Summary
The hallmark of chronic viral infections is a progressive exhaustion of antigen specific CD8+ T cells that leads to persisting viral replication. It is generally believed that exhaustion is a consequence of the accumulation of multiple inhibitory receptors on CD8+ T cells that makes them dysfunctional. Here we show that during human chronic HIV-1 infection a CD8+ T cell positive costimulatory pathway mediated by DNAM-1 is also disrupted. Thus, DNAM-1 downregulation on CD8+ T cells aggravates the impairment of CTL effector function in chronic HIV-1 infection.
Keywords: HIV-1, exhaustion, co-stimulation
Introduction
During chronic viral infections CD8+ T cells undergo a process of progressive immune dysfunction known as exhaustion [1, 2]. Recent studies have shown that multiple inhibitory receptors are frequently upregulated on antigen-specific exhausted CD8+ T cells [3, 4]. As a consequence, simultaneous blockade of several inhibitory pathways is more effective in correcting CD8 immune dysfunction than blockade of a single pathway [4]. In addition to increased expression of inhibitory receptors, one alternative mechanism that could contribute to T cell exhaustion during chronic infection is downregulation of positive costimulatory pathways, such as CD28-B7 interactions. Recent work from our group and others [5, 6] has shown that DNAM-1 (also known as CD226) – PVR (Poliovirus Receptor) (also known as CD155) interactions are important for CD8+ T cell proliferation and antigen- specific CD8+ T cell responses. This costimulatory pathway becomes essential when the cells providing cognate antigen are non-professional antigen presenting cells, such as epithelial or tumor cells. Chronic HIV-1 infection is the prototype clinically-important example of a situation where CD8+ T cell exhaustion perpetuates viral spreading and replication. Several studies have established that upregulation of the inhibitory receptor PD1 on HIV-1 specific T cells associates with T-cell exhaustion and disease progression [7–10]. Here, we evaluated whether impairment of the DNAM-1/PVR costimulatory pathway also contributes to HIV-1-induced the T-cell exhaustion.
Results and discussion
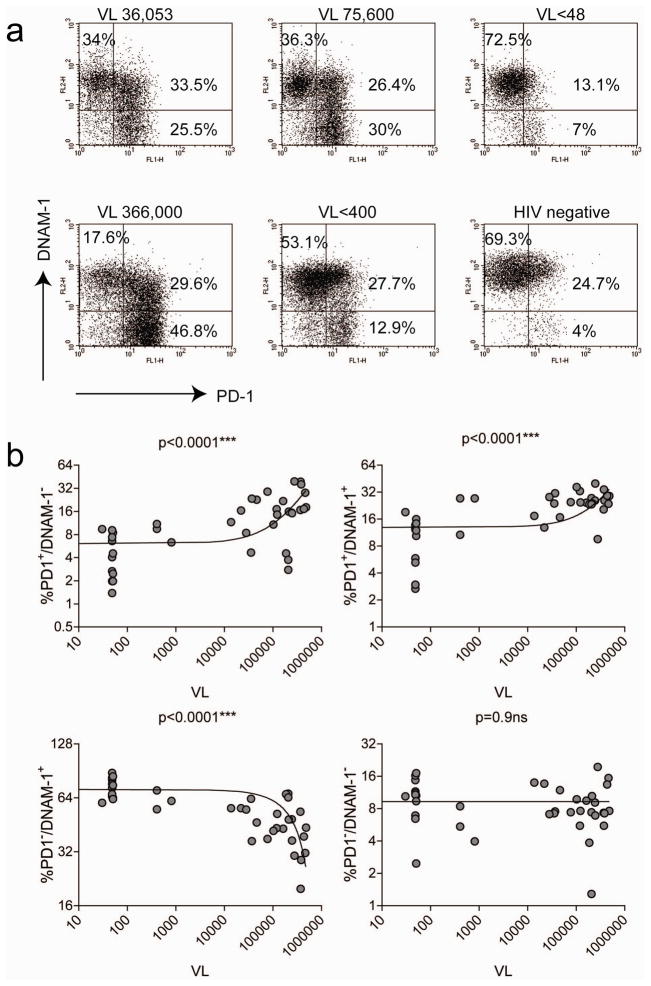
To establish whether the DNAM-1 costimulatory pathway is involved in chronic HIV-1 immune dysfunction, we examined the expression of DNAM-1 on exhausted CD8+ T cells that express the inhibitory receptor PD1. We found that a subset of CD8+ T cells with high expression of PD1 lost expression of DNAM-1 (Fig. 1A). The presence of PD1+/DNAM-1− CD8+ T cells highly correlated with HIV-1 viral load, while the percentage of DNAM-1+/PD1− CD8+ T cells inversely correlated with viral loads (Fig. 1B). Since it was previously shown that PD1 is preferentially expressed on HIV-1 specific CD8+ T cells [7, 8, 10], most likely DNAM-1 expression is lost on HIV-1 specific CD8+ T cells. Accordingly, we observed that a significant proportion of HIV-1-specific T cells downregulates