Abstract
Summary: Cytoscape is a popular bioinformatics package for biological network visualization and data integration. Version 2.8 introduces two powerful new features—Custom Node Graphics and Attribute Equations—which can be used jointly to greatly enhance Cytoscape's data integration and visualization capabilities. Custom Node Graphics allow an image to be projected onto a node, including images generated dynamically or at remote locations. Attribute Equations provide Cytoscape with spreadsheet-like functionality in which the value of an attribute is computed dynamically as a function of other attributes and network properties.
Availability and implementation: Cytoscape is a desktop Java application released under the Library Gnu Public License (LGPL). Binary install bundles and source code for Cytoscape 2.8 are available for download from http://cytoscape.org.
Contact: msmoot@ucsd.edu
1 INTRODUCTION
Networks are pervasive in biology. Vast datasets are being gathered to delineate networks of various types and levels, including networks of genetic and protein–protein interaction networks of transcriptional and post-transcriptional regulation, social networks and multiscale networks that span several of these domains (Fowler et al., 2009; Tan et al., 2007). These network data extend and compliment a great deal of other information available in the biomedical sciences. Although various datasets can appear quite different in quality and quantity, they all are reflections of the same underlying biological system and its responses. Thus data integration, along with integrated visualization, is a key.
Cytoscape, now in its eighth year of development, has become a standard tool for integrated analysis and visualization of biological networks (Cline et al., 2007; Shannon et al., 2003). Its central organizing principle is a network graph, with biological entities (e.g. genes, proteins, cells, patients) represented as nodes and biological interactions represented as edges between nodes. Data are integrated with the network using attributes, which map nodes or edges to specific data values such as gene expression levels or protein functions. Attribute values can be used to control visual aspects of nodes and edges (e.g. shape, color, size) as well as to perform complex network searches, filtering operations and other analysis.
Version 2.8 of Cytoscape has introduced two significant new features that improve its ability to integrate and visualize complex datasets. The first feature allows non-programmers to map graphical images onto nodes, which greatly increases the power and flexibility with which integrated data can be visualized. The second feature is the introduction of spreadsheet-like equations into Cytoscape's Attribute Browser to enable advanced transformation and combination of datasets directly within Cytoscape. Separately, each of these features provides useful new capabilities to Cytoscape. Taken together, however, these features provide a mechanism for expressing relationships between sets of data while simultaneously visualizing the integrated results.
2 INTEGRATED VISUALIZATION
2.1 Custom node graphics
A key function of Cytoscape is to allow diverse types of attribute data to be visualized on the nodes and edges of a biological network. Scalar data can be linked to simple visualization properties such as node color, shape or size as node and edge attributes. To represent multivariate data associated with a node, Cytoscape can control the visualization of each node using a custom graphical image (since version 2.3) through a programming API. Version 2.8 introduces the ability for non-programmers to specify images through the Cytoscape GUI and to map these images to nodes using Cytoscape's standard VizMapper interface.
Using this new feature, images are loaded using a standard file browser or by dragging and dropping them into a pool of available images maintained by Cytoscape. For cases involving large numbers of custom images, Cytoscape also allows images to be loaded as node attributes by providing a Uniform Resource Locator (URL) address for each image. Given a URL, Cytoscape will try to read that URL and generate an image for display.
Images can be mapped to nodes using Cytoscape's VizMapper framework, which connects the visualization properties of the network to attribute data. Placement and sizing of images can also be directly controlled within Cytoscape.
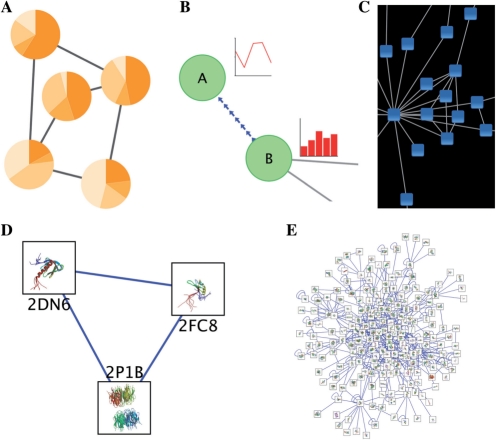
In addition to rich, multivariate visualizations (Fig. 1A and B), images enable shading, highlights and other aesthetic effects to be applied to nodes (Fig. 1C) to further enhance network visualization.
Fig. 1.
