenotype of Su silencing started appearing in the leaves of all the 25 inoculated plants. The white phenotype initially appeared in the veins and later scattered to the mesophyll. Eventually, almost all the leaves turned into white-yellow except a small proportion of mesophyll cells between the veins at 3–4 weeks post inoculation. Control plants, inoculated with the 2mDNA1 and TbCSV, remained green (Fig. 2a).
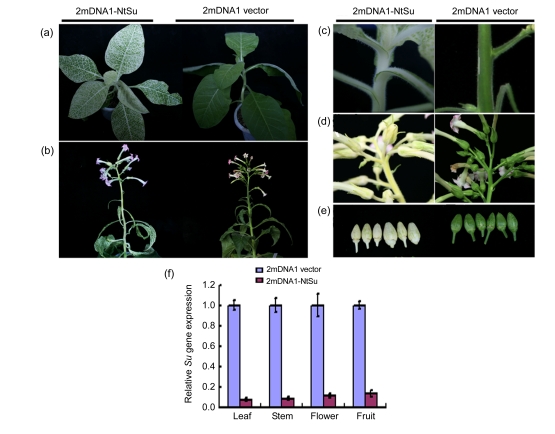
Fig. 2.
Development of the Su gene silencing in N. tabacum induced by 2mDNA1 and TbCSV
(a, b) Typical Su gene silencing in N. tabacum leaves at 30 dpi (a) and 120 dpi (b); (c–e) The Su-silenced N. tabacum stems (c), flowers (d), and fruits (e) with white-yellow phenotypes; (f) Silencing efficiency of Su gene in different plant organs was detected by real-time RT-PCR. Three samples were tested and error bars represent the standard deviation of the mean
In order to investigate whether the silencing phenotype can be persistent during plant growth, plants with silencing phenotype were kept from seedling to fructescence stage, and silencing was observed in newly developed tissues at more than 120 dpi (Fig. 2b). The typical white-yellow phenotype of the Su gene silencing was developed in stems, shoots, sepals, and even fruits (Figs. 2c–2e). SYBR real-time RT-PCR showed that the cognate Su mRNA level in Su-silenced plants was decreased more than 90.0% in leaf and stem tissues, and 89.1% and 85.8% in fruit and calyx of flowers, respectively (Fig. 2f). In contrast, the level of GAPDH mRNA, serving as an internal control, was similar in Su-silenced and non-silenced plants.
3.3. Induction of gene silencing in different N. tabacum cultivars by 2mDNA1 and TbCSV
To investigate whether 2mDNA1 system induces gene silencing in other tobacco cultivars, tobacco cultivars K236, NC89, Xanthi, Yun87, and Yunyan85 were agroinoculated with Agrobacterium suspensions containing 2mDNA1-NtSu and TbCSV. The yellowing phenotype developed in all tested cultivars in a similar temporal and spatial dynamics and VIGS efficiency in these cultivars was similar (Table 1). Initially, the veins in the newly emerged leaves turned to yellow at 10–12 dpi, and then the mesophyll tissues appeared yellow 2–3 weeks after agroinoculation, meanwhile the upper new leaves continuously obtained this phenotype as they grew up. This result indicates that the 2mDNA1 vector can efficiently induce gene silencing in all the five tested N. tabacum cultivars.
Table 1.
