Abstract
Background
Pseudohypoparathyroidism (PHP) defines a rare group of disorders whose common feature is resistance to the parathyroid hormone. Patients with PHP-Ia display additional hormone resistance, Albright hereditary osteodystrophy (AHO) and reduced Gsα activity in easily accessible cells. This form of PHP is associated with heterozygous inactivating mutations in Gsα-coding exons of GNAS, an imprinted gene locus on chromosome 20q13.3. Patients with PHP-Ib typically have isolated parathyroid hormone resistance, lack AHO features and demonstrate normal erythrocyte Gsα activity. Instead of coding Gsα mutations, patients with PHP-Ib display imprinting defects of GNAS, caused, at least in some cases, by genetic mutations within or nearby this gene.
Patients
Two unrelated PHP families, each of which includes at least one patient with a Gsα coding mutation and another with GNAS loss of imprinting, are reported here.
Results
One of the patients with GNAS imprinting defects has paternal uniparental isodisomy of chromosome 20q, explaining the observed imprinting abnormalities. The identified Gsα coding mutations include a tetranucleotide deletion in exon 7, which is frequently found in PHP-Ia, and a novel single nucleotide change at the acceptor splice junction of intron 11.
Conclusions
These molecular data reveal an interesting mixture, in the same family, of both genetic and epigenetic mutations of the same gene.
Pseudohypoparathyroidism (PHP; MIM 103580) consists of a heterogeneous group of endocrine diseases, the common feature of which is end-organ resistance to the parathyroid hormone (PTH).1–3 PHP type I, which is characterised by blunted urinary excretion of both cAMP and phosphate in response to exogenous PTH, is further subdivided based on the presence or the absence of additional endocrine abnormalities, deficiency of the α-subunit of the stimulatory G protein (Gsα) and the dysmorphic features of Albright hereditary osteodystrophy (AHO), which may include short stature, brachydactyly, ectopic ossifications and/or mental retardation. Patients with AHO and resistance to PTH, thyroid-stimulating hormone, and, often, additional hormones are referred to as having PHP-Ia. These patients typically carry maternally inherited heterozygous inactivating mutations in 1 of 13 GNAS exons that encode Gsα. Paternal inheritance of the same mutations leads to AHO without hormone resistance, a condition termed “pseudopseudohypoparathyroidism”.
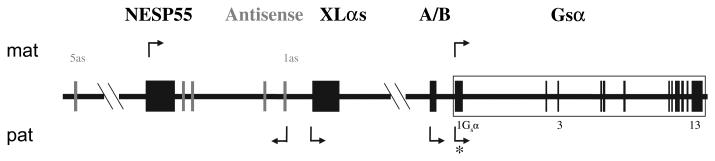
GNAS is a complex imprinted locus located on chromosome 20q13.3 (reviewed in refs1 2; figure 1). Upstream of exon 1 are three alternative first exons that individually splice onto exons 2–13 to form different transcripts: XLαs and A/B (also referred to as 1A) are paternally expressed, and NESP55 is maternally expressed. In addition, GNAS gives rise to a non-coding antisense transcript, which also shows paternal expression. Consistent with their monoallelic expression, the promoters of the imprinted GNAS transcripts are differentially methylated, with the active allele lacking methylation. In contrast, the promoter of the Gsα transcript lacks differential methylation, although it is silenced on the paternal allele in some tissues, including the renal cortex and the thyroid gland.
Figure 1.
General organisation and imprinting patterns of the GNAS locus (drawing not to scale). Arrows above and below represent maternally and paternally expressed transcripts, respectively. The boxes and connecting lines represent the exons and introns, respectively; grey boxes are used for the exons of the antisense transcripts. The asterisk indicates the silencing of Gsα from the paternal allele in some but not all tissues, such as renal proximal tubule.
