Abstract
Stuttering is a common speech disorder with substantial genetic contributions. To better understand the genetic factors involved in stuttering, we performed a genome-wide linkage study in a newly-ascertained consanguineous stuttering family from Pakistan. A linkage scan in this family using parametric linkage analysis revealed significant linkage only on chromosome 3q13.2–3q13.33, with a maximum two-point LOD score of 4.23 under an autosomal recessive model of inheritance.
Stuttering is a disorder of the flow of speech characterized by involuntary repetitions or prolongations of sounds or syllables, and by interruptions of speech known as blocks. While twin and family studies have suggested substantial heritability for this disorder, linkage studies have identified few highly significant linkage signals, and numerous suggestive linkage signals (Riaz et al. 2005; Shugart et al. 2004; Suresh et al. 2006; Wittke-Thompson et al. 2007). Because of marginally significant linkage scores and difficulties reproducing linkage observations across studies, we sought to increase the power to detect linkage using highly consanguineous families.
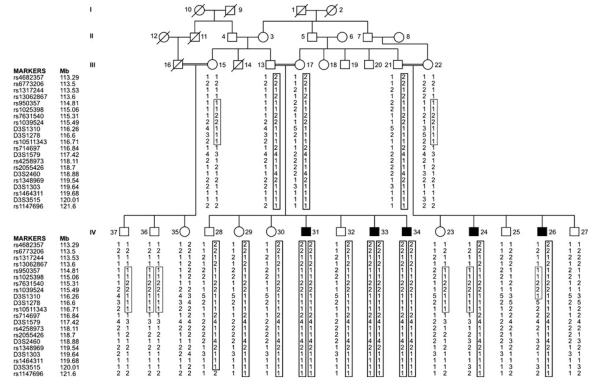
This study was approved by the Institutional Review Board (IRB) of the National Institutes of Health, USA (protocol number 97-DC-0057) and the IRB of the Center of Excellence in Molecular Biology, University of the Punjab, Lahore, Pakistan. Blood and speech samples were collected from all the available individuals of a consanguineous family (PKST77, Fig. 1), and written informed consent was obtained from each subject. Stuttering diagnosis was performed using the Stuttering Severity Instrument, 3rd edition (SSI-3) with individuals displaying severity scores above 16 (very mild) defined as affected. All individuals scored as affected were persistent stutterers, aged eight years or above who have stuttered for more than 6 months. A genome-wide linkage scan was performed using 6,090 SNPs assayed on the Illumina Human Linkage-12 Chip. Two-point parametric linkage analysis was performed using SuperLink v1.4 from the EasyLinkage v5.08 package (Hoffmann and Lindner 2005). The disease allele frequency was set at 0.01 and Caucasian allele frequencies provided by Illumina were used for the analysis. Due to the high degree of consanguinity in this family and in the Pakistani population, autosomal recessive inheritance was assumed. Significant evidence of linkage, with a LOD score of 3.23, was found at SNP rs7631540 on chromosome 3q13.31 (115.31 Mb) (Table 1). No marker showed significant linkage under either additive or dominant models of inheritance. The interval extending from markers D3S3044 (112.44 Mb) to D3S3636 (126.64 Mb) was genotyped with additional microsatellite and SNP markers. A two-point LOD score of 4.23 was obtained with the marker D3S1310 at 116.26 Mb (Table 1; Fig. 1). Multipoint analysis generated additional evidence for linkage, and an analysis including markers rs1317244, rs7631540, D3S1310, and D3S1303 under a recessive model generated a LOD score of 4.92.
Fig. 1.
Pedigree of Pakistani stuttering family PKST77. Filled symbols represent affected individuals. Double horizontal lines represent the consanguineous marriage. The disease associated haplotype is shown in vertical boxes
Table 1.
Two-point LOD score results for SNPs and microsatellite markers on chromosome 3q13.2–13.33
| Markers | hg18 Physical | Two-point LOD score results at recombination fraction theta (θ) |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Positions (Mb) | 0 | 0.01 | 0.03 | 0.05 | 0.07 | 0.09 | 0.11 | 0.13 | 0.15 | |
| rs4682357 | 113.29 | 1.57 | 1.68 | 1.76 | 1.77 | 1.73 | 1.67 | 1.59 | 1.5 | 1.41 |
| rs6773206 | 113.5 | 2.53 | 2.48 | 2.38 | 2.28 | 2.18 | 2.08 | 1.98 | 1.88 | 1.77 |
| rs1317244 | 113.53 | 3.13 | 2.75 | 2.65 | 2.54 | 2.43 | 2.32 | 2.2 | 2.09 | 1.98 |
| rs13062867a | 113.6 | 0.73 | 0.94 | 1.14 | 1.22 | 1.24 | 1.23 | 1.19 | 1.14 | 1.08 |
| rs950357 | 114.81 | 2.53 | 2.48 | 2.38 | 2.28 | 2.18 | 2.08 | 1.98 | 1.88 | 1.77 |
| rs1025398 | 115.06 | 2.57 | 2.44 | 2.34 | 2.25 | 2.15 | 2.05 | 1.95 | 1.85 | 1.75 |
| rs7631540 | 115.31 | 3.23 | 3.16 | 3.02 | 2.88 | 2.73 | 2.59 | 2.45 | 2.3 | 2.16 |
| rs1039524 | 115.49 | 2.86 | 2.8 | 2.69 | 2.58 | 2.47 | 2.36 | 2.24 | 2.13 | 2.01 |
| D3S1310 | 116.26 | 4.23 | 4.15 | 3.97 | 3.79 | 3.61 | 3.43 | 3.25 | 3.07 | 2.88 |