DNAM-1 expression, at least in the few patients of our cohort for which MHC-I tetramers were available to visualize virus-specific T cells (Supplementary Fig. 1). A similar loss of DNAM-1 expression on PD1+ CD8+ T cells was observed in mice during chronic but not acute infection with LCMV (Supplementary Fig. 2A–F). A significant proportion of the CD8+ T cells losing DNAM-1 expression were specific for the viral gp33 epitope gp33–41 (Supplementary Fig. 2B and data not shown).
Figure 1.
DNAM-1 downregulation on PD1+ CD8+ T cells correlates with HIV-1 viral load. (A) Staining of whole blood or PBMCs from representative chronically HIV-1-infected or uninfected donors for PD1 and DNAM-1 reveals that a subset of CD8+ T cells expressing high levels of PD1 also lose DNAM-1 expression. The dotplots shown are gated on CD3+CD8+ T cells. VL=viral load. (B) Correlation between percentages of PD1+/DNAM-1−, PD1+/DNAM-1+, PD1−/DNAM-1+ cell subsets and viral load. Correlations were calculated by Spearman rank-correlation using GraphPad Prism version 4.0a. All tests were two-tailed. The curve fit was calculated by linear regression. n=58.
In addition to PD1+DNAM-1− CD8+ T cells, we also observed the presence of PD1+DNAM-1+ CD8+ T cells in both HIV and LCMV chronic infections. DNAM-1 expression was also reduced on this cell subset in humans. However, in mice DNAM-1 expression was conserved or slightly upregulated. This discrepancy may be due to differences in the duration of HIV-1 and LCMV infections.
Previous analysis of the molecular signatures characteristic of exhausted CD8+ T cells did not reveal loss of DNAM-1 by gene expression profiling [11]. Therefore we hypothesized that downregulation of DNAM-1 was due to a post-transcriptional mechanism. It was earlier reported that another major NK/CD8-cell associated receptor, NKG2D, is regulated and its activation antagonized by the release of soluble NKG2D-ligands MIC-A and MIC-B during tumor progression [12]. Hence, we thought that release of soluble PVR into the serum of HIV-1 patients could be responsible for DNAM-1 downregulation or masking. Although we could detect soluble PVR in the serum of chronically HIV-1 infected patients, we did not observe any statistically significant correlation with viral load and/or DNAM-1 downregulation; furthermore, soluble PVR was also present in sera from uninfected individuals (Fig. 2A and data not shown). In an attempt to determine the mechanisms that caused loss of DNAM-1 on exhausted HIV-1-specific CD8+ T cells we generated HLA-A0201-restricted gag-specific CTLs and incubated them with PVR-bearing target cells in the presence or absence of the gag peptide p76-84. We chose as a target Colo205 colonic epithelial cells because they are HLA-A2+ and they express significant levels of PVR (data not shown). Incubation of gag-specific CTLs with Colo205 cells in the presence of antigen for 24 h induced downregulation of DNAM-1, while causing upregulation of the activation marker CD69 (Fig. 2B). In