Abstract
BRCA1 plays a critical role in the regulation of homologous recombination (HR)-mediated DNA double-strand break repair. BRCA1-deficient cancers have evolved to tolerate loss of BRCA1 function. This renders them vulnerable to agents, such as PARP inhibitors, that are conditionally ‘synthetic lethal' with their underlying repair defect. Recent studies demonstrate that BRCA1-deficient cells may acquire resistance to these agents by partially correcting their defect in HR-mediated repair, either through reversion mutations in BRCA1 or through ‘synthetic viable' loss of 53BP1. These findings and their clinical implications will be reviewed in this article.
Keywords: DNA repair, BRCA1, 53BP1, PARP
Introduction
BRCA1 is a critical component of the homologous recombination (HR)-mediated DNA repair pathway. Tumors that arise in BRCA1-mutation carriers show evidence of genomic instability and impaired DNA repair. The repair defect present in BRCA1-mutant cancers has been used to develop ‘synthetic lethal' treatment strategies that have been clinically validated. Recent data suggest that BRCA1-mutant cells can evade ‘synthetic lethal' treatment approaches and restore DNA repair capacity by several surprising mechanisms. In this article, the data behind synthetic lethal approaches to treatment of BRCA1-mutant cancers, and mechanisms of escape and ‘synthetic viability' will be reviewed.
The BRCA1 tumor syndrome
Women carrying a germline mutation in BRCA1 have a 50–80% lifetime risk of developing breast cancer and a 20–40% lifetime risk of developing ovarian cancer (King et al., 2003). The tumors that arise in these women have undergone loss of the wt BRCA1 allele, demonstrating that BRCA1 functions as a tumor suppressor (Neuhausen and Marshall, 1994). The breast cancers that arise tend to be of early onset, high grade, invasive ductal cancers that do not express the estrogen receptor or progesterone receptor, do not have amplification of HER2 (a ‘triple-negative' phenotype) and cluster with the ‘basal-like' subclass of breast cancers by gene expression profiling (Sorlie et al., 2003; Turner and Reis-Filho, 2006; Shakya et al., 2008). These tumors have high frequency of mutations in p53 and PTEN, and display a high level of genomic changes (Saal et al., 2008; Holstege et al., 2009; Manie et al., 2009).
BRCA1 and DNA repair
BRCA1 is a large protein that is expressed during the S and G2 phases of the cell cycle and mostly localized in discrete nuclear foci (Scully et al., 1996; 1997b). Upon DNA damage, BRCA1 is rapidly phosphorylated and redistributes to sites of DNA breaks, where it co-localizes with RAD51 and other proteins involved in DNA double-strand break (DSB) repair (DSBR) (Scully et al., 1997a). Cells lacking wt BRCA1 have a profound defect in DNA DSBR that can be partially complemented by re-introducing wt but not mutant BRCA1 (Scully et al., 1999). BRCA1-mutant cells also have a defect in DNA-damage induced S-phase and G2/M checkpoint function. The role of BRCA1 in DNA repair was further clarified when it was demonstrated that BRCA1 is specifically required for efficient HR-mediated DSBR (Moynahan et al., 1999; Snouwaert et al., 1999; Moynahan et al., 2001; Westermark et al., 2003). BRCA1 may also participate in other repair processes, including transcription-coupled repair, non-homologous end joining (NHEJ), and nucleotide-excision repair (Abbott et al., 1999; Hartman and Ford, 2002; Zhong et al., 2002; Bau et al., 2006). However, it is clear that BRCA1 function is critical for HR-mediated DSBR, and this pathway will the focus of this review.
The functional role of BRCA1 in DNA DSBR
Although BRCA1 is clearly required for efficient HR-mediated DSBR, its specific functional role in this process remains elusive. BRCA1 forms a stable heterodimer with BARD1, and the BRCA1–BARD1 complex has been shown to be a potent E3 ubiquitin ligase in vitro (Wu et al., 1996; Xia et al., 2003). This observation suggested that this enzymatic activity might be crucial to its function in DNA repair and tumorigenesis. Recent elegant mouse experiments have unexpectedly found that the ubiquitin ligase function of BRCA1 may be dispensable for its role in DNA repair (Reid et al., 2008). ES cells were generated that were homozygous for a BRCA1 mutation that rendered it inactive as an E3 ligase in vitro but retained its ability to form a stable heterodimer with BARD1. These E3-ligase dead BRCA1-mutant ES cells were found to be viable, did not accumulate spontaneous karyotypic abnormalities, and showed no measurable defect in homology-mediated repair of DNA breaks (Reid et al., 2008). The function role of the E3 ligase activity of the BRCA1–BARD complex remains uncertain at present.
BRCA1 interacts with multiple proteins involved in DNA repair and tumorigenesis, including, PALB2, BRIP1/BACH1, CtIP, TOPBP1, Abraxas, and RAP80 and is present in multiple distinct complexes (Wong et al., 1998; Yu et al., 1998; Cantor et al., 2001; Greenberg et al., 2006; Xia et al., 2006; Kim et al., 2007; Sobhian et al., 2007; Wang et al., 2007; Zhang et al., 2009b). Thus, BRCA1 may function partly as a scaffold protein coordinating the assembly of protein complexes involved in mediating HR-mediated DSBR and checkpoint function (Greenberg et al., 2006). Mutations that affect the BRCT domains of BRCA1 lead to increased cancer risk in humans, and when homozygous cause embryonic lethality in the mouse (Hohenstein et al., 2001; Williams et al., 2001). Thus, the BRCT domains of BRCA1 likely mediate critical interactions required for assembly of protein complexes required for proper DNA repair function and tumor suppression. This includes interaction of BRCA1 BRCT domains with: (i) BACH1/BRIP1 and TOBP1 (Cantor et al., 2001; Greenberg et al., 2006); (ii) Abraxas and RAP80 (Kim et al., 2007; Liu et al., 2007b; Wang et al., 2007); and (iii) CtIP and the MRE11/NBS/RAD50 complex (Wong et al., 1998; Sartori et al., 2007; Chen et al., 2008).
The interaction of BRCA1 with CtIP maybe especially critical for the regulation of processing of DNA ends at DSBs to promote HR (Sartori et al., 2007; Chen et al., 2008). BRCA1 also interacts with PALB2 linking it functionally to BRCA2 and its role in RAD51 loading during HR (Xia et al., 2006; Sy et al., 2009; Zhang et al., 2009a,b). However, much about the actual functional role of BRCA1 in the DNA repair process, and the regulation of its interaction with multiple DNA repair proteins remains unclear.
Targeting the role of BRCA1 in DNA repair in cancer therapy: interstrand cross-linking agents
Several studies have demonstrated that BRCA1-deficient cells are highly sensitive to interstrand cross-linking (ICL) agents such as cisplatin and mitomycin C (Moynahan et al., 2001; Tassone et al., 2003; 2009). The interstrand DNA crosslinks induced by these agents disrupt replication forks during S-phase and require efficient HR-mediated DSBR for viable S-phase progression and cell survival. Cell with defects in HR-mediated DSBR, including cells with mutations in BRCA1, BRCA2, or the Fanconi Anemia genes, are exquisitely sensitive to ICLs. This observation suggested that ICL might be utilized as targeted therapy for BRCA1-mutant breast and ovarian cancers. Breast cancers that arose in mice engineered to have tissue-specific knockout of Brca1 and p53 are exquisitely sensitive to treatment with platinum compounds (Rottenberg et al., 2007). Moreover, these BRCA1−/− tumors did not develop resistance to despite repeated treatments and late recurrences remained platinum sensitive (Rottenberg et al., 2007).
Clinical trials have validated the efficacy of ICL agents in the treatment of BRCA1-associated breast and ovarian cancers. Cisplatin has been shown to have a high rate of clinical–pathological complete responses when used as neoadjuvant treatment of breast cancer arising in BRCA1-mutation carriers (Byrski et al., 2009; 2010). Sporadic triple-negative breast cancers also had a significant rate of complete response to platinum therapy, suggesting that a subset of these cancers may be ‘BRCA1-like' and have a similar defect in HR-mediated repair (Silver et al., 2010). This has led to a great interest in defining the role of ICL compounds in the standard treatment of both BRCA1-associated cancers and sporadic triple-negative breast cancers.