PTH resistance without AHO and without other hormonal abnormalities is referred to as PHP-Ib1–3; however, some cases with PHP-Ib also demonstrate mild thyroid-stimulating hormone resistance.4 Few cases with the molecular diagnosis of PHP-Ib have also been recently reported to exhibit mild AHO features.5–7 Unlike patients with PHP-Ia and pseudopseudohypoparathyroidism, patients with PHP-Ib show normal Gsα activity in circulating blood cells and fibroblasts.1–3 Nevertheless, a loss of methylation at GNAS exon A/B, sometimes combined with epigenetic defects at other GNAS differentially methylated regions, is found in most patients with this disorder.8–10 Patients with an autosomal dominant form of PHP-Ib who display isolated loss of exon A/B methylation carry maternally inherited microdeletions within STX16,11 12 a neighbouring, non-imprinted gene encoding syntaxin-16. On the other hand, maternal deletions of the differentially methylated region comprising NESP55 and exons 3 and 4 of the GNAS antisense transcript have been identified in two unrelated PHP-Ib families in whom the disease is associated with broad GNAS epigenetic defects.13
We now report two unrelated PHP-I families, each of whom include at least one patient carrying Gsα coding mutations (ie, PHP-Ia) and another patient carrying GNAS imprinting defects (ie, PHP-Ib). One of the patients with GNAS imprinting defects was found to have paternal uniparental isodisomy of 20q (patUPD20q), thus explaining the observed imprinting abnormalities.
PATIENTS AND METHODS
Family 1
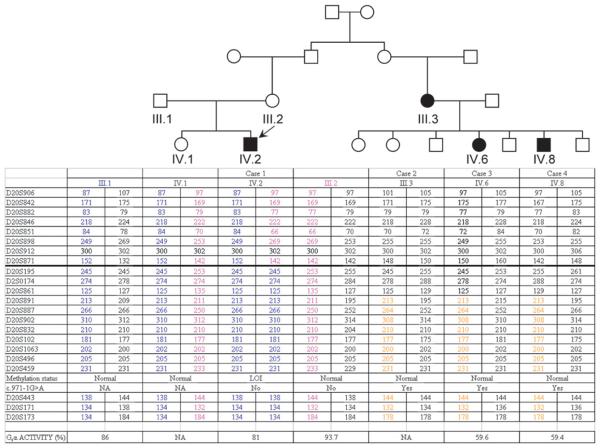
Case 1 is a 25-year-old male patient who presented with PTH and thyroid-stimulating hormone resistance along with physical features suggestive of mild AHO, including a round face, borderline short stature and cognitive impairment. He developed neonatal hyperbilirubinemia and was first studied at 10 months of age because of obesity (body mass index, 20; >p97), right cryptorchidism and psychomotor retardation. Case 2 is a 71-year-old woman with a clinical diagnosis of PHP-I. Cases 3 and 4 are 46-year-old female and 38 year-old male patients, respectively, who were diagnosed clinically as having PHP-Ia. Case 2 is the second cousin of case 1 and the mother of cases 3 and 4 (figure 2).
Figure 2.
Pedigree of family 1 showing the haplotypes for the analysed microsatellites and erythrocyte Gsα activity. Note that each branch has independent haplotypes carrying the genetic GNAS alteration. Moreover, IV.2 (case 1) shows homozygosity throughout 20q (markers below the thick line) and lacks the maternal allele for eight of the analysed markers. NA, not available.
Family 2
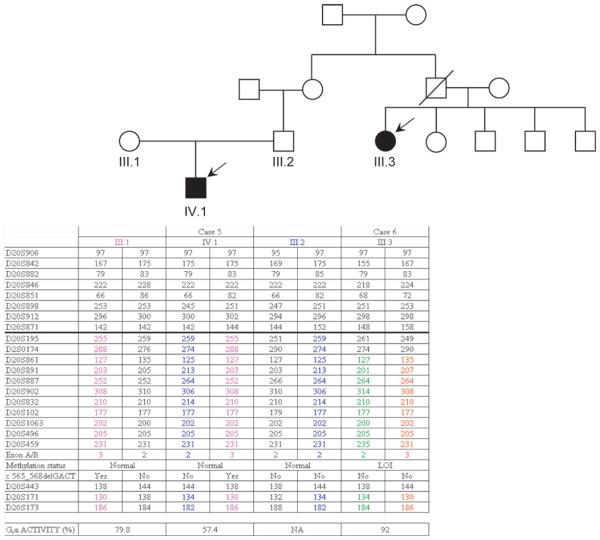
Case 5 is a 20-year-old man with a clinical diagnosis of PHP-Ia. Case 6 is a 53-year-old woman with a clinical diagnosis of PHP-Ib. Case 6 is the second cousin of case 5 (figure 3).
Figure 3.
