Abstract
Tumor suppressor p53 plays a central role in tumor prevention. As a transcription factor, p53 mainly exerts its function through transcription regulation of its target genes to initiate various cellular responses. To maintain its proper function, p53 is tightly regulated by a wide variety of regulators in cells. Thus, p53, its regulators and regulated genes form a complex p53 network which is composed of hundreds of genes and their products. microRNAs (miRNAs) are a class of endogenously expressed, small non-coding RNA molecules which play a key role in regulation of gene expression at the post-transcriptional level. Recent studies have demonstrated that miRNAs interact with p53 and its network at multiple levels. p53 regulates the transcription expression and the maturation of a group of miRNAs. On the other hand, miRNAs can regulate the activity and function of p53 through direct repression of p53 or its regulators in cells. These findings have demonstrated that miRNAs are important components in the p53 network, and also added another layer of complexity to the p53 network.
Keywords: p53, tumor suppression, microRNA
Introduction
Tumor suppressor p53 is the most frequently mutated gene in human tumors. p53 and its signaling pathway, which is composed of hundreds of genes and their products, play a central role in maintaining genomic stability and tumor suppression (Levine et al., 2004, 2006; Vousden and Prives, 2009; Feng and Levine, 2010). Since p53 was discovered 30 years ago, a tremendous amount of work has been done on this single protein and its signaling pathway, which makes p53 one of the most extensively studied protein, and at the same time leads to the growing complexity of the p53 network (Braithwaite and Prives, 2006; Levine et al., 2006; Vousden and Prives, 2009).
microRNAs (miRNAs) are a class of endogenously expressed, small (20–25 nucleotides) non-coding regulatory RNA molecules in cells. miRNAs bind to the 3′-untranslated regions (3′-UTRs) of protein-coding messenger RNAs (mRNAs) in a sequence-specific manner, reducing mRNA stability, inhibiting translation, and thereby negatively regulating the expression of genes at the post-transcriptional level (Pillai et al., 2007; Bartel, 2009). Recently, it has been reported that some miRNAs can also up-regulate translation (Vasudevan et al., 2007). Lin-4 is the first miRNA identified in 1993, which is required for the appropriate timing of post-embryonic development in Caenorhabditis elegans (Lee et al., 1993; Wightman et al., 1993). In 2000, let-7 was identified as the second miRNA, which controls developmental timing in C. elegans (Reinhart et al., 2000). Since then, miRNAs have attracted tremendous attention. miRNAs have been demonstrated to be a class of most abundant regulatory genes in humans; over 700 human miRNAs have been identified so far, and the total number of miRNAs is estimated to be close to 1000. It is estimated that over 30% of all genes and the majority of genetic pathways are subject to regulation by multiple miRNAs (Berezikov et al., 2005; Lim et al., 2005; Griffiths-Jones et al., 2006). However, relatively few miRNA-target interactions have been experimentally validated, and the biological functions of most miRNAs remain to be discovered (Griffiths-Jones et al., 2006; Kloosterman and Plasterk, 2006; Inui et al., 2010).
Recent studies have revealed the close interactions between p53 and miRNAs. p53 induces the transcription expression of several miRNAs and promotes the maturation of specific miRNAs, both of which contribute to the function of p53 in tumor suppression. On the other hand, miRNAs can negatively regulate p53 protein levels and function through direct repression of p53 expression, or positively regulate p53 activity and function through the repression of several negative regulators of p53. These findings have demonstrated that miRNAs are important components in the p53 network.
p53 and its signaling pathway
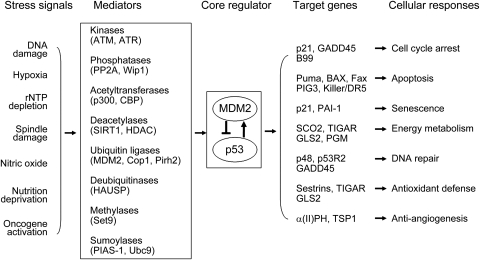
p53 plays a crucial role in maintaining genomic stability and tumor suppression (Levine et al., 2004, 2006; Vousden and Prives, 2009). Disruption of normal p53 function is in many circumstances a prerequisite for the initiation and/or progression of tumors (Donehower et al., 1992; Jacks et al., 1994). Over 50% of human tumors harbor DNA mutations in the p53 gene, and it is estimated that over 80% of tumors have dysfunctional p53 signaling (Levine et al., 2004, 2006; Olivier et al., 2004). In normal unstressed cells, p53 is maintained at a low level by rapid degradation through the ubiquitin–proteasome pathway. As an E3 ubiquitin ligase, MDM2 is a most critical negative regulator for p53, which binds to p53 and degrades p53 through ubiquitination (Harris and Levine, 2005; Brooks and Gu, 2006; Figure 1). p53 responds to a wide variety of intrinsic and extrinsic stress signals, including DNA damage, hypoxia, mitotic spindle damage, the inhibition of ribosome biogenesis, nutritional starvation, depletion of ribonucleotide triphosphates and even the activation of selected oncogenes (e.g. c-Myc, Ras and E2F-1; Figure 1). These signals are detected and communicated to p53 via a wide variety of enzymes that mediate the modifications of p53 protein, such as phosphorylation, acetylation, methylation, ubiquitination, summolation and neddylation, which lead to the increase of p53 protein half-life and therefore p53 protein accumulation in cells (Figure 1). As a transcription factor, the activated p53 protein binds to a specific DNA sequence, termed the p53-responsive element (RE) to regulate p53 target genes. A p53-RE is composed of RRRCWWGYYY(spacer of 0–21 nucleotides)RRRCWWGYYY, where R is a purine, W is A or T, and Y is a pyrimidine (el-Deiry et al., 1992). Thus, activated p53 selectively transcribes its target genes to start various cellular responses. Depending on cell type, environmental context, and the type and/or degree of stress, p53 induces cell cycle arrest, apoptosis or senescence to prevent the propagation of these damaged or mutant cells that could potentially become cancerous (Levine et al., 2006; Vousden and Prives, 2009). Recent studies have demonstrated that p53 exerts additional functions (Figure 1), including DNA repair (Hwang et al., 1999; Tanaka et al., 2000), antioxidant defense (Budanov et al., 2004; Sablina et al., 2005), energy metabolism (Bensaad et al., 2006; Matoba et al., 2006; Feng and Levine, 2010; Hu et al., 2010b) and anti-angiogenesis (Teodoro et al., 2006), which all contribute to the role of p53 in tumor suppression. To maintain the proper functions of p53, p53 protein levels and activity are tightly regulated by a wide variety of positive and negative regulators in cells (Harris and Levine, 2005; Levine et al., 2006). Thus, p53, its regulators and downstream target genes form a complex signaling network to respond to stress and prevent tumor formation (Figure 1).
