Summary of recent advances
The amenability of the zebrafish to in vivo imaging and genetic analysis has fueled expanded use of this vertebrate model to investigate the molecular and cellular foundations of host-microbe relationships. Study of microbial encounters in zebrafish hosts has concentrated on developing embryonic and larval stages, when the advantages of the zebrafish model are maximized. A comprehensive understanding of these host-microbe interactions requires appreciation of the developmental context into which a microbe is introduced, as well as the effects of that microbial challenge on host ontogeny. In this review, we discuss how in vivo imaging and genetic analysis in zebrafish has advanced our knowledge of host-microbe interactions in the context of a developing vertebrate host. We focus on recent insights into immune cell ontogeny and function, commensal microbial relationships in the intestine, and microbial pathogenesis in zebrafish hosts.
Introduction
Early zebrafish research was focused heavily on embryogenesis [1], however use of the zebrafish model has gradually expanded to include study of post-embryonic developmental and physiological processes [2]. This expanded scope of zebrafish research is exemplified by the efforts to characterize the zebrafish immune system and its interactions with pathogenic and commensal microbes [3,4]. Zebrafish have several key features that make it an attractive model for analyses of host-microbe interactions. First, the optical transparency of zebrafish embryos and larvae, together with availability of transgenic lines expressing fluorescent proteins in distinct immune cell lineages, permit high-resolution in vivo observation of developing host cells and resident microorganisms [5,6]. Second, the small size and rapid development of zebrafish embryos facilitates forward genetic screens using chemical or retroviral mutagenesis (Box 1), as well as screening of chemical libraries [2]. Furthermore, sequencing of the zebrafish genome (http://www.sanger.ac.uk/Projects/D_rerio/) has empowered functional genomic and reverse genetic techniques (Box 1 and reviewed in [2,7]). Finally, methods for rearing zebrafish under germ-free or gnotobiotic conditions have been established, thus allowing rigorous control of the animal's microbial environment [8]. These advantages of the zebrafish system are maximal during embryonic and larval stages, and analyses of host-microbe interactions in the zebrafish have consequently focused on this dynamic developmental period of the life cycle. This experimental platform therefore poses significant challenges and opportunities to understand host-microbe interactions in the context of a rapidly developing vertebrate host. Here we review recent progress using the zebrafish model to investigate host-microbe interactions, including interactions with pathogenic and commensal microorganisms. We highlight those studies that provide significant novel insights using in vivo imaging and genetic methods available in embryonic and larval zebrafish.
Box 1. Common methods for testing zebrafish gene function.
Morpholinos
Small modified oligonucleotides injected into zebrafish embryos to induce targeted gene knockdown during early development. Morpholinos can be designed to either block translation initiation of both maternal and zygotic transcripts, or correct splicing of zygotic transcripts of a target gene. Morpholino efficacy is limited to embryonic and early larval stages, and controls for non-specific effects must be included [99,100]
Ethylnitrosourea (ENU) mutagenesis
ENU is an alkylating agent often used as a zebrafish mutagen. Male zebrafish are treated with ENU and used to generate F1 animals containing random heterozygous germline mutations. To facilitate forward genetic screens, ENU-induced mutants can be identified by phenotypic screening, then the causative lesion identified by positional cloning [101,102]. Additionally, libraries of DNA and sperm from ENU-mutagenized fish can be stored and used in reverse genetic analysis to identify mutations in a selected gene of interest through direct sequencing or CEL1 nuclease assays in a process known as targeting induced local lesions in genomes (TILLING) [103].
Insertional mutagenesis
Pseudotyped retroviral particles are injected into blastula-stage embryos where they integrate into the host genome. These founder animals are then outcrossed to generate F1 germline mutants. For forward genetic analysis, retrovirus-induced mutants can then be identified by phenotypic screening. Retroviral insertion sites can be easily identified using PCR primers specific to the retroviral vector. For reverse genetic analysis, libraries of DNA and sperm from injected founders or F1 progeny can be stored and used to screen by DNA sequencing for insertions in or near a selected gene of interest [104,105].
Zinc-finger nucleases
This reverse genetic technique exploits double-strand break repair pathways to generate small targeted mutations in a gene of interest. Fusion proteins constructed from the Fok1 restriction enzyme and three (or more) zinc-finger motifs are designed to recognize specific DNA target sequences in a selected gene of interest. Matched pairs of zinc finger nuclease RNAs are injected into zebrafish blastulas resulting in germline mutations in the target gene that can be propagated into subsequent generations [7].
Alterations in gene expression
Wild-type, mutant, or dominant-negative forms of a selected gene can be misexpressed either by injection of mRNA or DNA expression constructs into zebrafish embryos. Whereas mRNA injections are only efficacious through embryonic stages, injection of expression constructs permit mosaic analysis through adult stages as well as establishment of stable transgenic lines. Transgenesis efficiency in zebrafish has been recently improved through the use of the Tol2 transposon system [106]. Tissue-specific or ubiquitous expression can be achieved by placing the selected gene under control of a defined cis-regulatory sequence. Conditional gene expression is possible by using inducible promoter systems (e.g., GAL4/UAS, Cre/lox, and heat-shock promoters)[107].
Mechanisms of immune cell development
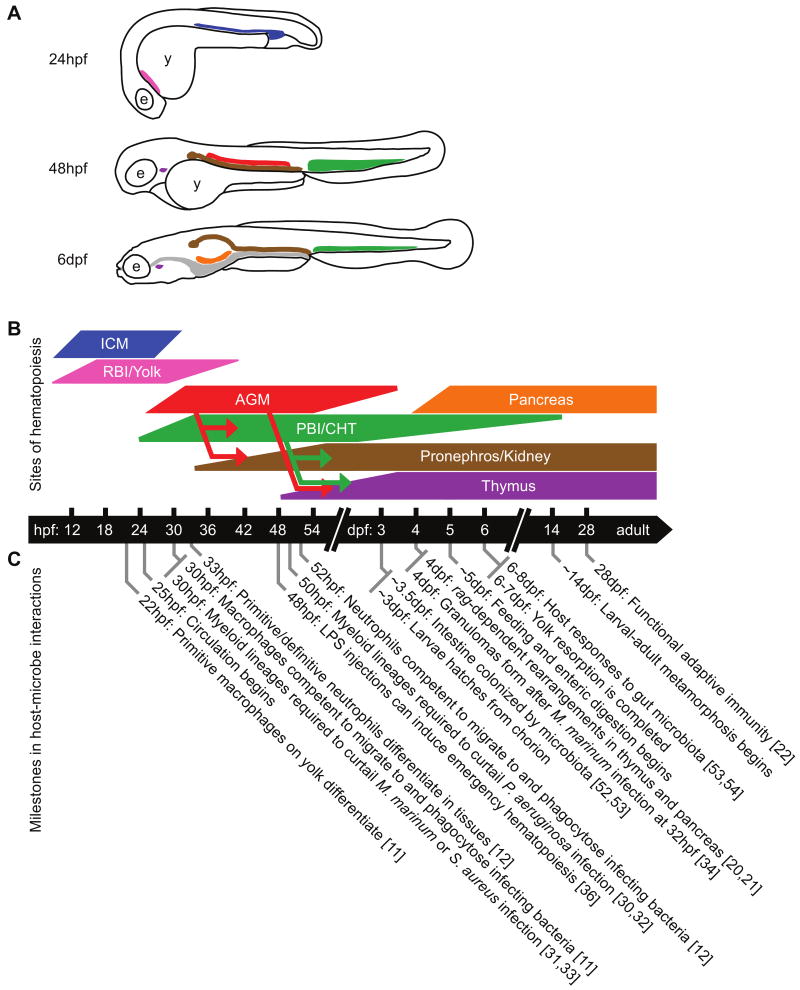
Hematopoiesis produces multiple immune cell types that perceive and respond to microbial stimuli. Zebrafish, like mammals, have distinct waves of hematopoiesis that occur in discrete yet functionally-analogous sites [3,6,9,10]. Recent in vivo imaging and lineage tracing studies have disclosed a dynamic pattern of cell seeding events that underlie the transfer of hematopoiesis between distinct anatomical sites during development (Fig. 1A,B). The ‘primitive’ wave of zebrafish hematopoiesis occurs in two separate locations in the embryo. Primitive myeloid precursors arise in the anterior mesoderm (also called rostral blood island or RBI) and migrate to the yolk, where they differentiate into primitive macrophages by 22 hours post-fertilization (hpf)[11] and primitive neutrophils by 33hpf [12]. Primitive erythroblasts arise in bilateral stripes of the ventral mesoderm and migrate to the midline to form the intermediate cell mass (ICM, analogous to the mammalian yolk sac), from where they enter circulation ∼24hpf (reviewed in [10]). ‘Definitive’, or multilineage, hematopoiesis begins ∼24hpf with the formation of a transient population of erythromyeloid progenitors (EMPs) within the posterior blood island (PBI) which later expands to become the caudal hematopoietic tissue (CHT; the PBI/CHT is analogous to the mammalian fetal liver)[13]. As early as 26hpf, cells within the presumptive zebrafish aorta-gonad-mesonephros (AGM) begin expressing markers of definitive hematopoietic stem cells (HSCs; e.g., c-myb, runx1, and cd41) [3,9,10]. Lineage tracing studies show these hematopoietic stem cells (HSCs) have multiple potential destinations. A subset of HSCs leave the AGM as early as 32hpf and migrate through the blood to colonize the CHT. After arriving in the CHT, these AGM-derived HSCs produce erythroid and myeloid cells, and migrate yet again beginning ∼50hpf from the CHT to colonize the sites of definitive hematopoietic tissues in adults, the pronephros and thymus [14,15]. Other HSCs enter circulation from the AGM after 40hpf to colonize the thymus, while others migrate anteriorly along the pronephric tubules to seed the developing kidney [16,17]. Other HSCs within the AGM can produce definitive myeloid cells in situ as early as 48hpf [18]. These early events are followed by initiation of definitive hematopoiesis in the kidney marrow and lymphopoiesis in the thymus starting ∼4dpf and continuing into adult stages [9]. Therefore, multiple waves of hematopoiesis in zebrafish occur at distinct anatomical sites analogous to mammalian hematopoiesis.