2mDNA1-induced gene silencing on different N. tabacum cultivars
| N. tabacum cultivar | Numbera |
Average VIGS efficiency (%) | ||
| Experiment 1 | Experiment 2 | Experiment 3 | ||
| Yun87 | 12/12/12 | 19/20/20 | 10/12/12 | 92.7±7.5b |
| Yunyan85 | 11/12/12 | 19/20/20 | 10/12/12 | 91.7±8.3 |
| K326 | 10/12/12 | 16/20/20 | 9/12/12 | 79.4±4.2 |
| NC89 | 12/12/12 | 20/20/20 | 12/12/12 | 100 |
| Xanthi | 12/12/12 | 20/20/20 | 12/12/12 | 100 |
Silenced plants/infected plants/inoculated plants
Data are expressed as mean±SD (n=3)
3.4. Induction of effective gene silencing in a wide range of temperatures by 2mDNA1 and TbCSV
To ascertain the effect of the growth temperature on 2mDNA1-induced gene silencing, tobacco plants after agroinoculation were grown at 15, 18, 20, 25, 30, 32, and 35 °C, respectively. The plants grown at 18–32 °C developed the yellowing phenotype in a similar temporal and spatial dynamics, with bright yellow in the newly emerged leaves at two weeks after inoculation (Table 2). Development of silencing in the new tissues continued as they grown up. However, the silencing phenotype was not found at 15 and 35 °C (Table 2). When plants without silencing phenotype were tested for presence of vectors, we found plants at 15 °C were all infected by the silencing vector and TbCSV while only a few plants at 35 °C were infected by the silencing vector and TbCSV. When infected plants kept at 15 and 35 °C were moved to 25 °C, silencing phenotype was observed at 5 d later (Table 2). This result demonstrates that 2mDNA1 vector can induce gene silencing at a wide range of temperatures.
Table 2.
Effect of the growth temperature on 2mDNA1-induced gene silencing in N. tabacum NC89
| Growth temperature (°C) | Numberb |
Average VIGS efficiency (%) | ||
| Experiment 1 | Experiment 2 | Experiment 3 | ||
| 15a | 0/6/6 | 0/10/10 | 0/10/10 | 0 |
| 18 | 5/6/6 | 8/10/10 | 4/5/5 | 84.4±5.1c |
| 20 | 6/6/6 | 9/10/10 | 5/5/5 | 96.7±5.7 |
| 25 | 5/6/6 | 10/10/10 | 5/5/5 | 94.4±9.6 |
| 30 | 6/6/6 | 8/9/9 | 5/5/5 | 96.3±6.4 |
| 32 | 4/6/6 | 6/8/8 | 4/4/5 | 73.9±6.7 |
| 35a | 0/2/6 | 0/4/10 | 0/3/10 | 0 |
When inoculated plants are moved to 25 °C growth chamber, all of them exhibit silencing phenotype 5 d later
Silenced plants/infected plants/inoculated plants
Data are expressed as mean±SD (n=3)
3.5. Essentiality of NtEDS1 for N gene-mediated resistance against TMV
To test whether NtEDS1 is required for N gene-mediated resistance against TMV, 320 bp of the cDNA of N. tabacum NtEDS1 was inserted into 2mDNA1 (2mDNA1-NtEDS1), and the construct and TbCSV were co-inoculated into N. tabacum cv. Xanthi. At three weeks post inoculation, NtEDS1 gene transcript in plants was reduced by more than 80% using SYBR real-time RT-PCR (data not shown). NtEDS1-silenced plants and control plants inoculated with 2mDNA1 vector and TbCSV were subsequently challenge-inoculated with wild-type TMV. Five days after inoculation, necrotic lesions were observed to resemble those produced in inoculated leaves of wild-type control plants, but the numbers of necrotic lesions induced by TMV were significantly reduced on inoculated leaves of NtEDS1-silenced plants (Fig. 3b), and typical mosaic symptoms appeared in the newly developed leaves (Fig. 3c).
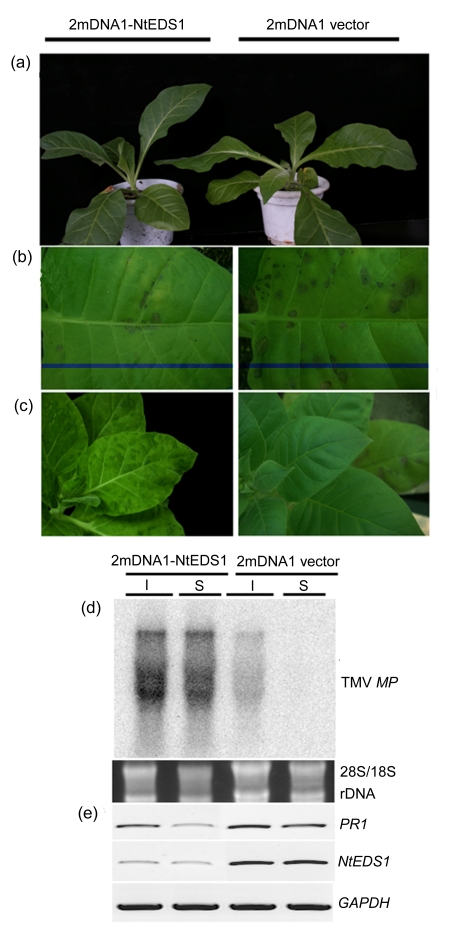
Fig. 3.
