Abstract
MicroRNAs (miRNAs) play essential roles in biological processes ranging from cellular proliferation to apoptosis. Recently, miRNAs have also been implicated in a number of diseases including cancers. However, the targets of most miRNAs remain unknown. The majority of reports describing identification of miRNA targets are based on computational approaches or detection of altered mRNA levels despite the fact that most miRNAs are thought to regulate their targets primarily at the level of translational inhibition in animals. miR-21 is a miRNA with oncogenic activity that is involved in various cancer related processes such as invasion and migration. Given the importance of miR-21 in tumorigenesis, we employed a quantitative proteomic strategy to systematically identify potential targets of miR-21. By knocking down the expression of endogenous miR-21 in MCF-7 breast cancer cells, we observed an increase in the abundance of 58 proteins signifying that they could be potential targets of miR-21. Validation of 12 of these candidate targets in luciferase assays showed that 6 of them were likely direct targets of miR-21. Importantly, the mRNA of the majority of the candidate targets tested did not show a concomitant increase in abundance. Overall, our results demonstrate that miR-21 affects the expression of many of its targets through translational inhibition and highlights the utility of proteomic approaches for identifying miRNA targets.
Keywords: miRNA, quantitative analysis, Proteomics
Introduction
miRNAs are non-coding, single-stranded RNAs of ~22 nucleotides that constitute a novel class of gene regulators [1, 2]. In animals, miRNAs are known to control a wide range of biological functions such as cellular proliferation, differentiation, migration and apoptosis [3–6]. miRNAs are initially transcribed by RNA Polymerase II as long primary transcripts (pri-miRNAs). Pri-miRNAs are processed by Drosha and Pasha, producing 60–70-nt stem-loop precursor miRNAs (pre-miRNAs). Pre-miRNAs are then exported to the cytoplasm by the nuclear export factor exportin 5 and cleaved further by Dicer, giving rise to transient ~22-nucleotide duplexes containing mature miRNAs and their complementary nucleotides. Mature miRNAs are then loaded onto the RNA-induced silencing complex (RISC), where they interact with target mRNA transcripts. When a miRNA and its cognate mRNA interact with perfect complementarity, RISC directly cleaves the target mRNA [7–9]. However, in animals, miRNAs generally interact with target mRNAs with only partial complementarity. The highly conserved 6 or 7 nucleotides at the 5' end of the miRNA are referred to as the ‘seed region’ for binding activities [10]. This mode of recognition results in translational repression of the target mRNA with much smaller effects at the level of target mRNA abundance [11–14].
Thus far, relatively few miRNA targets have been experimentally validated [15], although there are numerous miRNA targets that have been predicted through bioinformatics approaches [16–18]. Most computational algorithms predict miRNA targets based primarily on sequence complementarity between the 5’ end of the mature miRNA and the 3’untranslated region (3’UTR) of target genes. Given the relatively high rates of both false-positives and false-negatives from current miRNA target prediction programs, it is critically important to experimentally identify miRNA targets [10]. High-throughput approaches based on DNA microarrays that rely on changes in target mRNA abundance have been used to experimentally identify miRNA targets [19–21]. However, this approach has limitations as most miRNAs are thought to regulate gene expression by translational inhibition rather than mRNA degradation in animals [1].
In the present study, we sought to identify targets of miR-21 using quantitative tandem mass spectrometry to directly identify miRNA targets at the protein level. miR-21 is a miRNA that has been reported to be associated with multiple cancer-related processes including proliferation, apoptosis, invasion, and metastasis. For example, Si et al. found that inhibition of miR-21 suppressed both cell growth in vitro and tumor growth in the xenograft mouse model [22]. In another study, Corsten et al found that combined miR-21 inhibition and cytotoxic tumor treatment led to complete eradication of gliomas in the murine brain [23]. Recently, Asangani et al. showed that miR-21 inhibition significantly reduced cell intravasation and lung metastasis in chicken embryos [24]. Other studies showed similar results in human hepatocytes, breast cancer and glioblastomas cells [25–27].
Although miR-21 is clearly an important miRNA, only four targets of miR-21 have been described thus far. Zhu et al. identified tropomyosin1 (TPM1) as a potential target of miR-21 using two-dimensional gel electrophoresis [28]. While inhibition of miR-21 increased TPM1 protein expression about 2-fold in breast cancer cells, the expression of TPM1 mRNA remained unchanged. Phosphatase and tensin homolog (PTEN) was discovered as a potential target of miR-21 through a bioinformatics approach [25]. PTEN protein expression level was increased about 2 to 3 fold upon miR-21 inhibition in human hepatocellular carcinoma cells, while again there was no direct effect of miR-21 on PTEN mRNA abundance. Recently, three different groups identified programmed cell death 4 (PDCD4) as a target of miR-21. Asangani et al. used a computational approach to select PDCD4 as a potential target of miR-21 and demonstrated that miR-21 significantly decreased PDCD4 protein expression without affecting the level of this mRNA in colorectal cancer cells [24]. Frankel and colleagues used DNA microarrays to identify PDCD4 as a potential target of miR-21 targets in MCF-7 cells [21]. They further showed that miR-21 inhibition significantly increased PDCD4 expression at both the mRNA and protein level in MCF-7 cells. Zhu et al. identified PDCD4 and maspin as candidate miR-21 targets using a genetic screen/selection system [27]. They observed that inhibition of miR-21 significantly upregulated protein expression of PDCD4 and maspin in the metastatic breast cancer cell line MDA-MB-231.
We carried out global proteomic profiling to identify targets of miR-21 in MCF-7 breast cancer cells. Using an iTRAQ based proteomics strategy in combination with strong cation exchange (SCX) chromatography, we have identified 58 putative targets of miR-21. Using luciferase assays, we demonstrate that a subset of these targets identified from the proteomic screen are direct targets of miR-21. Further studies on the same subset of these putative targets confirm that the many of them are regulated through translational inhibition without affecting the mRNA levels.