Figure 1.
Tumor suppressor p53 and its signaling pathway. A wide variety of intracellular and extracellular stress signals are detected by the cell and communicated to the p53 protein by numerous mediators (shown are representative mediators). Stress signals result in the degradation of the MDM2 protein and the increase in the levels and activity of the p53 protein. Depending on the cell type, environmental context and the type and/or degree of stress, activated p53 selectively transcribes a group of its target genes (shown are representative genes) and initiates various cellular responses to exert its function in tumor suppression.
miRNAs
Most miRNAs localize within introns of either protein-coding or non-coding transcriptional units. These miRNAs either have the same orientation as their host genes and are coordinately expressed with their host genes, or have their own promoters independent of their host genes (Ozsolak et al., 2008). miRNAs can also localize within exons of mRNA-like non-coding RNAs. The maturation of miRNAs is a complex process, which includes the transcription of primary (pri)-miRNAs, the cleavage of pri-miRNAs to form precursor (pre)-miRNAs and the cleavage of pre-miRNAs to form mature miRNAs (Lee et al., 2006; Bartel, 2009; Brodersen and Voinnet, 2009). First, miRNAs are transcribed as primary transcripts (pri-miRNAs) in the nucleus by RNA polymerase II. Recently it was also reported that some miRNAs are transcribed by RNA polymerase III (Borchert et al., 2006; Ozsolak et al., 2008). The size of pri-miRNAs is variable, which is usually between 1 and 3 kb. Pri-miRNAs contain hairpin structures that are recognized and cleaved by the RNase III endonuclease Drosha in concert with its partner DGCR8. This leads to the generation of 60–100 bp stem loops known as pre-miRNAs. Pre-miRNAs are then exported from the nucleus to the cytoplasm by exportin 5. In the cytoplasm, pre-miRNAs are recognized by the RNase III endonuclease Dicer. Dicer cleaves pre-miRNAs and generates 20–25 bp mature miRNA duplexes. Normally, one strand of this duplex is degraded, whereas the other strand accumulates as the mature miRNA. The mature miRNAs are incorporated into an RNA-induced silencing complex (RISC), which allows the RISC to recognize target mRNAs through partial sequence complementarity with the 3′-UTRs of target mRNAs. The 5′ sequence of miRNAs, particularly those at nucleotide positions 2–7 relative to the 5′ end of the miRNA, are important for binding to target mRNAs and are known as miRNA ‘seed’. While the seed is the major determinant of mRNA target choice, the surrounding miRNA sequence also contributes to the strength of binding (Griffiths-Jones et al., 2006; Grimson et al., 2007). miRNAs regulate the expression of the target mRNA through two main mechanisms: blockade of translation at the initiation step or at the elongation step; and removal of the polyA tail (deadenylation) by fostering the activity of deadenylases (such as CCR4–NOT), followed by mRNA degradation (Seitz and Zamore, 2006; Bartel, 2009; Brodersen and Voinnet, 2009). Thus, by regulating the expression of genes involved in different genetic pathways through the post-transcriptional regulation, miRNAs have been demonstrated to play important roles in a wide array of biological processes, including development and cell differentiation, cell proliferation, apoptosis and metabolism (Wightman et al., 1993; Brennecke et al., 2003; Giraldez et al., 2005; Chen et al., 2006; Wilfred et al., 2007).
miRNAs in cancer
Results from numerous studies have revealed the important roles of miRNAs in tumorigenesis. miRNAs play important roles in regulating development and differentiation, cell proliferation, apoptosis and metabolism, all of which are often perturbed in tumors. By knocking down the endogenous expression of 90 different miRNAs with anti-sense oligonucleotides, a study has shown that cell proliferation and apoptosis can be regulated by multiple miRNAs in cancer cells, which suggests that miRNAs may promote tumorigenesis by modulating the expression of critical genes involved in tumor cell proliferation and survival (Cheng et al., 2005). Global alterations in miRNA expression patterns are a common feature of tumors; aberrant miRNA expression, and amplification or deletion of miRNAs have been frequently observed in various human tumors (Caldas and Brenton, 2005; Lu et al., 2005; Calin and Croce, 2006; Kent and Mendell, 2006). Alteration of maturation and biogenesis of miRNAs also greatly impacts upon tumorigenesis. It has been shown that global repression of miRNA maturation by suppression of key components of the miRNA biogenesis machinery in cancer cells promotes cell growth, transformation and tumorigenesis (Kumar et al., 2007). In addition, more than 50% of human miRNA genes are found to be located in cancer-associated genomic regions or at fragile sites, which further suggests the important role of miRNAs in tumorigenesis (Calin et al., 2004).