Figure 1. Developmental milestones in zebrafish host-microbe interactions.
Schematic depiction of anatomical sites (A) and approximate durations (B) of hematopoietic activity in developing zebrafish. The eye (e), yolk (y) and gastrointestinal tract (gray) are indicated in A. Relocation of definitive hematopoietic stem cells (HSCs) between sites is represented by arrows in B. Primitive erythropoiesis occurs in the intermediate cell mass (ICM, blue) which is active ∼11-30hpf [59], whereas primitive myelopoiesis begins in the rostral blood island (RBI) and later the yolk (pink) from ∼12-40hpf [11]. HSCs appear in the aorta-gonad-mesonephros region (AGM, red) ∼26hpf until ∼3dpf [15,16]. These HSCs are mobilized to seed the caudal hematopoietic tissue (PBI/CHT; green) and pronephros (brown) as early as 32hpf, and the thymus (purple) as early as 48hpf [14-17]. Definitive hematopoiesis in the CHT begins de novo ∼24hpf [13] and continues until at least 14dpf [14]. Cells from the CHT contribute to the pronephros and thymus as early as 48hpf [14,15]. B cell development initiates in the pancreas (orange) starting 4dpf [21], although the hematopoietic origins of these cells remain unknown. The thymus, pancreas, and pronephros/kidney subsequently serve as sites of definitive hematopoiesis into adult stages [9,10]. (C) Within this dynamic developmental context, important milestones relevant to zebrafish immunity and microbial interactions are indicated and referenced.
Zebrafish hematopoiesis produces largely the same differentiated cell types observed in mammals. Within the zebrafish myeloid lineage, monocytes, macrophages, neutrophils, and eosinophils have all been described (reviewed in [3,9]). In addition, mast cells [19] and cells with cytotoxic properties similar to mammalian natural killer (NK) cells (J. Yoder, personal communication) have recently been identified in the zebrafish. The innate immune functions of these cells provide a robust defense beginning early in development (Fig. 1C). Macrophages and neutrophils can be recruited to a site of infection and phagocytose invading bacteria as early as 30hpf [11] and 52hpf respectively, though the phagocytic activity of neutrophils is relatively low [12]. By 4dpf, T and B lymphocyte progenitors begin undergoing rag-dependent rearrangements within the thymus [20] and pancreas [21], respectively. However, zebrafish appear to be incapable of mounting an antibody response until early adulthood ∼4 weeks post fertilization [22], before which they rely exclusively on innate defense mechanisms. An important challenge for future research is to define the developmental origin, potency, and sensitivity of HSC populations in these distinct anatomic sites during zebrafish development, as well as the functions of their derivative lineages.
Mechanisms of pathogenic host-microbe relationships
The genetic tractability of zebrafish has prompted its use as a host organism for an expanding number of bacteria and virus pathogenic to fish and mammals (Table 1; reviewed in [3,4,23]). The majority of these pathogen studies have used microinjection to deliver pathogens at controlled doses into zebrafish embryos, while others have used static immersion to mimic natural infection routes [24-26]. Genetic analysis of pathogens in zebrafish hosts has focused almost entirely on known virulence factors, although systematic bacterial forward genetic analyses in adult zebrafish hosts have identified novel virulence determinants in the zoonotic pathogen Streptococcus iniae [27] and the human pathogen S. pyogenes [28]. Together, these studies are revealing bacterial and host factors mediating virulence, and in some cases, defining their mechanism of action (Table 1). Although a discussion of all recent zebrafish microbial pathogenesis studies is beyond the scope of this review, several emerging themes are discussed below.
Table 1. Microorganisms used for in vivo experimental analysis in zebrafish hosts.
| Bacterium | Reference |
|---|---|
| Aeromonas hydrophilaa,b | [54,55,60] |
| Aeromonas salmonicida b | [61] |
| Aeromonas veronii b | [53] |
| Bacillus subtilusa | [11] |
| Burkholderia cenocepaciaa | [62] |
| Edwardsiella ictaluria | [63] |
| Edwardsiella tarda a,b | [26,64,65] |
| Escherichia coli MG1655 b | [52,55] |
| Escherichia coli O157:H7 b | [66] |
| Flavobacterium columnare a,b | [67] |
| Flavobacterium johnsoniae a,b | [67] |
| Francisella spp. a | [68] |
| Leptospira interrogansa | [69] |
| Listeria monocytogenesa | [70,71] |
| Listeria spp. a | [70] |
| Listonella anguillarum a,b | [72] |
| Mycobacterium haemophiluma | [73] |
| Mycobacterium marinum a,b | [24,33,34,41-46,74-77] (D.Tobin et al., personal communication) |
| Pseudomonas aeruginosa a,b | [30,32,37,52,54,55,78] |
| Pseudomonas fluorescens b | [53] |
| Salmonella arizonaea | [24] |
| Salmonella typhimuriuma | [38,79,80] |
| Staphylococcus aureusa | [31,61] |
| Steptococcus ininaea | [27,81,82] |
| Streptococcus pyogenesa | [28,81,83-85] |
| Vibrio anguillarum b | [25] |
| Virus | Reference |
| Herpes simplex virus type 1 (HSV-1) a | [86] |
| Infectious hematopoietic necrosis virus (IHNV) a | [87,88] |
| Infectious pancreatic necrosis virus (IPNV) a | [87,89] |
| Infectious spleen and kidney necrosis virus (ISKNV) a | [90,91] |
| Nervous necrosis virus (NNV) a | [92] |
| Snakehead rahabdovirus (SHRV) a,b | [64,93-97] |
| Spring viremia of carp virus (SVCV) a | [88,98] |
| Viral hemorrhagic septicemia virus (VHSV) a,b | [99] |
| Microbial consortia | Reference |
| Intestinal microbiota b | [52-55, 58**] (M.Kanther et al., unpublished) |
Microbe introduced by injection.