Poly(ADP-ribose)polymerase and HR-mediated DNA repair pathways: conditional synthetic lethality
A major breakthrough in targeted treatment of BRCA1-mutant cancers was heralded by the finding that BRCA1 and BRCA2-mutant cells are exquisitely sensitive to poly(ADP-ribose)polymerase (PARP) inhibitors (Farmer et al., 2005; Helleday et al., 2005). PARP activity plays a critical role in mediating some of the chromatin changes required for efficient DNA repair. PARP1 (and the closely related PARP2) is rapidly activated at sites of DNA breaks, where it catalyzes the formation of poly(ADP-ribose) (PAR) polymers both on itself (autoPARylation) and local substrates (Satoh and Lindahl, 1992; Ame et al., 1999; Allinson et al., 2003; Woodhouse and Dianov, 2008). DNA damage associated PARylation, either directly or through the recruitment of proteins such as APLF and the chromatin remodeling enzyme ALC1, induces changes in local chromatin structure that lead to chromatin relaxation and recruitment of other DNA repair proteins (Tulin and Spradling, 2003; Ahel et al., 2008; 2009; Rulten et al., 2008; Gottschalk et al., 2009).
The role of PARP1 is best characterized in single-strand break (SSB) repair. PARP1 and PARP2 activity are critical for the efficient processing of DNA SSBs, and recruitment of XRCC1 (Masson et al., 1998; Sanderson and Lindahl, 2002; El-Khamisy et al., 2003; Woodhouse and Dianov, 2008; Figure 1). In the setting of PARP inhibition, unrepaired SSBs accumulate and can cause replication fork collapse and DNA DSB equivalents upon entry into S-phase (Oikawa et al., 1980; Wang et al., 1997; Simbulan-Rosenthal et al., 1999). These lesions must be repaired, and replication forks restarted using HR-mediated DSBR pathways, or the cell will accumulate lethal levels of DNA breaks. An increased level of HR-mediated DSBR during S-phase can allow cells to tolerate loss of PARP1 activity. Consistent with this model, PARP1-deficient mice are viable and have a relatively mild phenotype. However, both PARP1−/− cells and normal cell treated with PARP inhibitors show an increased level of sister-chromatid exchange and increased HR-mediated repair of DNA DSBs during S-phase (Oikawa et al., 1980; Wang et al., 1997; Simbulan-Rosenthal et al., 1999). The standard model to explain the sensitivity of BRCA1−/− cells to PARP inhibitors invokes the effect on PARP inhibitors on SSB repair and subsequent generation of large amounts of stalled replication forks and DSB equivalents which are lethal to HR-defective cells (Helleday et al., 2005; Ashworth, 2008b; Figure 1).
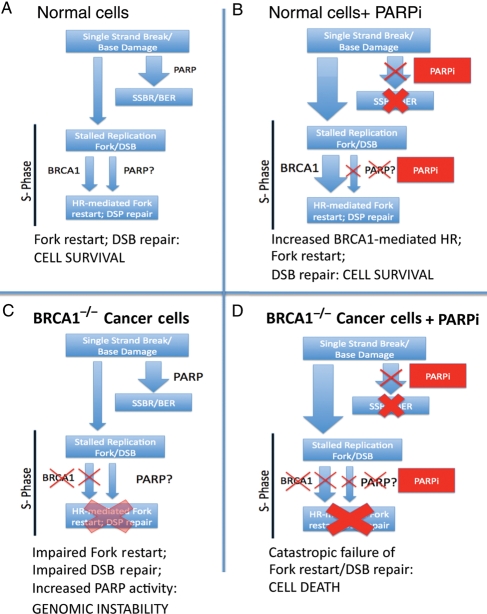
Figure 1.
Conditional synthetic lethality of PARP inhibitors with BRCA1−/− cells. (A) In normal cells, single-strand breaks and base lesions are repaired efficiently by PARP mediated single-strand break repair (SSBR)/base excision repair (BER). The few unrepaired breaks that persist into S-phase and lead to replication fork arrest and double-strand breaks (DSBs) are efficiently handled by BRCA1-dependent HR-mediated repair and fork restart. PARP may also play a role in fork restart. (B) In the setting of PARP inhibitor (PARPi), SSBR is impaired leading to accumulation of unrepaired single-strand breaks. This results in an great increase in the amount of replication fork lesions that must be repaired by a BRCA1-dependent HR-mediated repair pathway. (C) In BRCA1−/− cells, HR mediated repair of stalled replication forks and DSBs is impaired; there may be increased reliance of PARP-dependent fork-restart mechanisms in S-phase and increased PARP activity. (D) In BRCA1−/− cells treated with PARPi, an increased burden of unrepaired single-strand breaks enter S-phase. The resulting lesions cannot be repaired, as HR-mediated repair is completely disabled, leading to catastrophic failure of replication fork restart, accumulation of unrepaired DNA DSBs and cell arrest/death.
Although there has been much focus on the role of PARP1/2 in SSB repair, PARP also has other important roles that may contribute to the sensitivity of BRCA1−/− cells to PARP inhibitors. PARP1/2 binds to and is activated by stalled replication forks during S-phase and PARP1/2 activity is required for the efficient HR-mediated restart of stalled replication forks (Yang et al., 2004; Sugimura et al., 2008). Depletion of PARP1 or PARP2 results in reduced recruitment of MRE11, RPA, and RAD51 to hydroxyurea-induced collapsed replication forks, and reduces hydroxyurea-induced recombination and fork restart (Yang et al., 2004). These data support a model in which PARP1/2 activity is required for efficient recruitment of MRE11-dependent end resection activity to stalled replication forks. Of note, PARP1−/− cells and PARP inhibitor treated cells do not have drastic defects in HR-mediated repair of DNA DSBs induced by restriction enzymes and PARP1−/− ES cells do not have a dramatic defect in gene targeting (Schultz et al., 2003; Yang et al., 2004). These data together imply that PARP1/2 may have a specific role in HR-mediated replication fork restart, but not an essential role in HR-mediated DSBR in general.
BRCA1 is also recruited to stalled replication forks during S-phase, and interacts with CtIP and the MRE11/Rad50/NBS complex to regulate end resection activity (Scully et al., 1997a; Yu et al., 1998; Sartori et al., 2007; Chen et al., 2008). These data taken together suggest there may be a functional interaction between BRCA1 and PARP1/2 in regulating DNA end-processing activity at stalled replication forks. BRCA1 has a broader role that PARP1/2 in HR-mediated repair of DNA DSBs, as loss of BRCA1 has a much greater effect on multiple assays of HR. However, in the absence of BRCA1, there may be greater reliance on PARP-mediated pathways regulating restart of stalled replication forks. Thus, PARP inhibitors may deal a double blow to BRCA1-deficient cells, by both increasing the number of stalled replication forks, and by inhibiting PARP-dependent recruitment of end-resection activity to these lesions (Figure 1). PARP activity has been reported to be increased in HR-defective cells; however, this activity was not localized to stalled replication forks, suggesting that compensatory PARP activity may be directed to yet another function in HR-defective cells (Gottipati et al., 2010). PARP has broad roles in transcriptional regulation, chromatin dynamics, epigenetic regulation, and metabolism. It is possible that these other functions of PARP may contribute to PARP inhibitor sensitivity of BRCA1−/− cells.
The landmark pre-clinical studies with PARP inhibitors cited above have led to the rapid clinical development of these agents as targeted therapy for BRCA1 and BRCA2-mutant cancers. Early phase clinical trials showed impressive single agent activity for PARP inhibitors in heavily pre-treated BRCA1 and BRCA2-associated cancers (Fong et al., 2009). Recent Phase II studies testing the activity of single agent olaparib in heavily pre-treated breast cancers arising in BRCA1 or BRCA2 mutation carriers demonstrated an overall response rate of ∼41% in patients, with a very tolerable side effect profile (Tutt et al., 2010). A similar study in BRCA1/2-mutant women with advanced ovarian cancer demonstrated an overall response rate of 33% (Audeh et al., 2010). These studies demonstrate that PARP inhibitors will play a significant role in the future treatment of cancers arising in BRCA1-mutation carriers. There is also much interest in developing these agents for the treatment of sporadic cancers, such as ‘triple-negative' breast cancers, that may have defects in HR-mediated DSBR.