Materials and Methods
Real time RT-PCR analysis
Breast cancer cell line MCF-7 were cultured in Eagle’s Minimum Essential Medium (ATCC, Manassas, VA) supplemented with 10% FBS, 0.01mg/ml bovine insulin, 100U/ml penicillin and 100 μg/ml streptomycin. MCF-7 cells were seeded in 150 mm dishes and transfected with 50 nM miR-21 antisense oligonucleotides (anti-miR-21 oligo) or control oligo (Dharmacon, Lafayette, CO) using DharmaFECT 1 (Dharmacon). Total RNA from transfected cells was isolated with miRNeasy Mini kit (Qiagen, Valencia, CA) according to the manufacturer’s protocol. Real time RT-PCR of miR-21 was performed using TaqMan® MicroRNA Reverse Transcription Kit, TaqMan Universal PCR Master Mix and TaqMan MicroRNA Assay (Applied Biosystems, Foster City, CA). CT value of miR-21 was normalized to the CT value of U6B (a small RNA) in the same sample. Real time RT-PCR of miR-21 candidate targets was performed using QuantiTect® reverse transcription kit (Qiagen) and SybrGreen 2X qPCR master mix (Roche, Basel, Switzerland). The sequences of primers are provided in supplemental data (Supplementary Table 1). CT value from each gene is normalized to the CT value of β-actin in the same sample. Relative expression of miR-21 or miR-21 candidate targets in anti-miR-21 oligo transfections was normalized to control oligos and a p-value was calculated with a two-tailed t-test.
iTRAQ and SCX fractionation
MCF-7 cells were seeded in 150 mm dishes and transfected with 50 nM anti-miR-21 oligo or control oligo using DharmaFECT 1 (Dharmacon). The cells were harvested and resuspended in 0.05% SDS and lysed by sonication. 100 μg of each sample was digested using trypsin as previously described [29]. The tryptic peptides were labeled with iTRAQ reagents (Applied Biosystems) having 114, 115, 116 or 117 reporter ions. Subsequently, the labeled peptides were mixed equally and the mixture was fractionated using SCX chromatography on PolySULFOETHYL A column (PolyLC, Columbia, MD) (100 × 2.1 mm, 5qm particles with 300Å pores). SCX chromatography was performed using Ultimate HPLC system (LC Packing) and fractions were collected using a Probot fraction collector. Twenty-eight SCX fractions (0.5ml) were collected using a nonlinear gradient of 0 to 350 mM KCl in the presence of 10 mM potassium phosphate buffer (pH 2.85), containing 20% acetonitrile. The fractions were then dried and reconstituted in 2% trifluoroacetic acid and samples were analyzed using LC-MS/MS as described below.
Mass spectrometry and protein quantification
LC-MS/MS analysis of the sample was carried out using a RP-LC interfaced with a quadrupole time-of-flight mass spectrometer (QSTAR/Pulsar, Applied Biosystems). RP-LC system (LC Packing) consisted of a trap column (75qm × 3 cm, C18 material 5-10qm, 120Å) and an analytical column (75qm × 10 cm, C18 material 5qm, 120Å). The peptides were separated by acetonitrile gradient (0-60%) containing 0.1% formic acid. The MS spectra were acquired in a data dependent manner targeting the three most abundant ions in the survey scan. ProteinPilot software version 2.0.1 (Applied Biosystems MDS SCIEX) was used for identification and quantitation of proteins. The data from 28 LC-MS/MS analyses were processed together and search results were merged. The data were searched against human RefSeq database version 26 containing 39,380 protein entries. Search parameters included iTRAQ labeling at N-terminus and lysine residues, cysteine modification by methyl methanethiosulfonate (MMTS) and methionine oxidation. Protein identification was based on criteria of “unused” ProtScore. The total “unused” ProtScore is a measurement of all the peptide evidence for a protein that is not better explained by a higher ranking protein. Proteins identified with >95% confidence (Protscore >1.3) were used for further quantitation. Paragon Algorithm was used to quantitate relative protein levels between control and anti-miR-21 oligo transfected samples. Quantitation was carried out whenever there are one or more iTRAQ reagent ratios for a peptide.
Plasmid constructs and Luciferase assays
3’UTR fragments of selected genes were PCR amplified from human cDNA and cloned into pGL3-control vector (Promega, Madison, WI) at Bgl II restriction site. The primer sequences used for PCR amplification are listed in Supplementary Table 1. 24 hours prior transfection, 1× 105 cells were plated per well in a 24-well plate. pGL3 constructs (150 ng) plus 10 ng of the Renilla luciferase plasmid phRL-SV40 (Promega) were co-transfected with anti-miR-21 oligo or control oligo using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) to MCF-7 [30]. 24 hours after transfection, luciferase assays were performed using the dual luciferase reporter assay system (Promega). Firefly luciferase activity was normalized to Renilla luciferase activity for each transfected well. For each experimental trial, cells were transfected in triplicate and each well was assayed in triplicate. For each construct the values from anti-miR-21 oligo are normalized to control oligo. A p-value was calculated with the two-tailed t-test to compare relative luciferase activities in each construct with empty vector.
Bioinformatics analysis
For determining of the frequency of miR-21 seed regions, genes identified in the present study with annotated 3’UTR, 5’UTR and coding sequences were selected. The sequences were downloaded from March 2006 build of the human genome. The enrichment of motifs complementary to 7 mer (2 to 8 nucleotide) of the 5’ end of miR-21 was evaluated by chi square test. Pictar (http://pictar.bio.nyu.edu/) [17], Targetscan4.1 (http://www.targetscan.org/) [16, 31] and miRanda (http://www.microrna.org/microrna/home.do) [18] were used to computationally predict targets of miR-21.