Microbe introduced by static immersion, with or without wounding
The central role of myeloid leukocytes in controlling pathogen infections is one such emerging theme. Myeloid lineages can be transiently depleted in zebrafish embryos via morpholino knock-down of the myeloid transcription factor pu.1 [29]. Recent studies have shown that injection of Pseudomonas aeruginosa, Staphylococcus aureus, or Mycobacterium marinum into embryos lacking myeloid leukocytes (pu.1 ‘morphant’ embryos) results in increased virulence associated with elevated bacterial burden [30-33]. Moreover, loss of myeloid leukocytes results in restored virulence of several attenuated pathogen mutants, indicating that those respective virulence factors may function in phagocyte evasion in zebrafish embryos [30-34]. This requirement for myeloid lineages in bacterial clearance appears to be relatively specific for pathogens, as wild-type and pu.1 morphants are similarly competent to clear non-pathogenic Escherichia coli infections [33]. Importantly, the role of Pu.1 (and perhaps other host factors) in pathogen clearance may depend on the site of infection, as virulence of S. aureus was elevated in pu.1 morphants when injected into the bloodstream, but not into the yolk [31]. Although the mechanisms by which zebrafish myeloid leukocytes are recruited to sites of infection are not well understood, a recent study identified a novel role for H2O2 in the process of leukocyte recruitment to a wound. Niethammer and colleagues used a genetically encoded H2O2 sensor to reveal a local increase in H2O2 production at the margin of a new wound in 3dpf zebrafish, which stimulated recruitment of leukocytes to the wound site. The authors used morpholino knock-down and pharmacological inhibitors to show that dual oxidase, a NADPH oxidase, is required for the increase in H2O2 production and inflammation at wound sites [35]. We speculate that similar mechanisms might mediate leukocyte recruitment to sites of infection.
Another emerging theme suggests that microbial cells and their products can influence zebrafish immune cell production and function. Many microbial pathogenesis studies in zebrafish have focused on embryonic stages when hematopoietic development is especially dynamic (Fig. 1), raising the possibility that immune challenges during these stages could alter the proliferation, differentiation, and/or maintenance of hematopoietic immune cells. For example, injection of bacterial lipopolysaccharide (LPS) into 48hpf zebrafish embryos results in elevated expression of granulocyte colony-stimulating factor (gcsf) and its receptor (gcsfr), as well as production of a previously-unappreciated gcsfr-dependent hematopoietic wave (‘demand-driven’ or ‘emergency’ hematopoiesis) [36]. Microbial challenge can also affect the maintenance of immune cells, as illustrated by the observed decrease in neutrophil number and subsequent death following injection with S. aureus [31]. In addition to production and maintenance of hematopoietic cells, microbial products might also influence their circulation indirectly by affecting cardiovascular development. For example, the P. aeruginosa extracellular phospholipase PlcHR can inhibit angiogenesis in developing zebrafish, resulting in reduced numbers of circulating blood cells [37]. These studies indicate that distinct microbial challenges in developing zebrafish might have specific effects on hematopoietic immune cell formation and function. Conversely, the virulence of a given pathogen can vary as a function of the developmental stage at which zebrafish are infected. For example, virulence of P. aeruginosa mutants defective for quorum sensing or type III secretion is indistinguishable from wild-type parent strains when injected into zebrafish at 28hpf when only primitive myeloid cells are present. However, these same mutants are significantly attenuated when injected into zebrafish at 50hpf after definitive hematopoiesis has begun [32]. Together, these results indicate that a comprehensive understanding of any zebrafish-microbe interaction must include appreciation of the hematopoietic context into which the microbe is introduced, as well as the effects of that microbial challenge on hematopoietic lineages. Although the importance of developmental context has been best illustrated in the study of immune cell interactions with pathogenic bacteria, it is likely that this theme will also be relevant for microbial encounters with other host cell lineages and tissues.
Several classes of receptors for conserved microbe-associated molecular patterns (MAMPs) have been identified in the zebrafish. Additionally several downstream signal transduction cascades appear to be conserved between zebrafish and mammals (reviewed in [3,4,23]). The zebrafish genome encodes 24 putative variants of the Toll-like receptor (TLR) family, including homologs of 10 mammalian TLRs [4]. In some cases, the ligand and downstream signaling cascades of zebrafish TLRs are conserved with mammals. Stockhammer and co-workers used reverse genetic tests to show that the MAMP flagellin signals via zebrafish TLR5 homologs (tlr5a and tlr5b) and the MyD88 adaptor protein to induce both pro- and anti-inflammatory genes [38]. However, recent evidence indicates that TLR sequence homology may not always equate to functional conservation. Whereas mammalian TLR4 recognizes the MAMP LPS, functional analysis of two zebrafish genes that are paralogous to mammalian TLR4 (tlr4a and tlr4b) indicated that these receptors do not recognize LPS [39,40]. These studies underscore that the evolutionary divergence between zebrafish and mammals has resulted in conservation as well as divergence in immune gene function.
The unique combination of features available in the zebrafish model provides the potential to define the molecular and cellular mechanisms underlying host-microbe interaction. At present, this potential has been best realized in the zebrafish model of M. marinum infection. M. marinum is a natural pathogen of fish and a close relative of M. tuberculosis, the causative agent of human tuberculosis (TB). Similar to human TB, infecting M. marinum recruit and are phagocytosed by host macrophages. Infected macrophages subsequently migrate to deeper tissues, where they recruit additional macrophages and other immune cells to form cellular aggregates called granulomas. Injection of M. marinum into zebrafish embryos results in granuloma formation by 3 days post-infection (dpi) and does not require adaptive immunity [24]. The effect of M. marinum infection on larval and adult zebrafish gene expression has been analyzed extensively using both microarray and RNA-seq methods [41-43]. These studies have identified host genes and pathways responsive to infection that can serve as useful biomarkers and candidate host resistance/virulence determinants.
M. marinum pathogenesis was the recent focus of the first implementation of forward genetic analysis to identify zebrafish genes involved in a host-microbe relationship. Tobin and colleagues conducted an ENU mutagenesis screen to identify zebrafish mutations that cause hyper-susceptibility or hyper-resistance to M. marinum infection. One hyper-susceptible mutation was mapped to the leukotriene A4 hydrolase (lta4h) locus, which encodes an enzyme involved in the production of the chemoattractant and pro-inflammatory eicosanoid, Leukotriene B4. Loss of lta4h function causes a redirection of eicosanoid substrates to anti-inflammatory lipoxins, resulting in a decrease of TNFα production and consequential increases in bacterial growth and macrophage necrosis. The authors subsequently discovered that polymorphisms at the human LTA4H locus are strongly associated with susceptibility to TB, demonstrating the potential of forward genetic analysis in zebrafish to provide novel models and mechanisms of human infectious disease (D.M. Tobin et al., personal communication).
In vivo imaging in the zebrafish-M. marinum model has provided novel insights into the function of macrophages and granulomas during mycobacterial infection. Clay and co-workers found that macrophages were required to both curtail bacterial growth during early infection of zebrafish embryos, and to disseminate infecting bacteria into deeper tissues [33]. This indicated that infecting mycobacteria tolerate sub-optimal growth conditions within macrophages in exchange for access to deeper tissues where granulomas are formed. Granulomas have long been considered to serve as host protective structures that physically isolate the infecting bacteria. Davis and Ramakrishnan recently used the zebrafish-M. marinum model to show that the granuloma can benefit the infecting mycobacteria by facilitating their expansion and dissemination. M. marinum-infected macrophages residing in granulomas recruit uninfected macrophages which then phagocytose dying infected cells and consequently become infected. These newly infected macrophages have the potential to seed new granulomas [44]. Insights into the mechanism by which mycobacteria promote granuloma formation have been provided by analysis of a M. marinum strain lacking the ESX-1/RD1 locus (ΔRD1) encoding a specialized secretion system that promotes virulence in both M. marinum and M. tuberculosis [45]. ΔRD1 M. marinum can survive within macrophages during early infection of zebrafish embryos, but have attenuated initial granuloma formation [45], and recruitment and infection of new macrophages to existing granulomas [44]. These results indicate that infecting mycobacteria use RD1-dependent mechanisms to promote granuloma formation and also to exploit the granuloma environment to enhance pathogenesis. To identify host mechanisms co-opted by RD1-competent M. marinum, Volkman and co-workers compared host gene expression in zebrafish infected with wild-type or ΔRD1 M. marinum. Infection of zebrafish embryos induced RD1-dependent expression of host matrix metalloproteinase 9 (mmp9), and wild-type M. marinum infection of mmp9 morphant embryos caused a phenotype similar to ΔRD1 infection of wild-type hosts. Although mmp9 is expressed by multiple cell types including leukocytes, RD1-dependent mmp9 induction surprisingly occurred only in epithelial cells proximal to infected macrophages. Moreover, RD1-dependent induction of epithelial mmp9 expression did not require myeloid cells, and could be stimulated by injection of a single secreted product of the RD1 locus ESAT-6 [46]. These results establish that M. marinum residing within infected macrophages secrete ESAT-6 to induce mmp9 expression in neighboring epithelial cells, which in turn promotes granuloma formation to facilitate further bacterial dissemination. Together, these studies have defined novel molecular and cellular mechanisms underlying mycobacterial pathogenesis, and demonstrate the potential of the zebrafish for elucidating similar mechanisms in other host-microbe relationships.