Effect of NtEDS1 on N. tabacum cv. Xanthi resistance against TMV
(a) NtEDS1-silenced plant does not express any visible phenotype; (b) TMV induces abundant necrosis lesions in inoculated leaf of NtEDS1 non-silenced plants but few in NtEDS1-silenced plants; (c) TMV symptoms are displayed in the systemic leaves of NtEDS1-silenced plant but not on NtEDS1 non-silenced plant; (d) Northern blot analysis shows TMV accumulation in inoculated (I) and systemic (S) leaves at 5 dpi using TMV movement protein (MP) gene as a probe; (e) RT-PCR analysis shows the effect of NtEDS1-silencing on PR1 transcription
The accumulation level of TMV was analyzed by TAS-ELISA using TMV monoclonal antibody and Northern blot using a TMV MP gene-specific probe. As expected, high levels of TMV accumulation on 2mDNA1-NtEDS1 and TbCSV inoculated plants were observed by TAS-ELISA and Northern blot analyses at 5 d after TMV inoculation, but no TMV was detected in control plants inoculated with 2mDNA1 and TbCSV (Fig. 3d, Table 3). We also tested the transcript level of pathogenesis-related protein 1 (PR1), which is a marker gene for resistance responses that depend on salicylic acid (SA), and an overexpression of PR1 with TMV infection was found in NtEDS1 non-silenced control plant. However, the PR1 mRNA existed at low level in NtEDS1-silenced plant (Fig. 3e). Taken together, these data show that silencing of NtEDS1 mRNA caused loss of N gene-mediated resistance against TMV and led to enhanced susceptibility to TMV.
Table 3.
Detection of TMV accumulation in NtEDS1-silenced N. tabacum cv. Xanthi plants by TAS-ELISA
| Leaf | OD405 |
|
| 2mDNA1-NtEDS1 | 2mDNA1 vector | |
| Inoculated | 0.56±0.09 | 0.26±0.08 |
| Systemic | 1.12±0.14 | 0 |
Data are expressed as mean±SD (n=3)
4. Discussion
The application of VIGS as a tool for gene function studies and high-throughput functional genomics in plants has so far led to the development of many VIGS vectors derived from plant RNA and DNA viruses (Burch-Smith et al., 2004; Purkayastha and Dasgupta, 2009). Most of these vectors work well in Solanaceae family, especially in tomato and N. benthamiana, but few in another economically important crop, N. tabacum. Four vectors, derived from Apple latent spherical virus (ALSV), Satellite tobacco mosaic virus (STMV), Tobacco rattle virus (TRV), and 2mDNA1 with TYLCCNV, were reported to be functional in the N. tabacum plant (Gossele et al., 2002; Ryu et al., 2004; Huang et al., 2009; Igarashi et al., 2009). However, their use has been limited by the difficulties encountered in gene delivery to N. tabacum plants. In the case of ALSV-based VIGS, it is necessary to prepare silencing vector infected Chenopodium quinoa leaves as inocula for inducing VIGS in target plants and the reported STMV vector requires RNA transcription in vitro, which is neither reliable nor amenable to high-throughput applications, and silencing efficiency of 2mDNA1 with TYLCCNV in N. tabacum is low. We demonstrate that 2mDNA1 vector and TbCSV can induce high efficient gene silencing in N. tabacum following simple agroinoculation. Compared with other currently used strategies, the attractive features of the TbCSV with 2mDNA1 include: (1) linearization or in vitro transcription of plasmid DNA is not required; (2) delivery is obtained upon conventional agroinoculation, without any requirement for vacuum infiltration, spraying infection, or particle bombardment; (3) high silencing efficiency can be reached in all the five tested N. tabacum cultivars; (4) persistent silencing can be induced at different growth stages; and, (5) 2mDNA1 silencing vector produces mild downward leaf curling (Fig. 2) (Li et al., 2005), which allows silencing effects to be recognized easily.