Results
Identification of putative miR-21 targets using a quantitative proteomic approach
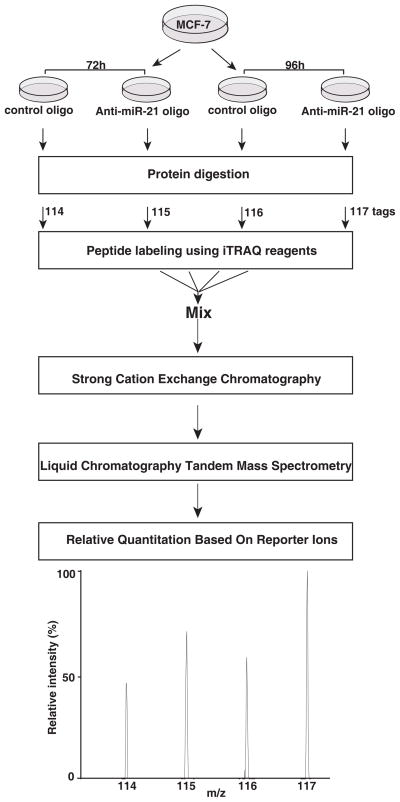
Most miRNAs are thought to regulate their targets through translational inhibition more potently than through mRNA degradation in animals [1]. However, most studies thus far have used strategies based on mRNA changes instead of protein changes to identify miRNA targets. Here, we describe an iTRAQ-based quantitative proteomics strategy to identify putative targets of miR-21 (Figure 1). Because miR-21 is highly expressed in the breast cancer cell line MCF-7, we reasoned that miR-21 inhibition would increase the expression of its cognate targets. We chose to inhibit the expression of miR-21 instead of overexpressing miR-21 because specific inhibition of endogenous miRNAs is less likely to cause off-site target effects than its overexpression. In order to repress miR-21 expression, we transfected MCF-7 cells with an anti-miR-21 oligo. Cells transfected with an oligo without any specificity to mammalian miRNAs were used as a control. To assess the efficiency of miR-21 knockdown, the expression of mature miR-21 was measured by real time RT-PCR. miR-21 expression in MCF-7 cells was dramatically decreased by anti-miR-21 oligo as compared to control oligo after 72 and 96 hours post-transfection (Figure 2). Subsequently, equal amounts of protein lysates from cells transfected with anti-miR-21 and control oligos were used for proteomic analysis as outlined in Figure 1. The proteins were subjected to trypsin digestion and the resulting peptides were labeled with iTRAQ isobaric reagents as per manufacturer’s protocol. Essentially, peptides from the 72 hour time point were labeled with 114 and 115 reporter ions while the peptides from the 96 hour time point were labeled with 116 and 117 reporter ions (Figure 1). Relative changes in protein abundance between anti-miR-21 and control oligo transfected cells were calculated based on the intensity of reporter ions generated during tandem mass spectrometry analysis.
Figure 1. Strategy for iTRAQ labeling.
MCF-7 cells were transfected with anti-miR-21 oligo or control oligo. After 72 or 96 hours post-transfection, total proteins were harvested and protein lysates (100qg from each sample) were digested with trypsin and labeled with iTRAQ reagents. Labeled peptides were combined and fractionated by strong cation exchange chromatography (SCX). Twenty-eight fractions were obtained and analyzed by LC-MS/MS. The fold changes were calculated from the ratio of intensity of iTRAQ reporter ions obtained from samples with anti-miR-21 oligos to those with control oligos.
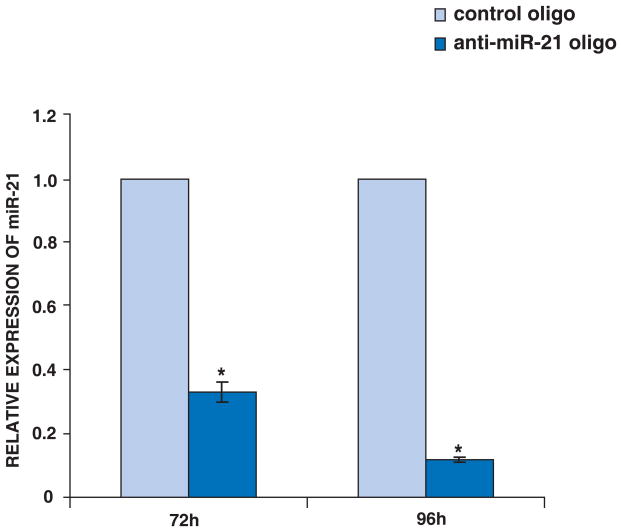
Figure 2. The expression of miR-21 in MCF-7 cells.
MCF-7 cells were transfected with anti-miR-21 oligo or control oligo. After 72 or 96 hours transfection, total RNA were isolated from the transfections. The relative expression of miR-21 in MCF-7 was significantly decreased by anti-miR-21 oligo compared with control oligo as detected by Real Time RT-PCR. Data are shown as the mean ± SD of 3 replicates and are representative of 3 independent experiments. *: p<0.05 using two-tailed t-test (anti-miR-21 oligo vs control oligo).
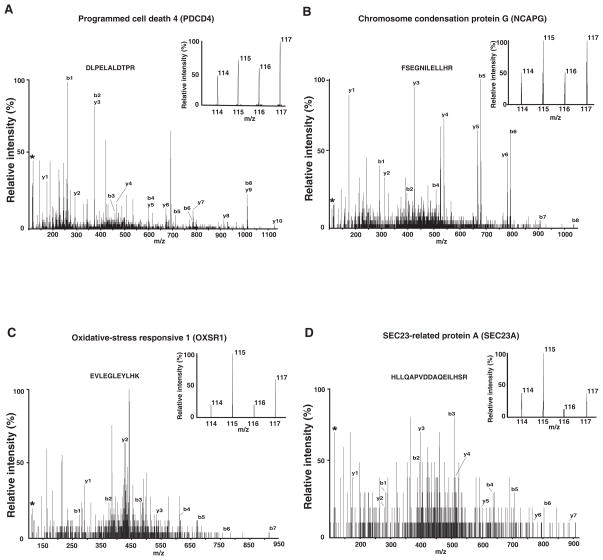
The data from a total of 31,437 MS/MS spectra generated by LC-MS/MS analysis from 28 SCX fractions were searched against the human RefSeq database. We identified a total of 1,151 proteins (Supplementary table 2). Using 1.5-fold as a cut-off to designate up or down-regulated proteins, we found 58 (5.0%) upregulated and 13 (1.1%) downregulated proteins in MCF-7 cells 72 hours following transfection of the anti-miR-21 oligo. The expression of many of these proteins at 96 hours post-transfection time point was found to be similar (Supplementary table 2). Representative MS/MS spectra and corresponding reporter ions of 4 putative targets are shown in Figure 3. Programmed cell death 4 (PDCD4) has been identified as a target of miR-21 by three different groups [21, 24, 27]. They demonstrated that PDCD4 protein was increased around 2-fold following miR-21 inhibition by Western blot. Our results show that PDCD4 protein was increased ~ 1.6 fold by anti-miR-21 oligo, which is consistent with the previous reports. Chromosome condensation protein G (NCAPG), Oxidative-stress responsive 1 (OXSR1), and SEC23-related protein A (SEC23A) are three of the novel targets of miR-21 identified in the present study.
