Abstract
A large number of plant microRNAs (miRNAs) are now documented in the miRBase, among which only 30 are for Solanum lycopersicum (tomato). Clearly, there is a far-reaching need to identify and profile the expression of miRNAs in this important crop under various physiological and pathological conditions. In this study, we used an in situ synthesized custom microarray of plant miRNAs to examine the expression and temporal presence of miRNAs in the leaves of tomato plants infected with Cucumber mosaic virus (CMV). Following computational sequence homology search and hairpin structure prediction, we identified three novel tomato miRNA precursor genes. Our results also show that, in accordance with the phenotype of the developing leaves, the tomato miRNAs are differentially expressed at different stages of plant development and that CMV infection can induce or suppress the expression of miRNAs as well as up-regulate some star miRNAs (miRNA*s) which are normally present at much lower levels. The results indicate that developmental anomalies elicited by virus infection may be caused by more complex biological processes.
Keywords: Solanum lycopersicum, Plant miRNA, Cucumber mosaic virus, miRNA array, miRNA response
1. Introduction
Solanum lycopersicum (tomato) is an economically important vegetable crop worldwide. The biology of tomato growth, fruit bearing, disease-resistance, and environmental damage-resistance has been the subject of extensive investigation. Advancement in these areas would aid in the development of high-yield crops and improve the quality of tomato fruits globally. Recently, microRNAs (miRNAs) have emerged as a new family of regulator molecules involved in plant development, signal transduction, transcription factor accumulation, protein degradation, environmental stress response, and pathogen invasion (Jones-Rhoades et al., 2006; Wang and Li, 2007). A large number of plant miRNAs from 37 plant species are now registered in a widely-used public registry miRBase (Release 15.0, Sept. 2009), of which 30 have been reported for tomato (Moxon et al., 2008; Zhang J. et al., 2008). Besides that, a few others have been reported in (Pilcher et al., 2007; Itaya et al., 2008; Yin et al., 2008).
Cucumber mosaic virus (CMV) is a plant virus in the Cucumovirus genus of the Bromoviridae family (Palukaitis et al., 1992). Its genome structure consists of three single-stranded sense RNAs (RNAs 1, 2, and 3), and two subgenomic RNAs (RNAs 4 and 4A), which contain five genes encoding proteins designated 1a, 2a, 2b, 3a, and 3b. Recently, protein 2b was found to be active in an anti-silencing mechanism of CMV in a host system (Guo and Ding, 2002). CMV can infect more than 1 000 plant species and numerous strains of CMV have been described, which can be classified into subgroups I and II according to molecular analyses of the viral genomic RNAs (Palukaitis et al., 1992). Tomato exhibits no natural resistance or tolerance towards CMV infection, and thus the infection causes catastrophic crop loss, as recorded for the CMV epidemic in Spain in 1986 (García-Arenal et al., 2000) and in northern Alabama, USA in 1992 (Sikora et al., 1998). Therefore, it is of direct economic interest to reveal the molecular signatures of the CMV infection and the plant response, and to eventually control this unfortunate event.
Newly revealed insights into plant regulatory mechanisms emphasize the key roles of small, non-coding RNAs in RNA silencing (Jones-Rhoades et al., 2006; Wang and Li, 2007). In particular, there has been mounting evidence to show that miRNAs, a particular class of endogenous molecules consisting of approximately 21 nucleotides (nt) and presenting in almost all life kingdoms, regulate plant development, flowering, and fruit production. In plants, precursors or genes of these miRNAs (pre-miRNAs) are distributed mostly in intergenic regions; these precursors are stem-loop RNA hairpins of approximately 50–350 nt. Upon activation, the pre-miRNAs are processed into RNA duplexes with a two-base extension at the 3′-end of each strand by the Dicer protein complex; post-transcriptional gene silencing (PTGS) then occurs via an assembly, i.e., RNA-induced silencing complex (RISC), involving the aforementioned small RNA duplexes and several proteins. The complex is involved in producing mature miRNA, a single strand of the hairpin RNA, with sufficient complementarity to a target gene (in plants) and the cleaving of the messenger RNA (mRNA) recruited by a mature miRNA. The counter strand of the mature miRNA, referred to as miRNA*, is degraded in a normal biogenic process and thus is rarely detected by Northern blotting or miRNA microarray profiling experiments. The detection of miRNA*, on the other hand, may indicate dysfunction of the normal miRNA processing.
miRNA microarray profiling is typically used for obtaining differential gene expression at different stages and/or under different conditions of plant growth. These studies make use of probes which are complementary to the known plant miRNAs for hybridization detection of expressed miRNAs, and the results provide the basis for investigation of the molecular mechanisms of plants (Axtell and Bartel, 2005). For plant species like tomato, whose miRNAs are largely unknown, a necessary step involves miRNA identification which can potentially be accomplished by ways of cloning (Sunkar et al., 2005), microarray profiling using computationally predicted probes (He et al., 2008), and deep sequencing, which has recently become the method of choice (Moxon et al., 2008). Our rationale was that, since mature miRNAs are highly conserved among plant species (Axtell and Bartel, 2005; Zhang B. et al., 2006) and a large number of miRNAs are already known, a microarray built on such a basis would allow discovery of conserved tomato miRNAs. We have constructed a custom miRNA microarray carrying heterologous probes to profile tomato miRNAs and miRNA*s in normal and CMV-infected leaf tissues at 3, 7, 14, and 20 d post-inoculation (dpi) (Gao et al., 2004). We showed that these documented miRNAs are a valuable information source not only for studying plant species known in the miRNA database but also for uncovering miRNAs and common gene regulatory pathways in species whose miRNAs are presently less well-characterized, such as the tomato. Our experiments identified some miRNAs and miRNA*s and their expression levels in tomato at different plant developmental stages in healthy and CMV-infected plants. These results represented a comprehensive survey of the miRNAs in tomato and proposed that miRNAs are not only regulated by viral silencing suppressors but also may be caused by more complex biological processes.