The specific sensitivity of BRCA1−/− (and BRCA2−/−) cells to PARP inhibitor has been cited as an example of therapeutic ‘synthetic lethality'. Synthetic lethality classically describes a state where two mutations, each having a viable phenotype, when combined, produce a lethal phenotype. However, acute loss of BRCA1 alone leads to proliferation arrest in most primary diploid cells, and BRCA1 null mutants are non-viable in mice. It is postulated that cancers that arise in the setting of BRCA1-mutation must acquire other mutations that allow proliferation in the setting of BRCA1 loss. Thus, BRCA1-mutant tumor cells have acquired a state in which repeated transit through S-phase can be accomplished without genomic catastrophe, despite loss of BRCA1 function. This compensated state remains highly dependent on PARP activity, as PARP inhibitors can induce cell death. Thus, the sensitivity of BRCA1-mutant cancer cells to PARP inhibitors is a more complicated or ‘conditional synthetic lethality' that occurs in cancer cells that have already acquired the ability to proliferate in the setting of BRCA1 loss. Indeed there are several setting in which BRCA1-null cells can tolerate PARP inhibitors and overcome this ‘conditional synthetic lethality'.
Development of resistance to PARP inhibitors
Although there is a high response rate of BRCA1-mutant cancers to treatment with PARP inhibitors, a substantial fraction of advanced BRCA1-mutant cancers are resistant to these agents (Fong et al., 2009; 2010; Tutt et al., 2010). Tumors that are initially sensitive to PARP inhibitor treatment also ultimately progress despite treatment with PARP inhibitors. These data demonstrate that de novo and acquired resistance to PARP will be a significant clinical problem. Experiment in cell culture and mouse models of BRCA1-associated cancers have led to new insight into several mechanisms of acquired resistance to PARP inhibitors.
In both cell culture and mouse models, increased expression of a p-glycoprotein efflux transporter mediating multiple drug resistance (MDR) can lead to acquired resistance to the PARP inhibitor olaparib (Rottenberg et al., 2008). Olaparib is a substrate for multiple MDR efflux transporters, and MDR inhibitors can reverse this mechanism of resistance in mouse models of BRCA1−/− breast cancer. As MDR up-regulation is a common mechanism of acquired resistance to many chemotherapeutic agents, it is likely to play a part in the development of clinical resistance to olaparib. Development of PARP inhibitors that are not substrates for MDR-mediating efflux pumps may decrease this potential mechanism of resistance. 6-Thioguanine has also been reported to be effective in killing PARP-resistant BRCA1−/− tumor cells that have p-glycoprotein mediated resistance to olaparib (Issaeva et al., 2010).
Resistance to PARP inhibitors: reversion mutations of BRCA1/2
Several elegant studies have demonstrated that BRCA1 and BRCA2-mutant cells can develop resistance to PARP inhibitors by acquiring secondary mutations in BRCA1 or BRCA2 that result in restoration of a partially functional gene (Ashworth, 2008a; Sakai et al., 2008; Swisher et al., 2008; Wang and Figg, 2008). Many germline BRCA1 and BRCA2 mutations are frameshift mutations resulting in a truncated transcript that is degraded by non-sense mediated decay. Excision of genomic region containing the mutation can lead to an in-frame deletion of one or more exons, and restoration of an open reading frame with production of a hypomorphic BRCA1 or BRCA2 protein. These novel hypomorphic BRCA1 or BRCA2 alleles are sufficient to drive resistance to PARP inhibitors and platinum compounds. These data suggest that BRCA1 and BRCA2 reversion mutations can partially restore HR-mediated repair function in these cells. As reversion mutations can lead to resistance to both PARP inhibitors and platinum, this suggests potential for cross-resistance to these agents. Poor response to prior platinum therapy is associated with poor response in BRCA1/2 mutation carriers treated with PARP inhibitors in recent clinical trials, supporting this concept of cross resistance (Fong et al., 2010).
The development of reversion mutations of BRCA1 is a vivid demonstration that while loss of BRCA1 function likely facilitates tumorigenesis, this defect is not required to maintain the malignant phenotype of an established tumor. Thus, restoration of BRCA1 function by reversion mutations is well-tolerated in tumors that develop in BRCA1-mutation carriers. These observations imply that methods to restore BRCA1 function in BRCA1-mutant tumors are unlikely to succeed as therapeutic strategies.
53BP1 and BRCA1: synthetic viability
Although BRCA1 functions as a tumor suppressor, mice homozygous for null BRCA1 mutations show early embryonic lethality (Hakem et al., 1996; Liu et al., 1996; Ludwig et al., 2001). ES-cells containing homozygous null mutations of BRCA1 are not viable, although some hypomorphic alleles, such as the BRCA1Δ11 mutation, result in viable ES cells (Gowen et al., 1996; Ludwig et al., 1997). Thus, loss of BRCA1 leads to significant defects in genomic stability that normally inhibit proliferation. Cells must acquire additional checkpoint or and other mutations to allow proliferation and tumorigenesis in the setting of BRCA1 loss. Almost all BRCA1-mutation associated cancers have acquired p53 mutations. However, p53 mutation alone is not enough to overcome the growth defect associated with BRCA1 loss. Loss of p53 only delays embryonic lethality in full null BRCA1-mutant mice by a few days (Hakem et al., 1997; Ludwig et al., 1997). Similarly, there is long latency of tumor development in mouse models with tissue-specific conditional BRCA1 mutations even when combined with p53 deletion (Cressman et al., 1999b; Xu et al., 2001; Liu et al., 2007a; Shakya et al., 2008).
These observations have led to great interest in identifying mutations that can allow cells to tolerate Brca1 loss. In screens aimed at identifying what additional mutations will allow cells to overcome the proliferation defect associated with loss of wt BRCA1, we and others have found that loss of 53BP1 is able to reverse many aspects of the phenotype associated with BRCA1 loss (Cao et al., 2009; Bouwman et al., 2010; Bunting et al., 2010). This result was striking and may lead to new insight into the role of BRCA1 and 53BP1 in regulating DNA repair.
53BP1 is the human ortholog of yeast DNA damage checkpoint proteins Rad9p/Crb2, with a key role in DNA repair response and checkpoint control. 53BP1 is a nuclear protein that contains Tudor domains and paired BRCT repeats, and is found in all mammalian cells. Upon the induction of DNA DSBs, 53BP1 rapidly redistributes from a diffuse nuclear localization to discrete foci that co-localize with phosphorylated histone H2AX and other repair proteins including BRCA1 (Schultz et al., 2000; Wang et al., 2002; Ward et al., 2003). This localization is dependent upon interaction of the Tudor domains with methylated histone residues (Huyen et al., 2004; Sanders et al., 2004). Mice lacking 53BP1 are viable and fertile, but are small, and very sensitive to ionizing radiation. Cells lacking 53BP1 are sensitive to DNA damaging agents and have defects in both S-phase and G2M checkpoints (most noticeable after low but not high doses of ionizing radiation) (Fernandez-Capetillo et al., 2002; Mochan et al., 2003). 53BP1 function is also required for class-switch recombination and has been implicated in NHEJ in a non-classical V(D)J recombination pathway (Ward et al., 2004; Xie et al., 2007; Difilippantonio et al., 2008). Overexpression of a dominant-negative 53BP1 construct suppressed NHEJ and increased frequency of HR-mediated repair, suggesting 53BP1 is involved in regulating the choice between NHEJ and HR-mediated repair of DNA DSBs (Xie et al., 2007).
New insight into the functional interaction between 53BP1 and BRCA1 was found by Cao et al. They investigated how mutations in other DNA repair and checkpoint proteins would affect the proliferative defect seen in cells homozygous for the Brca1−Δ11 mutation (Cao et al., 2009). The Brca1−Δ11 mutation has an in-frame deletion of exon 11 that results in production of a hypomorphic protein resulting from in-frame splicing from exon 10 to exon 12. This BRCA1Δ11 variant has an intact RING domain, tandem BRCT-repeats and localizes to the nucleus, but is associated with a defect in HR-mediated repair (Huber et al., 2001). Unlike homozygous null-BRCA1 mutants, which show early embryonic lethality that is only modestly delayed by being placed in a p53−/− background, homozygous Brca1Δ11/Δ11 mutants show mid-gestational embryonic lethality that can be reversed in certain strain backgrounds or by breeding to a p53+/− genotype (Cressman et al., 1999a; Xu et al., 2001). However, BRCA1Δ11/Δ11; p53+/− MEFs showed early senescence in culture and BRCA1Δ11/Δ11;p53+/− animals demonstrate rapid aging and are tumor prone (Cao et al., 2003). Cao et al. found that placing BRCA1Δ11/Δ11 mice in a 53BP1−/− background rescued the mid-gestational embryonic lethality and reversed the early senescence phenotype of BRCA1Δ11/Δ11 MEFs (Cao et al., 2009). Moreover, BRCA1Δ11/Δ11;53BP1−/− mice were viable, showed normal aging and did not have an increased tumor phenotype. Thus, loss of 53BP1 could reverse much of the phenotype associated found in Brca1Δ11/Δ11 mice and cells.