Figure 3. MS/MS spectra of selected proteins identified by proteomics.
Panels A-D show the MS/MS spectra of representative peptides from programmed cell death 4 (PDCD4), chromosome condensation protein G (NCAPG), Oxidative-stress responsive 1 (OXSR1), and SEC23-related protein A (SEC23A), respectively. The inset in each case shows the corresponding relative intensity of reporter ions generated during MS/MS fragmentation and indicates upregulation of protein in samples with anti-miR-21 oligo compared to control oligo. (*: the spectrum of reporter ions)
Sequence analysis of candidate targets identified through proteomics
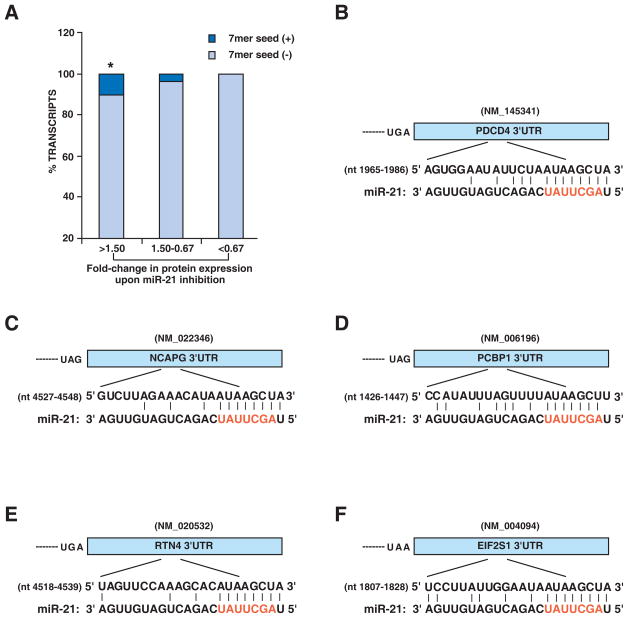
We searched nucleotide sequence databases for the transcripts of all genes identified by proteomics and obtained their 3’UTR, 5’UTR and coding sequences. For some of the transcripts, we were unable to obtain any 3’UTR, 5’UTR or coding sequence information and these were not analyzed further. The 3’UTR, 5’UTR and coding sequences were next searched for miR-21 seed matches, respectively. The distribution of perfect 7 mer seed matches (complementary to 2 - 8 nucleotides at the 5’end of miR-21) is shown in Table 2. We found that 3’UTRs of 5 out of 53 upregulated genes (9.4 %) bore the perfect 7 mer seed matches (Figure 4A). These genes were PDCD4, reticulon 4 isoform A (RTN4), chromosome condensation protein G (NCAPG), poly (rC) binding protein 1 (PCBP1) and eukaryotic translation initiation factor 2 (EIF2S1). The complementarity of these five 3’UTR sequences to the sequence of miR-21 is shown in Figure 4B-F. In contrast, the 3’UTRs from only 4.3 % genes whose protein levels were unchanged contained a sequence corresponding exactly to the 7 mer seed sequence. Our results demonstrate that matches to the 7 mer seed sequence of miR-21 are significantly enriched in upregulated genes compared with unchanged genes identified through the proteomic approach (Figure 4A). The same seed match was further searched in the 5’UTRs and the coding sequences, which did not show the enrichment of the seed match in 5’UTR or coding sequences of upregulated genes (Table 2). None of the downregulated genes had the seed matches in the 3’UTR, 5’UTR or coding sequences. Noticeably, many upregulated genes did not contain miR-21 seed matches, which may due to two possible reasons. First, miR-21 may regulate those genes through indirect mechanisms without directly bind to those targets. Second, sequences outside of seed region might play a role in binding specificity and stability.
Table 2.
The distribution of miR-21 7 mer seed matches in transcripts from proteins identified by proteomics
| Fold-change | 3′UTR | 5′UTR | Coding sequences |
|---|---|---|---|
| >1.5 | 5 (9.4%) | 0 (0%) | 1 (1.9%) |
| 1.5 – 0.67 | 44 (4.3%) | 1 (0.1%) | 47 (4.6%) |
| < 0.67 | 0 (0%) | 0 (0%) | 0 (0%) |
Figure 4. Sequence complementary analysis of miR-21 with 3’-UTR of candidate targets.
A: The transcripts of all genes identified by proteomics were searched for annotated 3’UTRs. The frequency of motifs in the 3’UTRs complementary to perfect 7 mer seed region of miR-21 (2 to 8 nucleotide at 5’ end) was evaluated. * p<0.05 using chi square statistic when upregulated genes (protein fold-change: >1.5) were compared with unchanged genes (protein fold-change: 1.5-0.67). 3’UTRs of 5 out of 53 upregulated genes (9.4%) bore the 7 mer miR-21 seed matches. Panels B-F show the complementarity of miR-21 sequence to the five upregulated genes bearing perfect seed matches. The seed sequence of miR-21 is shown in red. Vertical lines denote identity between miR-21 sequence and the corresponding 3’UTR sequence. Nucleotide accession numbers from RefSeq database are indicated.