2. Materials and methods
2.1. Plant, virus inoculation, and RNA extraction
Forty-eight tomato plants (cv. Hezuo903) at the four true leaves stages were used in this experiment, and plants were grown under greenhouse conditions with a 14-h photo-period at 23–26 °C a day. The cotyledons of 24 plants were dusted with carborundum and were mechanically inoculated with 2 μg purified virions of CMV-Phy (GenBank accession Nos. DQ402477, DQ412731, and DQ412732), a severe strain cloned in our lab. Simultaneously, another 24 mock plants were dusted with carborundum and inoculated with 20 mmol/L sodium phosphate buffer (pH 7.2). At 3, 7, 14, and 20 dpi, the upper systemically leaves of mock or virus-inoculated plants were harvested from six plants. They were split into two subgroups with three plants each, which stands for the biological replicates. Total RNAs were extracted from leaf tissues using the mirVana™ miRNA Isolation Kit (Ambion, USA).
2.2. miRNA microarray assay
The array design was based on 513 well-characterized miRNAs and 511 miRNA*s from Arabidopsis, rice, maize, sorghum, medick, sugarcane, and soybean. The design included 165 sequence-unique mature miRNAs and 365 sequence-unique miRNA*s. They were employed on a custom microarray platform, a µParaflo® microfluidic microchip (LC Sciences, USA), using in situ parallel synthesis and RNA hybridization optimized probes (LC Sciences, USA). This microarray platform has been commercialized and successfully employed in diverse species’ miRNA microarray experiments (Shiboleth et al., 2007; He et al., 2008; Zhang Z. et al., 2008; Meng et al., 2009; Wilson et al., 2009). For redundant miRNAs, the sequence IDs were used according to the priority of Arabidopsis (ath-miR), rice (osa-miR), maize (zma-miR), sorghum (sbi-miR), medick (mtr-miR), sugarcane (sof-miR), and soybean (gma-miR). The final set of probes was the result of two generations of iteration of the array design (Table S1) and consisted of probes for hybridization specificity evaluation based on 13 miRNA and 11 miRNA* mismatch probes. Additionally, the array contained probes for evaluation of synthesis quality and hybridization quality, and a number of probes for hybridization with other non-coding RNAs. Each miRNA probe was repeated at least three times. Detailed information on the microarray design and performance is provided in File S1.
2.3. miRNA microarray experiment and data analysis
The microarray assays were performed according to the protocol provided by the LC Sciences (USA). For each hybridization experiment, 5 μg total RNAs were size-fractionated using a YM-100 microcon centrifugal filter (Millipore, USA) and the small RNAs (<300 nt) isolated were 3′-extended with a poly(A) tail using poly(A) polymerase. An oligonucleotide tag was then ligated to the poly(A) tail for later fluorescent dye staining; two different tags, Cy3 and Cy5, were used for our dual-sample experiments. Hybridization used a µParaflo® microfluidic chip station and 100 µl 6× SSPE (saline sodium phosphate ethylene diamine tetraacetic acid) buffer (0.90 mol/L NaCl, 60 mmol/L Na2HPO4, 6 mmol/L ethylene diamine tetraacetic acid (EDTA); pH 6.8) containing 25% (v/v) formamide at 34 °C overnight. Stringency wash using 0.1× SSPE buffer was carried out before detection using a fluorescence image scanner GenePix 4000B (Axon/Molecular Device, Sunnyvale, CA, USA) to detect the tag-specific staining Cy3 and Cy5 dyes. The biological replicate samples for each time point were separately labeled with either Cy3 or Cy5 fluorescent dye, and color-reversal hybridization experiments were carried out using a pair of Cy3 and Cy5 samples of either mock versus CMV-inoculated or vice versa. These color reversal experiments were used to exclude the data points which had inconsistent hybridization results.
The hybridization images were digitized using the Array-Pro image analysis software Media Cybernetics. Data were analyzed by first subtracting the background and then normalizing the signals using a locally weighted regression (LOWESS) filter (Bolstad et al., 2003). The reported detected signals met with two criteria: greater than the background value plus three times the background standard deviation (SD) and spot co-variance (SCV=SD×100%/intensity) of less than 50%. The mean and the spot-to-spot co-variance (SSCV=SD×100%/replicate mean) of each probe having a detectable signal were calculated, and those having an SSCV less than 50% were used in further analyses. Generation of comparison data sets (Z-values) from detected signals included log2 transformation, gene centering, and normalization (Bolstad et al., 2003). Hierarchical clustering was performed by average linkage and Euclidean distance metric.