Further insight into this remarkable observation was put forth by Bunting et al. (2010) and Bouwman et al. (2010). Bunting et al. found that loss of 53BP1 could reverse the defect in HR-mediated DSBR found in BRCA1Δ11/Δ11 cells. Loss of 53BP1 also alleviates the level of spontaneous chromosomal abnormalities, and checkpoint mediated arrest that is seen in BRCA1Δ11/Δ11 cells. Moreover loss of 53BP1 also reverses the sensitivity of BRCA1Δ11/Δ11 cells to PARP inhibitors and restored RAD51 focus formation. This effect of 53BP1 was specific to BRCA1 as loss of 53BP1 could not alleviate the phenotype associated with XRCC1 loss (Bunting et al., 2010).
Bouwman et al. (2010) worked with a conditional null-allele of Brca1 that is not viable in ES cells. A piggyBac retrotransposon-based insertional mutagenesis screen (Ding et al., 2005) was used to search for insertion events that would allow ES cells to tolerate a null allele of BRCA1. Loss of 53BP1 was found to allow ES cells to survive after acute deletion of BRCA1. This demonstrated that loss of 53BP1 could rescue ES cells with a null-mutation of BRCA1, extending the prior findings with the BRCA1Δ11 mutation. Loss of 53BP1 also alleviated the spontaneous DNA damage, chromosomal abnormalities, and G2/M checkpoint activation associated with loss of BRCA1 in this model. Loss of 53BP1, but not p53, reversed the sensitivity of BRCA1−/− cells to cisplatin and mitomycin C. Moreover loss of 53BP1 restored RAD51 foci formation after ionizing radiation in BRCA1−/− cells, and partly restored HR function in BRCA1−/− as measured by gene targeting.
These studies show that loss of 53BP1 is ‘synthetically viable' with BRCA1 loss in ES cells. Loss of 53BP1 function can rescue the severe proliferation defect of BRCA1-mutant cells and restore functional capacity to perform HR mediated DNA repair. Insight into the mechanism behind this observation was further elucidated by Bunting et al. (2010). End resection of DNA breaks is a critical step in allow HR-mediated repair and is in part mediated by CtIP and the MRE11 complex. Loss of BRCA1 is associated with failure of end-resection at DNA doubles-strand breaks, thus leading to impaired HR-mediated repair, and promotion of NHEJ mediated repair of DNA DSBs. Bunting et al. (2010) found that loss of 53BP1 restores end-resection in Brca1Δ11/Δ11 cells, and that this rescue was dependent on both CtIP and ATM. These observations can lead to a model (Figure 2) in which during G0/G1, where BRCA1 is not normally present, 53BP1 inhibits the end-resection function of CtIP. During S phase, BRCA1, by an as yet unknown mechanism, inhibits this function of 53BP1 at DNA breaks, allowing activation of CtIP mediated end-resection and promoting HR-mediated repair. Loss of BRCA1 leads to unregulated 53BP1-mediated inhibition of CtIP function at breaks, leading to failure of HR-mediated repair and promotion of NHEJ during S-phase. If 53BP1 is also lost, CtIP is now released from inhibition and end-resection and the capacity to perform HR-mediated repair is restored (Figure 3).
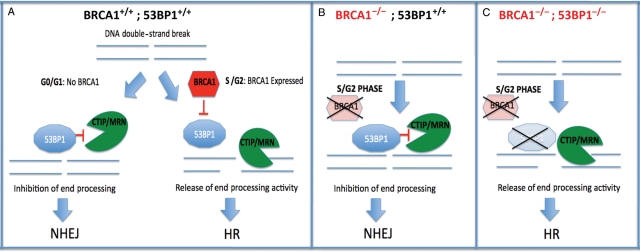
Figure 2.
Collaboration of BRCA1 loss and 53BP1 loss results in synthetic viability and restoration of HR-mediated repair. (A) Normal cells: in G0/G1 there is no BRCA1 expression, and 53BP1 is recruited to DNA double-strand breaks (DSBs) where it restrains CtIP activity, inhibits end processing and promotes NHEJ. In S/G2 phases of the cell cycle, BRCA1 is normally expressed and recruited to sites of breaks, where it inhibits the action of 53BP1 on CtIP, allowing end processing and promoting HR mediated repair. (B) BRCA1−/− cells: BRCA1 is not present in S-phase, and 53BP1 remains free to inhibit CtIP function, leading to impaired end processing of breaks, suppression of HR and promotion of error-prone NHEJ. (C) BRCA1−/−; 53BP1−/− cells: during S-phase CtIP is recruited to sites of DNA breaks. In the absence of 53BP1, its activity is unrestrained allowing end processing and HR mediated repair.
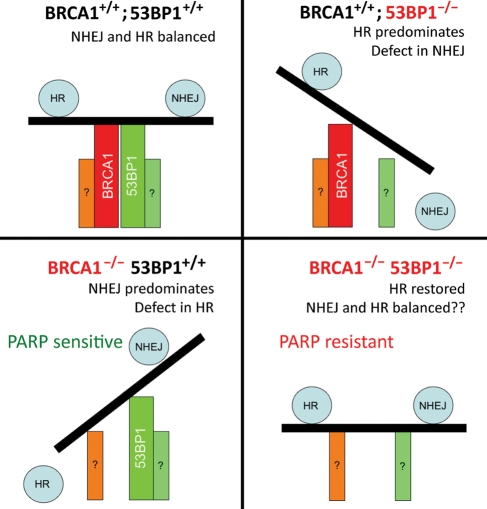
Figure 3.
53BP1 and BRCA1 as regulators of the balance between NHEJ and HR. In normal cells (upper left), 53BP1 and BRCA1 together maintain an overall balance between NHEJ and HR. In BRCA1+/+; 53BP1−/− cells (upper right), 53BP1 loss inhibits NHEJ and promotes HR, resulting in increased frequency of HR-mediated repair. In BRCA1−/−; 53BP1+/+ cells (lower left), loss of BRCA1 results in a profound defect in HR, with 53BP1 now promoting NHEJ as the predominant repair pathway, resulting in genomic instability. In BRCA1−/−; 53BP1−/− cells (lower right), the loss of both BRCA1 and 53BP1 restores HR-capacity. The factors that now regulate choice between HR and NHEJ in this setting are unknown.
This model does leave many unanswered questions. For example, BRCA1 interacts with PALB2 and is required for efficient recruitment of BRCA2 to DNA breaks (Zhang et al., 2009a,b). Whether and how 53BP1 restores PALB2 and BRCA2 recruitment and function in BRCA1−/− cells remain unclear. How loss of 53BP1 affects the function of the different distinct BRCA1 associated complexes that are assembled during S-phase (Greenberg et al., 2006) and whether this contributes to the ‘synthetic viable' phenotype is also uncertain.
This model highlights the role of BRCA1 and 53BP1 in the functional competition between the NHEJ and HR pathways in mediating the repair of DNA DSBs (Kass and Jasin, 2010; Figure 2). BRCA1 and 53BP1 may be a ‘master regulator' of the repair choice that occurs at DNA breaks (Figure 3). When 53BP1 is absent, end processing is not inhibited and DNA DSBs are preferentially repaired by HR-mediated repair. When BRCA1 is absent, DSBs do not undergo end processing, resulting in suppression of HR and preferential repair by NHEJ. When both BRCA1 and 53BP1 are absent, end-resection at DNA DSBs and subsequent HR-mediated repair can once again take place.