Temperature is one of the most important factors for efficient silencing. In virus-infected plants, high temperatures frequently result in attenuated symptoms and low virus accumulation (Chellappan et al., 2005). By contrast, low temperatures often induce a rapid spread of virus diseases by the control of small interfering RNA (siRNA) generation (Szittya et al., 2003). For example, a growth temperature lower than 22 °C is required for efficient TRV-induced gene silencing in tomato, and endogenous gene silencing in cotton is more efficient by Cotton leaf crumple virus (CLCrV) at a relatively cool temperature (22/18 °C) for cotton growth (Liu et al., 2002b; Tuttle et al., 2008). We found that gene silencing based on 2mDNA1 and TbCSV can work in a wide range of temperatures, and no difference in silencing efficiency was observed between 18–32 °C. The silencing phenotype in all inoculated plants, however, could not be observed at extreme temperatures (15 or 35 °C). We also found that viral replication was detected at 15 °C but not at 35 °C. It is possible that some enzymes in the pathway involved in virus-induced siRNA production might be passivated at extremely low temperatures, while some host factor(s) involved in geminivirus replication, transcription, and/or movement may be less effective at high temperature. Similar results were found previously with lower temperatures inhibiting silencing and higher temperatures eliminating virus replication (Szittya et al., 2003; Chellappan et al., 2005).
TMV, a member of the α-like virus supergroup of positive-strand RNA viruses, is a model for virus function research. TMV infection induces hypersensitive response (HR) in tobacco plants containing the N gene. The interaction between TMV and tobacco harbouring the N gene is a classical system for studying gene-for-gene interactions in disease resistance (Whitham et al., 1994). To examine the effectiveness of silencing N. tabacum disease resistance related genes, we chose NtEDS1 gene, an important positive regulator of SA synthesis and an essential factor in resistance response, which plays an important role in N gene-mediated TMV resistance. EDS1 encodes a protein with homology to lipases, and is necessary for resistance (including, HR) mediated by Toll/interleukin 1 receptor (TIR)-containing R-proteins (Falk et al., 1999; Liu et al., 2002a; Peart et al., 2002; Wiermer et al., 2005). In a previous study, researchers cloned the NtEDS1 gene and predicted its function in N. tabacum; however, they only confirmed NtEDS1 gene function through gene silencing in N. benthamiana because of lack of a VIGS system in N. tabacum (Liu et al., 2002a; Peart et al., 2002). Using our silencing system, we found that NtEDS1 is required for N gene-mediated TMV resistance. When NtEDS1 was silenced by using an alphasatellite-based vector, TMV infection elicited less necrotic lesions and produced systemic infection. From a previous study in A. thaliana and N. benthamiana, EDS1 plays an important role in Toll-like/interleukin-1 receptor-nucleotide binding-leucine-rich repeat (TIR-NB-LRR) proteins-induced cell death, but is not required for Pto (a protein kinase) or Rx (a coiled-coil (CC)-NB-LRR protein) signaling pathways (Falk et al., 1999; Peart et al., 2002). Our data suggest that TMV-induced HR also requires NtEDS1. Therefore, we can conclude that recruitment of EDS1 by TIR-NB-LRR proteins is evolutionarily conserved between dicotyledonous plant species. We also found high-level accumulation of PR1 transcript in plants inoculated with empty vector, but not in the NtEDS1-silenced controls. These results support the conclusion that NtEDS1 is involved in a point upstream of PR1 expression.
In conclusion, we have shown the effectiveness of a vector based on geminivirus alphasatellite in N. tabacum and demonstrated that the vector can be used to study gene-for-gene interactions in disease resistance.