2.4. Bioinformatics analysis
The detected miRNA and miRNA* sequences from microarray experiments were mapped to corresponding regions in SOL Genomics Network (SGN) bacterial artificial chromosome (BAC) (bacs.v439), and BAC-end sequences (http://sgn.cornell.edu/) by BLASTN algorithm. The sequences that matched at least 90% of a given small RNA (with up to one mismatch) were extracted with flanking sequences (120 nt on both sides) and further screened by RNAFold prediction program for their potential hairpin structure(s) (Hofacker, 2003). More specifically, a gene was considered as a potential miRNA precursor candidate only if it met the following criteria: (1) number of base pairs (bp) in a stem ≥16; (2) number of allowed errors in one bulge in a stem ≤12; (3) free energy ΔG≤−15 kcal/mol (1 cal=4.184 J); (4) length of hairpin (up and down stems plus hairpin loop) ≥50 nt; and, (5) number of base pairs appearing in mature miRNA or miRNA* region ≥12. Target prediction was performed using psRNA Target (http://bioinfo3.noble.org/psRNATarget/) (Zhang, 2005) based on the SGN unigene sequences (http://sgn.cornell.edu/) and expressed sequence tag (EST) sequences (http://www.ncbi.nlm.nih.gov/).
2.5. miRNA Northern blotting
Small RNAs from a 10 µg total RNA sample were separated on a 15% (v/v) denaturing polyacrylamide gel containing 8 µmol/L urea and the loading was visualized using ethidium bromide staining. The RNA was transferred to a Hybond N nylon membrane (Amersham, USA) by electroblotting, and was then fixed by ultraviolet (UV) cross-linking at 1200 µJ× 100 in a Stratalinker 1800 (Stratagene, USA). To detect miRNA and miRNA* sequences, antisense oligonucleotide probes were synthesized. Probe sequences were: 5′-GTGCTCTCTATCTTCTGTCAA-3′ for miR156, 5′-AGCACGTGCCCTGCTTCTCCA-3′ for miR164, 5′-TCGGCAAGTCATCCTTGGCTG-3′ for miR169, 5′-TCGAGCCAGACAACATTCCCC-3′ for miR166*, and 5′-TCAGTTGATGCAAGGCGGGAC-3′ for miR168*. Probes were prepared by end labeling with 32P-γATP using T4 polynucleotide kinase (TaKaRa, Japan) according to the manufacturer′s instructions. Hybridization was performed at 42 °C in hybridization solution (0.2% sodium dodecyl sulfate (SDS), 6× saline sodium citrate (SSC), 5× Denhart’s solution) for 24 h, and membranes were washed three times with 0.2% SDS and 6× SSC at 42 °C.
3. Results
Our experiments involved: (1) miRNA microarray profiling of tomato plant leaves at 3, 7, 14, and 20 dpi using mock and CMV-inoculated plants grown under identical conditions; (2) prediction of tomato miRNA precursor genes and their targets; and (3) Northern blotting verification of the selected differentially expressed miRNAs and miRNA*s.
3.1. miRNA expression profiling
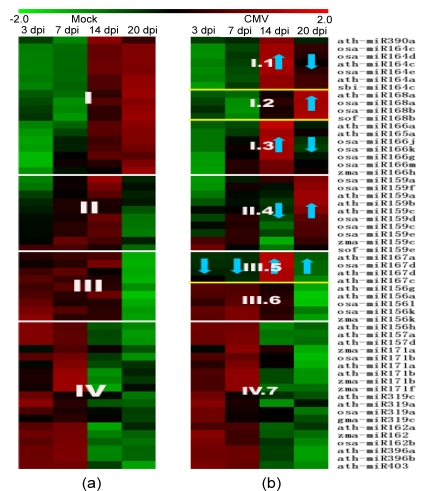
According to the detectivity determination mentioned in Section 2, the microarray experiments revealed that, out of the 165 unique miRNA probes (Table S1), 56 probes were found to express across all time points, both in mock and CMV-infected tomatoes. A hierarchical clustering of the data from these probes, based on the Z-values of the log2 data (Table S2), which were averaged from the two color reversal hybridization experiments, is shown in Fig. 1. The miRNA expression profiling revealed four major distinctly different expression patterns, labeled as I, II, III, and IV in Fig. 1a, over the time course in healthy plants. The miRNA response to CMV infection was also surveyed for the same probes, labeled as I.1, I.2, I.3, II.4, III.5, III.6, and IV.7 in Fig. 1b. Group I (miR165, miR390, and the miR164, miR166, and miR168 families) exhibited generally increasing up-regulation in healthy plants, but two kinds of expression patterns after CMV infection. The miR168 expression levels of Group I.2 remained unaffected during 14 dpi and were up-regulated at 20 dpi in the CMV-infected plants. Groups I.1 (miR390 and miR164) and I.3 (miR165 and miR166) showed few differences after CMV infection when compared with mock inoculation at 7 dpi, but they exhibited more strongly induced expression at 14 dpi, while the expression levels were subsequently reduced at 20 dpi. Group II miRNAs (the miR159 family) exhibited increasing expression levels up to 14 dpi, and then drastic decreased in expression at 20 dpi in healthy plants. However, in the CMV-infected plants, they exhibited relatively constant expression levels up to 7 dpi, sharp decreased at 14 dpi, then drastic increased in expression at 20 dpi (Group II.4). Group III miRNAs (part of the miR156 family and the miR167 family) exhibited higher expression levels up to 14 dpi but reduced expression at 20 dpi in healthy plants. The miR156 family (Group III.6) showed an expression pattern in CMV-infected plants that was very similar to the pattern in healthy plants. On the other hand, the miR167 family (Group III.5) showed lower expression in the presence of CMV infection up to 7 dpi, and clearly up-regulated expression at 14 and 20 dpi. The Group III.5 pattern is similar to those of Groups I.1 and I.3 in the CMV-infected plants. Group IV miRNAs (miR403, part of the miR156 family, and the miR157, miR162, miR171, miR319, and miR396 families) showed the opposite pattern of expression to that of Group I in healthy plants, and their expression was unaffected by CMV infection.