Emerging evidence has suggested that miRNAs can act as oncogenes or tumor suppressor genes in tumorigenesis. For example, the miR-17-92 cluster miRNAs, which are transcribed as a polycistronic unit, are highly expressed in many tumors, including lymphoma and colorectal cancer. They can augment the oncogenic effect of the c-Myc and therefore function as an oncogene (He et al., 2005; Diosdado et al., 2009; Mu et al., 2009). miR-221 and miR-222 are frequently overexpressed in lung and liver cancers. Their overexpression has been demonstrated to enhance tumorigenicity through the down-regulation of PTEN and TIMP3 tumor suppressors (Garofalo et al., 2009). On the other hand, some miRNAs have been shown to act as tumor suppressors. Let-7 family miRNAs can directly suppress the expression of oncogenes, including Ras and c-Myc, and therefore show tumor suppressive functions (Johnson et al., 2005; Roush and Slack, 2008). Consistent with their functions in tumor suppression, their expressions are frequently down-regulated in various cancers, including lung and colorectal cancers (Johnson et al., 2005; Roush and Slack, 2008). miR-15a and miR-16-1 are found to be deleted or down-regulated in more than 65% of B-cell chromic lymphocytic leukemia (B-CLL). They can negatively regulate anti-apoptotic protein BCL2. Therefore, decreased expression of miR-15a and miR-16-1 up-regulates BCL2 levels and reduces apoptosis, thereby contributing to malignant transformation (Calin et al., 2002, 2008).
The transcription regulation of miRNAs by p53
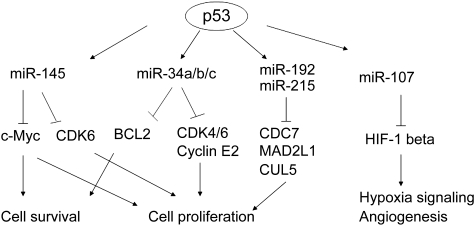
The interactions between p53 and miRNAs have been first demonstrated through the identification of several miRNAs as direct p53 target genes. As a transcription factor, p53 mainly exerts its function through its transcriptional regulation of protein-coding target genes to initiate cellular responses. Recent studies have revealed a novel mechanism for p53 in tumor suppression: p53 induces the expression of specific miRNAs which show tumor suppressive functions. In 2007, several groups reported that p53 can directly regulate the expression of the miR-34 family members (Chang et al., 2007; Corney et al., 2007; He et al., 2007; Raver-Shapira et al., 2007; Tazawa et al., 2007). miR-34 family consists of miR-34a, miR-34b and miR-34c, which are encoded by two different genes. miR-34a is encoded by an individual transcript and expressed in a majority of tissues, and miR-34b and miR-34c share a common primary transcript and are mainly expressed in lung tissues. p53 binds to p53 REs in miR-34a and miR-34b/c promoters and activates transcription of the miR-34 family. Ectopic expression of miR-34a promotes p53-mediated apoptosis, cell cycle arrest and senescence, whereas inactivation of endogenous miR-34a strongly inhibits p53-dependent apoptosis in cells. Studies have shown that miR-34 family members directly repress the expression of several targets involved in the regulation of cell cycle and in the promotion of cell proliferation and survival. These targets involve cyclin E2, cyclin-dependent kinases 4 and 6 (CDK4 and CDK6), and BCL2. Decreased expression of miR-34 has been frequently observed in various tumors (Bommer et al., 2007; Chang et al., 2007; Tazawa et al., 2007), suggesting that loss of miR-34 could promote tumorigenesis. These findings, for the first time, strongly suggest that in addition to many protein-encoding genes, miRNAs, the non-coding genes, can also be regulated by p53 as a new class of p53 target genes. Thus, miRNAs are involved in the composition of the complex p53 signaling pathway and contribute to the role of p53 in tumor suppression (Figure 2).
Figure 2.
The transcription regulation of specific miRNAs by p53. p53 induces the expression of a set of miRNAs, including miR-34a/b/c, miR-145, miR-107, miR-192 and miR-215, which can all contribute to the role of p53 in tumor suppression as a new group of p53 target genes. miR-34a/b/c down-regulate CDK4 and CDK6 to induce cell cycle arrest, and down-regulate BCL2 to promote apoptosis. miR-145 down-regulates c-Myc to reduce cell proliferation. miR-192 and miR-215 down-regulate a group of genes which regulate DNA synthesis and cell cycle checkpoints, including CDC7, MAD2L1 and CUL5, to induce cell cycle arrest and reduce tumor cell growth. miR-107 down-regulates HIF-1 beta to negatively regulate hypoxia signaling and suppress angiogenesis.
In addition to the miR-34 family, p53 also directly regulates the transcriptional expression of several additional miRNAs through binding to the p53 REs in their promoters, including miR-145, miR-107, miR-192 and miR-215 (Figure 2). For example, miR-145 was identified as a direct target for p53; p53 binds to a p53 RE in the miR-145 promoter and induces the expression of miR-145. miR-145 negatively regulates oncogene c-Myc; overexpression of miR-145 reduces c-Myc expression, whereas inactivation of endogenous miR-145 enhances c-Myc expression. This negative regulation of c-Myc by miR-145 accounts partially for the miR-145-mediated inhibition of tumor cell growth both in vitro and in vivo (Sachdeva et al., 2009). miR-107 is an additional target gene of p53, which contributes to the role of p53 in the regulation of hypoxic signaling and anti-angiogenesis (Yamakuchi et al., 2010). miR-107 can repress the expression of hypoxia inducible factor-1β (HIF-1β), which interacts with HIF-1α subunits to form a HIF-1 complex, which is a transcription factor that mediates the transcriptional response to hypoxia and plays an important role in tumorigenesis. Overexpression of miR-107 inhibits HIF-1β expression and hypoxic signaling, whereas inactivation of endogenous miR-107 enhances HIF-1β expression and hypoxic signaling in cells. Consistently, miR-107 levels are inversely associated with the expression levels of HIF-1β in human colon cancer. Ectopic expression of miR-107 in colon cancer cells suppresses angiogenesis and tumor growth in mice. Recently, p53 was also reported to induce the expression of miR-192 and miR-215 (Braun et al., 2008; Georges et al., 2008). The expression of these two miRNAs is severely reduced in many colon cancers. A set of downstream targets of miR-192 and miR-215 were identified, which include a number of regulators of DNA synthesis and the G1 and G2 cell cycle checkpoints in cells, such as CDC7, MAD2L1 and CUL5 (Georges et al., 2008). Overexpression of miR-192 and miR-215 in tumor cell lines can all result in the increased p21 protein levels, cell cycle arrest and suppression of tumor cell colony formation. Furthermore, these effects were partially p53-dependent. Taken together, these findings suggest an important role of p53 in the regulation of miRNA expression, and at the same time, also suggest an important role of miRNAs in mediating the role of p53 in tumor suppression as a new class of p53 target genes.