Pedigree of family 2 showing the haplotypes for the analysed microsatellites and erythrocyte Gsα activity. It can be seen that each branch has independent haplotypes carrying the genetic GNAS alteration. Markers from 20p and 20q are indicated above or below the thick line, respectively. NA, not available.
Biochemical features of these cases are shown in table 1, and detailed clinical descriptions of cases can be found as supplementary information.
Table 1.
Clinical and biochemical features of the pseudohypoparathyroidism (PHP) cases
| Family 1 |
Family 2 |
|||||
|---|---|---|---|---|---|---|
| Case 1‡ (IV.2) | Case 2 (III.3) | Case 3 (IV.6) | Case 4 (IV.8) | Case 5 (IV.1) | Case 6 (III.3) | |
| Sex | Male | Female | Female | Male | Male | Female |
| Age at diagnosis (years) | 9 | 54 | 20 | 11 | 5 | 23 |
| Rounded face | Yes | Yes | Yes | Yes | Yes | No |
| Weight (kg) | 26 (−1SD) | 66.8 (+2.5SD) | 55 (−0.5SD) | 30 (−1SD) | 21.6 (+1.5SD) | 51.8 (−1.5SD) |
| Height (cm) | 124.7 (−2SD) | 146.5 (−3SD) | 137 (−3SD) | 127 (−3SD) | 107 (−0.5SD) | 149.3 (−3SD) |
| Midparental height (cm) | 162.2 | NA | NA | NA | 157 | NA |
| Macrocephaly | Yes | Yes | Yes | Yes | Yes | No |
| Subcutaneous ossifications | No | Yes§ | No | No | Yes§ | No |
| Intracranial calcifications | Yes | Yes | Yes | Yes | Yes | Yes |
| Brachydactyly | No | Yes | Yes | Yes | Yes | No |
| Cataracts | No | Yes | Yes | Yes | No | Yes |
| Cognitive impairment | Yes | Yes | Yes | Yes | Yes | No |
| Calcium (2.2–2.6 mmol/L)* | 2.18 | 2.72 | 1.42 | 1.45 | 1.32 | 1.79 |
| Phosphate (mmol/L) | 2.09 (1.3–2.26) | 1.16 (1–1.5) | 1.67 (1–1.5) | 2.55 (1–1.5) | 3.32 (1.3–2.26) | 1.61 (1–1.5) |
| PTH (10–65 pg/ml) | 940 | 54 | 113 | 633 | 434 | 244 |
| Free T4 (0.7–1.9 ng/dl) | 1.2 | 1.16 | 1.04 | 0.88 | 0.7 | NA |
| TSH (0.5–5 mU/L) | 2 | 1.11 | 7.22 | 5.71 | 16 | 13 |
| Anti-thyroglobulin antibodies (0–60 IU/ml) | Negative | Negative | Negative | Negative | Negative | 192.6 |
| FSH/LH (IU/L)† | 3.35/9.85* | 33.7/67.2 | 37/11 | 1.6/5.8 | 1.9/0.5 | NA |
| Twenty-four-hour urinary calcium excretion (100–250 mg/24 h) |
131.8 | 159.6 | 70 | 100 | 172.8 | 165.6 |
| GNAS molecular defect | Loss of imprinting | Gsα coding mutation | Gsα coding mutation | Gsα coding mutation | Gsα coding mutation | Loss of imprinting |
FSH, follicle-stimulating hormone; LH, luteinising hormone; NA, not available; PTH, parathyroid hormone; TSH, thyroid-stimulating hormone.
All data were measured at the time of PHP diagnosis except gonadotropin levels of case 1, which were measured at 18 years of life.
Total serum calcium.
FSH and LH normal ranges in premenopausal women 1.4–21 and 0.8–57 IU/L, postmenopausal women 34–96 and 40–140 IU/L, and men 0.5–15 and 1.3–13 IU/L.
The patient was already on levothyroxine treatment at the time of diagnosis.
Radiograms reveal subcutaneous calcified lesions and mass; however, no biopsy information is available for determining ossification.
Molecular studies
GNAS gene analysis
The institutional review board approved the study. Genetic analyses were performed after informed consent of the patient or parents was obtained. Genomic DNA was extracted from peripheral blood leucocytes using a commercial kit, following the manufacturer's instructions (NucleoSpin Blood, Macherey-Nagel, Germany). The 13 coding exons of the GNAS gene as well as the exon A/B region were amplified, and both strands of amplicons were sequenced (primers and conditions are available on request).