contrast, when gag-specific CTLs were incubated with a PVR−, HLA-A2+ target, such an EBV transformed B cell line, DNAM-1 expression increased significantly (Fig. 2C). These data suggest that DNAM-1 downregulation in chronic HIV-1 infection is a specific consequence of CD8+ T cells interacting with PVR-expressing HIV-1-replicating cells. The most common cell types permissive to HIV-1 replication are CD4+ T cells and monocytes, macrophages or dendritic cells. Indeed PVR expression was upregulated on peripheral blood CD4+ T cells stimulated with mitogens (Fig. 2D). PVR expression was transient, peaking at day 2 and almost entirely lost by day 5 post-stimulation (data not shown), suggesting that in vivo recently activated CD4+ T cells infected by HIV-1 could become a target responsible for the loss of DNAM-1 on virus-specific CTLs. Moreover, in vivo in lymph nodes of patients chronically infected with HIV-1, PVR+CD3+ T cells were detected (Fig. 2E). Strong PVR expression was also detected on interdigitating dendritic cells of the T cell area, a cell type suspected to play a role in delivering HIV-1 to secondary lymphoid organs upon migration from mucosal sites (Fig. 2F), although there are no in vivo studies that directly demonstrate that interdigitating DC are a major reservoir for HIV-1 replication during the chronic phase. It is also possible that other cell types expressing PVR may contribute to downregulation of DNAM-1 on CD8+ T cells. Accordingly, HIV-1 replication was reported to occur to some extent in intestinal epithelial cells [13] and endothelial cells of brain vessels [14].
Figure 2.
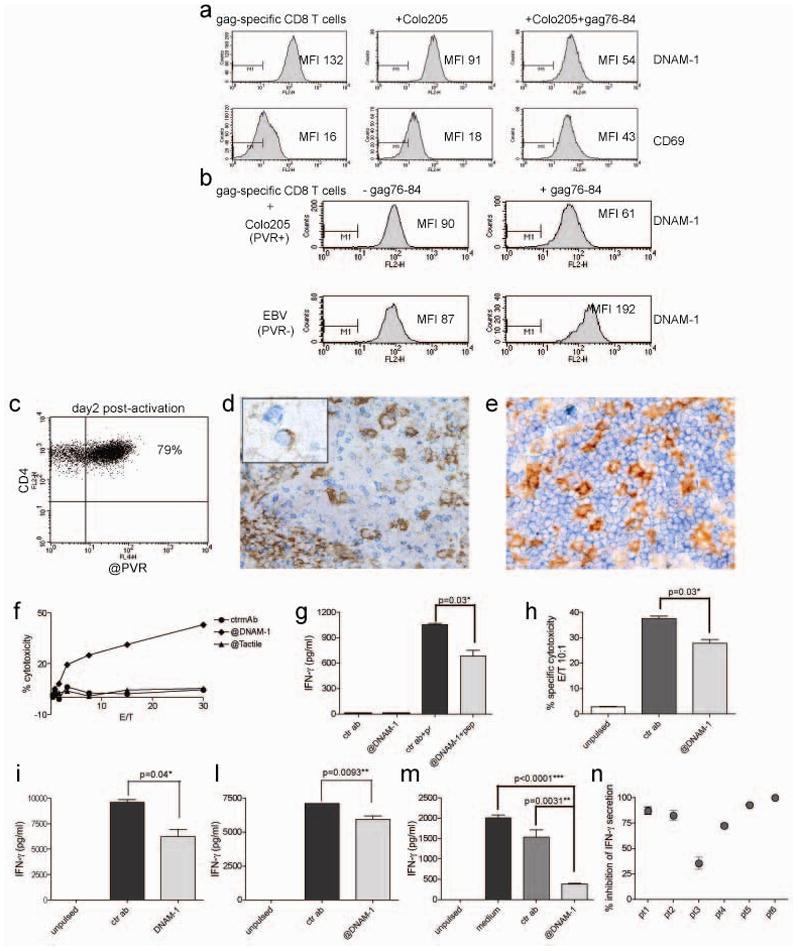
DNAM-1 expression is decreased when CD8+ T cells encounter antigen-bearing cells expressing PVR. (A) Levels of soluble PVR were determined by ELISA as described in the Materials and methods. Correlations were calculated by Spearman rank-correlation using GraphPad Prism version 4.0a. All tests were two-tailed. No correlation was found. n=32. (B, C) DNAM-1 and CD69 expression was assessed on gag-specific HLA-A2-restricted CTLs cultured for 24h alone or in the presence of either PVR-expressing Colo205 cells or PVR- APC (EBV-transformed B cell line), pulsed or notwith the gag-derived peptide 76-84 (100ng/ml). MFI=mean fluorescence intensity. Data are representative of 3 experiments. (D) Expression of PVR on CD4+ T cells assessed by flow cytometry two days after in vitro stimulation with the mitogen phytohemagglutinin (0.5μg/ml). PBMCs (106 cells/ml/well) where cultured in medium supplemented with rIL-2 .(E) Staining of reactive lymph nodes of HIV-1 patients shows that PVR (brown) is expressed in the germinal center by follicular dendritic cells and also by some cells with lymphoid morphology that co-express CD3 (blue) (see inset). (F) Staining of a normal lymph node shows that PVR (brown) is highly expressed by interdigitating dendritic cells surrounded by CD3+ (blue) T cells in the T cell area. Magnification, 200X (inset, 400X).