| D3S1278 | 116.6 | 2.25 | 2.47 | 2.6 | 2.61 | 2.57 | 2.49 | 2.4 | 2.3 | 2.19 |
| rs10511343 | 116.71 | −0.68 | −0.55 | −0.37 | −0.26 | −0.18 | −0.13 | −0.08 | −0.05 | −0.02 |
| rs714697a | 116.84 | 1.53 | 1.74 | 1.87 | 1.87 | 1.83 | 1.76 | 1.67 | 1.57 | 1.46 |
| D3S1579 | 117.42 | 1.43 | 1.66 | 1.83 | 1.88 | 1.88 | 1.85 | 1.8 | 1.74 | 1.66 |
| rs4258973 | 118.11 | 1.11 | 1.32 | 1.48 | 1.52 | 1.51 | 1.48 | 1.42 | 1.35 | 1.28 |
| rs2055426 | 118.7 | 0.87 | 1.1 | 1.28 | 1.34 | 1.35 | 1.33 | 1.29 | 1.25 | 1.19 |
| D3S2460 | 118.88 | 2.71 | 2.7 | 2.65 | 2.56 | 2.46 | 2.35 | 2.23 | 2.1 | 1.97 |
| rs1348969 | 119.54 | 2.45 | 2.4 | 2.31 | 2.21 | 2.12 | 2.02 | 1.92 | 1.82 | 1.73 |
| D3S1303 | 119.64 | 3.74 | 3.66 | 3.5 | 3.33 | 3.17 | 3.01 | 2.84 | 2.68 | 2.51 |
| rs1464311 | 119.68 | 0.09 | 0.09 | 0.08 | 0.07 | 0.07 | 0.06 | 0.05 | 0.05 | 0.04 |
| D3S3515 | 120.01 | −0.87 | −0.74 | −0.57 | −0.45 | −0.36 | −0.3 | −0.25 | −0.21 | −0.18 |
| rs1147696 | 121.6 | −0.46 | −0.33 | −0.17 | −0.07 | 0 | 0.05 | 0.08 | 0.1 | 0.12 |
Markers in bold show significant two-point LOD scores
hg18 Human Genome Assembly 18
Markers rs13062867 and rs714697 show the proximal and distal boundaries of the refined linkage region
One individual in PKST77 (#36) carries the stuttering-associated diplotype across part of this interval but he does not stutter. The genetics of stuttering are known to be complex (Suresh et al. 2006), and we hypothesize that this represents a case of non-penetrance, perhaps due to recovery from early childhood stuttering, both of which have previously been documented in this disorder (Kang et al. 2010).
A previous linkage study reported a nominal linkage on chromosome 3q, although somewhat more distally, at 192.02 Mb (Wittke-Thompson et al. 2007). A previous candidate gene association study of stuttering found a suggestive association with variants in the DRD2 dopamine receptor gene (located on chromosome 11) (Lan et al. 2009). Another member of the dopamine receptor gene family (DRD3) is found in our linkage region. We sequenced all coding exons of this gene in all family members, however, no genetic variation was observed. In addition to the DRD3 gene, there are 46 known and predicted genes in this region of chromosome 3q (3.24 Mb). Consanguineous families may provide a generally powerful resource for identifying linkage for this complex trait.
Acknowledgments
We thank Ms. Bushra Raza for assistance with stuttering diagnosis, and the research subjects for their generous cooperation with this study.
Footnotes
Present Address: S. Riazuddin Allama Iqbal Medical College, Lahore, Pakistan
References
- Hoffmann K, Lindner TH. easyLINKAGE-Plus—automated linkage analyses using large-scale SNP data. Bioinformatics. 2005;21(17):3565–3567. doi: 10.1093/bioinformatics/bti571. [DOI] [PubMed] [Google Scholar]
- Kang C, Riazuddin S, Mundorff J, Krasnewich D, Friedman P, Mulikan J, Drayna D. Mutations in the lysosomal enzyme-targeting pathway and persistent stuttering. N Engl J Med. 2010;362:677–685. doi: 10.1056/NEJMoa0902630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan J, Song M, Pan C, Zhuang G, Wang Y, Ma W, Chu Q, Lai Q, Xu F, Li Y, Liu L, Wang W. Association between dopaminergic genes (SLC6A3 and DRD2) and stuttering among Han Chinese. J Hum Genet. 2009;54(8):457–460. doi: 10.1038/jhg.2009.60. [DOI] [PubMed] [Google Scholar]
- Riaz N, Steinberg S, Ahmad J, Pluzhnikov A, Riazuddin S, Cox NJ, Drayna D. Genomewide significant linkage to stuttering on chromosome 12. Am J Hum Genet. 2005;76(4):647–651. doi: 10.1086/429226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shugart YY, Mundorff J, Kilshaw J, Doheny K, Doan B, Wanyee J, Green ED, Drayna D. Results of a genome-wide linkage scan for stuttering. Am J Med Genet A. 2004;124A(2):133–135. doi: 10.1002/ajmg.a.20347. [DOI] [PubMed] [Google Scholar]
- Suresh R, Ambrose N, Roe C, Pluzhnikov A, Wittke-Thompson JK, Ng MC, Wu X, Cook EH, Lundstrom C, Garsten M, Ezrati R, Yairi E, Cox NJ. New complexities in the genetics of stuttering: significant sex-specific linkage signals. Am J Hum Genet. 2006;78(4):554–563. doi: 10.1086/501370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittke-Thompson JK, Ambrose N, Yairi E, Roe C, Cook EH, Ober C, Cox NJ. Genetic studies of stuttering in a founder population. J Fluency Disord. 2007;32(1):33–50. doi: 10.1016/j.jfludis.2006.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]