p53 regulates miRNA processing and maturation
A recent study revealed an additional mechanism for p53 in the regulation of miRNAs; p53 promotes the post-transcriptional maturation of miRNAs (Suzuki et al., 2009). Drosha plays a critical role in the processing of pri-miRNA transcripts into pre-miRNAs. For the maturation of some miRNAs (but not all), Drosha requires the involvement of RNA-associated proteins such as the DEAD box RNA helicases p68 and p72 (also known as DDX17) to carry out its function. It was recently reported that p53 promotes the Drosha-mediated processing of certain miRNAs with growth-suppressive functions, including miR-16-1, miR-143 and miR-145, in cells in response to DNA damage, such as doxorubicin treatment. These miRNAs are decreased in various human cancers, and overexpression of these miRNAs reduces tumor cell proliferation. These miRNAs negatively regulate some important regulators of the cell cycle and cell proliferation, such as K-Ras (as a target of miR-143) and CDK6 (as a target of miR-16-1 and miR-145). The study further shows that p53 interacts with Drosha in doxorubicin-treated cells and this interaction requires p68 and p72. p53 also mediates the interaction of pri-miRNAs with Drosha in response to doxorubicin and is required for the increased pri-miRNA processing activity that is associated with Drosha. Furthermore, DNA mutations in the p53 gene, such as R175H and R273H which are frequently observed in tumors, can lead to the decreased processing of pri-miRNAs by Drosha and decreased levels of pri-miRNAs and mature miRNAs in cells, including miR-16-1, miR-143 and miR-145. These findings suggest that transcription-independent modulation of miRNA biogenesis is intrinsically embedded in a tumor suppressive program governed by p53, which could be a new mechanism by which p53 mutation contributes to cancer. Further investigation would establish the full repertoire of miRNAs whose maturation is regulated by p53 through the aforementioned mechanism, and furthermore, would provide insights into how the specificities of recognition of pri-miRNAs are determined.
While p53 promotes the maturation of miRNAs, p53 also monitors the maturation of miRNAs. Loss of maturation of miRNAs can activate the p53 signaling and induce p53-mediated senescence (Mudhasani et al., 2008). The ablation of Dicer and loss of mature miRNAs in embryonic fibroblasts activate p53 and induce senescence which could be rescued by deletion of p53 (Mudhasani et al., 2008). Since p53-mediated senescence is an important mechanism by which p53 exerts its function in tumor suppression, loss of p53 function may greatly facilitate the tumorigenic potential of cells with reduced levels of mature miRNAs.
The regulation of the p53 and its pathway by miRNAs
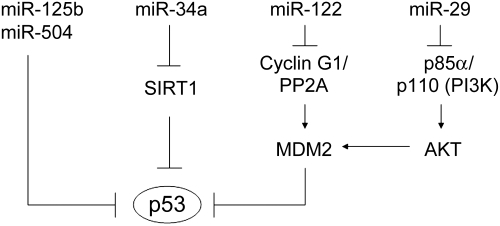
Computational analyses estimate that over 30% of all genes and the majority of genetic pathways are subject to regulation by multiple miRNAs (Bentwich et al., 2005; Lim et al., 2005; Griffiths-Jones et al., 2006). This raises a possibility that some specific miRNAs could regulate p53 and its pathway. To test this possibility, we screened for miRNAs which could potentially regulate p53 expression by performing an in silico search for putative binding sites of miRNAs in the 3′-UTR of human p53 gene. Through this approach, we identified miR-504 as a novel miRNA that can negatively regulate p53 expression through its binding to two binding sites in human p53 3′-UTR (Hu et al., 2010a). Our results demonstrate that ectopic expression of miR-504 reduces p53 protein levels and impairs p53 functions, especially p53-mediated apoptosis and G1 cell cycle arrest in cells in response to stress. Furthermore, miR-504 overexpression promotes tumorigenicity of colon cancer cells in vivo (Hu et al., 2010a). Our results demonstrate that p53 is subject to the negative regulation of specific miRNAs, which is a novel mechanism for cells to regulate p53 protein levels and functions (Figure 3). In addition to our study, a recent study reported that miR-125b is another novel miRNA targeting p53 (Le et al., 2009). miR-125b is a brain-enriched miRNA, which acts as a negative regulator of p53 in both zebrafish and humans. Overexpression of miR-125b represses the endogenous levels of p53 protein and reduces apoptosis in cells, whereas knockdown of miR-125b increases the levels of p53 protein and induces apoptosis in human cells and zebrafish brain. These results demonstrate that miR-125b is an important negative regulator of p53 and p53-induced apoptosis during development and during the stress response.
Figure 3.