Thus, BRCA1 is not required for the actual mechanics of HR-mediated repair, but plays a crucial role, together with 53BP1, in mediating repair choice. At present it is unclear what repair factors are regulating end resection and choice of repair pathways in the absence of both 53BP1 and BRCA1. ATM is activated by PARP inhibition, and BRCA1Δ11/Δ11;53BP1−/− cells are very sensitive to ATM inhibitors, suggesting that ATM may play a crucial role in mediating repair in this setting (Bryant and Helleday, 2006; Bunting et al., 2010).
Bouwman et al. (2010) found that a subset of BRCA1-associated human breast cancers have lost 53BP1 protein expression. This loss of 53BP1 in BRCA1-associated cancers may result in resistance to PARP inhibitors and platinum agents. Similarly, a subset of sporadic basal-like breast cancers, which are postulated to be ‘BRCA1-like', also show loss of 53BP1 expression (Bouwman et al., 2010). Thus, loss of 53BP1 may functionally affect the repair capacity of both BRCA1 mutant, and ‘BRCA1-like' human breast cancers (Turner et al., 2007). As PARP inhibitors and platinum agents will likely play a significant role for in treatment of BRCA1-mutant cancers and ‘BRCA1-like' cancers, understanding of the mechanisms of resistance to these agents is very important. It is quite possible that abnormalities in 53BP1 may contribute to both de novo and acquired resistance to of these tumors to PARP inhibitors and platinum.
Summary
BRCA1−/− cancers have acquired a state where they can proliferate in the setting of a profound defect in HR-mediated DNA repair and associated genomic instability. BRCA1-mutant cells are now critically dependent on other parts of the repair pathway to avoid repair catastrophe during replication. This renders them vulnerable to agents that target these pathways to induce ‘conditional synthetic lethality' (Figure 4). The PARP inhibitors are a highly powerful and clinically validated example of this approach. Other potential ‘synthetic lethal' pathways may also exist. It has been reported that BRCA1-mutant cells are also specifically sensitive to EZH2 inhibitors (Puppe et al., 2009). As polycomb group proteins have been recently identified as being involved in DNA repair pathways (Chou et al., 2010; Ismail et al., 2010), this may be another example of repair-associated synthetic lethality in BRCA1−/− cells.
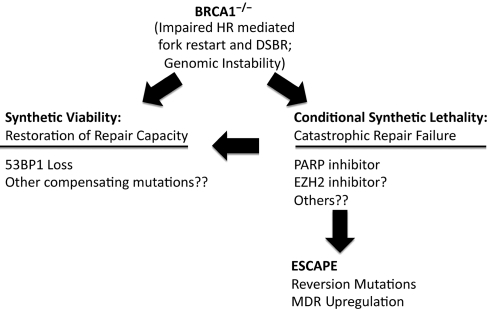
Figure 4.
Schema showing roles of synthetic lethality, escape and synthetic viability affecting survival of BRCA1−/− cells.
BRCA1-mutant cells can escape synthetic lethality by several mechanisms (Figure 4). The genomic instability associated with BRCA1 loss may enable the development of reversion mutations in BRCA1 that restore wildtype function and allow escape from synthetic lethal therapies. Alternatively, compensating mutations in other genes that regulate repair choice, such as 53BP1, may allow functional restoration of HR-mediated repair pathways and result in ‘synthetic viability'. Identification of other genes and pathways that collaborate with BRCA1 to produce either synthetic lethality or synthetic viability may lead to better insight into the role of BRCA1 in the repair process, and provide opportunities for novel therapeutic interventions.
Funding
The authors received support from the National Institutes of Health (S.G.), the Department of Defense (A.A. and S.G.) and the New Jersey Commission on Cancer Research (S.G.).
Acknowledgements
We are indebted to Bing Xia, Zhiyuan Shen, and Mark Brenneman, Jos Jonkers, Jiri Bartek, Amal Aly, Jay Oza, Atul Kulkarni, Vasudeva Ginjala, and Ming Yao for many helpful discussions. We apologize to those investigators whose primary work was not cited due to space considerations.
Conflict of interest: none declared.
References
- Abbott D.W., Thompson M.E., Robinson-Benion C., et al. BRCA1 expression restores radiation resistance in BRCA1-defective cancer cells through enhancement of transcription-coupled DNA repair. J. Biol. Chem. 1999;274:18808–18812. doi: 10.1074/jbc.274.26.18808. [DOI] [PubMed] [Google Scholar]
- Ahel I., Ahel D., Matsusaka T., et al. Poly(ADP-ribose)-binding zinc finger motifs in DNA repair/checkpoint proteins. Nature. 2008;451:81–85. doi: 10.1038/nature06420. [DOI] [PubMed] [Google Scholar]
- Ahel D., Horejsi Z., Wiechens N., et al. Poly(ADP-ribose)-dependent regulation of DNA repair by the chromatin remodeling enzyme ALC1. Science. 2009;325:1240–1243. doi: 10.1126/science.1177321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allinson S.L., Dianova, Dianov G.L. Poly(ADP-ribose) polymerase in base excision repair: always engaged, but not essential for DNA damage processing. Acta Biochim. Pol. 2003;50:169–179. [PubMed] [Google Scholar]
- Ame J.C., Rolli V., Schreiber V., et al. PARP-2, a novel mammalian DNA damage-dependent poly(ADP-ribose) polymerase. J. Biol. Chem. 1999;274:17860–17868. doi: 10.1074/jbc.274.25.17860. [DOI] [PubMed] [Google Scholar]
- Ashworth A. Drug resistance caused by reversion mutation. Cancer Res. 2008a;68:10021–10023. doi: 10.1158/0008-5472.CAN-08-2287. [DOI] [PubMed] [Google Scholar]
- Ashworth A. A synthetic lethal therapeutic approach: poly(ADP) ribose polymerase inhibitors for the treatment of cancers deficient in DNA double-strand break repair. J. Clin. Oncol. 2008b;26:3785–3790. doi: 10.1200/JCO.2008.16.0812. [DOI] [PubMed] [Google Scholar]
- Audeh M.W., Carmichael J., Penson R.T., et al. Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and recurrent ovarian cancer: a proof-of-concept trial. Lancet. 2010;376:245–251. doi: 10.1016/S0140-6736(10)60893-8. [DOI] [PubMed] [Google Scholar]
- Bau D.T., Mau Y.C., Shen C.Y. The role of BRCA1 in non-homologous end-joining. Cancer Lett. 2006;240:1–8. doi: 10.1016/j.canlet.2005.08.003. [DOI] [PubMed] [Google Scholar]
- Bouwman P., Aly A., Escandell J.M., et al. 53BP1 loss rescues BRCA1 deficiency and is associated with triple-negative and BRCA-mutated breast cancers. Nat. Struct. Mol. Biol. 2010;17:688–695. doi: 10.1038/nsmb.1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant H.E., Helleday T. Inhibition of poly (ADP-ribose) polymerase activates ATM which is required for subsequent homologous recombination repair. Nucleic Acids Res. 2006;34:1685–1691. doi: 10.1093/nar/gkl108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunting S.F., Callen E., Wong N., et al. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell. 2010;141:243–254. doi: 10.1016/j.cell.2010.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrski T., Huzarski T., Dent R., et al. Response to neoadjuvant therapy with cisplatin in BRCA1-positive breast cancer patients. Breast Cancer Res. Treat. 2009;115:359–363. doi: 10.1007/s10549-008-0128-9. [DOI] [PubMed] [Google Scholar]
- Byrski T., Gronwald J., Huzarski T., et al. Pathologic complete response rates in young women with BRCA1-positive breast cancers after neoadjuvant chemotherapy. J. Clin. Oncol. 2010;28:375–379. doi: 10.1200/JCO.2008.20.7019. [DOI] [PubMed] [Google Scholar]
- Cantor S.B., Bell D.W., Ganesan S., et al. BACH1, a novel helicase-like protein, interacts directly with BRCA1 and contributes to its DNA repair function. Cell. 2001;105:149–160. doi: 10.1016/s0092-8674(01)00304-x. [DOI] [PubMed] [Google Scholar]