Mechanisms of commensal host-microbe relationships
Although individual pathogens can have salient effects on vertebrate biology, the vast majority of host-microbe interactions are non-pathogenic. The epithelial surfaces of zebrafish and all other vertebrates are colonized at birth by large communities of microorganisms (microbiota) that form commensal or mutualistic relationships with their hosts [47,48]. The majority of these microbes reside in digestive tract communities, where they influence a broad range of host biological processes. The zebrafish digestive system shares extensive homology with that of mammals, including a liver, gall bladder, endocrine and exocrine pancreas, and an intestine with proximal-distal functional specification. Initial morphogenesis is completed by 3dpf, however further differentiation and remodeling continue into later stages (reviewed in [49-51]). Zebrafish larvae hatch from the axenic environment within their protective chorion around 3dpf, and the intestine is colonized by microbes within 12-24 hours [52,53]. 16S rRNA sequence-based surveys have revealed that the zebrafish gut microbiota is dominated at all ages by members of the bacterial phylum Proteobacteria, with Firmicutes and Fusobacteria comprising additional dominant phyla at larval and adult stages respectively [54-56] (E. Mittge et al., unpublished). This is in marked contrast to the mammalian gut microbiota, which is dominated by members of the Fimicutes and Bacteroidetes phyla [57]. A recent comparison of the gut microbiota from adult zebrafish raised in different aquaculture facilities or collected in the wild disclosed minimal compositional differences, suggesting that the microbial community assembly in the zebrafish intestine is strongly shaped by deterministic forces within the intestinal habitat (E. Mittge et al., unpublished).
Comparison of zebrafish larvae (6-8dpf) raised in the absence of microorganisms (germ-free or GF) and age-matched controls colonized with a normal zebrafish microbiota, revealed that the zebrafish gut microbiota impacts upon a wide range of host biological processes, including many that are similarly influenced in rodent models [8,53,54]. Conserved host responses include fortification of innate immune defenses, enhancement of nutrient digestion, regulation of intestinal glycan expression, and stimulation of epithelial cell renewal [53-55]. Intriguingly, the presence of a gut microbiota in zebrafish larvae stimulates expression of the neutrophil biomarker myeloperoxidase [54,55] and increased neutrophil infiltration into the intestine [58], suggesting that production and/or activity of immune cells can be regulated by commensal as well as pathogenic microbes. Consistent with this notion, disease severity in a zebrafish model of oxazolone-induced enterocolitis was recently shown to be sensitive to the composition of the gut microbiota [56]. The roles of the gut microbiota on host biology are therefore similar between zebrafish and mammals, despite salient differences in the bacterial composition of their intestinal microbiotas.
To facilitate host tolerance to the gut microbiota, animals have evolved mechanisms to prevent detection of MAMPS as illustrated by the activity of intestinal alkaline phosphatase (Iap). Iap is a brush border enzyme expressed by enterocytes that acts to dephosphorylate and thereby detoxify the lipid A moiety of LPS, the source of LPS endotoxic effects. Bates and colleagues showed that colonization of GF zebrafish with a gut microbiota stimulates intestinal epithelial expression of the iap gene. Morpholino knock-down of iap in zebrafish results in LPS hypersensitivity and neutrophilic inflammation of the gut. Genetic tests revealed that microbial induction of iap and its anti-inflammatory activity are dependent on the TLR adapter protein Myd88 as well as TNFα signaling [58]. Iap therefore promotes mucosal tolerance to commensal gut microbes by reducing the pro-inflammatory potential of LPS in the gut lumen.
Host responses to microbial encounters frequently involve communication between multiple host cell lineages and tissues. It is therefore important to not only identify the signaling mechanisms that facilitate a host response, but also the temporal and spatial pattern in which those signaling events occur. The NF-κB pathway integrates a variety of microbial and physiological stimuli to regulate gene expression. We recently exploited the amenability of the zebrafish to in vivo imaging and transgenesis to define the temporal and spatial pattern of zebrafish NF-κB activity. We generated transgenic reporter zebrafish expressing GFP under control of a NF-κB-responsive cis-element, and used it to reveal that the zebrafish microbiota induces NF-κB activity in multiple tissues including intestine, liver, and dorsal root ganglia. Microbial stimulation of NF-κB activity was associated with NF-κB-dependent induction of acute phase and complement genes in those same tissues, indicating that the gut microbiota can regulate innate immune responses in intestinal as well as extraintestinal sites (M. Kanther et al., unpublished).
Gnotobiotic zebrafish provide opportunities for reductionist analysis of host-bacterial interactions. To this end, we have shown that colonization of GF zebrafish with individual bacterial species can induce host responses normally evoked by the commensal gut microbiota. Among several primary bacterial isolates and laboratory strains representing the normal zebrafish or mouse gut microbiota, P. aeruginosa was the most robust inducer of innate immune and nutrient metabolic responses [55]. Although P. aeruginosa is pathogenic when injected into zebrafish embryos, immersion of zebrafish larvae in P. aeruginosa results in a robust colonization of the intestinal lumen without overt pathogenesis [32,52]. In vivo imaging of GFP-labeled P. aeruginosa in the zebrafish intestine revealed features of intestinal bacterial community assembly, as well as an unanticipated diversity of bacterial behaviors within the intestine. We found that P. aeruginosa strains harboring mutations in genes required for flagella assembly or rotation elicited attenuated innate immune responses in zebrafish hosts, but elicited normal nutrient metabolic responses similar to wild-type P. aeruginosa [52]. These results demonstrate that genetic analysis in model gut bacteria can be conducted in gnotobiotic zebrafish hosts to reveal bacterial factors regulating host-microbe relationships.
Prospectus
The developing zebrafish continues to provide attractive opportunities to investigate the mechanisms underlying host-microbe relationships. With the production of new transgenic reporters of specific host cell lineages and host signaling events, in vivo imaging in transparent developing zebrafish will provide improved spatial and temporal resolution of immune system development and function as well as dynamic host-microbe interactions. Expanded use of forward and reverse genetic analysis in zebrafish hosts and individual microbes can be expected to reveal novel factors regulating distinct aspects of host-microbe interaction. Likewise, the zebrafish provides useful platforms for small molecule screens to identify chemical regulators of specific host-microbe interactions. These approaches should be similarly applicable to the analysis of individual pathogens as well as representative members of the commensal microbiota. As research on host-microbe interactions in the zebrafish continues to grow, it will become increasingly important to appreciate the developmental context in which a given microbial encounter occurs, as well as the effects of that microbial challenge on host ontogeny and physiology.
Acknowledgments
We regret that many excellent references relevant to the topics discussed in this review could not be included due to space limitations. We are grateful to Jeff Yoder, David Tobin, and Lalita Ramakrishnan for communicating data prior to publication, and to Gray Camp, James Minchin, and Suk-Won Jin for helpful comments on this manuscript. Work in the authors' laboratory is supported by NIH grants DK073695 and DK081426, the Center for Gastrointestinal Biology and Disease (NIH grant P30 DK034987), and a Pew Scholar Award to J.F.R.
Footnotes
Annotated references
* Niethammer et al., 2009 (ref. 35): Utilizing the optical transparency and genetic tractability of the zebrafish, Niethammer et al. describe a novel H2O2 gradient responsible for wound to leukocyte signaling created by dual oxidase (duox).
This is the first description of a role for H2O2 in leukocyte signaling.
* Liongue et al., 2009 (ref. 36): Liongue et al. reveal roles for the zebrafish granylocyte colony-stimulating factor and its receptor (gcsf/gcsfr) in production of myeloid cells during primitive hematopoiesis, as well as a previously unappreciated role in production of myeloid cells in response to LPS injection (emergency or demand-driven hematopoiesis).