Multiple miRNAs regulate the activity and function of p53. miR-125b and miR-504 directly down-regulate p53 protein levels and functions in apoptosis and cell cycle arrest through their direct binding to p53 3′-UTR. miR-34a up-regulates p53 activity and function by down-regulating SIRT1, which is a negative regulator of p53 through deacetylating p53. miR-122 enhances p53 activity through its down-regulation of cyclin G1, which forms a complex with PP2A phosphatase and enhances MDM2 activity to inhibit p53. miR-29 down-regulates p85α, a regulatory subunit of PI3K, and thereby enhances p53 activity through the negative loop between PI3K-AKT-MDM2 and p53.
In addition to the direct negative regulation of the p53 protein by miRNAs, recent studies also show that p53 can be indirectly regulated by several miRNAs through the regulation of regulators of p53 in cells, including miR-34a, miR-29 and miR-122 (Figure 3). miR-34a, a transcription target of the p53 protein, was found to positively regulate p53 activity and function in apoptosis through its direct negative regulation of SIRT1 (Yamakuchi et al., 2008). SIRT1 is a negative regulator of p53, which physically interacts with p53 and deacetylates Lys382 of p53 (Luo et al., 2001). Overexpression of miR-34a decreases SIRT1 expression, leading to the increase in acetylated p53 levels and p53 activity, which in turn induces apoptosis in a p53-dependent manner in cells (Yamakuchi et al., 2008). Recently, miR-29 was identified as another positive regulator of p53 (Park et al., 2009). miR-29 family members (miR-29a, miR-29b and miR-29c) up-regulate p53 protein levels and induce p53-mediated apoptosis through repression of p85α, a regulatory subunit of PI3 kinase (PI3K). The PI3K/AKT pathway can negatively regulate p53 activity through the direct phosphorylation and activation of MDM2 by AKT (Zhou et al., 2001). As a positive upstream regulator of AKT, PI3K is composed of a catalytic subunit, p110, and a regulatory subunit, p85α, which binds to p110 and stabilizes it. By targeting p85α, miR-29 family members reduce PI3K/AKT activity and this results in the reduced phosphorylation of AKT and MDM2, which in turn leads to the activation of p53. Thus, miR-29 up-regulates p53 through the negative loop between PI3K–AKT–MDM2 and p53. Down-regulation of miR-29 miRNAs has been reported in various cancers, including leukemia, and lung and breast cancers. Furthermore, miR-122 was recently shown to increase p53 protein levels and activity through its negative regulation of cyclin G1 (Fornari et al., 2009). MiR-122 is a liver-specific miRNA; its down-regulation is a common feature of both human and mouse liver cancer. Cyclin G1 is transcriptionally regulated by p53. Cyclin G1 protein forms a complex with the PP2A phosphatase, and removes a phosphate residue from MDM2, which is added to the MDM2 protein by cyclin A/cdk2 (Okamoto et al., 2002). The phosphorylation of MDM2 by cyclin A/cdk2 inhibits MDM2 activity, and the cyclin G1/PP2A enhances MDM2 activity and inhibits p53. Thus, cyclin G1 forms a negative feedback loop with p53 to negatively regulate p53. Cyclin G1 is overexpressed in several human cancers, including breast cancers. By directly repressing the expression of cyclin G1, miR-122 increases p53 protein levels and activity and inhibits tumorigenesis in liver cancer.
Conclusion
These findings from recent studies have demonstrated that p53 cross-talks with miRNAs at multiple levels. First, p53 induces the transcription expression of a set of miRNAs, including miR-34a/b/c, miR-145, miR-107, miR-192 and miR-215. These miRNAs have been shown to be able to mediate the role of p53 in tumor suppression through inducing apoptosis, cell cycle arrest and/or senescence (Figure 2). Second, p53 promotes the Drosha-mediated processing and maturation of certain miRNAs with growth-suppressive function, including miR-16-1, miR-143 and miR-145. At the same time, p53 monitors the maturation of miRNAs; loss of miRNA maturation activates p53 and induces p53-mediated senescence. Third, miRNAs can negatively regulate p53 function through direct repression of the p53 protein, or can positively regulate p53 activity and function through the regulation of some negative regulators of the p53 protein. For example, p53 can be negatively regulated by specific miRNAs, such as miR-504 and miR-125b, which can directly down-regulate p53 protein levels and function. At the same time, some specific miRNAs positively regulate p53 through the direct repression of SIRT1 (by miR-34a), p85α (by miR-29) or cyclin G1 (by miR-122) (Figure 3). These findings demonstrated that through the regulation of transcription expression and/or maturation of specific miRNAs by p53, miRNAs could be a new group of p53 target genes to mediate the function of p53 in tumor suppression. At the same time, miRNAs could be a new group of regulators for p53, joining a panel of kinases (e.g. ATM, ATR), ubiquitin ligases (e.g. MDM2, Cop1, Pirh2) and phosphatases (e.g. Wip1, PP2A) to tightly regulate the levels and activity of p53 (Figure 1).
miRNAs have emerged as a class of most abundant regulatory genes in humans. Over 700 miRNAs have been identified, and more miRNAs are expected to be identified. Each miRNA is predicted to have numerous putative targets that have disparate functions. A tremendous amount of work has been done on the miRNAs since their discovery, which makes miRNAs one of the most rapidly growing fields. However, the miRNA world remains largely uncharted since the biological functions and target genes of most miRNAs remain unknown. With the identification of more targets of miRNAs and biological functions of miRNAs, we are expecting that more miRNAs will be identified to regulate one or more gene products involved in the p53 signaling pathway which consists of hundreds of proteins. These miRNAs can positively or negatively regulate the activity and function of the p53 signaling pathway. Furthermore, with the identification of more miRNAs, more miRNAs could be identified as direct p53 target genes. Therefore, we are only beginning to see how miRNAs join the p53 network, and many questions remain to be addressed. One of future challenges is to identify systematically all the miRNAs that regulate the p53 signaling pathway or are regulated by the p53 pathway in different cellular contexts and, furthermore, to study the physiological and pathological (e.g. tumor) significance of these regulations.