To characterise the chromosome 20 haplotype, eight polymorphic markers located at 20p and 14 microsatellites at 20q were typed by fluorescent PCR.
Mutational data analysis
The possible effects on mRNA splicing were evaluated in silico using the GeneSplicer (http://www.tigr.org/tdb/GeneSplicer/gene_spl.html) and Gene Finding and Intron Splice Site Prediction (http://www.cbs.dtu.dk/biolinks/pserve2.php) web interfaces. In addition, the CRYP-SKIP software (http://www.dbass.org.uk/cryp-skip/) was used to predict the overall probability of cryptic splice-site activation (pCR-E =1) versus exon skipping (pCR-E =0).
Methylation-specific multiplex-ligation-dependent probe amplification
Dosage and methylation analyses were studied by methylation-specific multiplex-ligation-dependent probe amplification (MS-MLPA) using the ME031A kit (MRC-Holland, Amsterdam, The Netherlands). The protocol was performed following the manufacturer's recommendations. Analysis of the MS-MLPA PCR products was performed on an ABI3130XL genetic analyser and the GeneMapper software (Applied Biosystems Foster City, California, USA). A more detailed description of this method is provided as supplementary information.
Gsα activity measurement
In heparinised blood samples, the activity of Gsα protein from erythrocyte membranes was analysed in vitro as previously described.14 Results obtained in triplicate were expressed as the percentage of the mean of healthy controls (normal range 85–115%). PHP-Ia patients with Gsα coding mutations were used as positive controls.
RESULTS
Family 1
We analysed the 13 GNAS exons encoding Gsα in genomic DNA from case 1 but could not identify any nucleotide alterations. Instead, epigenetic evaluation of the region by MS-MLPA revealed a loss of methylation at the maternal A/B and XLαs exons and a gain of methylation at NESP55 exon (figure 2). At the same time, two second cousins of the patient's mother (cases 3 and 4) were independently analysed, and instead of epigenetic alterations, a single nucleotide change was identified at the acceptor splice site of GNAS intron 11 (c.971-1G>A) (figure 2). The analysis of their affected mother (case 2) also revealed the same mutation, which was predicted, by three different genome analysis softwares, to affect the acceptor splicer and to promote exon skipping (data not shown).
To rule out a loss of one GNAS allele in case 1, which would account for the observed epigenetic abnormalities, 22 micro-satellites in chromosome 20 were typed, revealing a loss of the maternal haplotype in case 1 throughout the long arm of chromosome 20 (figure 2). However, MS-MLPA analysis of the GNAS locus argued against a copy number loss, and taken together with his normal karyotype, these findings suggested that case 1 had patUPD20q. Consistent with this conclusion, this patient demonstrated nearly normal erythrocyte Gsα activity, while his two cousins with the splice-site mutation were shown to have diminished Gsα activity (figure 2). The haplotype analysis of the entire family also showed that the mother of case 1 and her cousin (case 2) had inherited different parental haplotypes near the GNAS locus based on microsatellites D20S832, D20S459 and D20S173. The mother of case 1 also lacked the c.971-1G>A mutation, consistent with the absence of AHO features and hormone resistance in that individual.
Family 2
Nucleotide sequence analysis of the 13 Gsα coding GNAS exons in the index patient's DNA (case 5) revealed a heterozygous deletion of GACT tetranucleotide in exon 7 (c.565_568delGACT) (figure 3). The genetic analysis of the whole family revealed that, whereas the mother carried the same mutation, his second cousin (case 6), who also displayed PTH resistance, did not have any coding mutations. Methylation analysis of GNAS in this patient revealed loss of methylation at the maternal A/B and XLαs DMR and an apparent gain of methylation at NESP55 DMR. Typing the microsatellites at chromosome 20 ruled out a large deletion within GNAS that would explain the observed epigenetic defects; particularly, the patient was heterozygous for the pentanucleotide repeat polymorphism located within GNAS exon A/B (figure 3).
Similar to the results obtained in family 1, while erythrocyte Gsα activity was diminished in case 5, who had a Gsα coding mutation, it was normal in case 6, who showed GNAS methylation defects (figure 3).