Fig. 1.
Comparison of the expression patterns of miRNAs in mock (a) and CMV-infected (b) tomato leaves
The color scale is based on the Z-value of the log2 detected signal, from green relatively-low to red relatively-high expression of miRNAs in samples. The heat maps presented here summarize four distinct white line expression patterns in (a), over the time course in healthy plants. The expression patterns of miRNA’s response to the CMV infection [yellow lines in (b)] were also surveyed and characterized by the regulation blue arrow, as compared with their expression in healthy plants
3.2. miRNA* accumulation in CMV-infected plants
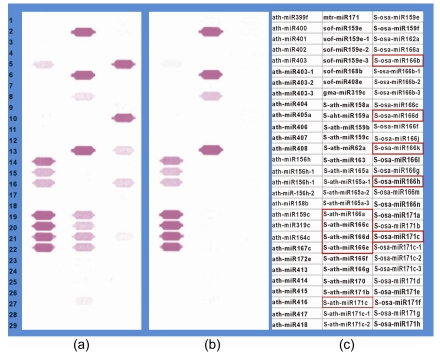
miRNA* probes (365 probes on array; Table S1) were those complementary to the strand opposite to the miRNA mature strand. Signals were detected for 16 miRNA* probes in co-hybridization experiments of the CMV-infected samples at 20 dpi, and a representative region of microarray image is shown in Fig. 2. These signals were not present in the hybridizations of the mock tomatoes and the 3, 7, and 14 dpi of CMV-infected plants. The detected probes of miRNA*s include one member of miR168* (S-ath-miR168a), two members of miR171* (S-ath-miR171c, S-osa-miR171c) and miR396* (S-ath-miR396a, S-osa-miR396a), and 11 members of miR166* (S-ath-miR166a, S-ath-miR166c, S-ath-miR166d, S-ath-miR166e, S-osa-miR166b, S-osa-miR166d, S-osa-miR166h, S-osa-miR166k, S-zma-miR166e, S-zma-miR166k, and S-gma-miR166b). While most of the detected miRNA* probes only appeared during CMV infection, we note that the probe of miR162* (S-ath-miR162a) can be detected in both mock and CMV-infected tomatoes (Fig. 2), and we would not be able to rule out that the detection may be an artifact of cross-hybridization.
Fig. 2.
Microarray image of miRNA*s accumulation in CMV-infected plants at 20 dpi
Image in a region containing probes for miRNA*s for (a) CMV-inoculated samples at 20 dpi with Cy3 labeling and (b) mock samples at 20 dpi with Cy5 labeling; (c) Probe layout for the image regions shown in (a) and (b). For example, ath-miR399f annotates the probes complementary to the mature miRNAs; S-ath-miR158a annotates the probes complementary to miRNA*s; ath-miR403-1 and S-ath-miR165a-1 annotate the mismatch probes with one different mismatched nucleotide. The red boxes annotate the miRNA* probes only appeared during CMV-inoculated samples. The hybridization images were obtained using the Array-Pro image analysis software Media Cybernetics
3.3. Predicted precursor genes and putative tomato miRNAs targets
We searched for miRNA precursor genes in the SGN database (http://sgn.cornell.edu/) according to the 72 detected sequences (56 miRNAs and 16 miRNA*s). We predicted 16 putative precursor genes (Table 1; Fig. S1). As shown in Table 1, 13 precursor genes were previously reported (Pilcher et al., 2007; Itaya et al., 2008; Moxon et al., 2008; Yin et al., 2008; Zhang J. et al., 2008), but three new precursor genes are reported for the first time in this study. The hairpin folding forms of these three precursor genes, sly-MIR156d, sly-MiR168, and sly-MIR396, are show in Fig. S1. Experimentally, when we considered the cross-hybridization of the microarray (Table S3) in CMV-infected samples, we detected the accumulation of five miRNA*s (sly-miR166b*, sly-miR168*, sly-miR171b*, sly-miR171c*, and sly-miR396*), which can be mapped to the corresponding predicted precursor genes (Table 1; Fig. S1). Moxon et al. (2008) detected both miR171 and miR171* in normal tomato leaves using deep sequencing, but in our case, we only detected miR171* accumulation in virus-infected tomato leaves. This comparison confirms the presence of miR171* in both mock and CMV-infected samples. Since the expression level of miR171* in healthy tomatoes is significantly lower than that of miR171 by deep sequencing (Moxon et al., 2008), it suggests that the absence of miR171* in the mock sample by our microarray hybridization is possibly due to the lower detection limit of deep sequencing. miR164 and miR390 were recently cloned and sequenced (Itaya et al., 2008; Moxon et al., 2008) and they can both be detected in our experiments. We can map the two miRNAs to the genome (Table 1), but the mapped “precursors” fail to fold into the hairpin structures, which need to satisfy the five criteria mentioned in Section 2.4.