miRNA dysregulation has been suggested to be an important mechanism for the aberrant expression of a list of oncogenes and/or tumor suppressor genes which are not affected directly by genetic mutations or transcriptional regulation in tumorigenesis. The activity and function of p53 are frequently down-regulated in a number of cancers without mutations in the p53 gene itself, and miRNA dysregulation could be an important mechanism. Considering the relative chemical simplicity of miRNA molecules, introduction of miRNAs or miRNA antagonists into tumor cells to restore the tumor-suppressive effects of p53 raises exciting possibilities for novel tumor therapeutic approaches. It would be of interest to investigate whether we can restore p53 function and induce tumor regression by introducing specific p53-induced miRNAs and miRNAs positively regulating p53 function which are down-regulated in tumors, or by introducing antagonists for specific miRNAs which negatively regulate p53 functions and are up-regulated in tumors. These future studies will provide new insights into the interactions between miRNAs and the p53 pathway and open a new window for therapeutic intervention in cancer.
Conflict of interest: none declared.
Funding
Z.F. is supported by the grant from National Institutes of Health (1R01CA143204-01) and New Jersey Commission on Cancer Research. W.H. is supported by the grant from National Institutes of Health (1P30CA147892-01).
References
- Bartel D.P. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensaad K., Tsuruta A., Selak M.A., et al. TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell. 2006;126:107–120. doi: 10.1016/j.cell.2006.05.036. [DOI] [PubMed] [Google Scholar]
- Bentwich I., Avniel A., Karov Y., et al. Identification of hundreds of conserved and nonconserved human microRNAs. Nat. Genet. 2005;37:766–770. doi: 10.1038/ng1590. [DOI] [PubMed] [Google Scholar]
- Berezikov E., Guryev V., van de Belt J., et al. Phylogenetic shadowing and computational identification of human microRNA genes. Cell. 2005;120:21–24. doi: 10.1016/j.cell.2004.12.031. [DOI] [PubMed] [Google Scholar]
- Bommer G.T., Gerin I., Feng Y., et al. p53-mediated activation of miRNA34 candidate tumor-suppressor genes. Curr. Biol. 2007;17:1298–1307. doi: 10.1016/j.cub.2007.06.068. [DOI] [PubMed] [Google Scholar]
- Borchert G.M., Lanier W., Davidson B.L. RNA polymerase III transcribes human microRNAs. Nat. Struct. Mol. Biol. 2006;13:1097–1101. doi: 10.1038/nsmb1167. [DOI] [PubMed] [Google Scholar]
- Braithwaite A.W., Prives C.L. p53: more research and more questions. Cell Death Differ. 2006;13:877–880. doi: 10.1038/sj.cdd.4401938. [DOI] [PubMed] [Google Scholar]
- Braun C.J., Zhang X., Savelyeva I., et al. p53-responsive microRNAs 192 and 215 are capable of inducing cell cycle arrest. Cancer Res. 2008;68:10094–10104. doi: 10.1158/0008-5472.CAN-08-1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J., Hipfner D.R., Stark A., et al. Bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell. 2003;113:25–36. doi: 10.1016/s0092-8674(03)00231-9. [DOI] [PubMed] [Google Scholar]
- Brodersen P., Voinnet O. Revisiting the principles of microRNA target recognition and mode of action. Nat. Rev. Mol. Cell Biol. 2009;10:141–148. doi: 10.1038/nrm2619. [DOI] [PubMed] [Google Scholar]
- Brooks C.L., Gu W. p53 ubiquitination: Mdm2 and beyond. Mol. Cell. 2006;21:307–315. doi: 10.1016/j.molcel.2006.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budanov A.V., Sablina A.A., Feinstein E., et al. Regeneration of peroxiredoxins by p53-regulated sestrins, homologs of bacterial AhpD. Science. 2004;304:596–600. doi: 10.1126/science.1095569. [DOI] [PubMed] [Google Scholar]
- Caldas C., Brenton J.D. Sizing up miRNAs as cancer genes. Nat. Med. 2005;11:712–714. doi: 10.1038/nm0705-712. [DOI] [PubMed] [Google Scholar]
- Calin G.A., Croce C.M. MicroRNA signatures in human cancers. Nat. Rev. Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- Calin G.A., Dumitru C.D., Shimizu M., et al. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. USA. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin G.A., Sevignani C., Dumitru C.D., et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc. Natl. Acad. Sci. USA. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin G.A., Cimmino A., Fabbri M., et al. MiR-15a and miR-16-1 cluster functions in human leukemia. Proc. Natl. Acad. Sci. USA. 2008;105:5166–5171. doi: 10.1073/pnas.0800121105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang T.C., Wentzel E.A., Kent O.A., et al. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol. Cell. 2007;26:745–752. doi: 10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J.F., Mandel E.M., Thomson J.M., et al. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat. Genet. 2006;38:228–233. doi: 10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng A.M., Byrom M.W., Shelton J., et al. Antisense inhibition of human miRNAs and indications for an involvement of miRNA in cell growth and apoptosis. Nucleic Acids Res. 2005;33:1290–1297. doi: 10.1093/nar/gki200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corney D.C., Flesken-Nikitin A., Godwin A.K., et al. MicroRNA-34b and microRNA-34c are targets of p53 and cooperate in control of cell proliferation and adhesion-independent growth. Cancer Res. 2007;67:8433–8438. doi: 10.1158/0008-5472.CAN-07-1585. [DOI] [PubMed] [Google Scholar]