- Cao L., Li W., Kim S., et al. Senescence, aging, and malignant transformation mediated by p53 in mice lacking the Brca1 full-length isoform. Genes Dev. 2003;17:201–213. doi: 10.1101/gad.1050003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao L., Xu X., Bunting S.F., et al. A selective requirement for 53BP1 in the biological response to genomic instability induced by Brca1 deficiency. Mol. Cell. 2009;35:534–541. doi: 10.1016/j.molcel.2009.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Nievera C.J., Lee A.Y., et al. Cell cycle-dependent complex formation of BRCA1.CtIP.MRN is important for DNA double-strand break repair. J. Biol. Chem. 2008;283:7713–7720. doi: 10.1074/jbc.M710245200. [DOI] [PubMed] [Google Scholar]
- Chou D.M., Adamson B., Dephoure N.E., et al. A chromatin localization screen reveals poly (ADP ribose)-regulated recruitment of the repressive polycomb and NuRD complexes to sites of DNA damage. Proc. Natl. Acad. Sci. USA. 2010;107:18475–18480. doi: 10.1073/pnas.1012946107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cressman V.L., Backlund D.C., Avrutskaya A.V., et al. Growth retardation, DNA repair defects, and lack of spermatogenesis in BRCA1-deficient mice. Mol. Cell. Biol. 1999a;19:7061–7075. doi: 10.1128/mcb.19.10.7061. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- Cressman V.L., Backlund D.C., Hicks E.M., et al. Mammary tumor formation in p53- and BRCA1-deficient mice. Cell Growth Differ. 1999b;10:1–10. [PubMed] [Google Scholar]
- Difilippantonio S., Gapud E., Wong N., et al. 53BP1 facilitates long-range DNA end-joining during V(D)J recombination. Nature. 2008;456:529–533. doi: 10.1038/nature07476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding S., Wu X., Li G., et al. Efficient transposition of the piggyBac (PB) transposon in mammalian cells and mice. Cell. 2005;122:473–483. doi: 10.1016/j.cell.2005.07.013. [DOI] [PubMed] [Google Scholar]
- El-Khamisy S.F., Masutani M., Suzuki H., et al. A requirement for PARP-1 for the assembly or stability of XRCC1 nuclear foci at sites of oxidative DNA damage. Nucleic Acids Res. 2003;31:5526–5533. doi: 10.1093/nar/gkg761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer H., McCabe N., Lord C.J., et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- Fernandez-Capetillo O., Chen H.T., Celeste A., et al. DNA damage-induced G2-M checkpoint activation by histone H2AX and 53BP1. Nat. Cell Biol. 2002;4:993–997. doi: 10.1038/ncb884. [DOI] [PubMed] [Google Scholar]
- Fong P.C., Boss D.S., Yap T.A., et al. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N. Engl. J. Med. 2009;361:123–134. doi: 10.1056/NEJMoa0900212. [DOI] [PubMed] [Google Scholar]
- Fong P.C., Yap T.A., Boss D.S., et al. Poly(ADP)-ribose polymerase inhibition: frequent durable responses in BRCA carrier ovarian cancer correlating with platinum-free interval. J. Clin. Oncol. 2010;28:2512–2519. doi: 10.1200/JCO.2009.26.9589. [DOI] [PubMed] [Google Scholar]
- Gottipati P., Vischioni B., Schultz N., et al. Poly(ADP-ribose) polymerase is hyperactivated in homologous recombination-defective cells. Cancer Res. 2010;70:5389–5398. doi: 10.1158/0008-5472.CAN-09-4716. [DOI] [PubMed] [Google Scholar]
- Gottschalk A.J., Timinszky G., Kong S.E., et al. Poly(ADP-ribosyl)ation directs recruitment and activation of an ATP-dependent chromatin remodeler. Proc. Natl. Acad. Sci. USA. 2009;106:13770–13774. doi: 10.1073/pnas.0906920106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowen L.C., Johnson B.L., Latour A.M., et al. Brca1 deficiency results in early embryonic lethality characterized by neuroepithelial abnormalities. Nat. Genet. 1996;12:191–194. doi: 10.1038/ng0296-191. [DOI] [PubMed] [Google Scholar]
- Greenberg R.A., Sobhian B., Pathania S., et al. Multifactorial contributions to an acute DNA damage response by BRCA1/BARD1-containing complexes. Genes Dev. 2006;20:34–46. doi: 10.1101/gad.1381306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakem R., de la Pompa J.L., Sirard C., et al. The tumor suppressor gene Brca1 is required for embryonic cellular proliferation in the mouse. Cell. 1996;85:1009–1023. doi: 10.1016/s0092-8674(00)81302-1. [DOI] [PubMed] [Google Scholar]
- Hakem R., de la Pompa J.L., Elia A., et al. Partial rescue of Brca1 (5–6) early embryonic lethality by p53 or p21 null mutation. Nat. Genet. 1997;16:298–302. doi: 10.1038/ng0797-298. [DOI] [PubMed] [Google Scholar]
- Hartman A.R., Ford J.M. BRCA1 induces DNA damage recognition factors and enhances nucleotide excision repair. Nat. Genet. 2002;32:180–184. doi: 10.1038/ng953. [DOI] [PubMed] [Google Scholar]
- Helleday T., Bryant H.E., Schultz N. Poly(ADP-ribose) polymerase (PARP-1) in homologous recombination and as a target for cancer therapy. Cell Cycle. 2005;4:1176–1178. doi: 10.4161/cc.4.9.2031. [DOI] [PubMed] [Google Scholar]
- Hohenstein P., Kielman M.F., Breukel C., et al. A targeted mouse Brca1 mutation removing the last BRCT repeat results in apoptosis and embryonic lethality at the headfold stage. Oncogene. 2001;20:2544–2550. doi: 10.1038/sj.onc.1204363. [DOI] [PubMed] [Google Scholar]
- Holstege H., Joosse S.A., van Oostrom C.T., et al. High incidence of protein-truncating TP53 mutations in BRCA1-related breast cancer. Cancer Res. 2009;69:3625–3633. doi: 10.1158/0008-5472.CAN-08-3426. [DOI] [PubMed] [Google Scholar]
- Huber L.J., Yang T.W., Sarkisian C.J., et al. Impaired DNA damage response in cells expressing an exon 11-deleted murine Brca1 variant that localizes to nuclear foci. Mol. Cell. Biol. 2001;21:4005–4015. doi: 10.1128/MCB.21.12.4005-4015.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huyen Y., Zgheib O., Ditullio R.A., Jr., et al. Methylated lysine 79 of histone H3 targets 53BP1 to DNA double-strand breaks. Nature. 2004;432:406–411. doi: 10.1038/nature03114. [DOI] [PubMed] [Google Scholar]
- Ismail I.H., Andrin C., McDonald D., et al. BMI1-mediated histone ubiquitylation promotes DNA double-strand break repair. J. Cell Biol. 2010;191:45–60. doi: 10.1083/jcb.201003034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Issaeva N., Thomas H.D., Djureinovic T., et al. 6-thioguanine selectively kills BRCA2-defective tumors and overcomes PARP inhibitor resistance. Cancer Res. 2010;70:6268–6276. doi: 10.1158/0008-5472.CAN-09-3416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kass E.M., Jasin M. Collaboration and competition between DNA double-strand break repair pathways. FEBS Lett. 2010;584:3703–3708. doi: 10.1016/j.febslet.2010.07.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H., Huang J., Chen J. CCDC98 is a BRCA1-BRCT domain-binding protein involved in the DNA damage response. Nat. Struct. Mol. Biol. 2007;14:710–715. doi: 10.1038/nsmb1277. [DOI] [PubMed] [Google Scholar]
- King M.C., Marks J.H., Mandell J.B. Breast and ovarian cancer risks due to inherited mutations in BRCA1 and BRCA2. Science. 2003;302:643–646. doi: 10.1126/science.1088759. [DOI] [PubMed] [Google Scholar]
- Liu C.Y., Flesken-Nikitin A., Li S., et al. Inactivation of the mouse Brca1 gene leads to failure in the morphogenesis of the egg cylinder in early postimplantation development. Genes Dev. 1996;10:1835–1843. doi: 10.1101/gad.10.14.1835. [DOI] [PubMed] [Google Scholar]