Table 1.
Predicted precursor genes and potential targets of detected tomato miRNAs and miRNA*s
| Genea | Predicted precursorb | Gene ID | Predicted target gened |
| sly-miR156d | sly-MIR156dc | LE_HBa0125A23_SP6_300719 | SPL2 (SGN-U#324312) |
| sly-miR156a–c | sly-MIR156b | C02HBa0194N24.1 | SPL3 (SGN-U#317176) |
| sly-miR156a–c | sly-MIR156c | C02SLe0022J22.1 | SPL3 (SGN-U#317176) |
| sly-miR159 | sly-MIR159 | C03HBa0029M12.1 | GAMyb-like1 (EF175474) |
| sly-miR162 | sly-MIR162 | SL_EcoRI0035M01_T7_295421 | |
| sly-miR166a | sly-MIR166a | C06HBa0073H07.1 | REV (SGN-U#321033) |
| sly-miR166b/sly-miR166b* | sly-MIR166b | C08HBa0005L01.1 | REV (SGN-U#321033) |
| sly-miR167 | sly-MIR167 | C06HBa0028D14.1 | ARF8 (SGN-U#327976) |
| sly-miR168/sly-miR168* | sly-MIR168c | C12HBa0067C22.2 | AGO1 (SGN-U#338285) |
| sly-miR171a | sly-MIR171a | C02HBa0060J03.2 | SCL6 (SGN-U#333058) |
| sly-miR171b/sly-miR171b* | sly-MIR171b | C02HBa0167J21.2 | SCL6 (SGN-U#333058) |
| sly-miR171c/sly-miR171c* | sly-MIR171c | C12HBa0090D09.1 | SCL6 (SGN-U#333058) |
| sly-miR171d | sly-MIR171d | C12HBa0146I19.1 | SCL6 (SGN-U#333058) |
| sly-miR319 | sly-MIR319 | LE_HBa0015P16_SP6_130147 | TCP (SGN-U#324434) |
| sly-miR396/sly-miR396* | sly-MIR396c | C12HBa0211M21.1 | Transcription activator GRL1 (SGN-U#323341) |
| sly-miR403 | sly-MIR403 | SL_EcoRI0010F07 | PAZ domain-containing protein (SGN-U#315036) |
| sly-miR390 | C07HBa0229H10.2 | PP2C (SGN-U#325391) | |
| sly-miR164 | C07HBa0179K09.1 | CUC2 (SGN-U#326326) |
The predicted precursor structures are indicated in Fig. S1
Precursor genes that are first predicted in this study
SPL: squamosa promoter-binding protein-like; REV: homeodomain-leucine zipper protein Revoluta; ARF: auxin-responsive factor; AGO: argonaute protein; SCL: scarecrow-like transcription factor; PP2C: protein phosphatase 2C; CUC2: cup-shaped cotyledon CUC2
A total of 12 potential target genes predicted by psRNA target (http://bioinfo3.noble.org/psRNATarget/) (Zhang, 2005) are listed in Table 1. Among them, seven miRNA targets were validated in tomato using a 5′-rapid amplification of cDNA ends (RACE) method (Moxon et al., 2008).
3.4. Northern blot validation
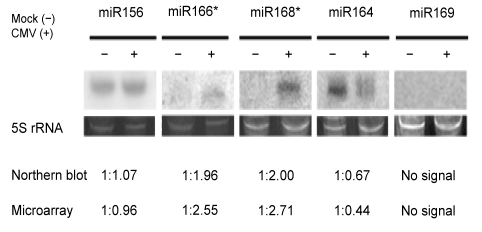
To confirm the microarray hybridization results and our putative prediction results, five representative miRNAs and miRNA*s were selected for Northern blots in tomato leaf samples at 20 dpi. Among them, sly-miR156a–c and sly-miR166b* can be found in miRBase, sly-miR168* was first reported in this study and sly-miR164 was recently cloned and sequenced (Itaya et al., 2008; Moxon et al., 2008) but failed to fold into the hairpin structures. In addition, miR169, which was deemed as highly conserved miRNAs in plants (Zhang B. et al., 2006), but not detected by microarray in this study, was also selected for Northern blot validation. The results confirmed the microarray results that the relative levels of miR156 expression remained nearly unchanged in mock (−) and CMV (+) samples, but a greater accumulation of miR166* and miR168* and reduced level of miR164 were detected in CMV-infected samples, and miR169 was undetected in all samples (Fig. 3).
Fig. 3.