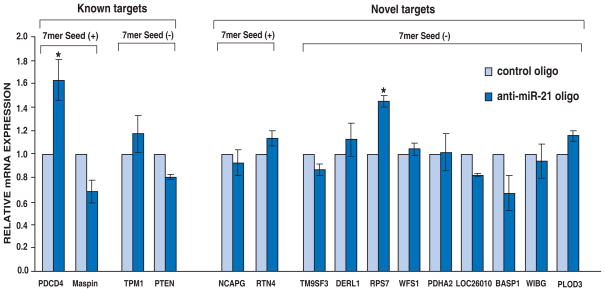
Measurement of mRNA expression of known and candidate miR-21 targets
We used real time RT-PCR to examine any changes in mRNA abundance for the identified miR-21 targets. We chose 12 targets identified by proteomics, including 3 genes with perfect 7 mer matches of the miR-21 seed region (this includes PDCD4 which was recently described to as a target of miR-21). Among the 11 novel candidate targets, only one gene showed significantly increased mRNA expression induced by miR-21 inhibition (Figure 5). The remaining 10 genes were unchanged at the mRNA levels. Interestingly, RTN4 and NCAPG genes contain perfect 7 mer seed matches but no change in the mRNA levels was observed indicating that although they might be direct targets, they could not be identified by approaches designed to identify targets through differentially expressed mRNAs. We also validated mRNA expression of four known targets in order to compare the results to our proteomic data. Two of them, PDCD4 and maspin contain perfect 7 mer matches. The results showed that only one out of the four known targets (PDCD4) had increased mRNA expression induced by miR-21 inhibition (Figure 5). Taken together, our results show that it is likely that these targets were regulated by miR-21 through translational inhibition instead of mRNA degradation.
Figure 5. mRNA expression of miR-21 targets.
The expression of selected miR-21 targets was performed by real time RT-PCR. Data are shown as the mean ± SD of 3 replicates. * p<0.05 using two-tailed t-test (anti-miR-21 oligo vs control oligo).
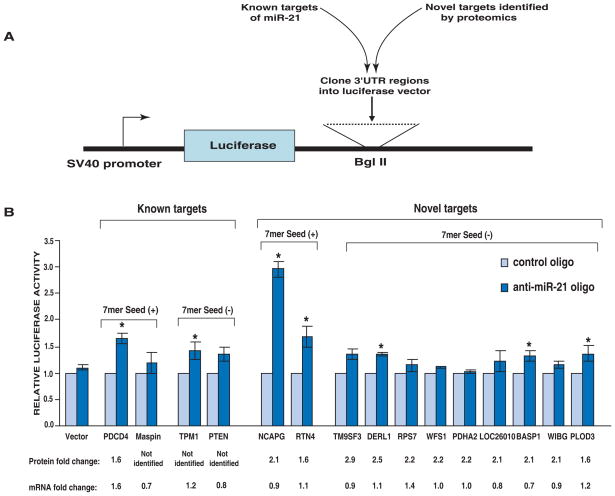
Verification of direct miR-21 targets using luciferase assay
We performed luciferase assays to determine if miR-21 directly regulates putative targets that we have identified. We chose the same genes that were used for mRNA expression for these experiments. The 3’UTR regions of these targets were cloned downstream of the luciferase open reading frame as indicated in Figure 6A. The constructs were co-transfected with anti-miR-21 or control oligos into MCF-7 cells. A significantly increased luciferase activity was observed in both genes (RTN4 and NCAPG) containing perfect 7 mer seed matches upon miR-21 inhibition (Figure 6B). Among the remaining nine potential targets without perfect 7 mer matches, three had less pronounced but significantly increased activity (Figure 6B). They are Der1-like domain family member 1 (DERL1), procollagen-lysine 2-oxoglutarate 5-dioxygenase 3 precursor (PLOD3) and brain abundant membrane attached signal protein 1 (BASP1). Both DERL1 and PLOD3 are involved in cellular protein metabolic process [32, 33]. BASP1 gene encodes a membrane bound protein with several phosphorylation sites and PEST motifs [34]. We also validated 4 known targets of miR-21 by luciferase assays. All of them showed increased luciferase activity upon miR-21 inhibition, although only two of them reached statistical significance (Figure 6B). Differences between our results and those reported by others may be due to different cell types or cell conditions when using the same cell line.
Figure 6. Verification of direct miR-21 targets using luciferase assay.
A: 3’UTR fragments of known and selected novel targets of miR-21 were cloned downstream of the luciferase open reading frame at Bgl II restriction site of pGL3-control vector. B: The above constructs and anti-miR-21 oligo or control oligo were co-transfected along with Renilla luciferase plamid. Shown are relative luciferase activity normalized to corresponding transfections with control oligo. Data are shown as the mean ± SD of 3 replicates and are representative of 3 independent experiments. * p<0.05 using two-tailed t-test (pGL3-3’UTR construct vs pGL3-control). The protein and mRNA fold change for each gene are also indicated.
Discussion
The association between dysregulated miRNAs and cellular signaling pathways involved in cancer has been proposed. However, the molecular mechanisms by which miRNA can modulate tumor growth or metastases remain largely unknown. This may due to our limited knowledge of miRNA target recognition. mRNA changes based strategy including gene expression array have been used to identify miRNA targets. Because a miRNA may regulate the expression of its targets only through translational inhibition, we demonstrate the utility of proteomic approaches to discover targets of miRNAs in an unbiased manner. Our mRNA based validation experiments clearly show that miR-21 regulates many of its targets without affecting mRNA abundance. We employed miR21 anti-sense oligonucletides to inhibit endogenous expression of miR-21 because exogenous miRNAs could lead to the identification of false positives.
Pictar, Targetscan and miRanda predictions were used to predict potential targets of miR-21 and the prediction results were compared with our iTRAQ results. Only few putative targets identified by proteomics were predicted by these three prediction methods (Data not shown). This is consistent with our sequence analysis results for 3’UTR of candidate targets and miR-21 seed region (Figure 4). It is possible that some of candidate targets identified by proteomics may be indirect targets of miR-21, which cannot be predicted by the prediction programs. Also the computational prediction systems based on miRNA seed regions have relative high false-positives and negatives. Thus, additional experimental data for different miRNAs are needed for increasing the accuracy of prediction programs.