To investigate the functional role of DNAM-1 engagement on HIV-1-specific T cells we tested redirected cytotoxicity of gag76-84 specific CTLs on FcR+ P815 cells in the absence or presence of an anti-DNAM-1 antibody. Indeed engagement of DNAM-1 alone was able to induce strong redirected cytotoxicity (Fig. 3A). Moreover, blocking of DNAM-1 on CTLs co-cultured with gag76-84-pulsed Colo205 cells significantly reduced both IFN-γ production(Fig. 3B) and cytotoxicity (Fig. 3C). Notably, blockade of DNAM-1 on HIV-1-specific CD8+ T cells co-cultured with peptide-pulsed HLA-A2+ immature (Fig. 3D) or resiquimod-stimulated (Fig. 3E) dendritic cells significantly reduced IFN-γ release (but not cytotoxicity, data not shown). Finally, addition of the anti-DNAM-1 antibody to total PBMCs from HIV-1 infected patients stimulated with a mixture of 15 mer gag-derived peptides that cover different HLA specificities dramatically reduced IFN-γ secretion in all the patients who responded to the peptides in a 18h in vitro assay (Fig. 3F,G).
Figure 3.
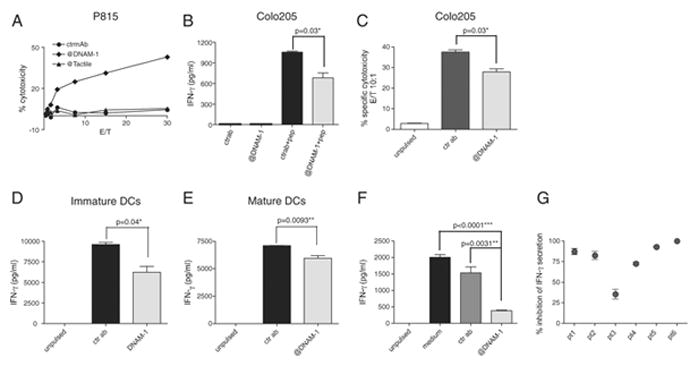
DNAM-1 engagement regulates cytotoxicity and IFN-γ production by HIV-1-specific CD8+ T cells. (A) Redirected cytotoxicity with gag-specific HLA-A2 restricted CTLs was performed against anti-DNAM-1/Tactile(CD96)/isotype matched control Ab-coated P815 cells at different effector to target (E/T) ratios. One representative experiment of two is shown. (B,C) Unpulsed or peptide-pulsed (100ng/ml) Colo205 cells were incubated with gag-specific HLA-A2 restricted CTLs in the presence of anti-DNAM-1 or isotype-matched control antibody. (B) IFN-γ was measured in cell culture supernatants after 6h of co-culture. (C) Cytotoxicity was evaluated after 4h of co-incubation. One representative experiment of four (B) or two (C) is shown. Bars indicate means ± s.d; n=3 replicates. (D, E) Unpulsed or peptide-pulsed (100ng/ml) (D) immature or (E) resiquimod-stimulated monocyte-derived dendritic cells (DCs) were incubated with gag-specific HLA-A2 restricted CTLs (10:1 E/T ratio) in the presence of anti-DNAM-1 or isotype-matched control antibody. IFN-γ was measured in cell culture supernatants after 6h of co-culture One representative experiment of three is shown. Bars indicate means ± s.d n=3. (F) PBMCs (5 × 105/well) from HIV infected patients were left unstimulated or stimulated with a pool of gag-derived 15-mer peptides in the presence of medium, anti-DNAM-1 or isotype-matched control antibody. IFN-γ was measured in cell culture supernatants after 18h of co-culture. Data from one representative donor is shown. Bars indicate means ± s.d.; n=2. (B–F) Significance was calculated by two-tailed unpaired t test using GraphPad Prism version 4.0a. (G) PBMCs from 20 HIV infected patients were stimulated as in (F) and IFN-γ measured in cell culture supernatants. The percentage of inhibition of IFN-γrelease by the anti-DNAM-1 antibody against isotype control is shown for the 6 patients that produced detectable amounts of IFN-γ in this assay.
