Abstract
Summary: Sequencing reads generated by RNA-sequencing (RNA-seq) must first be mapped back to the genome through alignment before they can be further analyzed. Current fast and memory-saving short-read mappers could give us a quick view of the transcriptome. However, they are neither designed for reads that span across splice junctions nor for repetitive reads, which can be mapped to multiple locations in the genome (multi-reads). Here, we describe a new software package: ABMapper, which is specifically designed for exploring all putative locations of reads that are mapped to splice junctions or repetitive in nature.
Availability and Implementation: The software is freely available at: http://abmapper.sourceforge.net/. The software is written in C++ and PERL. It runs on all major platforms and operating systems including Windows, Mac OS X and LINUX.
Contact: tf.chan@cuhk.edu.hk
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
The advent of RNA-seq technology has dramatically transformed transcriptomics within just a few years (Wang et al., 2009). The next-generation sequencing platform can generate far more data per experiment than traditional sequencing technology. Millions of short, sometimes paired-end reads have brought about new challenges: to map them back onto the genome for determining gene expression level and for detecting splicing events.
Two groups, Li et al. and Langmead et al. have developed short-read alignment tools based on the Burrow–Wheeler transformation algorithm (Burrows and Wheeler, 1994), which runs fast and with low memory requirement (Langmead et al., 2009; Li and Durbin, 2009). A major drawback of these fast aligners is that they require reads to map to contiguous genomic sequence, and thus are not appropriate for reads spanning splice junctions. SplitSeek (Ameur et al., 2010), TopHat (Trapnell et al., 2009) and SpliceMap (Au et al., 2010) are three widely known software packages written to make up for the lack of ability of these short-read aligners for splice site identification. However, these tools have to rely on output from existing aligners: TopHat requires bowtie; SpliceMap requires ELAND or bowtie; and SplitSeek depends on the SOLiD Whole Transcriptome analysis pipeline and is therefore restricted only to SOLiD dataset.
In this work, we present ABMapper, which has been developed as a stand-alone tool for ab initio mapping of sequencing reads that span across splicing junctions or reads that have multiple putative locations within the genome. It adopts a fast suffix-array algorithm and a dual-seed strategy to find all putative locations of reads.
2 METHODS
2.1 Overview
ABMapper is a portable, easy-to-use package for spliced alignment, splice site detection and read mapping. The core module was written in C++ and wrapped in PERL scripts, and the executable file can be called by: Perl runABMapper.pl—ref chromosome list—input reads (.fa or .fq)
Full documentation is available from the associated web site.
2.2 Algorithm
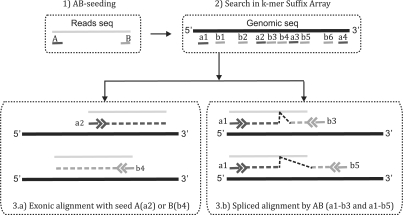
Reference genomic sequences are first indexed into kmers with a user-defined k, i.e. the respective suffix array consisting of all the kmers of the genome is built. After indexing, for every read being mapped, two seeds (namely A and B, by which the name of this tool was derived) with each at length k from each end of the read are retrieved and extended toward each other through alignment by steps as illustrated in Figure 1.
Fig. 1.
Workflow of ABMapper. (1) ‘A’ and ‘B’ refer to the two seeds from each end of a read. (2) Hits that are found within the genome (black line) matching the two seeds are represented by bars with the corresponding labels (A: a1, a2, a3; similarly for B). Finally, seeds are extended during (3a) exonic alignment; and (3b) spliced alignment.
Two possible scenarios could arise during a seed extension: without or with splice junctions. For the first case, the fully matched alignment without splicing (3.a in Fig. 1), which is termed ‘exonic alignment’, only extends from one seed. For the second case, which is termed ‘spliced alignment’, one seed from each end of a read (3.b in Fig. 1) will be extended toward each other until a stop point is reached. A good spliced alignment is defined as two extended fragments that could form a complete read. However, a seed could sometimes be extended beyond its real stop point, especially when errors are allowed. In such cases, an overlapping tolerance would be used, which allows the sum of the lengths of the two extensions to be larger than the length of the read. The two overlapped fragments would then be subjected to canonical splicing motifs search, i.e. ‘GT-AG’, ‘GC-AG’ and ‘AT-AC’, to determine a stop point for spliced alignment. The above steps are performed for one chromosome at a time. Finally, outputs including alignment details, junction sites, putative locations of repetitive reads, etc., which depend on user's parameter, will be reported.