Among the 11 selected novel candidate targets of miR-21 for luciferase assays, two targets (NCAPG and RTN4) contain perfect 7 mer miR-21 seed matches. Both of these showed significantly increased luciferase activities upon miR-21 inhibition. NCAPG is involved in precise separation of two replicated sister chromatids during mitosis. It is also essential for chromosome condensation in metaphase of single, unreplicated sister chromatids [35]. In the present study, NCAPG protein was increased 2.1-fold following miR-21 inhibition as detected by proteomics. Luciferase assay showed that anti-miR-21 oligo dramatically increased luciferase activity of constructs containing the 3’UTR of NCAPG (Figure 6B). These results uncover an unexpected link between miR-21 and chromatin condensation, providing a mechanism by which the functional activation of miR-21 in tumor cells may contribute to genome instability. Reticulons (RTNs) are a large family of transmembrane proteins present throughout the eukaryotic domain in virtually every cell type. RTN4, also termed Nogo, comes in three isoforms, Nogo-A, -B, and –C [36]. Nogo-A has turned out to be a neuronal protein involved in diverse processes that go from axonal fasciculation to apoptosis [37]. RTN4 (isoform A) was identified as a putative target of miR-21 by proteomics, and further confirmed as a direct target by luciferase assays in our study. It is consistent with the function of miR-21 as an oncogenic miRNA involved in apoptosis.
Knockdown of miR-21 in cultured glioblastoma cells leads to increased apoptotic cell death [26]. Anti-miR-21 oligo mediated cell growth inhibition has been associated with increased apoptosis and decreased cell proliferation in breast cancer [22]. Among the putative targets identified by proteomics, three are apoptosis-related genes including PDCD4, RTN4, and tetratricopeptide repeat domain 11 (FIS1). Both PDCD4 and RTN4 contain perfect 7 mer seed matches of miR-21 and were confirmed as direct targets of miR-21 by luciferase assay. Our results indicate that miR-21 may regulate apoptosis by inhibition of apoptosis-related genes.
Although miRNAs are largely known to downregulate gene expression, Vasudevan et al have recently shown that miRNAs can also upregulate translation under certain circumstances [38]. In our study, we also observed 13 proteins to be similarly regulated by miR-21. However, further experiments will be required to determine if any of these are indeed direct targets of miR-21.
In conclusion, we have identified 58 genes as candidate targets of miR-21 using a global quantitative proteomic approach. Six out of 12 tested target genes were likely to be direct targets of miR-21 as shown by luciferase assays - 3 of these contain a perfect 7 mer miR-21 seed match. Overall, our mRNA based validation experiments clearly show that miR-21 regulates many of its targets at translational level without affecting mRNA abundance.
Supplementary Material
The sequences of primers used in the present study
The entire list of proteins identified in this study
Table 1.
Candidate targets of miR-21 identified by proteomics
| Accession | Gene Symbol | Protein Name | Ratio |
|---|---|---|---|
| gi|33859833 | TM9SF3 | Transmembrane 9 superfamily member 3 | 2.92 |
| gi|62912457 | ALDH18A1 | Pyrroline-5-carboxylate synthetase isoform 2 | 2.80 |
| gi|13236516 | DERL1 | Der1-like domain family, member 1 | 2.49 |
| gi|89035017 | LOC646195 | PREDICTED: similar to 40S ribosomal protein S28 isoform 2 | 2.33 |
| gi|4885543 | PDHA2 | Pyruvate dehydrogenase (lipoamide) alpha 2 | 2.24 |
| gi|4506741 | RPS7 | Ribosomal protein S7 | 2.21 |
| gi|4826878 | OXSR1 | Oxidative-stress responsive 1 | 2.16 |
| gi|5174749 | WFS1 | Wolframin | 2.15 |
| gi|21359945 | NCAPG | Chromosome condensation protein G | 2.14 |
| gi|7661598 | LOC26010 | Hypothetical protein LOC26010 | 2.13 |
| gi|30795231 | BASP1 | Brain abundant, membrane attached signal protein 1 | 2.12 |
| gi|14150139 | WIBG | Within bgcn homolog | 2.08 |
| gi|6912586 | PGLS | 6-phosphogluconolactonase | 1.94 |
| gi|88998889 | LOC643936 | PREDICTED: hypothetical protein | 1.93 |
| gi|31543140 | USHBP1 | Usher syndrome 1C binding protein 1 | 1.91 |
| gi|5174529 | MAT2A | Methionine adenosyltransferase II, alpha | 1.87 |
| gi|25777602 | PSMD2 | Proteasome 26S non-ATPase subunit 2 | 1.83 |
| gi|5031573 | ACTR3 | ARP3 actin-related protein 3 homolog | 1.81 |
| gi|116812600 | STARD10 | START domain containing 10 | 1.81 |
| gi|63252888 | P4HA1 | Prolyl 4-hydroxylase, alpha I subunit isoform 2 precursor | 1.78 |
| gi|24308271 | DC2 | DC2 protein | 1.78 |
| gi|40548408 | KIAA1967 | P30 DBC protein | 1.74 |
| gi|21735598 | PDCD4 | Programmed cell death 4 isoform 2 | 1.73 |
| gi|38202214 | SEC23A | SEC23-related protein A | 1.71 |
| gi|5454158 | VARS | Valyl-tRNA synthetase | 1.71 |
| gi|4885379 | HIST1H1E | Histone cluster 1, H1e | 1.71 |
| gi|5902102 | SNRPD1 | Small nuclear ribonucleoprotein D1 polypeptide 16kDa | 1.68 |
| gi|89026818 | RPL21 | PREDICTED: similar to 60S ribosomal protein L21 | 1.63 |
| gi|24431935 | RTN4 | Reticulon 4 isoform A | 1.62 |
| gi|7705632 | FIS1 | Tetratricopeptide repeat domain 11 | 1.62 |