DISCUSSION
We report, for the first time, two unrelated PHP-I families with two different molecular defects of GNAS: each family includes at least one patient with a Gsα coding mutation and one patient with imprinting abnormalities. One of the Gsα coding mutations is the frequently found 4 bp deletion in exon 7, and the other is a novel splice site mutation. In family 1, the methylation defects in the proband are the result of paternal UPD. In contrast, the coding Gsα mutations in this family are present on the maternal side of the proband. Likewise, the coding and imprinting GNAS defects in family 2 are located on different branches of the family. While cases 5 and 6 are related through their fathers, the GNAS defect in each of these cases is located on the maternal allele. It appears that the presence of these two distinct GNAS defects in the same family is coincidental, even if the probability of this event to occur, according to the estimated PHP-I prevalence of 0.79/100 000 (Orphanet Report Series, November 2008), is 6.2×10−11.
In addition to these genetic findings, our results indicate that the definitive diagnosis of a PHP subtype cannot rely solely on clinical and biochemical information, and that a complete molecular study of the GNAS gene and its methylation status should be carried out. For example, our case 1 demonstrated clinical features consistent with mild AHO, and his erythrocyte Gsα activity was slightly below the normal range. While the diagnosis of PHP-Ia was therefore considered, molecular characterisations showed a lack of maternal-specific GNAS methylation pattern, an epigenetic defect observed in PHP-Ib patients. In addition, the molecular analysis of case 6, who also had short stature besides PTH resistance, revealed GNAS imprinting defects of the maternal allele without any deletion of this genomic region. Coexistence of GNAS methylation defects and AHO-like features have been previously reported.5–7 Our new findings provide further evidence that an overlap can exist between the clinical features of different PHP subtypes. However, mild AHO features and GNAS imprinting defects may be unrelated in some cases. For example, it is plausible that some of the subtle AHO findings in our case 1 are due to the documented patUPD20q.
There is only one previously reported case of PHP caused by patUPD20q.10 Similar to that case (K-1), case 1 had a normal karyotype and developed neonatal hyperbilirubinemia. On the other hand, our patient did not present with craniosynostosis, unlike K-1. The coincidence of neonatal hyperbilirubinemia in the two described patients with patUPD20q may suggest that this feature is due to impaired expression of other imprinted genes located on chromosome 20q, as opposed to the unmasking of recessive traits present in the paternally inherited DNA, which may perhaps explain the craniosynostosis phenotype observed in K-1.10
In conclusion, the presented cases highlight the importance of a complete clinical and biochemical characterisation of patients with PHP that belong to the same family, and of a careful molecular and epigenetic analysis of GNAS and the chromosomal region comprising this gene for establishing the correct diagnosis and, thereby, providing appropriate genetic counselling.
Supplementary Material
Acknowledgements
We thank all members of the affected families for their collaborative participation in this study.
Funding GPN is supported by Miguel Servet contracts from the Instituto de Salud Carlos III of the Spanish Ministry of Health (grant CP03/0064). This work was partially funded by grant GV2008/111035 from the Department of Health of the Basque Government and BIO08/ER/001. This group is also supported by the Centro de Investigación Biomédica en Red de Enfermedades Raras (CIBERER), ISCIII. This work was also funded, in part, by grant number R01DK073911 from the National Institute of Diabetes and Digestive and Kidney Diseases (to MB) and from the German Ministry of Research and Education (GMG01GM0315 to OH). Other Funders: National Institutes of Health.
Footnotes
Competing interests None to declare.
Patient consent Obtained
Ethics approval This study was conducted with the approval of the Hospital de Cruces Ethics Committee.
Provenance and peer review Not commissioned; externally peer reviewed.