Concluding remarks
Together, our data indicate that DNAM-1/PVR interactions may play a critical role in the effector function of HIV-1-specific CD8+ T cells. This costimulatory pathway may become even more crucial in the case of HIV-1-specific CD8+ T cell clones with low avidity TCRs. Downregulation of DNAM-1 coupled to upregulation of inhibitory receptors, such as PD1 and Lag3, may generate a subset of deeply dysfunctional cytotoxic T cells that are highly impaired in their capacity to eradicate the viral infection. Strategies aimed at avoiding loss or regaining expression of DNAM-1 may be instrumental in the treatment of chronic HIV-1 infection.
Materials and methods
Patients
Blood samples were obtained from three cohorts of HIV-1-infected individuals: (i) patients at the Clinical Trial Unit at Washington University, (ii) chronically-infected subjects recruited into the CHAVI 001 study in Africa and (iii) patients recruited at the Mortimer Market Centre for Sexual Health and HIV Research (London, UK). In each case, study approval was obtained from local ethics committees, and blood samples were withdrawn with written informed consent.
Flow cytometry
Flow cytometry was performed on whole blood or PBMCs of uninfected individuals or patients chronically infected with HIV-1 by four-color staining with anti-CD3-PerCP (Biolegend), anti-CD8-APC (Beckmann-Coulter), anti-DNAM-1 PE (Miltenyi Biotech) and anti-PD1-FITC (Pharmingen). Samples were analyzed on a BD FacsCalibur with the CellQuest software.
Generation of gag-specific HLA-A2 restricted CTLs
An HIV-1-infected patient known to be HLA-A2+ due to positive reactivity with the anti-HLA-A2 antibody BB7.2 (Hb-82, ATCC) was stained with a HLA-A0201 gag76-84 RPE-conjugated tetramer (Beckman Coulter). Upon detection of antigen specific T cells, RPE+ CD8+ T cells were purified by anti-PE microbeads (Miltenyi Biotech). Two consecutive rounds of purification were necessary to generate a stable cell line with >99% of tetramer+ cells. Cells were expanded and maintained by restimulating them every 20–25 days with irradiated (3,000 Rads) allogeneic PBMCs in the presence of phytohaemagglutinin (HA16, Murex) and recombinant IL–2 (R&D), as previously described [15].
Redirected cytotoxicity
Antibody-coated P815 cells were used as target cells. Antibodies to DNAM-1 (11A8), Tactile (NK92.39) and PVR (SKII.4) were generated in our laboratory as previously described [16]. Lysis of target cells was measured by standard Chromium release assay.
In vitro downregulation of DNAM-1
Antigen presenting cells (Colo205 or the EBV-transformed HLA-A2+ B cell line CA) (5 × 105) were left unpulsed or pulsed with the gag-derived peptide 76–84 for three hours. Free peptide was removed by extensive washing and the cells were co-incubated with gag-specific CTLs for 18 h. DNAM-1 staining was performed on ice, gating on CD8+ T cells.