| gi|57242766 | FLJ36031 | Hypothetical protein LOC168455 | 1.60 |
| gi|56181387 | STUB1 | STIP1 homology and U-box containing protein 1 | 1.60 |
| gi|4759046 | POLR1C | RNA polymerase I subunit isoform 2 | 1.59 |
| gi|50658065 | SMC4 | SMC4 structural maintenance of chromosomes 4-like 1 | 1.59 |
| gi|4503377 | DPYSL2 | Dihydropyrimidinase-like 2 | 1.59 |
| gi|10835025 | NDUFV2 | NADH dehydrogenase (ubiquinone) 24kDa flavoprotein 2, | 1.58 |
| gi|73427803 | LLGL2 | Lethal giant larvae homolog 2 isoform c | 1.58 |
| gi|27262649 | ATXN2L | Ataxin 2 related protein isoform C | 1.57 |
| gi|20070228 | NUCB1 | Nucleobindin 1 | 1.57 |
| gi|89886460 | INF2 | Hypothetical protein LOC64423 isoform 2 | 1.57 |
| gi|5174449 | H1FX | H1 histone family, member X | 1.56 |
| gi|7661734 | DCPS | mRNA decapping enzyme | 1.56 |
| gi|4505891 | PLOD3 | Procollagen-lysine, 2-oxoglutarate 5-dioxygenase 3 precursor | 1.55 |
| gi|88955151 | LOC647153 | PREDICTED: similar to voltage-dependent anion channel 2 | 1.55 |
| gi|34452703 | ASNS | Asparagine synthetase | 1.55 |
| gi|24586675 | SSH3 | Slingshot homolog 3 | 1.55 |
| gi|40254924 | LRRC59 | Leucine rich repeat containing 59 | 1.54 |
| gi|89026256 | LOC653888 | PREDICTED: similar to Actin-related protein 2/3 complex subunit 1B (ARP2/3 complex 41 kDa subunit) (p41-ARC) | 1.54 |
| gi|87196351 | DDX3X | DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 3 | 1.54 |
| gi|24308295 | GRPEL1 | GrpE-like 1, mitochondrial | 1.54 |
| gi|5453854 | PCBP1 | Poly(rC) binding protein 1 | 1.54 |
| gi|4758256 | EIF2S1 | Eukaryotic translation initiation factor 2, subunit 1 alpha, 35kDa | 1.53 |
| gi|4885381 | HIST1H1B | Histone cluster 1, H1b | 1.53 |
| gi|62241015 | AKT1 | V-akt murine thymoma viral oncogene homolog 1 | 1.53 |
| gi|27436901 | MRPL12 | Mitochondrial ribosomal protein L12 | 1.52 |
| gi|13376617 | PTGES2 | Prostaglandin E synthase 2 isoform 1 | 1.51 |
| gi|4504251 | HIST2H2AA3 | H2A histone family, member O | 1.51 |
| gi|4502303 | ATP5O | Mitochondrial ATP synthase, O subunit precursor | 1.50 |
Acknowledgments
This study was supported by Department of Defense Era of Hope Scholar award (W81XWH-06-1-0428) and NIH roadmap grant (U54RR020839) for “Technology Centers for Networks and Pathways”. We thank Marjan Gucek for his expert assistance. Special thanks go to Hun-Way Hwang for helping in luciferase assays. We are grateful to H. C. Harsha and Jun Zhong for fruitful discussions in manuscript preparation. We also thank Kumaran Kandasamy for helping in sequence analysis.
Abbreviations
- miRNA
microRNA
- 3’UTR
3’untranslated region
- SCX
strong cation exchange
Footnotes
All authors have declared no financial/commercial conflict of interest.
References
- 1.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Bagga S, Bracht J, Hunter S, Massirer K, et al. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell. 2005;122:553–563. doi: 10.1016/j.cell.2005.07.031. [DOI] [PubMed] [Google Scholar]
- 3.Lee YS, Kim HK, Chung S, Kim KS, Dutta A. Depletion of human micro-RNA miR-125b reveals that it is critical for the proliferation of differentiated cells but not for the down-regulation of putative targets during differentiation. J Biol Chem. 2005;280:16635–16641. doi: 10.1074/jbc.M412247200. [DOI] [PubMed] [Google Scholar]
- 4.Ivanovska I, Ball AS, Diaz RL, Magnus JF, et al. MicroRNAs in the miR-106b family regulate p21/CDKN1A and promote cell cycle progression. Mol Cell Biol. 2008 doi: 10.1128/MCB.01977-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kobayashi T, Lu J, Cobb BS, Rodda SJ, et al. Dicer-dependent pathways regulate chondrocyte proliferation and differentiation. Proc Natl Acad Sci U S A. 2008 doi: 10.1073/pnas.0707900105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang Q, Gumireddy K, Schrier M, le Sage C, et al. The microRNAs miR-373 and miR-520c promote tumour invasion and metastasis. Nat Cell Biol. 2008;10:202–210. doi: 10.1038/ncb1681. [DOI] [PubMed] [Google Scholar]
- 7.Hannon GJ. RNA interference. Nature. 2002;418:244–251. doi: 10.1038/418244a. [DOI] [PubMed] [Google Scholar]
- 8.Palatnik JF, Allen E, Wu X, Schommer C, et al. Control of leaf morphogenesis by microRNAs. Nature. 2003;425:257–263. doi: 10.1038/nature01958. [DOI] [PubMed] [Google Scholar]
- 9.Tang G, Reinhart BJ, Bartel DP, Zamore PD. A biochemical framework for RNA silencing in plants. Genes Dev. 2003;17:49–63. doi: 10.1101/gad.1048103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martin G, Schouest K, Kovvuru P, Spillane C. Prediction and validation of microRNA targets in animal genomes. J Biosci. 2007;32:1049–1052. doi: 10.1007/s12038-007-0106-0. [DOI] [PubMed] [Google Scholar]
- 11.Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 12.Olsen PH, Ambros V. The lin-4 regulatory RNA controls developmental timing in Caenorhabditis elegans by blocking LIN-14 protein synthesis after the initiation of translation. Dev Biol. 1999;216:671–680. doi: 10.1006/dbio.1999.9523. [DOI] [PubMed] [Google Scholar]
- 13.Mathonnet G, Fabian MR, Svitkin YV, Parsyan A, et al. MicroRNA inhibition of translation initiation in vitro by targeting the cap-binding complex eIF4F. Science. 2007;317:1764–1767. doi: 10.1126/science.1146067. [DOI] [PubMed] [Google Scholar]