Northern blots of small RNAs extracted from mock and CMV-inoculated tomato plants at 20 dpi
5S rRNA stained with ethidium bromide was used as loading control. The graph shows the calculated absolute fold changes between mock and CMV-inoculated samples at 20 dpi deduced from Northern blotting after quantification by a Phosphor-Imager and microarray data analysis
4. Discussion
4.1. Conserved tomato miRNAs
We detected 56 miRNA signals and 16 miRNA* signals by miRNA microarray expression profiling. They were grouped into 12 families (miR156/157, miR159, miR162, miR164, miR165/166, miR167, miR168, miR171, miR319, miR390, miR396, and miR403) and each family contains one or more sequence analogs (Table 1; Table S3). Our test had indicated that the chip hybridization specificity lied at the borderline of single nucleotide mismatch located in a central position. Sometimes additional mismatches located toward the ends of a sequence can be tolerated (File S1). Therefore, it is likely that the detection of analog probes is due to cross-hybridizations (Table S3) resulting in similar expression patterns (Fig. 1). However, in the miR156/miR157 family, different expression patterns were detected in healthy plants. These miRNAs belonged to either Group III (osa-miR156k, ath-miR156a, zma-miR156k, ath-miR156g, and osa-miR156l) or Group IV (ath-miR157a, ath-miR157d, and ath-miR156h) (Fig. 1a; Fig. 4). The Group IV (miR157a and miR157d, homologous to sly-miR156a-c) may have tolerated the mismatch at the 12th position to detect tomato miRNAs (Fig. 4), or tomato might have analogue miRNAs that are similar to miR157a and miR157d from the other species. Therefore, the detection of multiple miR156 probes indicates that in tomato, miR156 may have larger family members than reported in miRBase. Interestingly, the microarray results were validated by our prediction of their precursor genes (Table 1; Fig. S1). A novel miR156 precursor gene was found, whose mature sequence is analogous to the Group III, with a tolerated mismatch at the 15th position (Fig. 4).
Fig. 4.
Homologous analysis of miR156/miR157 family sequences and their expression patterns
A total of eight different miR156/157 analog probes on the array, and eight probes of the two expression groups (shaded in yellow) had detectable hybridization signals. They may be homologous to the sequence we predicted (sly-miR156d, shaded in blue) or the sequence which was reported in miRBase (sly-miR156a–c shaded in gray). Red letters are the mismatched nucleotides
The detected sequences were distributed in 12 miRNA families. According to Zhang B. et al. (2006), plant miRNAs are classified into four classes based on their conservation across 71 plant species. miRNAs have been grouped into highly conserved (conserved among 10–20 plant families), moderately conserved (among 5–9 plant families), lowly conserved (among 2–4 plant families), or non-conserved (one plant family). Our results are highly consistent with the classification of Zhang B. et al. (2006). We identified eight highly conserved miRNA families (miR156, miR159, miR166, miR168, miR171, miR319, miR390, and miR396) and three moderately conserved miRNA families (miR162, miR164, and miR167). Notably, miR403, a lowly conserved miRNA, was detected in our experiments. Further analysis revealed that it is a eudicot-specific miRNA that may play roles in eudicot-specific characteristics of plant development (Sunkar and Zhu, 2004). However, miR169, which was deemed as highly conserved miRNA in plants, was not detected by microarray and northern blots and we hypothesized that different hybridization efficiencies may cause some low-expressed miRNAs escape from detection. Further analysis revealed that miR169 expressed at a higher level in fruit and flower stages and it was almost undetectable in leaves (Moxon et al., 2008). Such results confirmed that the certain miRNAs were expressed in a spatial or temporal specific pattern.
4.2. miRNA/miRNA* responses during CMV infection
Previous studies have revealed that different isolates of CMV have different effects on miRNA pathways in Arabidopsis (Chapman et al., 2004; Zhang X. et al., 2006). The protein 2b suppressors from a mild CMV-Q strain exhibited little or no influence on miRNA pathways (Chapman et al., 2004), while those from the severe strain CMV-Fny dramatically interrupted the miRNA pathways (Zhang X. et al., 2006). Such results indicated that CMV-Fny blocks argonaute-1 (AGO1) activity through the protein 2b, causing an increased accumulation of miRNA and miRNA*, and thus impairs miRNA-guided mRNA cleavage (Zhang X. et al., 2006). However, previous work on the effects of CMV infection on the miRNA pathway were mainly limited to analysis of a single time point after CMV infection (Cillo et al., 2009; Feng et al., 2009), or focused on protein 2b transgenic plants (Chapman et al., 2004; Zhang X. et al., 2006). By contrast, our study explored the effects of virus infection on miRNA expression over the time course of infection with a severe strain CMV-Phy, whose protein 2b sequence shares 78.4% similarity with the Fny but only 52% similarity with the CMV-Q 2b, and produced some original results.
The results revealed that CMV infection initially caused a down-regulation of miR167 expression during the stage of 3–7 dpi, and that plants infected with CMV were visually indistinguishable from healthy plants with respect to leaf morphology and height (Fig. S2). At 14 dpi, three miRNA families (miR164, miR165/166, and miR167) were up-regulated in their expression, while the miR159 family was down-regulated. miR159, miR164, miR166, and miR167 are known to take part in the regulation of plant hormone-signaling pathways by targeting MYB, NAC, homeodomain leucine zipper (HD-ZIP), and auxin response factor (ARF). Plant hormones, such as auxin, have been implicated in the development of serrations, lobes, and leaflets in different species (Wang et al., 2005; Hay and Tsiantis, 2006; Barkoulas et al., 2008). This may explain why mild alterations in leaf phenotype, such as leaf area, were observed at this stage of CMV infection (Fig. S2). At 20 dpi, miR164, miR165/166, miR167, and miR159 showed altered expression patterns compared with the 14 dpi stage, while miR168 showed up-regulated expression. This miRNA expression profile was accompanied by the accumulation of some miRNA*s (miR168*, miR396*, miR166*, and miR171*). Interestingly, at 20 dpi, the accumulation was highly consistent with a severe symptom of CMV infection which results in severely stunted plants with some reduction in aerial tissue weight and leaf area, and in mosaic symptoms seen in newly developed leaves (Fig. S2). It appears that the severity of morphological alterations generally correlated with the perturbation of related miRNA pathways as observed by expression profiling of miRNAs and miRNA*s (Zhang X. et al., 2006).