Acknowledgments
We want to thank Prof. David BAULCOMBE (University of Cambridge, UK) for providing the T19 seeds.
Footnotes
Project supported by the National Science and Technology Major Projects of China (No. 2009ZX08009-026B), the China Postdoctoral Science Foundation (No. 20090461375), and the National Basic Research Program (973) of China (No. 2006CB101903)
References
- 1.Becker A, Lange M. VIGS—genomics goes functional. Trends Plant Sci. 2010;15(1):1–4. doi: 10.1016/j.tplants.2009.09.002. [DOI] [PubMed] [Google Scholar]
- 2.Briddon RW, Stanley J. Subviral agents associated with plant single-stranded DNA viruses. Virology. 2006;344(1):198–210. doi: 10.1016/j.virol.2005.09.042. [DOI] [PubMed] [Google Scholar]
- 3.Burch-Smith TM, Anderson JC, Martin GB, Dinesh-Kumar SP. Applications and advantages of virus-induced gene silencing for gene function studies in plants. Plant J. 2004;39(5):734–746. doi: 10.1111/j.1365-313X.2004.02158.x. [DOI] [PubMed] [Google Scholar]
- 4.Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55(4):611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 5.Cai XZ, Wang CC, Xu YP, Xu QF, Zheng Z, Zhou XP. Efficient gene silencing induction in tomato by a viral satellite DNA vector. Virus Res. 2007;125(2):169–175. doi: 10.1016/j.virusres.2006.12.016. [DOI] [PubMed] [Google Scholar]
- 6.Carrillo-Tripp J, Shimada-Beltran H, Rivera-Bustamante R. Use of geminiviral vectors for functional genomics. Curr Opin Plant Biol. 2006;9(2):209–215. doi: 10.1016/j.pbi.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 7.Chellappan P, Vanitharani R, Ogbe F, Fauquet CM. Effect of temperature on geminivirus-induced RNA silencing in plants. Plant Physiol. 2005;138(4):1828–1841. doi: 10.1104/pp.105.066563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cogoni C, Macino G. Isolation of quelling-defective (qde) mutants impaired in posttranscriptional transgene-induced gene silencing in Neurospora crassa . PNAS. 1997;94(19):10233–10238. doi: 10.1073/pnas.94.19.10233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cogoni C, Macino G. Post-transcriptional gene silencing across kingdoms. Curr Opin Genet Dev. 2000;10(6):638–643. doi: 10.1016/S0959-437X(00)00134-9. [DOI] [PubMed] [Google Scholar]
- 10.Cui XF, Tao XR, Xie Y, Fauquet CM, Zhou XP. A DNAβ associated with Tomato yellow leaf curl China virus is required for symptom induction. J Virol. 2004;78(24):13966–13974. doi: 10.1128/JVI.78.24.13966-13974.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ding CJ, Qing L, Li ZH, Liu Y, Qian YJ, Zhou XP. Genetic determinants of symptoms on viral DNA satellites. Appl Environ Microbiol. 2009;75(16):5380–5389. doi: 10.1128/AEM.01193-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ding XS, Schneider WL, Chaluvadi SR, Mian MA, Nelson RS. Characterization of a Brome mosaic virus strain and its use as a vector for gene silencing in monocotyledonous hosts. Mol Plant Microbe Interact. 2006;19(11):1229–1239. doi: 10.1094/MPMI-19-1229. [DOI] [PubMed] [Google Scholar]
- 13.English JJ, Mueller E, Baulcombe DC. Suppression of virus accumulation in transgenic plants exhibiting silencing of nuclear genes. Plant Cell. 1996;8(2):179–188. doi: 10.1105/tpc.8.2.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Falk A, Feys BJ, Frost LN, Jones JD, Daniels MJ, Parker JE. EDS1, an essential component of R gene-mediated disease resistance in Arabidopsis has homology to eukaryotic lipases. PNAS. 1999;96(6):3292–3297. doi: 10.1073/pnas.96.6.3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fauquet CM, Briddon RW, Brown JK, Moriones E, Stanley J, Zerbini M, Zhou XP. Geminivirus strain demarcation and nomenclature. Arch Virol. 2008;153(4):783–821. doi: 10.1007/s00705-008-0037-6. [DOI] [PubMed] [Google Scholar]