Histology
Specimens included reactive lymph nodes from three HIV-1+ patients (tissue bank of the Department of Pathology, Spedali Civili di Brescia) removed for diagnostic purposes. Cryostat sections obtained from fresh-frozen tissue blocks were air- dried overnight at room temperature and fixed in acetone for 10 minutes. By immunohistochemistry, PVR was detected using mAb SKII.4 followed by Dako Real Detection System Alkaline Phosphatase using New Fucsin as chromogen (Dako). For double immunohistochemistry SKII.4 was detected by NovoLink Polymer Detection System (Novocastra Laboratories) HRP-linked followed by DAB whereas CD3 (rabbit monoclonal Sp7, NeoMarkers) was revealed by Mach 4 AP-linked (Biocare Medical) followed by Ferangi Blue as chromogen (Biocare Medical).
Detection of IFN-γ in cell culture supernatants
Unpulsed or peptide-pulsed (100ng/ml) Colo205 (104/well), immature or resiquimod-stimulated monocyte-derived dendritic cells (5 × 103/well) were co-incubated with gag-specific CTLs (5 × 104/well). After 6 h of co-culture supernatants were collected and IFN-γ measured by the human Th1/2 CBA kit (Pharmingen, BD). Anti-DNAM-1 antibody and an isotype-matched control antibody (anti-myc, CRL-1729, ATCC) were used at a final concentration of 20μg/ml. For stimulation of whole PBMCs 5 × 105 cells were seeded/well into a 96-well plate and left unstimulated or stimulated with 400ng/ml of HXB2 gag1-122 15-mer peptide pool (Anaspec).
Infection of mice with LCMV
Mice were infected intraperitoneally with 2 × 105 pfu of LCMV Arm (kindly provided by Dr. R. Ahmed, Emory Vaccine Center, Emory University School of Medicine, Atlanta) or intravenously with 2 × 106 PFU of LCMV clone 13 (kindly provided by Dr. M. Oldstone, Department of Immunology and Microbial Science, The Scripps Research Institute, La Jolla, California). Nine days or one month after infection mice were sacrificed and splenocytes stained with anti-DNAM-1-biotin or Alexa 647 (mab480, previously described) followed by Streptavidin-APC (Molecular Probes), CD3-PerCP (Biolegend), anti-PD1-PE (eBioscience) and CD8-FITC (Pharmingen). Staining with H2Db gp33-41 tetramers (Beckmann-Coulter) was performed on purified CD8+ T cells (CD8 T cell isolation kit, Miltenyi Biotech).
Detection of soluble PVR by ELISA
ELISA plates (Maxisorp) were coated with purified anti-CD155 (Beckman-Coulter) at 5μg/ml overnight at 4° C in carbonate buffer pH 9.5. After washing plates were blocked for 1h at RT with PBS/10%FCS. Dilutions of sera were incubated for 2 h at RT. After additional washing we used biotinylated SKII.4 as detection antibody at a concentration of 0.5μg/ml, followed by Avidin-HRP (Sigma). As a standard we used purified soluble PVR-hFc (3 domains) [16].
Supplementary Material
Acknowledgments
This work was supported by the NIAID Center for HIV/AIDS Vaccine Immunology grant A1067854 and JDRF#24-2008-938. We thank CHAVI and DUKE management and support teams for study coordination, and all the patients who generously donated blood for this study.
Abbreviations
- DNAM-1
DNAX Activating Molecule-1
- PVR
Poliovirus Receptor
- CBA
cytometric bead array
- ATCC
American Tissue Cell Culture Collection
Footnotes
Conflict of interest: The authors declare no financial or commercial conflict of interest.