These results reveal some interesting characteristics. Firstly, not all miRNAs exhibited differential expression upon CMV infection, and those related to development regulation, hormone signaling, and miRNA biogenesis exhibited either induced or suppressed expression (Fig. 1). Secondly, hormone signaling response miRNAs, such as miR167, started to show reduced expression at an early stage of CMV infection. Thirdly, the accumulation of miRNA*s in the infected tomato occurred in later infection stage accompanied by an aggravated disease symptom. Therefore, we concluded that miRNAs are not only regulated by viral silencing suppressors but also may be caused by more complex biological processes. This observation was supported by Bazzini et al. (2009) in which virus infection elevates transcriptional activity of miR164a promoter in plants.
In conclusion, our results suggested that plant developmental anomalies elicited by virus infection are caused by a synchronized network. miRNAs, target mRNAs, and other regulatory factors including the plant hormone signaling may be involved. This work points new sights to recognize virus infection in hosts through a better understanding of the relationships between viral infection, the miRNA expression, and disease symptoms in plants.
List of electronic supplementary materials
Footnotes
Project supported by the National High-Tech Research and Development Program (863) of China (No. 2008AA10Z129), and the National Natural Science Foundation of China (No. 30800716)
Electronic supplementary materials: The online version of this article (doi:10.1631/jzus.B1000278) contains supplementary materials, which are available to authorized users
References
- 1.Axtell MJ, Bartel DP. Antiquity of microRNAs and their targets in land plants. Plant Cell. 2005;17(6):1658–1673. doi: 10.1105/tpc.105.032185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barkoulas M, Hay A, Kougioumoutzi E, Tsiantis M. A developmental framework for dissected leaf formation in the Arabidopsis relative Cardamine hirsuta . Nat Genet. 2008;40(9):1136–1141. doi: 10.1038/ng.189. [DOI] [PubMed] [Google Scholar]
- 3.Bazzini A, Almasia N, Manacorda C, Mongelli V, Conti G, Maroniche G, Rodriguez M, Distefano A, Hopp HE, del Vas M, et al. Virus infection elevates transcriptional activity of miR164a promoter in plants. BMC Plant Biol. 2009;9(1):152. doi: 10.1186/1471-2229-9-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19(2):185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 5.Chapman EJ, Prokhnevsky AI, Gopinath K, Dolja VV, Carrington JC. Viral RNA silencing suppressors inhibit the microRNA pathway at an intermediate step. Genes Dev. 2004;18(10):1179–1186. doi: 10.1101/gad.1201204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cillo F, Mascia T, Pasciuto MM, Gallitelli D. Differential effects of mild and severe cucumber mosaic virus strains in the perturbation of microRNA-regulated gene expression in tomato map to the 3′ sequence of RNA 2. Mol Plant-Microbe Interact. 2009;22(10):1239–1249. doi: 10.1094/MPMI-22-10-1239. [DOI] [PubMed] [Google Scholar]
- 7.Feng J, Wang K, Liu X, Chen S, Chen J. The quantification of tomato microRNAs response to viral infection by stem-loop real-time RT-PCR. Gene. 2009;437(1-2):14–21. doi: 10.1016/j.gene.2009.01.017. [DOI] [PubMed] [Google Scholar]
- 8.Gao X, Gulari E, Zhou X. In situ synthesis of oligonucleotide microarrays. Biopolymers. 2004;73(5):579–596. doi: 10.1002/bip.20005. [DOI] [PubMed] [Google Scholar]
- 9.García-Arenal F, Escriu F, Aranda MA, Alonso-Prados JL, Malpica JM, Fraile A. Molecular epidemiology of Cucumber mosaic virus and its satellite RNA. Virus Res. 2000;71(1-2):1–8. doi: 10.1016/S0168-1702(00)00183-0. [DOI] [PubMed] [Google Scholar]
- 10.Guo HS, Ding SW. A viral protein inhibits the long range signaling activity of the gene silencing signal. EMBO J. 2002;21(3):398–407. doi: 10.1093/emboj/21.3.398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hay A, Tsiantis M. The genetic basis for differences in leaf form between Arabidopsis thaliana and its wild relative Cardamine hirsuta. Nat Genet. 2006;38(8):942–947. doi: 10.1038/ng1835. [DOI] [PubMed] [Google Scholar]
- 12.He PA, Nie Z, Chen J, Lv Z, Sheng Q, Zhou S, Gao X, Kong L, Wu X, Jin Y, et al. Identification and characteristics of microRNAs from Bombyx mori . BMC Genom. 2008;9(1):248. doi: 10.1186/1471-2164-9-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hofacker IL. Vienna RNA secondary structure server. Nucleic Acids Res. 2003;31(13):3429–3431. doi: 10.1093/nar/gkg599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Itaya A, Bundschuh R, Archual AJ, Joung JG, Fei Z, Dai X, Zhao PX, Tang Y, Nelson RS, Ding B. Small RNAs in tomato fruit and leaf development. Biochim Biophys Acta. 2008;1779(2):99–107. doi: 10.1016/j.bbagrm.2007.09.003. [DOI] [PubMed] [Google Scholar]