* Sullivan et al., 2009 (ref. 40): Sullivan et al. demonstrate that zebrafish TLR4 paralogs (tlr4a and tlr4b) are not responsive to LPS as in mammals. This study highlights the evolutionary divergence of immune components between zebrafish and mammals, and the necessity of functional examination of conserved gene candidates.
** Tobin et al., personal communication: This is the first report of a forward genetic screen to identify zebrafish genes involved in microbial pathogenesis. Tobin et al. identify leukotriene A4 hydrolase (lta4h) as an important host hyper-susceptibilty locus in zebrafish M. marinum infection, and discover that polymorphisms at the human LTA4H locus are strongly associated with susceptibility to TB.
** Volkman et al., 2010 (ref. 46): Volkman et al. show that infecting M. marinum use a single secreted factor, ESAT-6, to induce host production of matrix metalloproteinase 9 (mmp9) in neighboring epithelial cells in order to promote granuloma formation in zebrafish hosts. Epithelial expression of mmp9 enhances recruitment of uninfected macrophages to initiate granuloma formation to facilitate bacterial dissemination.
** Bates et al., 2007 (ref. 58): Bates et al. identify a role for intestinal alkaline phosphatase (iap) in preventing detection of MAMPS and promoting intestinal bacterial tolerance. They show that iap is responsible for dephosphorylation and detoxification of lipopolysaccharide produced by the gut microbiota to control homeostatic levels of neutrophils and prevent neutrophilic inflammation of the gut.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Grunwald D, Eisen J. Headwaters of the zebrafish -- emergence of a new model vertebrate. Nat Rev Genet. 2002;3:717–724. doi: 10.1038/nrg892. [DOI] [PubMed] [Google Scholar]
- 2.Lieschke GJ, Currie PD. Animal models of human disease: zebrafish swim into view. Nat Rev Genet. 2007;8:353–367. doi: 10.1038/nrg2091. [DOI] [PubMed] [Google Scholar]
- 3.Meeker N, Trede N. Immunology and zebrafish: spawning new models of human disease. Dev Comp Immunol. 2008;32:745–757. doi: 10.1016/j.dci.2007.11.011. [DOI] [PubMed] [Google Scholar]
- 4.Sullivan C, Kim CH. Zebrafish as a model for infectious disease and immune function. Fish Shellfish Immunol. 2008;25:341–350. doi: 10.1016/j.fsi.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 5.Hall CJ, Flores MV, C KE, Crosier PS. Live imaging early immune cell ontogeny and function in zebrafish Danio rerio. Journal of Fish Biology. 2008:1833–1871. [Google Scholar]
- 6.Bertrand JY, Traver D. Hematopoietic cell development in the zebrafish embryo. Curr Opin Hematol. 2009;16:243–248. doi: 10.1097/MOH.0b013e32832c05e4. [DOI] [PubMed] [Google Scholar]
- 7.Amacher SL. Emerging gene knockout technology in zebrafish: zinc-finger nucleases. Brief Funct Genomic Proteomic. 2008;7:460–464. doi: 10.1093/bfgp/eln043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pham LN, Kanther M, Semova I, Rawls JF. Methods for generating and colonizing gnotobiotic zebrafish. Nat Protoc. 2008;3:1862–1875. doi: 10.1038/nprot.2008.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carradice D, Lieschke GJ. Zebrafish in hematology: sushi or science? Blood. 2008;111:3331–3342. doi: 10.1182/blood-2007-10-052761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davidson AJ, Zon LI. The ‘definitive’ (and ‘primitive’) guide to zebrafish hematopoiesis. Oncogene. 2004;23:7233–7246. doi: 10.1038/sj.onc.1207943. [DOI] [PubMed] [Google Scholar]
- 11.Herbomel P, Thisse B, Thisse C. Ontogeny and behaviour of early macrophages in the zebrafish embryo. Development. 1999;126:3735–3745. doi: 10.1242/dev.126.17.3735. [DOI] [PubMed] [Google Scholar]
- 12.Le Guyader D, Redd MJ, Colucci-Guyon E, Murayama E, Kissa K, Briolat V, Mordelet E, Zapata A, Shinomiya H, Herbomel P. Origins and unconventional behavior of neutrophils in developing zebrafish. Blood. 2008;111:132–141. doi: 10.1182/blood-2007-06-095398. [DOI] [PubMed] [Google Scholar]
- 13.Bertrand J, Kim A, Violette E, Stachura D, Cisson J, Traver D. Definitive hematopoiesis initiates through a committed erythromyeloid progenitor in the zebrafish embryo. Development. 2007;134:4147–4156. doi: 10.1242/dev.012385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Murayama E, Kissa K, Zapata A, Mordelet E, Briolat V, Lin H, Handin R, Herbomel P. Tracing hematopoietic precursor migration to successive hematopoietic organs during zebrafish development. Immunity. 2006;25:963–975. doi: 10.1016/j.immuni.2006.10.015. [DOI] [PubMed] [Google Scholar]
- 15.Jin H, Xu J, Wen Z. Migratory path of definitive hematopoietic stem/progenitor cells during zebrafish development. Blood. 2007;109:5208–5214. doi: 10.1182/blood-2007-01-069005. [DOI] [PubMed] [Google Scholar]
- 16.Bertrand J, Kim A, Teng S, Traver D. CD41+ cmyb+ precursors colonize the zebrafish pronephros by a novel migration route to initiate adult hematopoiesis. Development. 2008;135:1853–1862. doi: 10.1242/dev.015297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kissa K, Murayama E, Zapata A, Cortés A, Perret E, Machu C, Herbomel P. Live imaging of emerging hematopoietic stem cells and early thymus colonization. Blood. 2008;111:1147–1156. doi: 10.1182/blood-2007-07-099499. [DOI] [PubMed] [Google Scholar]
- 18.Jin H, Sood R, Xu J, Zhen F, English MA, Liu PP, Wen Z. Definitive hematopoietic stem/progenitor cells manifest distinct differentiation output in the zebrafish VDA and PBI. Development. 2009;136:647–654. doi: 10.1242/dev.029637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dobson JT, Seibert J, Teh EM, Da'as S, Fraser RB, Paw BH, Lin TJ, Berman JN. Carboxypeptidase A5 identifies a novel mast cell lineage in the zebrafish providing new insight into mast cell fate determination. Blood. 2008;112:2969–2972. doi: 10.1182/blood-2008-03-145011. [DOI] [PubMed] [Google Scholar]
- 20.Willett CE, Cherry JJ, Steiner LA. Characterization and expression of the recombination activating genes (rag1 and rag2) of zebrafish. Immunogenetics. 1997;45:394–404. doi: 10.1007/s002510050221. [DOI] [PubMed] [Google Scholar]
- 21.Danilova N, Steiner LA. B cells develop in the zebrafish pancreas. Proc Natl Acad Sci U S A. 2002;99:13711–13716. doi: 10.1073/pnas.212515999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lam SH, Chua HL, Gong Z, Lam TJ, Sin YM. Development and maturation of the immune system in zebrafish, Danio rerio: a gene expression profiling, in situ hybridization and immunological study. Dev Comp Immunol. 2004;28:9–28. doi: 10.1016/s0145-305x(03)00103-4. [DOI] [PubMed] [Google Scholar]
- 23.Phelps H, Neely M. Evolution of the zebrafish model: from development to immunity and infectious disease. Zebrafish. 2005;2:87–103. doi: 10.1089/zeb.2005.2.87. [DOI] [PubMed] [Google Scholar]
- 24.Davis JM, Clay H, Lewis JL, Ghori N, Herbomel P, Ramakrishnan L. Real-time visualization of mycobacterium-macrophage interactions leading to initiation of granuloma formation in zebrafish embryos. Immunity. 2002;17:693–702. doi: 10.1016/s1074-7613(02)00475-2. [DOI] [PubMed] [Google Scholar]
- 25.O'Toole R, Von Hofsten J, Rosqvist R, Olsson PE, Wolf-Watz H. Visualisation of zebrafish infection by GFP-labelled Vibrio anguillarum. Microb Pathog. 2004;37:41–46. doi: 10.1016/j.micpath.2004.03.001. [DOI] [PubMed] [Google Scholar]