- Diosdado B., van de Wiel M.A., Terhaar Sive Droste J.S., et al. MiR-17-92 cluster is associated with 13q gain and c-myc expression during colorectal adenoma to adenocarcinoma progression. Br. J. Cancer. 2009;101:707–714. doi: 10.1038/sj.bjc.6605037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donehower L.A., Harvey M., Slagle B.L., et al. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature. 1992;356:215–221. doi: 10.1038/356215a0. [DOI] [PubMed] [Google Scholar]
- el-Deiry W.S., Kern S.E., Pietenpol J.A., et al. Definition of a consensus binding site for p53. Nat. Genet. 1992;1:45–49. doi: 10.1038/ng0492-45. [DOI] [PubMed] [Google Scholar]
- Feng Z., Levine A.J. The regulation of energy metabolism and the IGF-1/mTOR pathways by the p53 protein. Trends Cell Biol. 2010;20:427–434. doi: 10.1016/j.tcb.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornari F., Gramantieri L., Giovannini C., et al. MiR-122/cyclin G1 interaction modulates p53 activity and affects doxorubicin sensitivity of human hepatocarcinoma cells. Cancer Res. 2009;69:5761–5767. doi: 10.1158/0008-5472.CAN-08-4797. [DOI] [PubMed] [Google Scholar]
- Garofalo M., Di Leva G., Romano G., et al. miR-221&222 regulate TRAIL resistance and enhance tumorigenicity through PTEN and TIMP3 downregulation. Cancer Cell. 2009;16:498–509. doi: 10.1016/j.ccr.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Georges S.A., Biery M.C., Kim S.Y., et al. Coordinated regulation of cell cycle transcripts by p53-inducible microRNAs, miR-192 and miR-215. Cancer Res. 2008;68:10105–10112. doi: 10.1158/0008-5472.CAN-08-1846. [DOI] [PubMed] [Google Scholar]
- Giraldez A.J., Cinalli R.M., Glasner M.E., et al. MicroRNAs regulate brain morphogenesis in zebrafish. Science. 2005;308:833–838. doi: 10.1126/science.1109020. [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones S., Grocock R.J., van Dongen S., et al. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimson A., Farh K.K., Johnston W.K., et al. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol. Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris S.L., Levine A.J. The p53 pathway: positive and negative feedback loops. Oncogene. 2005;24:2899–2908. doi: 10.1038/sj.onc.1208615. [DOI] [PubMed] [Google Scholar]
- He L., Thomson J.M., Hemann M.T., et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L., He X., Lim L.P., et al. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W., Chan C.S., Wu R., et al. Negative regulation of tumor suppressor p53 by microRNA miR-504. Mol. Cell. 2010a;38:689–699. doi: 10.1016/j.molcel.2010.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W., Zhang C., Wu R., et al. Glutaminase 2, a novel p53 target gene regulating energy metabolism and antioxidant function. Proc. Natl. Acad. Sci. USA. 2010b;107:7455–7460. doi: 10.1073/pnas.1001006107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang B.J., Ford J.M., Hanawalt P.C., et al. Expression of the p48 xeroderma pigmentosum gene is p53-dependent and is involved in global genomic repair. Proc. Natl. Acad. Sci. USA. 1999;96:424–428. doi: 10.1073/pnas.96.2.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inui M., Martello G., Piccolo S. MicroRNA control of signal transduction. Nat. Rev. Mol. Cell Biol. 2010;11:252–263. doi: 10.1038/nrm2868. [DOI] [PubMed] [Google Scholar]
- Jacks T., Remington L., Williams B.O., et al. Tumor spectrum analysis in p53-mutant mice. Curr. Biol. 1994;4:1–7. doi: 10.1016/s0960-9822(00)00002-6. [DOI] [PubMed] [Google Scholar]
- Johnson S.M., Grosshans H., Shingara J., et al. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- Kent O.A., Mendell J.T. A small piece in the cancer puzzle: microRNAs as tumor suppressors and oncogenes. Oncogene. 2006;25:6188–6196. doi: 10.1038/sj.onc.1209913. [DOI] [PubMed] [Google Scholar]
- Kloosterman W.P., Plasterk R.H. The diverse functions of microRNAs in animal development and disease. Dev. Cell. 2006;11:441–450. doi: 10.1016/j.devcel.2006.09.009. [DOI] [PubMed] [Google Scholar]
- Kumar M.S., Lu J., Mercer K.L., et al. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat. Genet. 2007;39:673–677. doi: 10.1038/ng2003. [DOI] [PubMed] [Google Scholar]
- Le M.T., Teh C., Shyh-Chang N., et al. MicroRNA-125b is a novel negative regulator of p53. Genes Dev. 2009;23:862–876. doi: 10.1101/gad.1767609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee R.C., Feinbaum R.L., Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- Lee Y., Han J., Yeom K.H., et al. Drosha in primary microRNA processing. Cold Spring Harb. Symp. Quant. Biol. 2006;71:51–57. doi: 10.1101/sqb.2006.71.041. [DOI] [PubMed] [Google Scholar]
- Levine A.J., Finlay C.A., Hinds P.W. P53 is a tumor suppressor gene. Cell. 2004;116:S67–S69. doi: 10.1016/s0092-8674(04)00036-4. 61 p following S69. [DOI] [PubMed] [Google Scholar]
- Levine A.J., Hu W., Feng Z. The P53 pathway: what questions remain to be explored? Cell Death Differ. 2006;13:1027–1036. doi: 10.1038/sj.cdd.4401910. [DOI] [PubMed] [Google Scholar]