- 15.Jones-Rhoades MW, Bartel DP, Bartel B. MicroRNAs and their regulatory roles in plants. Ann Rev Plant Biol. 2006;57(1):19–53. doi: 10.1146/annurev.arplant.57.032905.105218. [DOI] [PubMed] [Google Scholar]
- 16.Meng Y, Huang F, Shi Q, Cao J, Chen D, Zhang J, Ni J, Wu P, Chen M. Genome-wide survey of rice microRNAs and microRNA-target pairs in the root of a novel auxin-resistant mutant. Planta. 2009;230(5):883–898. doi: 10.1007/s00425-009-0994-3. [DOI] [PubMed] [Google Scholar]
- 17.Moxon S, Jing R, Szittya G, Schwach F, Rusholme Pilcher RL, Moulton V, Dalmay T. Deep sequencing of tomato short RNAs identifies microRNAs targeting genes involved in fruit ripening. Genome Res. 2008;18(10):1602–1609. doi: 10.1101/gr.080127.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Palukaitis P, Roossinck MJ, Dietzgen RG, Francki RI. Cucumber mosaic virus. Adv Virus Res. 1992;41:281–348. doi: 10.1016/S0065-3527(08)60039-1. [DOI] [PubMed] [Google Scholar]
- 19.Pilcher RL, Moxon S, Pakseresht N, Moulton V, Manning K, Seymour G, Dalmay T. Identification of novel small RNAs in tomato (Solanum lycopersicum) Planta. 2007;226(3):709–717. doi: 10.1007/s00425-007-0518-y. [DOI] [PubMed] [Google Scholar]
- 20.Shiboleth YM, Haronsky E, Leibman D, Arazi T, Wassenegger M, Whitham SA, Gaba V, Gal-On A. The conserved FRNK box in HC-Pro, a plant viral suppressor of gene silencing, is required for small RNA binding and mediates symptom development. J Virol. 2007;81(23):13135–13148. doi: 10.1128/JVI.01031-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sikora EJ, Gudauskas RT, Murphy JF, Porch DW, Andrianifahanana M, Zehnder GW, Bauske EM, Kemble JM, Lester DF. A multivirus epidemic of tomatoes in Alabama. Plant Dis. 1998;82(1):117–120. doi: 10.1094/PDIS.1998.82.1.117. [DOI] [PubMed] [Google Scholar]
- 22.Sunkar R, Zhu JK. Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell. 2004;16(8):2001–2019. doi: 10.1105/tpc.104.022830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sunkar R, Girke T, Jain PK, Zhu JK. Cloning and characterization of microRNAs from rice. Plant Cell. 2005;17(5):1397–1411. doi: 10.1105/tpc.105.031682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang H, Jones B, Li Z, Frasse P, Delalande C, Regad F, Chaabouni S, Latche A, Pech JC, Bouzayen M. The tomato Aux/IAA transcription factor IAA9 is involved in fruit development and leaf morphogenesis. Plant Cell. 2005;17(10):2676–2692. doi: 10.1105/tpc.105.033415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang QL, Li ZH. The functions of microRNAs in plants. Front Biosci. 2007;12:3975–3982. doi: 10.2741/2364. [DOI] [PubMed] [Google Scholar]
- 26.Wilson KD, Venkatasubrahmanyam S, Jia F, Sun N, Butte AJ, Wu JC. MicroRNA profiling of human-induced pluripotent stem cells. Stem Cells Devel. 2009;18(5):749–758. doi: 10.1089/scd.2008.0247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yin Z, Li C, Han X, Shen F. Identification of conserved microRNAs and their target genes in tomato (Lycopersicon esculentum) Gene. 2008;414(1-2):60–66. doi: 10.1016/j.gene.2008.02.007. [DOI] [PubMed] [Google Scholar]
- 28.Zhang B, Pan X, Cannon CH, Cobb GP, Anderson TA. Conservation and divergence of plant microRNA genes. Plant J. 2006;46(2):243–259. doi: 10.1111/j.1365-313X.2006.02697.x. [DOI] [PubMed] [Google Scholar]
- 29.Zhang J, Zeng R, Chen J, Liu X, Liao Q. Identification of conserved microRNAs and their targets from Solanum lycopersicum Mill. Gene. 2008;423(1):1–7. doi: 10.1016/j.gene.2008.05.023. [DOI] [PubMed] [Google Scholar]
- 30.Zhang X, Yuan YR, Pei Y, Lin SS, Tuschl T, Patel DJ, Chua NH. Cucumber mosaic virus-encoded 2b suppressor inhibits Arabidopsis Argonaute1 cleavage activity to counter plant defense. Genes Devel. 2006;20(23):3255–3268. doi: 10.1101/gad.1495506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Y. miRU: an automated plant miRNA target prediction server. Nucleic Acids Res. 2005;33:W701–W704. doi: 10.1093/nar/gki383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Z, Wei L, Zou X, Tao Y, Liu Z, Zheng Y. Submergence-responsive microRNAs are potentially involved in the regulation of morphological and metabolic adaptations in maize root cells. Ann Bot. 2008;102(4):509–519. doi: 10.1093/aob/mcn129. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.