3 RESULTS
We chose 213 893 reads, each with the size of 75 bp that cover all human known junction sites, and the same number of reads from the exons of human genome to compare the results of ABMapper (version 2.0.3; seed length = 11; maximum output m = 500 and output format t = 1) with SpliceMap (version 3.1.1) and TopHat (version 1.0.13). Performance of the three software packages are summarized in Table 1. Additional performance data of ABMapper, as well as resource usage of the three programs can be found in the Supplementary Material.
Table 1.
Comparison between TopHat (TH), SpliceMap (SM) and ABMapper (ABM)
| Total read = 427 786 | sp.fa |
sp_e1.fa |
sp_e2.fa |
||||||
|---|---|---|---|---|---|---|---|---|---|
| TH | SM | ABM | TH | SM | ABM | TH | SM | ABM | |
| Found | 366 918 | 354 502 | 424 532 | 367 249 | 320 949 | 424 385 | 372 274 | 277 369 | 409 676 |
| Perfect hit | 326 510 | 334 241 | 420 338 | 326 422 | 279 146 | 421 543 | 326 033 | 222 907 | 407 636 |
| Accuracy (%) | 88.99 | 94.28 | 99.01 | 88.88 | 86.98 | 99.33 | 87.58 | 80.36 | 99.50 |
| Recall (%) | 76.33 | 78.13 | 98.26 | 76.30 | 65.25 | 98.54 | 76.21 | 52.11 | 95.29 |
sp.fa is the benchmark dataset, with sp_e1.fa and sp_e2.fa having 1 and 2 random errors, respectively. We defined accuracy as %(Perfect hit/Found) and recall as %(Perfect hit/Total).
Support for multiple input formats: ABMapper supports BWA SAM file, reads file in raw, FASTA and FASTQ formats. Reads that are mapped to multiple locations (multi-reads) or unmapped reads could be extracted from BWA's SAM/BAM file and mapped by ABMapper.
User-defined seed length: the seed length is a key parameter that would affect the searching speed, sensitivity and accuracy. The shorter the seed length is, the more sensitive and accurate the mapping becomes, but at the expense of running time. Users could define it according to sample size (i.e. number of reads to be processed) and computation power available. The default seed length is 10. (See Supplementary Figure S1 for resource usage based on seed lengths.)
Dynamic error tolerance and overlapping extension exploration: ABMapper can give more chance to reads that would be extended exceeding a cutoff, to find out whether it could be mapped in full length by allowing one additional error. This is most useful in the case of paired-end reads, where it is a commonly known phenomenon that the second read (or p2) always has a lower quality (higher error rate) than the first read (p1). Another important feature of ABMapper is the processing of overlapping sequence of two fragments of a spliced read. Due to error tolerance, the two parts of a read could be extended from both ends beyond their corresponding mapped positions toward each other to an overlapping region, which can be further analyzed [see (4)]. This is more favorable than tools such as SpliceMap, which aligns and extends a read only from one end.
Splice site detection: canonical motifs, ‘GT-AG’, ‘GC-AG’ ‘AT-AC’ are used to determine a probable junction, especially in the case where overlapping fragments are found.
Repetitive reads filtering: repetitive elements are discovered and filtered in the RNA content by ABMapper according to the number of repeats specified by a user. This feature is also meaningful in research (Xu et al., 2010). Filtered repetitive reads in FASTA format would be used for RepeatMasker analysis.
Unbiased searching and support for multiple output formats: by default, ABMapper will provide as much useful information as possible, which includes alignment details, splicing junction sites and repetitive reads. In addition, ABMapper supports multiple formats including SAM and BED.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the anonymous reviewers for their comments.
Funding: This study is supported by a Focused Investment Scheme of the Chinese University of Hong Kong granted to S.K.-W.T.; and a General Research Fund (GRF461708) from the Research Grant Committee, Hong Kong SAR Government granted to T.-F.C.
Conflict of Interest: none declared.
REFERENCES
- Ameur A, et al. Global and unbiased detection of splice junctions from RNA-seq data. Genome Biol. 2010;11:R34. doi: 10.1186/gb-2010-11-3-r34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Au KF, et al. Detection of splice junctions from paired-end RNA-seq data by SpliceMap. Nucleic Acids Res. 2010;38:4570–4578. doi: 10.1093/nar/gkq211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrows M, Wheeler DJ. Technical Report 124. Palo Alto, CA: Digital Equipment Corporation; 1994. A block-sorting lossless data compression algorithm. [Google Scholar]
- Langmead B, et al. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, et al. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, et al. RNA-Seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu AG, et al. Intergenic and repeat transcription in human, chimpanzee and macaque brains measured by RNA-Seq. PLoS Comput. Biol. 2010;6:e1000843. doi: 10.1371/journal.pcbi.1000843. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.