Rich network visualizations enabled by the new Cytoscape features. Simple networks are shown with custom node images based on (A) pie chart displays or (B) line plots and bar charts generated using Google's Chart API. (C) Nodes have a transparent custom graphic to give the appearance of shading. (D and E) Protein–protein interaction networks in which each node contains a 3D image of the protein structure of the protein represented by the node.
2.2 Equations
The Cytoscape Attribute Browser provides a tabular view of data that have been loaded into Cytoscape and attached to nodes, edges or networks. To improve the ability of users to manipulate attribute data within Cytoscape, we have added Attribute Equations to version 2.8. Attribute Equations extend the existing spreadsheet-like behavior of the Cytoscape Attribute Browser (rows and columns of editable cells) by allowing users to write equations to populate attribute values, much like the functions and formulas offered by spreadsheet software such as Microsoft Excel or Open Office.
Attribute Equations simplify data analysis, by permitting direct manipulation of data, and allow for very complex analysis given the wide variety of functions provided. Example functions include conditionals (IF), list manipulation (FIRST) and mathematical transformations (SIN, LOG). We also provide support for basic mathematical operators such as plus (+), minus (−), multiply (*), divide (/) and exponent (ˆ) along with the string concatenation operator (&).
For navigation of the built-in functions (66 total), the Cytoscape Attribute Browser provides a Formula Builder, which displays a list of available functions with usage descriptions and guides the user in the construction of valid formulas. Attribute formulas may reference other attributes, constant values or other functions. A public programming API is available that allows programmers to write custom functions and to integrate them alongside the native functions provided by Cytoscape.
2.3 Two illustrative examples: integration of custom graphics with equations
As a first example illustrating the joint use of Custom Node Graphics and Equations, we integrate networks of protein interactions with corresponding visualizations of the known protein structures. This can be accomplished in Cytoscape v2.8 by constructing a URL that points to an image of the 3D structure of a protein and then mapping that image to a node in the network. We begin by creating a data attribute called ‘pdbID’ and, for each node, we populate the value of this attribute with a valid protein name in the Protein Data Bank (http://www.pdb.org). We then create a new string attribute, which we name ‘imageURL’. We then write the following equation for the imageURL attribute:
![]()
This equation is copied to all nodes via an option in the right-click menu of the Attribute Browser. For each node, Cytoscape interprets the $pdbID variable using the corresponding pdbID attribute value and concatenates the resulting strings into a valid URL. We then use the Cytoscape VizMapper to map the imageURL attribute to a Custom Graphics visual property. As described in Section 2.1, this visual property interprets the URL and loads the image it points to. Following this procedure produces rich network visualizations such as those seen in Figure 1D and E.
A second example that demonstrates the use of both new features is the GLINECHART function available from the GoogleChartFunctions plugin (available in the Plugin Manager). This function uses the Google Visualization API (http://code.google.com/apis/visualization) to dynamically generate a URL to an image. For instance, if a user has time course data (values {1.1, 2.3, 1.7, 0.6}) for an experiment stored in an attribute named ‘timeCourse’, the following function call will produce a URL of a line chart image of the data with a Y-axis range of 0–3:
![]()
This function evaluates to a URL, which in turn is interpreted by the VizMapper as a node image, resulting in integrated network visualizations such as those shown in Figure 1A and B.
3 CONCLUSION
Custom node graphics provide a new tool for non-programmers that allows rich new network visualizations to be created that integrate large and complex datasets. Equations provide a powerful mechanism for data transformation within Cytoscape. Together, these features can be used to embed rich visualizations of data within the nodes of large networks.
ACKNOWLEDGEMENTS
Our team would like to acknowledge the the Ideker Laboratory and the core Cytoscape development team.
Funding: National Institutes of Health (grant numbers: GM070743, P01HG005062).
Conflict of Interest: none declared.
REFERENCES
- Cline MS, et al. Integration of biological networks and gene expression data using Cytoscape. Nat. Protoc. 2007;2:2366–2382. doi: 10.1038/nprot.2007.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler JH, et al. Model of genetic variation in human social networks. Proc. Natl Acad. Sci. 2009;106:1720–1724. doi: 10.1073/pnas.0806746106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan K, et al. Transcriptional regulation of protein complexes within and across species. Proc. Natl Acad. Sci. USA. 2007;104:1283. doi: 10.1073/pnas.0606914104. [DOI] [PMC free article] [PubMed] [Google Scholar]