- Lim L.P., Lau N.C., Garrett-Engele P., et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- Lu J., Getz G., Miska E.A., et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- Luo J., Nikolaev A.Y., Imai S., et al. Negative control of p53 by Sir2alpha promotes cell survival under stress. Cell. 2001;107:137–148. doi: 10.1016/s0092-8674(01)00524-4. [DOI] [PubMed] [Google Scholar]
- Matoba S., Kang J.G., Patino W.D., et al. p53 regulates mitochondrial respiration. Science. 2006;312:1650–1653. doi: 10.1126/science.1126863. [DOI] [PubMed] [Google Scholar]
- Mu P., Han Y.C., Betel D., et al. Genetic dissection of the miR-17∼92 cluster of microRNAs in Myc-induced B-cell lymphomas. Genes Dev. 2009;23:2806–2811. doi: 10.1101/gad.1872909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudhasani R., Zhu Z., Hutvagner G., et al. Loss of miRNA biogenesis induces p19Arf-p53 signaling and senescence in primary cells. J. Cell Biol. 2008;181:1055–1063. doi: 10.1083/jcb.200802105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto K., Li H., Jensen M.R., et al. Cyclin G recruits PP2A to dephosphorylate Mdm2. Mol. Cell. 2002;9:761–771. doi: 10.1016/s1097-2765(02)00504-x. [DOI] [PubMed] [Google Scholar]
- Olivier M., Hussain S.P., Caron de Fromentel C., et al. TP53 mutation spectra and load: a tool for generating hypotheses on the etiology of cancer. IARC Sci. Publ. 2004;157:247–270. [PubMed] [Google Scholar]
- Ozsolak F., Poling L.L., Wang Z., et al. Chromatin structure analyses identify miRNA promoters. Genes Dev. 2008;22:3172–3183. doi: 10.1101/gad.1706508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.Y., Lee J.H., Ha M., et al. miR-29 miRNAs activate p53 by targeting p85 alpha and CDC42. Nat. Struct. Mol. Biol. 2009;16:23–29. doi: 10.1038/nsmb.1533. [DOI] [PubMed] [Google Scholar]
- Pillai R.S., Bhattacharyya S.N., Filipowicz W. Repression of protein synthesis by miRNAs: how many mechanisms? Trends Cell Biol. 2007;17:118–126. doi: 10.1016/j.tcb.2006.12.007. [DOI] [PubMed] [Google Scholar]
- Raver-Shapira N., Marciano E., Meiri E., et al. Transcriptional activation of miR-34a contributes to p53-mediated apoptosis. Mol. Cell. 2007;26:731–743. doi: 10.1016/j.molcel.2007.05.017. [DOI] [PubMed] [Google Scholar]
- Reinhart B.J., Slack F.J., Basson M., et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- Roush S., Slack F.J. The let-7 family of microRNAs. Trends Cell Biol. 2008;18:505–516. doi: 10.1016/j.tcb.2008.07.007. [DOI] [PubMed] [Google Scholar]
- Sablina A.A., Budanov A.V., Ilyinskaya G.V., et al. The antioxidant function of the p53 tumor suppressor. Nat. Med. 2005;11:1306–1313. doi: 10.1038/nm1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachdeva M., Zhu S., Wu F., et al. p53 represses c-Myc through induction of the tumor suppressor miR-145. Proc. Natl. Acad. Sci. USA. 2009;106:3207–3212. doi: 10.1073/pnas.0808042106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seitz H., Zamore P.D. Rethinking the microprocessor. Cell. 2006;125:827–829. doi: 10.1016/j.cell.2006.05.018. [DOI] [PubMed] [Google Scholar]
- Suzuki H.I., Yamagata K., Sugimoto K., et al. Modulation of microRNA processing by p53. Nature. 2009;460:529–533. doi: 10.1038/nature08199. [DOI] [PubMed] [Google Scholar]
- Tanaka H., Arakawa H., Yamaguchi T., et al. A ribonucleotide reductase gene involved in a p53-dependent cell-cycle checkpoint for DNA damage. Nature. 2000;404:42–49. doi: 10.1038/35003506. [DOI] [PubMed] [Google Scholar]
- Tazawa H., Tsuchiya N., Izumiya M., et al. Tumor-suppressive miR-34a induces senescence-like growth arrest through modulation of the E2F pathway in human colon cancer cells. Proc. Natl. Acad. Sci. USA. 2007;104:15472–15477. doi: 10.1073/pnas.0707351104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teodoro J.G., Parker A.E., Zhu X., et al. p53-mediated inhibition of angiogenesis through up-regulation of a collagen prolyl hydroxylase. Science. 2006;313:968–971. doi: 10.1126/science.1126391. [DOI] [PubMed] [Google Scholar]
- Vasudevan S., Tong Y., Steitz J.A. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318:1931–1934. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- Vousden K.H., Prives C. Blinded by the light: the growing complexity of p53. Cell. 2009;137:413–431. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- Wightman B., Ha I., Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- Wilfred B.R., Wang W.X., Nelson P.T. Energizing miRNA research: a review of the role of miRNAs in lipid metabolism, with a prediction that miR-103/107 regulates human metabolic pathways. Mol. Genet. Metab. 2007;91:209–217. doi: 10.1016/j.ymgme.2007.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamakuchi M., Ferlito M., Lowenstein C.J. miR-34a repression of SIRT1 regulates apoptosis. Proc. Natl. Acad. Sci. USA. 2008;105:13421–13426. doi: 10.1073/pnas.0801613105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamakuchi M., Lotterman C.D., Bao C., et al. P53-induced microRNA-107 inhibits HIF-1 and tumor angiogenesis. Proc. Natl. Acad. Sci. USA. 2010;107:6334–6339. doi: 10.1073/pnas.0911082107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B.P., Liao Y., Xia W., et al. HER-2/neu induces p53 ubiquitination via Akt-mediated MDM2 phosphorylation. Nat. Cell Biol. 2001;3:973–982. doi: 10.1038/ncb1101-973. [DOI] [PubMed] [Google Scholar]