- 16.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans . Nature. 1998;391(6669):806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 17.Gossele V, Fache I, Meulewaeter F, Cornelissen M, Metzlaff M. SVISS—a novel transient gene silencing system for gene function discovery and validation in tobacco plants. Plant J. 2002;32(5):859–866. doi: 10.1046/j.1365-313X.2002.01471.x. [DOI] [PubMed] [Google Scholar]
- 18.Huang CJ, Xie Y, Zhou XP. Efficient virus-induced gene silencing in plants using a modified geminivirus DNA1 component. Plant Biotechnol J. 2009;7(3):254–265. doi: 10.1111/j.1467-7652.2008.00395.x. [DOI] [PubMed] [Google Scholar]
- 19.Igarashi A, Yamagata K, Sugai T, Takahashi Y, Sugawara E, Tamura A, Yaegashi H, Yamagishi N, Takahashi T, Isogai M, et al. Apple latent spherical virus vectors for reliable and effective virus-induced gene silencing among a broad range of plants including tobacco, tomato, Arabidopsis thaliana, cucurbits, and legumes. Virology. 2009;386(2):407–416. doi: 10.1016/j.virol.2009.01.039. [DOI] [PubMed] [Google Scholar]
- 20.Jefferson RA, Kavanagh TA, Bevan MW. GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987;6(13):3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koncz C, Mayerhofer R, Koncz-Kalman Z, Nawrath C, Reiss B, Redei GP, Schell J. Isolation of a gene encoding a novel chloroplast protein by T-DNA tagging in Arabidopsis thaliana . EMBO J. 1990;9(5):1337–1346. doi: 10.1002/j.1460-2075.1990.tb08248.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kumagai MH, Donson J, Dellacioppa G, Harvey D, Hanley K, Grill LK. Cytoplasmic inhibition of carotenoid biosynthesis with virus-derived RNA. PNAS. 1995;92(5):1679–1683. doi: 10.1073/pnas.92.5.1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li ZH, Xie Y, Zhou XP. Tobacco curly shoot virus DNAβ is not necessary for infection but intensifies symptoms in a host-dependent manner. Phytopathology. 2005;95(8):902–908. doi: 10.1094/PHYTO-95-0902. [DOI] [PubMed] [Google Scholar]
- 24.Liu YL, Schiff M, Marathe R, Dinesh-Kumar SP. Tobacco Rar1, EDS1 and NPR1/NIM1 like genes are required for N-mediated resistance to Tobacco mosaic virus. Plant J. 2002;30(4):415–429. doi: 10.1046/j.1365-313X.2002.01297.x. [DOI] [PubMed] [Google Scholar]
- 25.Liu YL, Schiff M, Dinesh-Kumar SP. Virus-induced gene silencing in tomato. Plant J. 2002;31(6):777–786. doi: 10.1046/j.1365-313X.2002.01394.x. [DOI] [PubMed] [Google Scholar]
- 26.Mansoor S, Briddon RW, Zafar Y, Stanley J. Geminivirus disease complexes: an emerging threat. Trends Plant Sci. 2003;8(3):128–134. doi: 10.1016/S1360-1385(03)00007-4. [DOI] [PubMed] [Google Scholar]
- 27.Mansoor S, Zafar Y, Briddon RW. Geminivirus disease complexes: the threat is spreading. Trends Plant Sci. 2006;11(5):209–212. doi: 10.1016/j.tplants.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 28.Napoli C, Lemieux C, Jorgensen R. Introduction of a chimeric chalcone synthase gene into petunia results in reversible co-suppression of homologous genes in trans. Plant Cell. 1990;2(4):279–289. doi: 10.1105/tpc.2.4.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peart JR, Cook G, Feys BJ, Parker JE, Baulcombe DC. An EDS1 orthologue is required for N-mediated resistance against Tobacco mosaic virus. Plant J. 2002;29(5):569–579. doi: 10.1046/j.1365-313X.2002.029005569.x. [DOI] [PubMed] [Google Scholar]