- 26.Pressley ME, Phelan PE, 3rd, Witten PE, Mellon MT, Kim CH. Pathogenesis and inflammatory response to Edwardsiella tarda infection in the zebrafish. Dev Comp Immunol. 2005;29:501–513. doi: 10.1016/j.dci.2004.10.007. [DOI] [PubMed] [Google Scholar]
- 27.Miller JD, Neely MN. Large-scale screen highlights the importance of capsule for virulence in the zoonotic pathogen Streptococcus iniae. Infect Immun. 2005;73:921–934. doi: 10.1128/IAI.73.2.921-934.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kizy A, Neely M. First Streptococcus pyogenes signature-tagged mutagenesis screen identifies novel virulence determinants. Infect Immun. 2009;77:1854–1865. doi: 10.1128/IAI.01306-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rhodes J, Hagen A, Hsu K, Deng M, Liu TX, Look AT, Kanki JP. Interplay of pu.1 and gata1 determines myelo-erythroid progenitor cell fate in zebrafish. Dev Cell. 2005;8:97–108. doi: 10.1016/j.devcel.2004.11.014. [DOI] [PubMed] [Google Scholar]
- 30.Brannon M, Davis J, Mathias J, Hall C, Emerson J, Crosier P, Huttenlocher A, Ramakrishnan L, Moskowitz S. Pseudomonas aeruginosa type III secretion system interacts with phagocytes to modulate systemic infection of zebrafish embryos. Cell Microbiol. 2009 doi: 10.1111/j.1462-5822.2009.01288.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prajsnar T, Cunliffe V, Foster S, Renshaw S. A novel vertebrate model of Staphylococcus aureus infection reveals phagocyte-dependent resistance of zebrafish to non-host specialized pathogens. Cell Microbiol. 2008;10:2312–2325. doi: 10.1111/j.1462-5822.2008.01213.x. [DOI] [PubMed] [Google Scholar]
- 32.Clatworthy A, Lee J, Leibman M, Kostun Z, Davidson A, Hung D. Pseudomonas aeruginosa infection of zebrafish involves both host and pathogen determinants. Infect Immun. 2009;77:1293–1303. doi: 10.1128/IAI.01181-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Clay H, Davis J, Beery D, Huttenlocher A, Lyons S, Ramakrishnan L. Dichotomous role of the macrophage in early Mycobacterium marinum infection of the zebrafish. Cell Host Microbe. 2007;2:29–39. doi: 10.1016/j.chom.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Clay H, Volkman H, Ramakrishnan L. Tumor necrosis factor signaling mediates resistance to mycobacteria by inhibiting bacterial growth and macrophage death. Immunity. 2008;29:283–294. doi: 10.1016/j.immuni.2008.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Niethammer P, Grabher C, Look A, Mitchison T. A tissue-scale gradient of hydrogen peroxide mediates rapid wound detection in zebrafish. Nature. 2009;459:996–999. doi: 10.1038/nature08119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liongue C, Hall C, O'Connell B, Crosier P, Ward A. Zebrafish granulocyte colony-stimulating factor receptor signaling promotes myelopoiesis and myeloid cell migration. Blood. 2009;113:2535–2546. doi: 10.1182/blood-2008-07-171967. [DOI] [PubMed] [Google Scholar]
- 37.Vasil M, Stonehouse M, Vasil A, Wadsworth S, Goldfine H, Bolcome Rr, Chan J. A complex extracellular sphingomyelinase of Pseudomonas aeruginosa inhibits angiogenesis by selective cytotoxicity to endothelial cells. PLoS Pathog. 2009;5:e1000420. doi: 10.1371/journal.ppat.1000420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stockhammer O, Zakrzewska A, Hegedûs Z, Spaink H, Meijer A. Transcriptome profiling and functional analyses of the zebrafish embryonic innate immune response to Salmonella infection. J Immunol. 2009;182:5641–5653. doi: 10.4049/jimmunol.0900082. [DOI] [PubMed] [Google Scholar]
- 39.Sepulcre M, Alcaraz-Pérez F, López-Muñoz A, Roca F, Meseguer J, Cayuela M, Mulero V. Evolution of lipopolysaccharide (LPS) recognition and signaling: fish TLR4 does not recognize LPS and negatively regulates NF-kappaB activation. J Immunol. 2009;182:1836–1845. doi: 10.4049/jimmunol.0801755. [DOI] [PubMed] [Google Scholar]
- 40.Sullivan C, Charette J, Catchen J, Lage C, Giasson G, Postlethwait J, Millard P, Kim C. The gene history of zebrafish tlr4a and tlr4b is predictive of their divergent functions. J Immunol. 2009 doi: 10.4049/jimmunol.0803285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Meijer AH, Verbeek FJ, Salas-Vidal E, Corredor-Adamez M, Bussman J, van der Sar AM, Otto GW, Geisler R, Spaink HP. Transcriptome profiling of adult zebrafish at the late stage of chronic tuberculosis due to Mycobacterium marinum infection. Mol Immunol. 2005;42:1185–1203. doi: 10.1016/j.molimm.2004.11.014. [DOI] [PubMed] [Google Scholar]
- 42.van der Sar A, Spaink H, Zakrzewska A, Bitter W, Meijer A. Specificity of the zebrafish host transcriptome response to acute and chronic mycobacterial infection and the role of innate and adaptive immune components. Mol Immunol. 2009;46:2317–2332. doi: 10.1016/j.molimm.2009.03.024. [DOI] [PubMed] [Google Scholar]
- 43.Hegedus Z, Zakrzewska A, Agoston V, Ordas A, Rácz P, Mink M, Spaink H, Meijer A. Deep sequencing of the zebrafish transcriptome response to Mycobacterium infection. Mol Immunol. 2009;46:2918–2930. doi: 10.1016/j.molimm.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 44.Davis J, Ramakrishnan L. The role of the granuloma in expansion and dissemination of early tuberculous infection. Cell. 2009;136:37–49. doi: 10.1016/j.cell.2008.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Volkman HE, Clay H, Beery D, Chang JC, Sherman DR, Ramakrishnan L. Tuberculous granuloma formation is enhanced by a Mycobacterium virulence determinant. PLoS Biol. 2004;2:e367. doi: 10.1371/journal.pbio.0020367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Volkman HE, P T, Zheng J, Davis JM, Rawls JF, Ramakrishnan L. Tuberculous granuloma induction via interaction of bacterial secreted protein with host epithelium. Science. 2010 doi: 10.1126/science.1179663. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cheesman SE, Guillemin K. We know you are in there: conversing with the indigenous gut microbiota. Res Microbiol. 2007;158:2–9. doi: 10.1016/j.resmic.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 48.Camp JG, Kanther M, Semova I, Rawls JF. Patterns and scales in gastrointestinal microbial ecology. Gastroenterology. 2009;136:1989–2002. doi: 10.1053/j.gastro.2009.02.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pack M, Solnica-Krezel L, Malicki J, Neuhauss SC, Schier AF, Stemple DL, Driever W, Fishman MC. Mutations affecting development of zebrafish digestive organs. Development. 1996;123:321–328. doi: 10.1242/dev.123.1.321. [DOI] [PubMed] [Google Scholar]
- 50.Ng AN, de Jong-Curtain TA, Mawdsley DJ, White SJ, Shin J, Appel B, Dong PD, Stainier DY, Heath JK. Formation of the digestive system in zebrafish: III. Intestinal epithelium morphogenesis. Dev Biol. 2005;286:114–135. doi: 10.1016/j.ydbio.2005.07.013. [DOI] [PubMed] [Google Scholar]
- 51.Wallace KN, Akhter S, Smith EM, Lorent K, Pack M. Intestinal growth and differentiation in zebrafish. Mech Dev. 2005;122:157–173. doi: 10.1016/j.mod.2004.10.009. [DOI] [PubMed] [Google Scholar]