- Liu X., Holstege H., van der Gulden H., et al. Somatic loss of BRCA1 and p53 in mice induces mammary tumors with features of human BRCA1-mutated basal-like breast cancer. Proc. Natl. Acad. Sci. USA. 2007a;104:12111–12116. doi: 10.1073/pnas.0702969104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Wu J., Yu X. CCDC98 targets BRCA1 to DNA damage sites. Nat. Struct. Mol. Biol. 2007b;14:716–720. doi: 10.1038/nsmb1279. [DOI] [PubMed] [Google Scholar]
- Ludwig T., Chapman D.L., Papaioannou V.E., et al. Targeted mutations of breast cancer susceptibility gene homologs in mice: lethal phenotypes of Brca1, Brca2, Brca1/Brca2, Brca1/p53, and Brca2/p53 nullizygous embryos. Genes Dev. 1997;11:1226–1241. doi: 10.1101/gad.11.10.1226. [DOI] [PubMed] [Google Scholar]
- Ludwig T., Fisher P., Ganesan S., et al. Tumorigenesis in mice carrying a truncating Brca1 mutation. Genes Dev. 2001;15:1188–1193. doi: 10.1101/gad.879201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manie E., Vincent-Salomon A., Lehmann-Che J., et al. High frequency of TP53 mutation in BRCA1 and sporadic basal-like carcinomas but not in BRCA1 luminal breast tumors. Cancer Res. 2009;69:663–671. doi: 10.1158/0008-5472.CAN-08-1560. [DOI] [PubMed] [Google Scholar]
- Masson M., Niedergang C., Schreiber V., et al. XRCC1 is specifically associated with poly(ADP-ribose) polymerase and negatively regulates its activity following DNA damage. Mol. Cell. Biol. 1998;18:3563–3571. doi: 10.1128/mcb.18.6.3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochan T.A., Venere M., DiTullio R.A., Jr., et al. 53BP1 and NFBD1/MDC1-Nbs1 function in parallel interacting pathways activating ataxia-telangiectasia mutated (ATM) in response to DNA damage. Cancer Res. 2003;63:8586–8591. [PubMed] [Google Scholar]
- Moynahan M.E., Chiu J.W., Koller B.H., et al. Brca1 controls homology-directed DNA repair. Mol. Cell. 1999;4:511–518. doi: 10.1016/s1097-2765(00)80202-6. [DOI] [PubMed] [Google Scholar]
- Moynahan M.E., Cui T.Y., Jasin M. Homology-directed DNA repair, mitomycin-c resistance, and chromosome stability is restored with correction of a Brca1 mutation. Cancer Res. 2001;61:4842–4850. [PubMed] [Google Scholar]
- Neuhausen S.L., Marshall C.J. Loss of heterozygosity in familial tumors from three BRCA1-linked kindreds. Cancer Res. 1994;54:6069–6072. [PubMed] [Google Scholar]
- Oikawa A., Tohda H., Kanai M., et al. Inhibitors of poly(adenosine diphosphate ribose) polymerase induce sister chromatid exchanges. Biochem. Biophys. Res. Commun. 1980;97:1311–1316. doi: 10.1016/s0006-291x(80)80009-x. [DOI] [PubMed] [Google Scholar]
- Puppe J., Drost R., Liu X., et al. BRCA1-deficient mammary tumor cells are dependent on EZH2 expression and sensitive to polycomb repressive complex 2-inhibitor 3-deazaneplanocin A. Breast Cancer Res. 2009;11:R63. doi: 10.1186/bcr2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid L.J., Shakya R., Modi A.P., et al. E3 ligase activity of BRCA1 is not essential for mammalian cell viability or homology-directed repair of double-strand DNA breaks. Proc. Natl. Acad. Sci. USA. 2008;105:20876–20881. doi: 10.1073/pnas.0811203106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rottenberg S., Nygren A.O., Pajic M., et al. Selective induction of chemotherapy resistance of mammary tumors in a conditional mouse model for hereditary breast cancer. Proc. Natl. Acad. Sci. USA. 2007;104:12117–12122. doi: 10.1073/pnas.0702955104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rottenberg S., Jaspers J.E., Kersbergen A., et al. High sensitivity of BRCA1-deficient mammary tumors to the PARP inhibitor AZD2281 alone and in combination with platinum drugs. Proc. Natl. Acad. Sci. USA. 2008;105:17079–17084. doi: 10.1073/pnas.0806092105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rulten S.L., Cortes-Ledesma F., Guo L., et al. APLF (C2orf13) is a novel component of poly(ADP-ribose) signaling in mammalian cells. Mol. Cell. Biol. 2008;28:4620–4628. doi: 10.1128/MCB.02243-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saal L.H., Gruvberger-Saal S.K., Persson C., et al. Recurrent gross mutations of the PTEN tumor suppressor gene in breast cancers with deficient DSB repair. Nat. Genet. 2008;40:102–107. doi: 10.1038/ng.2007.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai W., Swisher E.M., Karlan B.Y., et al. Secondary mutations as a mechanism of cisplatin resistance in BRCA2-mutated cancers. Nature. 2008;451:1116–1120. doi: 10.1038/nature06633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders S.L., Portoso M., Mata J., et al. Methylation of histone H4 lysine 20 controls recruitment of Crb2 to sites of DNA damage. Cell. 2004;119:603–614. doi: 10.1016/j.cell.2004.11.009. [DOI] [PubMed] [Google Scholar]
- Sanderson R.J., Lindahl T. Down-regulation of DNA repair synthesis at DNA single-strand interruptions in poly(ADP-ribose) polymerase-1 deficient murine cell extracts. DNA Repair (Amst.) 2002;1:547–558. doi: 10.1016/s1568-7864(02)00054-x. [DOI] [PubMed] [Google Scholar]
- Sartori A.A., Lukas C., Coates J., et al. Human CtIP promotes DNA end resection. Nature. 2007;450:509–514. doi: 10.1038/nature06337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh M.S., Lindahl T. Role of poly(ADP-ribose) formation in DNA repair. Nature. 1992;356:356–358. doi: 10.1038/356356a0. [DOI] [PubMed] [Google Scholar]
- Schultz L.B., Chehab N.H., Malikzay A., et al. p53 binding protein 1 (53BP1) is an early participant in the cellular response to DNA double-strand breaks. J. Cell Biol. 2000;151:1381–1390. doi: 10.1083/jcb.151.7.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz N., Lopez E., Saleh-Gohari N., et al. Poly(ADP-ribose) polymerase (PARP-1) has a controlling role in homologous recombination. Nucleic Acids Res. 2003;31:4959–4964. doi: 10.1093/nar/gkg703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scully R., Ganesan S., Brown M., et al. Location of BRCA1 in human breast and ovarian cancer cells. Science. 1996;272:123–126. doi: 10.1126/science.272.5258.123. [DOI] [PubMed] [Google Scholar]
- Scully R., Chen J., Ochs R.L., et al. Dynamic changes of BRCA1 subnuclear location and phosphorylation state are initiated by DNA damage. Cell. 1997a;90:425–435. doi: 10.1016/s0092-8674(00)80503-6. [DOI] [PubMed] [Google Scholar]
- Scully R., Chen J., Plug A., et al. Association of BRCA1 with Rad51 in mitotic and meiotic cells. Cell. 1997b;88:265–275. doi: 10.1016/s0092-8674(00)81847-4. [DOI] [PubMed] [Google Scholar]
- Scully R., Ganesan S., Vlasakova K., et al. Genetic analysis of BRCA1 function in a defined tumor cell line. Mol. Cell. 1999;4:1093–1099. doi: 10.1016/s1097-2765(00)80238-5. [DOI] [PubMed] [Google Scholar]
- Shakya R., Szabolcs M., McCarthy E., et al. The basal-like mammary carcinomas induced by Brca1 or Bard1 inactivation implicate the BRCA1/BARD1 heterodimer in tumor suppression. Proc. Natl. Acad. Sci. USA. 2008;105:7040–7045. doi: 10.1073/pnas.0711032105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver D.P., Richardson A.L., Eklund A.C., et al. Efficacy of neoadjuvant cisplatin in triple-negative breast cancer. J. Clin. Oncol. 2010;28:1145–1153. doi: 10.1200/JCO.2009.22.4725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simbulan-Rosenthal C.M., Haddad B.R., Rosenthal D.S., et al. Chromosomal aberrations in PARP−/− mice: genome stabilization in immortalized cells by reintroduction of poly(ADP-ribose) polymerase cDNA. Proc. Natl. Acad. Sci. USA. 1999;96:13191–13196. doi: 10.1073/pnas.96.23.13191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snouwaert J.N., Gowen L.C., Latour A.M., et al. BRCA1 deficient embryonic stem cells display a decreased homologous recombination frequency and an increased frequency of non-homologous recombination that is corrected by expression of a Brca1 transgene. Oncogene. 1999;18:7900–7907. doi: 10.1038/sj.onc.1203334. [DOI] [PubMed] [Google Scholar]