References
- 1.Saibil SD, Deenick EK, Ohashi PS. The sound of silence: modulating anergy in T lymphocytes. Curr Opin Immunol. 2007;19:658–664. doi: 10.1016/j.coi.2007.08.005. [DOI] [PubMed] [Google Scholar]
- 2.Shin H, Wherry EJ. CD8 T cell dysfunction during chronic viral infection. Curr Opin Immunol. 2007;19:408–415. doi: 10.1016/j.coi.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 3.Barber DL, Wherry EJ, Masopust D, Zhu B, Allison JP, Sharpe AH, Freeman GJ, Ahmed R. Restoring function in exhausted CD8 T cells during chronic viral infection. Nature. 2006;439:682–687. doi: 10.1038/nature04444. [DOI] [PubMed] [Google Scholar]
- 4.Blackburn SD, Shin H, Haining WN, Zou T, Workman CJ, Polley A, Betts MR, et al. Coregulation of CD8+ T cell exhaustion by multiple inhibitory receptors during chronic viral infection. Nat Immunol. 2009;10:29–37. doi: 10.1038/ni.1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gilfillan S, Chan CJ, Cella M, Haynes NM, Rapaport AS, Boles KS, Andrews DM, et al. DNAM-1 promotes activation of cytotoxic lymphocytes by nonprofessional antigen-presenting cells and tumors. J Exp Med. 2008;205:2965–2973. doi: 10.1084/jem.20081752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iguchi-Manaka A, Kai H, Yamashita Y, Shibata K, Tahara-Hanaoka S, Honda S, Yasui T, et al. Accelerated tumor growth in mice deficient in DNAM-1 receptor. J Exp Med. 2008;205:2959–2964. doi: 10.1084/jem.20081611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Day CL, Kaufmann DE, Kiepiela P, Brown JA, Moodley ES, Reddy S, Mackey EW, et al. PD-1 expression on HIV-specific T cells is associated with T-cell exhaustion and disease progression. Nature. 2006;443:350–354. doi: 10.1038/nature05115. [DOI] [PubMed] [Google Scholar]
- 8.Trautmann L, Janbazian L, Chomont N, Said EA, Gimmig S, Bessette B, Boulassel MR, et al. Upregulation of PD-1 expression on HIV-specific CD8+ T cells leads to reversible immune dysfunction. Nat Med. 2006;12:1198–1202. doi: 10.1038/nm1482. [DOI] [PubMed] [Google Scholar]
- 9.Freeman GJ, Wherry EJ, Ahmed R, Sharpe AH. Reinvigorating exhausted HIV-specific T cells via PD-1-PD-1 ligand blockade. J Exp Med. 2006;203:2223–2227. doi: 10.1084/jem.20061800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Petrovas C, Casazza JP, Brenchley JM, Price DA, Gostick E, Adams WC, Precopio ML, et al. PD-1 is a regulator of virus-specific CD8+ T cell survival in HIV infection. J Exp Med. 2006;203:2281–2292. doi: 10.1084/jem.20061496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wherry EJ, Ha SJ, Kaech SM, Haining WN, Sarkar S, Kalia V, Subramaniam S, et al. Molecular signature of CD8+ T cell exhaustion during chronic viral infection. Immunity. 2007;27:670–684. doi: 10.1016/j.immuni.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 12.Gonzalez S, Groh V, Spies T. Immunobiology of human NKG2D and its ligands. Curr Top Microbiol Immunol. 2006;298:121–138. doi: 10.1007/3-540-27743-9_6. [DOI] [PubMed] [Google Scholar]
- 13.Meng G, Wei X, Wu X, Sellers MT, Decker JM, Moldoveanu Z, Orenstein JM, et al. Primary intestinal epithelial cells selectively transfer R5 HIV-1 to CCR5+ cells. Nat Med. 2002;8:150–156. doi: 10.1038/nm0202-150. [DOI] [PubMed] [Google Scholar]
- 14.Bussolino F, Mitola S, Serini G, Barillari G, Ensoli B. Interactions between endothelial cells and HIV-1. Int J Biochem Cell Biol. 2001;33:371–390. doi: 10.1016/s1357-2725(01)00024-3. [DOI] [PubMed] [Google Scholar]
- 15.Padovan E, Giachino C, Cella M, Valitutti S, Acuto O, Lanzavecchia A. Normal T lymphocytes can express two different T cell receptor beta chains: implications for the mechanism of allelic exclusion. J Exp Med. 1995;181:1587–1591. doi: 10.1084/jem.181.4.1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fuchs A, Cella M, Giurisato E, Shaw AS, Colonna M. Cutting edge: CD96 (tactile) promotes NK cell-target cell adhesion by interacting with the poliovirus receptor (CD155) J Immunol. 2004;172:3994–3998. doi: 10.4049/jimmunol.172.7.3994. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.