- 52.Rawls JF, Mahowald MA, Goodman AL, Trent CM, Gordon JI. In vivo imaging and genetic analysis link bacterial motility and symbiosis in the zebrafish gut. Proc Natl Acad Sci U S A. 2007;104:7622–7627. doi: 10.1073/pnas.0702386104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bates J, Mittge E, Kuhlman J, Baden K, Cheesman S, Guillemin K. Distinct signals from the microbiota promote different aspects of zebrafish gut differentiation. Dev Biol. 2006;297:374–386. doi: 10.1016/j.ydbio.2006.05.006. [DOI] [PubMed] [Google Scholar]
- 54.Rawls JF, Samuel BS, Gordon JI. Gnotobiotic zebrafish reveal evolutionarily conserved responses to the gut microbiota. Proc Natl Acad Sci U S A. 2004;101:4596–4601. doi: 10.1073/pnas.0400706101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rawls JF, Mahowald MA, Ley RE, Gordon JI. Reciprocal gut microbiota transplants from zebrafish and mice to germ-free recipients reveal host habitat selection. Cell. 2006;127:423–433. doi: 10.1016/j.cell.2006.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brugman S, Liu KY, Lindenbergh-Kortleve D, Samsom JN, Furuta GT, Renshaw SA, Willemsen R, Nieuwenhuis EE. Oxazolone-induced enterocolitis in zebrafish depends on the composition of the intestinal microbiota. Gastroenterology. 2009;137:1757–1767. e1751. doi: 10.1053/j.gastro.2009.07.069. [DOI] [PubMed] [Google Scholar]
- 57.Ley RE, Hamady M, Lozupone C, Turnbaugh PJ, Ramey RR, Bircher JS, Schlegel ML, Tucker TA, Schrenzel MD, Knight R, et al. Evolution of mammals and their gut microbes. Science. 2008 doi: 10.1126/science.1155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bates J, Akerlund J, Mittge E, Guillemin K. Intestinal alkaline phosphatase detoxifies lipopolysaccharide and prevents inflammation in zebrafish in response to the gut microbiota. Cell Host Microbe. 2007;2:371–382. doi: 10.1016/j.chom.2007.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Willett CE, Cortes A, Zuasti A, Zapata AG. Early hematopoiesis and developing lymphoid organs in the zebrafish. Dev Dyn. 1999;214:323–336. doi: 10.1002/(SICI)1097-0177(199904)214:4<323::AID-AJA5>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 60.Rodriguez I, Novoa B, Figueras A. Immune response of zebrafish (Danio rerio) against a newly isolated bacterial pathogen Aeromonas hydrophila. Fish Shellfish Immunol. 2008;25:239–249. doi: 10.1016/j.fsi.2008.05.002. [DOI] [PubMed] [Google Scholar]
- 61.Lin B, Chen S, Cao Z, Lin Y, Mo D, Zhang H, Gu J, Dong M, Liu Z, Xu A. Acute phase response in zebrafish upon Aeromonas salmonicida and Staphylococcus aureus infection: striking similarities and obvious differences with mammals. Mol Immunol. 2007;44:295–301. doi: 10.1016/j.molimm.2006.03.001. [DOI] [PubMed] [Google Scholar]
- 62.Deng Y, Boon C, Eberl L, Zhang LH. Differential modulation of Burkholderia cenocepacia virulence and energy metabolism by the quorum-sensing signal BDSF and its synthase. J Bacteriol. 2009;191:7270–7278. doi: 10.1128/JB.00681-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Petrie-Hanson L, Romano CL, Mackey RB, Khosravi P, Hohn CM, Boyle CR. Evaluation of zebrafish Danio rerio as a model for enteric septicemia of catfish (ESC) J Aquat Anim Health. 2007;19:151–158. doi: 10.1577/H06-026.1. [DOI] [PubMed] [Google Scholar]
- 64.Nayak AS, Lage CR, Kim CH. Effects of low concentrations of arsenic on the innate immune system of the zebrafish (Danio rerio) Toxicol Sci. 2007;98:118–124. doi: 10.1093/toxsci/kfm072. [DOI] [PubMed] [Google Scholar]
- 65.Phelan PE, Mellon MT, Kim CH. Functional characterization of full-length TLR3, IRAK-4, and TRAF6 in zebrafish (Danio rerio) Mol Immunol. 2005;42:1057–1071. doi: 10.1016/j.molimm.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 66.Szabady R, Lokuta M, Walters K, Huttenlocher A, Welch R. Modulation of neutrophil function by a secreted mucinase of Escherichia coli O157:H7. PLoS Pathog. 2009;5:e1000320. doi: 10.1371/journal.ppat.1000320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Moyer TR, Hunnicutt DW. Susceptibility of zebra fish Danio rerio to infection by Flavobacterium columnare and F. johnsoniae. Dis Aquat Organ. 2007;76:39–44. doi: 10.3354/dao076039. [DOI] [PubMed] [Google Scholar]
- 68.Vojtech LN, Sanders GE, Conway C, Ostland V, Hansen JD. Host immune response and acute disease in a zebrafish model of Francisella pathogenesis. Infect Immun. 2009;77:914–925. doi: 10.1128/IAI.01201-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Davis JM, Haake DA, Ramakrishnan L. Leptospira interrogans stably infects zebrafish embryos, altering phagocyte behavior and homing to specific tissues. PLoS Negl Trop Dis. 2009;3:e463. doi: 10.1371/journal.pntd.0000463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Menudier A, Rougier FP, Bosgiraud C. Comparative virulence between different strains of Listeria in zebrafish (Brachydanio rerio) and mice. Pathol Biol (Paris) 1996;44:783–789. [PubMed] [Google Scholar]
- 71.Levraud J, Disson O, Kissa K, Bonne I, Cossart P, Herbomel P, Lecuit M. Real-time observation of listeria monocytogenes-phagocyte interactions in living zebrafish larvae. Infect Immun. 2009;77:3651–3660. doi: 10.1128/IAI.00408-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rojo I, de Ilarduya OM, Estonba A, Pardo MA. Innate immune gene expression in individual zebrafish after Listonella anguillarum inoculation. Fish Shellfish Immunol. 2007;23:1285–1293. doi: 10.1016/j.fsi.2007.07.002. [DOI] [PubMed] [Google Scholar]
- 73.Whipps CM, Dougan ST, Kent ML. Mycobacterium haemophilum infections of zebrafish (Danio rerio) in research facilities. FEMS Microbiol Lett. 2007;270:21–26. doi: 10.1111/j.1574-6968.2007.00671.x. [DOI] [PubMed] [Google Scholar]
- 74.Gao LY, Pak M, Kish R, Kajihara K, Brown EJ. A mycobacterial operon essential for virulence in vivo and invasion and intracellular persistence in macrophages. Infect Immun. 2006;74:1757–1767. doi: 10.1128/IAI.74.3.1757-1767.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Harriff MJ, Bermudez LE, Kent ML. Experimental exposure of zebrafish, Danio rerio (Hamilton), to Mycobacterium marinum and Mycobacterium peregrinum reveals the gastrointestinal tract as the primary route of infection: a potential model for environmental mycobacterial infection. J Fish Dis. 2007;30:587–600. doi: 10.1111/j.1365-2761.2007.00839.x. [DOI] [PubMed] [Google Scholar]
- 76.Prouty MG, Correa NE, Barker LP, Jagadeeswaran P, Klose KE. Zebrafish-Mycobacterium marinum model for mycobacterial pathogenesis. FEMS Microbiol Lett. 2003;225:177–182. doi: 10.1016/S0378-1097(03)00446-4. [DOI] [PubMed] [Google Scholar]
- 77.Swaim LE, Connolly LE, Volkman HE, Humbert O, Born DE, Ramakrishnan L. Mycobacterium marinum infection of adult zebrafish causes caseating granulomatous tuberculosis and is moderated by adaptive immunity. Infect Immun. 2006;74:6108–6117. doi: 10.1128/IAI.00887-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Llamas M, van der Sar A, Chu B, Sparrius M, Vogel H, Bitter W. A Novel extracytoplasmic function (ECF) sigma factor regulates virulence in Pseudomonas aeruginosa. PLoS Pathog. 2009;5:e1000572. doi: 10.1371/journal.ppat.1000572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.van der Sar AM, Musters RJ, van Eeden FJ, Appelmelk BJ, Vandenbroucke-Grauls CM, Bitter W. Zebrafish embryos as a model host for the real time analysis of Salmonella typhimurium infections. Cell Microbiol. 2003;5:601–611. doi: 10.1046/j.1462-5822.2003.00303.x. [DOI] [PubMed] [Google Scholar]