- Sobhian B., Shao G., Lilli D.R., et al. RAP80 targets BRCA1 to specific ubiquitin structures at DNA damage sites. Science. 2007;316:1198–1202. doi: 10.1126/science.1139516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorlie T., Tibshirani R., Parker J., et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc. Natl. Acad. Sci. USA. 2003;100:8418–8423. doi: 10.1073/pnas.0932692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimura K., Takebayashi S., Taguchi H., et al. PARP-1 ensures regulation of replication fork progression by homologous recombination on damaged DNA. J. Cell. Biol. 2008;183:1203–1212. doi: 10.1083/jcb.200806068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swisher E.M., Sakai W., Karlan B.Y., et al. Secondary BRCA1 mutations in BRCA1-mutated ovarian carcinomas with platinum resistance. Cancer Res. 2008;68:2581–2586. doi: 10.1158/0008-5472.CAN-08-0088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sy S.M., Huen M.S., Chen J. PALB2 is an integral component of the BRCA complex required for homologous recombination repair. Proc. Natl. Acad. Sci. USA. 2009;106:7155–7160. doi: 10.1073/pnas.0811159106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tassone P., Tagliaferri P., Perricelli A., et al. BRCA1 expression modulates chemosensitivity of BRCA1-defective HCC1937 human breast cancer cells. Br. J. Cancer. 2003;88:1285–1291. doi: 10.1038/sj.bjc.6600859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tassone P., Di Martino M.T., Ventura M., et al. Loss of BRCA1 function increases the antitumor activity of cisplatin against human breast cancer xenografts in vivo. Cancer Biol. Ther. 2009;8:648–653. doi: 10.4161/cbt.8.7.7968. [DOI] [PubMed] [Google Scholar]
- Tulin A., Spradling A. Chromatin loosening by poly(ADP)-ribose polymerase (PARP) at Drosophila puff loci. Science. 2003;299:560–562. doi: 10.1126/science.1078764. [DOI] [PubMed] [Google Scholar]
- Turner N.C., Reis-Filho J.S. Basal-like breast cancer and the BRCA1 phenotype. Oncogene. 2006;25:5846–5853. doi: 10.1038/sj.onc.1209876. [DOI] [PubMed] [Google Scholar]
- Turner N.C., Reis-Filho J.S., Russell A.M., et al. BRCA1 dysfunction in sporadic basal-like breast cancer. Oncogene. 2007;26:2126–2132. doi: 10.1038/sj.onc.1210014. [DOI] [PubMed] [Google Scholar]
- Tutt A., Robson M., Garber J.E., et al. Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and advanced breast cancer: a proof-of-concept trial. Lancet. 2010;376:235–244. doi: 10.1016/S0140-6736(10)60892-6. [DOI] [PubMed] [Google Scholar]
- Wang W., Figg W.D. Secondary BRCA1 and BRCA2 alterations and acquired chemoresistance. Cancer Biol. Ther. 2008;7:1004–1005. doi: 10.4161/cbt.7.7.6409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z.Q., Stingl L., Morrison C., et al. PARP is important for genomic stability but dispensable in apoptosis. Genes Dev. 1997;11:2347–2358. doi: 10.1101/gad.11.18.2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B., Matsuoka S., Carpenter P.B., et al. 53BP1, a mediator of the DNA damage checkpoint. Science. 2002;298:1435–1438. doi: 10.1126/science.1076182. [DOI] [PubMed] [Google Scholar]
- Wang B., Matsuoka S., Ballif B.A., et al. Abraxas and RAP80 form a BRCA1 protein complex required for the DNA damage response. Science. 2007;316:1194–1198. doi: 10.1126/science.1139476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward I.M., Minn K., van Deursen J., et al. p53 binding protein 53BP1 is required for DNA damage responses and tumor suppression in mice. Mol. Cell. Biol. 2003;23:2556–2563. doi: 10.1128/MCB.23.7.2556-2563.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward I.M., Reina-San-Martin B., Olaru A., et al. 53BP1 is required for class switch recombination. J. Cell. Biol. 2004;165:459–464. doi: 10.1083/jcb.200403021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermark U.K., Reyngold M., Olshen A.B., et al. BARD1 participates with BRCA1 in homology-directed repair of chromosome breaks. Mol. Cell. Biol. 2003;23:7926–7936. doi: 10.1128/MCB.23.21.7926-7936.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams R.S., Green R., Glover J.N. Crystal structure of the BRCT repeat region from the breast cancer-associated protein BRCA1. Nat. Struct. Biol. 2001;8:838–842. doi: 10.1038/nsb1001-838. [DOI] [PubMed] [Google Scholar]
- Wong A.K., Ormonde P.A., Pero R., et al. Characterization of a carboxy-terminal BRCA1 interacting protein. Oncogene. 1998;17:2279–2285. doi: 10.1038/sj.onc.1202150. [DOI] [PubMed] [Google Scholar]
- Woodhouse B.C., Dianov G.L. Poly ADP-ribose polymerase-1: an international molecule of mystery. DNA Repair (Amst.) 2008;7:1077–1086. doi: 10.1016/j.dnarep.2008.03.009. [DOI] [PubMed] [Google Scholar]
- Wu L.C., Wang Z.W., Tsan J.T., et al. Identification of a RING protein that can interact in vivo with the BRCA1 gene product. Nat. Genet. 1996;14:430–440. doi: 10.1038/ng1296-430. [DOI] [PubMed] [Google Scholar]
- Xia Y., Pao G.M., Chen H.W., et al. Enhancement of BRCA1 E3 ubiquitin ligase activity through direct interaction with the BARD1 protein. J. Biol. Chem. 2003;278:5255–5263. doi: 10.1074/jbc.M204591200. [DOI] [PubMed] [Google Scholar]
- Xia B., Sheng Q., Nakanishi K., et al. Control of BRCA2 cellular and clinical functions by a nuclear partner, PALB2. Mol. Cell. 2006;22:719–729. doi: 10.1016/j.molcel.2006.05.022. [DOI] [PubMed] [Google Scholar]
- Xie A., Hartlerode A., Stucki M., et al. Distinct roles of chromatin-associated proteins MDC1 and 53BP1 in mammalian double-strand break repair. Mol. Cell. 2007;28:1045–1057. doi: 10.1016/j.molcel.2007.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Qiao W., Linke S.P., et al. Genetic interactions between tumor suppressors Brca1 and p53 in apoptosis, cell cycle and tumorigenesis. Nat. Genet. 2001;28:266–271. doi: 10.1038/90108. [DOI] [PubMed] [Google Scholar]
- Yang Y.G., Cortes U., Patnaik S., et al. Ablation of PARP-1 does not interfere with the repair of DNA double-strand breaks, but compromises the reactivation of stalled replication forks. Oncogene. 2004;23:3872–3882. doi: 10.1038/sj.onc.1207491. [DOI] [PubMed] [Google Scholar]
- Yu X., Wu L.C., Bowcock A.M., et al. The C-terminal (BRCT) domains of BRCA1 interact in vivo with CtIP, a protein implicated in the CtBP pathway of transcriptional repression. J. Biol. Chem. 1998;273:25388–25392. doi: 10.1074/jbc.273.39.25388. [DOI] [PubMed] [Google Scholar]
- Zhang F., Fan Q., Ren K., et al. PALB2 functionally connects the breast cancer susceptibility proteins BRCA1 and BRCA2. Mol. Cancer Res. 2009a;7:1110–1118. doi: 10.1158/1541-7786.MCR-09-0123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F., Ma J., Wu J., et al. PALB2 links BRCA1 and BRCA2 in the DNA-damage response. Curr. Biol. 2009b;19:524–529. doi: 10.1016/j.cub.2009.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Q., Boyer T.G., Chen P.L., et al. Deficient nonhomologous end-joining activity in cell-free extracts from Brca1-null fibroblasts. Cancer Res. 2002;62:3966–3970. [PubMed] [Google Scholar]