- 14.Fontana L, Pelosi E, Greco P, Racanicchi S, et al. MicroRNAs 17-5p-20a-106a control monocytopoiesis through AML1 targeting and M-CSF receptor upregulation. Nat Cell Biol. 2007;9:775–787. doi: 10.1038/ncb1613. [DOI] [PubMed] [Google Scholar]
- 15.Sethupathy P, Corda B, Hatzigeorgiou AG. TarBase: A comprehensive database of experimentally supported animal microRNA targets. Rna. 2006;12:192–197. doi: 10.1261/rna.2239606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 17.Krek A, Grun D, Poy MN, Wolf R, et al. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 18.John B, Enright AJ, Aravin A, Tuschl T, et al. Human MicroRNA targets. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lim LP, Lau NC, Garrett-Engele P, Grimson A, et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 20.Wang X, Wang X. Systematic identification of microRNA functions by combining target prediction and expression profiling. Nucleic Acids Res. 2006;34:1646–1652. doi: 10.1093/nar/gkl068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frankel LB, Christoffersen NR, Jacobsen A, Lindow M, et al. Programmed cell death 4 (PDCD4) is an important functional target of the microRNA miR-21 in breast cancer cells. J Biol Chem. 2008;283:1026–1033. doi: 10.1074/jbc.M707224200. [DOI] [PubMed] [Google Scholar]
- 22.Si ML, Zhu S, Wu H, Lu Z, et al. miR-21-mediated tumor growth. Oncogene. 2007;26:2799–2803. doi: 10.1038/sj.onc.1210083. [DOI] [PubMed] [Google Scholar]
- 23.Corsten MF, Miranda R, Kasmieh R, Krichevsky AM, et al. MicroRNA-21 knockdown disrupts glioma growth in vivo and displays synergistic cytotoxicity with neural precursor cell delivered S-TRAIL in human gliomas. Cancer Res. 2007;67:8994–9000. doi: 10.1158/0008-5472.CAN-07-1045. [DOI] [PubMed] [Google Scholar]
- 24.Asangani IA, Rasheed SA, Nikolova DA, Leupold JH, et al. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene. 2007 doi: 10.1038/sj.onc.1210856. [DOI] [PubMed] [Google Scholar]
- 25.Meng F, Henson R, Wehbe-Janek H, Ghoshal K, et al. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. 2007;133:647–658. doi: 10.1053/j.gastro.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chan JA, Krichevsky AM, Kosik KS. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005;65:6029–6033. doi: 10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- 27.Zhu S, Wu H, Wu F, Nie D, et al. MicroRNA-21 targets tumor suppressor genes in invasion and metastasis. Cell Res. 2008 doi: 10.1038/cr.2008.24. [DOI] [PubMed] [Google Scholar]
- 28.Zhu S, Si ML, Wu H, Mo YY. MicroRNA-21 targets the tumor suppressor gene tropomyosin 1 (TPM1) J Biol Chem. 2007;282:14328–14336. doi: 10.1074/jbc.M611393200. [DOI] [PubMed] [Google Scholar]
- 29.Amanchy R, Kalume DE, Pandey A. Stable isotope labeling with amino acids in cell culture (SILAC) for studying dynamics of protein abundance and posttranslational modifications. Sci STKE. 2005;2005:pl2. doi: 10.1126/stke.2672005pl2. [DOI] [PubMed] [Google Scholar]
- 30.Chang TC, Wentzel EA, Kent OA, Ramachandran K, et al. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol Cell. 2007;26:745–752. doi: 10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grimson A, Farh KK, Johnston WK, Garrett-Engele P, et al. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Katiyar S, Joshi S, Lennarz WJ. The retrotranslocation protein Derlin-1 binds peptide:N-glycanase to the endoplasmic reticulum. Mol Biol Cell. 2005;16:4584–4594. doi: 10.1091/mbc.E05-04-0345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rautavuoma K, Takaluoma K, Passoja K, Pirskanen A, et al. Characterization of three fragments that constitute the monomers of the human lysyl hydroxylase isoenzymes 1-3. The 30-kDa N-terminal fragment is not required for lysyl hydroxylase activity. J Biol Chem. 2002;277:23084–23091. doi: 10.1074/jbc.M112077200. [DOI] [PubMed] [Google Scholar]
- 34.Fitzgibbon J, Neat MJ, Foot N, Hill AS, et al. Assignment of brain acid-soluble protein 1 (BASP1) to human chromosome 5p15.1-->p14, differential expression in human cancer cell lines as a result of alterations in gene dosage. Cytogenet Cell Genet. 2000;89:147–149. doi: 10.1159/000015597. [DOI] [PubMed] [Google Scholar]
- 35.Dej KJ, Ahn C, Orr-Weaver TL. Mutations in the Drosophila condensin subunit dCAP-G: defining the role of condensin for chromosome condensation in mitosis and gene expression in interphase. Genetics. 2004;168:895–906. doi: 10.1534/genetics.104.030908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schweigreiter R, Stasyk T, Contarini I, Frauscher S, et al. Phosphorylation-regulated cleavage of the reticulon protein Nogo-B by caspase-7 at a noncanonical recognition site. Proteomics. 2007;7:4457–4467. doi: 10.1002/pmic.200700499. [DOI] [PubMed] [Google Scholar]
- 37.Mingorance A, Soriano-Garcia E, del Rio JA. Nogo-A functions during the development of the central nervous system and in the adult. Rev Neurol. 2004;39:440–446. [PubMed] [Google Scholar]
- 38.Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318:1931–1934. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The sequences of primers used in the present study
The entire list of proteins identified in this study