- 80.van der Sar A, Stockhammer O, van der Laan C, Spaink H, Bitter W, Meijer A. MyD88 innate immune function in a zebrafish embryo infection model. Infect Immun. 2006;74:2436–2441. doi: 10.1128/IAI.74.4.2436-2441.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Neely MN, Pfeifer JD, Caparon M. Streptococcus-zebrafish model of bacterial pathogenesis. Infect Immun. 2002;70:3904–3914. doi: 10.1128/IAI.70.7.3904-3914.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lowe B, Miller J, Neely M. Analysis of the polysaccharide capsule of the systemic pathogen Streptococcus iniae and its implications in virulence. Infect Immun. 2007;75:1255–1264. doi: 10.1128/IAI.01484-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bates CS, Toukoki C, Neely MN, Eichenbaum Z. Characterization of MtsR, a new metal regulator in group A streptococcus, involved in iron acquisition and virulence. Infect Immun. 2005;73:5743–5753. doi: 10.1128/IAI.73.9.5743-5753.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cho KH, Caparon MG. Patterns of virulence gene expression differ between biofilm and tissue communities of Streptococcus pyogenes. Mol Microbiol. 2005;57:1545–1556. doi: 10.1111/j.1365-2958.2005.04786.x. [DOI] [PubMed] [Google Scholar]
- 85.Montanez GE, Neely MN, Eichenbaum Z. The Streptococcal iron uptake (Siu) transporter is required for iron uptake and virulence in a zebrafish infection model. Microbiology. 2005;151:3749–3757. doi: 10.1099/mic.0.28075-0. [DOI] [PubMed] [Google Scholar]
- 86.Burgos JS, Ripoll-Gomez J, Alfaro JM, Sastre I, Valdivieso F. Zebrafish as a new model for herpes simplex virus type 1 infection. Zebrafish. 2008;5:323–333. doi: 10.1089/zeb.2008.0552. [DOI] [PubMed] [Google Scholar]
- 87.LaPatra SE, Barone L, Jones GR, Zon LI. Effects of infectious hematopoietic necrosis virus and infectious pancreatic necrosis virus infection on hematopoietic precursors of the zebrafish. Blood Cells Mol Dis. 2000;26:445–452. doi: 10.1006/bcmd.2000.0320. [DOI] [PubMed] [Google Scholar]
- 88.Wang L, Zhang HX, Zhang JH, Chen WH, Ruan XF, Xia C. In vitro effects of recombinant zebrafish IFN on spring viremia of carp virus and infectious hematopoietic necrosis virus. J Interferon Cytokine Res. 2006;26:256–259. doi: 10.1089/jir.2006.26.256. [DOI] [PubMed] [Google Scholar]
- 89.Garner JN, Joshi B, Jagus R. Characterization of rainbow trout and zebrafish eukaryotic initiation factor 2alpha and its response to endoplasmic reticulum stress and IPNV infection. Dev Comp Immunol. 2003;27:217–231. doi: 10.1016/s0145-305x(02)00096-4. [DOI] [PubMed] [Google Scholar]
- 90.Xu X, Zhang L, Weng S, Huang Z, Lu J, Lan D, Zhong X, Yu X, Xu A, He J. A zebrafish (Danio rerio) model of infectious spleen and kidney necrosis virus (ISKNV) infection. Virology. 2008;376:1–12. doi: 10.1016/j.virol.2007.12.026. [DOI] [PubMed] [Google Scholar]
- 91.Wang ZL, Xu XP, He BL, Weng SP, Xiao J, Wang L, Lin T, Liu X, Wang Q, Yu XQ, et al. Infectious spleen and kidney necrosis virus ORF48R functions as a new viral vascular endothelial growth factor. J Virol. 2008;82:4371–4383. doi: 10.1128/JVI.02027-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lu MW, Chao YM, Guo TC, Santi N, Evensen O, Kasani SK, Hong JR, Wu JL. The interferon response is involved in nervous necrosis virus acute and persistent infection in zebrafish infection model. Mol Immunol. 2008;45:1146–1152. doi: 10.1016/j.molimm.2007.07.018. [DOI] [PubMed] [Google Scholar]
- 93.Alonso M, Kim CH, Johnson MC, Pressley M, Leong JA. The NV gene of snakehead rhabdovirus (SHRV) is not required for pathogenesis, and a heterologous glycoprotein can be incorporated into the SHRV envelope. J Virol. 2004;78:5875–5882. doi: 10.1128/JVI.78.11.5875-5882.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Altmann SM, Mellon MT, Distel DL, Kim CH. Molecular and functional analysis of an interferon gene from the zebrafish, Danio rerio. J Virol. 2003;77:1992–2002. doi: 10.1128/JVI.77.3.1992-2002.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Altmann SM, Mellon MT, Johnson MC, Paw BH, Trede NS, Zon LI, Kim CH. Cloning and characterization of an Mx gene and its corresponding promoter from the zebrafish, Danio rerio. Dev Comp Immunol. 2004;28:295–306. doi: 10.1016/j.dci.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 96.Hermann AC, Kim CH. Effects of arsenic on zebrafish innate immune system. Mar Biotechnol (NY) 2005;7:494–505. doi: 10.1007/s10126-004-4109-7. [DOI] [PubMed] [Google Scholar]
- 97.Phelan PE, Pressley ME, Witten PE, Mellon MT, Blake S, Kim CH. Characterization of snakehead rhabdovirus infection in zebrafish (Danio rerio) J Virol. 2005;79:1842–1852. doi: 10.1128/JVI.79.3.1842-1852.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sanders GE, Batts WN, Winton JR. Susceptibility of zebrafish (Danio rerio) to a model pathogen, spring viremia of carp virus. Comp Med. 2003;53:514–521. [PubMed] [Google Scholar]
- 99.Novoa B, Romero A, Mulero V, Rodriguez I, Fernandez I, Figueras A. Zebrafish (Danio rerio) as a model for the study of vaccination against viral haemorrhagic septicemia virus (VHSV) Vaccine. 2006;24:5806–5816. doi: 10.1016/j.vaccine.2006.05.015. [DOI] [PubMed] [Google Scholar]
- 100.Nasevicius A, Ekker SC. Effective targeted gene ‘knockdown’ in zebrafish. Nat Genet. 2000;26:216–220. doi: 10.1038/79951. [DOI] [PubMed] [Google Scholar]
- 101.Eisen JS, Smith JC. Controlling morpholino experiments: don't stop making antisense. Development. 2008;135:1735–1743. doi: 10.1242/dev.001115. [DOI] [PubMed] [Google Scholar]
- 102.Driever W, Solnica-Krezel L, Schier AF, Neuhauss SC, Malicki J, Stemple DL, Stainier DY, Zwartkruis F, Abdelilah S, Rangini Z, et al. A genetic screen for mutations affecting embryogenesis in zebrafish. Development. 1996;123:37–46. doi: 10.1242/dev.123.1.37. [DOI] [PubMed] [Google Scholar]
- 103.Haffter P, Granato M, Brand M, Mullins MC, Hammerschmidt M, Kane DA, Odenthal J, van Eeden FJ, Jiang YJ, Heisenberg CP, et al. The identification of genes with unique and essential functions in the development of the zebrafish, Danio rerio. Development. 1996;123:1–36. doi: 10.1242/dev.123.1.1. [DOI] [PubMed] [Google Scholar]
- 104.Wienholds E, van Eeden F, Kosters M, Mudde J, Plasterk RH, Cuppen E. Efficient target-selected mutagenesis in zebrafish. Genome Res. 2003;13:2700–2707. doi: 10.1101/gr.1725103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Jao LE, Maddison L, Chen W, Burgess SM. Using retroviruses as a mutagenesis tool to explore the zebrafish genome. Brief Funct Genomic Proteomic. 2008;7:427–443. doi: 10.1093/bfgp/eln038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Amsterdam A, Hopkins N. Mutagenesis strategies in zebrafish for identifying genes involved in development and disease. Trends Genet. 2006;22:473–478. doi: 10.1016/j.tig.2006.06.011. [DOI] [PubMed] [Google Scholar]
- 107.Suster ML, Kikuta H, Urasaki A, Asakawa K, Kawakami K. Transgenesis in zebrafish with the tol2 transposon system. Methods Mol Biol. 2009;561:41–63. doi: 10.1007/978-1-60327-019-9_3. [DOI] [PubMed] [Google Scholar]
- 108.Esengil H, Chen JK. Gene regulation technologies in zebrafish. Mol Biosyst. 2008;4:300–308. doi: 10.1039/b718447f. [DOI] [PubMed] [Google Scholar]