- 30.Purkayastha A, Dasgupta I. Virus-induced gene silencing: a versatile tool for discovery of gene functions in plants. Plant Physiol Biochem. 2009;47(11-12):967–976. doi: 10.1016/j.plaphy.2009.09.001. [DOI] [PubMed] [Google Scholar]
- 31.Ramachandran V, Chen X. Small RNA metabolism in Arabidopsis. Trends Plant Sci. 2008;13(7):368–374. doi: 10.1016/j.tplants.2008.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ryu CM, Anand A, Kang L, Mysore KS. Agrodrench: a novel and effective agroinoculation method for virus-induced gene silencing in roots and diverse Solanaceous species. Plant J. 2004;40(2):322–331. doi: 10.1111/j.1365-313X.2004.02211.x. [DOI] [PubMed] [Google Scholar]
- 33.Szittya G, Silhavy D, Molnar A, Havelda Z, Lovas A, Lakatos L, Banfalvi Z, Burgyan J. Low temperature inhibits RNA silencing-mediated defence by the control of siRNA generation. EMBO J. 2003;22(3):633–640. doi: 10.1093/emboj/cdg74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tao XR, Zhou XP. A modified viral satellite DNA that suppresses gene expression in plants. Plant J. 2004;38(5):850–860. doi: 10.1111/j.1365-313X.2004.02087.x. [DOI] [PubMed] [Google Scholar]
- 35.Tuttle JR, Idris AM, Brown JK, Haigler CH, Robertson D. Geminivirus-mediated gene silencing from Cotton leaf crumple virus is enhanced by low temperature in cotton. Plant Physiol. 2008;148(1):41–50. doi: 10.1104/pp.108.123869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Voinnet O. RNA silencing as a plant immune system against viruses. Trends Genet. 2001;17(8):449–459. doi: 10.1016/S0168-9525(01)02367-8. [DOI] [PubMed] [Google Scholar]
- 37.Whitham S, Dinesh-Kumar SP, Choi D, Hehl R, Corr C, Baker B. The product of the Tobacco mosaic virus resistance gene N: similarity to toll and the interleukin-1 receptor. Cell. 1994;78(6):1101–1115. doi: 10.1016/0092-8674(94)90283-6. [DOI] [PubMed] [Google Scholar]
- 38.Wiermer M, Feys BJ, Parker JE. Plant immunity: the EDS1 regulatory node. Curr Opin Plant Biol. 2005;8(4):383–389. doi: 10.1016/j.pbi.2005.05.010. [DOI] [PubMed] [Google Scholar]
- 39.Wu PJ, Zhou XP. Interaction between a nanovirus-like component and the Tobacco curly shoot virus/satellite complex. Acta Biochim Biophys Sin. 2005;37(1):25–31. doi: 10.1093/abbs/37.1.25. [DOI] [PubMed] [Google Scholar]
- 40.Xie ZX, Johansen LK, Gustafson AM, Kasschau KD, Lellis AD, Zilberman D, Jacobsen SE, Carrington JC. Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2004;2(5):e104. doi: 10.1371/journal.pbio.0020104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou XP, Liu YL, Calvert L, Munoz C, Otim-Nape GW, Robinson DJ, Harrison BD. Evidence that DNA-A of a geminivirus associated with severe cassava mosaic disease in Uganda has arisen by interspecific recombination. J Gen Virol. 1997;78(8):2101–2111. doi: 10.1099/0022-1317-78-8-2101. [DOI] [PubMed] [Google Scholar]
- 42.Zhou XP, Xie Y, Zhang ZK. Molecular characterization of a distinct begomovirus infecting tobacco in Yunnan, China. Arch Virol. 2001;146(8):1599–1606. doi: 10.1007/s007050170081. [DOI] [PubMed] [Google Scholar]