REFERENCES
- 1.Weinstein LS, Yu S, Warner DR, Liu J. Endocrine manifestations of stimulatory G protein alpha-subunit mutations and the role of genomic imprinting. Endocr Rev. 2001;22:675–705. doi: 10.1210/edrv.22.5.0439. [DOI] [PubMed] [Google Scholar]
- 2.Bastepe M. The GNAS locus and pseudohypoparathyroidism. Adv Exp Med Biol. 2008;626:27–40. doi: 10.1007/978-0-387-77576-0_3. [DOI] [PubMed] [Google Scholar]
- 3.Levine MA. Hypoparathyroidism and pseudohypoparathyroidism. In: De Groot LJ, Jameson J, editors. Endocrinology. W.B.Saunders; Philadelphia: 2000. pp. 1133–53. [Google Scholar]
- 4.Liu J, Erlichman B, Weinstein LS. The stimulatory G protein alpha-subunit Gs alpha is imprinted in human thyroid glands: implications for thyroid function in pseudohypoparathyroidism types 1A and 1B. J Clin Endocrinol Metab. 2003;88:4336–41. doi: 10.1210/jc.2003-030393. [DOI] [PubMed] [Google Scholar]
- 5.de Nanclares GP, Fernandez-Rebollo E, Santin I, Garcia-Cuartero B, Gaztambide S, Menendez E, Morales MJ, Pombo M, Bilbao JR, Barros F, Zazo N, Ahrens W, Jüppner H, Hiort O, Castano L, Bastepe M. Epigenetic defects of GNAS in patients with pseudohypoparathyroidism and mild features of Albright's hereditary osteodystrophy. J Clin Endocrinol Metab. 2007;92:2370–73. doi: 10.1210/jc.2006-2287. [DOI] [PubMed] [Google Scholar]
- 6.Mariot V, Maupetit-Mehouas S, Sinding C, Kottler ML, Linglart A. A maternal epimutation of GNAS leads to Albright osteodystrophy and PTH resistance. J Clin Endocrinol Metab. 2008;93:661–5. doi: 10.1210/jc.2007-0927. [DOI] [PubMed] [Google Scholar]
- 7.Unluturk U, Harmanci A, Babaoglu M, Yasar U, Varli K, Bastepe M, Bayraktar M. Molecular diagnosis and clinical characterization of pseudohypoparathyroidism type-Ib in a patient with mild Albright's hereditary osteodystrophy-like features, epileptic seizures, and defective renal handling of uric acid. Am J Med Sci. 2008;336:84–90. doi: 10.1097/MAJ.0b013e31815b218f. [DOI] [PubMed] [Google Scholar]
- 8.Liu J, Litman D, Rosenberg MJ, Yu S, Biesecker LG, Weinstein LS. A GNAS1 imprinting defect in pseudohypoparathyroidism type IB. J Clin Invest. 2000;106:1167–74. doi: 10.1172/JCI10431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bastepe M, Pincus JE, Sugimoto T, Tojo K, Kanatani M, Azuma Y, Kruse K, Rosenbloom AL, Koshiyama H, Jüppner H. Positional dissociation between the genetic mutation responsible for pseudohypoparathyroidism type Ib and the associated methylation defect at exon A/B: evidence for a long-range regulatory element within the imprinted GNAS1 locus. Hum Mol Genet. 2001;10:1231–41. doi: 10.1093/hmg/10.12.1231. [DOI] [PubMed] [Google Scholar]
- 10.Bastepe M, Lane AH, Jüppner H. Paternal uniparental isodisomy of chromosome 20q d and the resulting changes in GNAS1 methylation d as a plausible cause of pseudohypoparathyroidism. Am J Hum Genet. 2001;68:1283–9. doi: 10.1086/320117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bastepe M, Frohlich LF, Hendy GN, Indridason OS, Josse RG, Koshiyama H, Korkko J, Nakamoto JM, Rosenbloom AL, Slyper AH, Sugimoto T, Tsatsoulis A, Crawford JD, Jüppner H. Autosomal dominant pseudohypoparathyroidism type Ib is associated with a heterozygous microdeletion that likely disrupts a putative imprinting control element of GNAS. J Clin Invest. 2003;112:1255–63. doi: 10.1172/JCI19159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Linglart A, Gensure RC, Olney RC, Jüppner H, Bastepe M. A novel STX16 deletion in autosomal dominant pseudohypoparathyroidism type Ib redefines the boundaries of a cis-acting imprinting control element of GNAS. Am J Hum Genet. 2005;76:804–14. doi: 10.1086/429932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bastepe M, Frohlich LF, Linglart A, Abu-Zahra HS, Tojo K, Ward LM, Jüppner H. Deletion of the NESP55 differentially methylated region causes loss of maternal GNAS imprints and pseudohypoparathyroidism type Ib. Nat Genet. 2005;37:25–7. doi: 10.1038/ng1487. [DOI] [PubMed] [Google Scholar]
- 14.Ahrens W, Hiort O. Determination of Gs alpha protein activity in Albright's hereditary osteodystrophy. J Pediatr Endocrinol Metab. 2006;19(Suppl 2):647–51. doi: 10.1515/jpem.2006.19.s2.647. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.