Abstract
Sensation seeking is a heritable personality trait that has been reliably linked to behavior disorders. The dopamine system has been hypothesized to contribute to individual differences in sensation seeking, and both experimental and observational studies in humans and non-human animals provide evidence for this relationship. We present here a candidate-system approach to genetic association analysis of sensation seeking, in which single nucleotide polymorphisms (SNPs) from a number of dopaminergic genes were analyzed. Using 273 SNPs from eight dopamine genes in a sample of 635 unrelated individuals, we examined the aggregate effects of those SNPs significantly associated with sensation seeking. Multiple SNPs in four dopamine genes accounted for significant variance in sensation seeking. These results suggest that aggregation of multiple SNPs within genes relevant to a specific neurobiological system into a “genetic risk score” may explain a nontrivial proportion of variance in human traits.
Keywords: sensation seeking, dopamine, candidate gene, association study
Sensation seeking is a personality trait of great importance to public health, in that it has been specifically associated with behavior disorders with high social costs, especially substance use disorders (Zuckerman & Neeb, 1979). Among individuals with substance use disorders, greater sensation seeking is associated with earlier age onset of use and abuse, more poly-substance use, a greater number of symptoms, and more severe impairment (Ball, Carroll, & Rounsaville, 1994). Higher sensation seeking levels are also associated with increased treatment drop-out rates and poorer treatment outcomes (Staiger, Kambouropoulos, & Dawe, 2007).
Heritability for sensation seeking ranges from 40% to 60% (Eysenck, 1983; Fulker, Eysenck, & Zuckerman, 1980; Hur & Bouchard, 1997; Koopmans, Boomsma, Heath, & van Doornen, 1995). Correlations among specific elements (facets) of sensation seeking are primarily accounted for by overlapping genetic factors (Hur & Bouchard, 1996; Koopmans, Boomsma, Heath, & van Doornen, 1995). Twin studies suggest no sex differences in the magnitude or nature of genetic effects on sensation seeking (Eysenck, 1983; Koopmans, Boomsma, Heath, & van Doornen, 1995). Further, behavioral undercontrol (similar to the disinhibition scale of sensation seeking) shares substantial genetic risk with alcohol dependence and conduct disorder (Slutske et al., 2002).
Sensation seeking therefore represents a promising endophenotypic aspect of externalizing problems (Benjamin, Ebstein, & Belmaker, 2001; Gottesman & Gould, 2003; Krueger, Markon, Patrick, Benning, & Kramer, 2007). It is also an important target phenotype for a theory-driven candidate neurogenetic system approach to linking molecular genetic polymorphisms with specific behavioral phenomena. Sensation seeking has a demonstrable neurobiological basis in humans (Joseph, Liu, Jiang, Lynam, & Kelly, 2009). The dopaminergic system has been long hypothesized to underlie individual variation in sensation seeking (Zuckerman, 1984), and recent research supports this hypothesis. A rodent model has demonstrated that availability of nucleus accumbens dopamine D2 and D3 receptors is negatively associated with impulsivity (similar to the disinhibition scale of sensation seeking; Dalley et al., 2007). A study in humans has shown a negative association between ventral midbrain dopamine D2 receptor availability and novelty seeking (similar to the experience seeking scale of sensation seeking; Zald et al., 2008). A pharmacological study in humans also suggested that dopamine stimulation increases nicotine craving in individuals who score highly on the experience seeking scale of sensation seeking (Netter, Henning, & Roed, 1996).
Candidate gene studies provide some evidence of a relationship between specific polymorphisms in genes involved in the dopaminergic system and sensation seeking. A commonly examined functional single nucleotide polymorphism (SNP; rs4680, also known as Val158Met) in the COMT gene has been associated with sensation seeking (though the effect was specific to females; Lang, Bajbouj, Sander, & Gallinat, 2007). A gene-gene interaction effect on sensation seeking has been demonstrated between rs1800497 (also known as DRD2 Taq1A or C32806T) located in the gene ANKK1 and the commonly studied variable-number-of-tandem-repeats (VNTR) polymorphism (48 base pairs that repeat a variable number of times) in the DRD4 gene (Eisenberg, Campbell, MacKillop, Lum, & Wilson, 2007). This DRD4 VNTR appears to be a developmentally stable predictor of experience seeking behaviors, and has been associated longitudinally with infant visual exploratory behavior and adolescent novelty seeking (Laucht, Becker, & Schmidt, 2006). However, as with many other traits and diseases, non-replication of specific candidate gene effects on sensation seeking and related traits is a common occurrence, and evidence for the involvement of any specific variant is typically modest at best (e.g. Heck et al., 2009).
As a continuous trait conferring risk for the development of externalizing disorders, and greater disorder severity, sensation seeking is an appealing target for human genetic research. Candidate gene studies of sensation seeking and personality traits more generally have previously focused on a small number of polymorphisms within a single gene (Ebstein, 2006), the individual effects of SNPs from multiple genes (Heck et al., 2009), or the aggregate effects of single polymorphisms from each of several genes (Beaver, 2009). The advent of dense whole-genome SNP genotyping allows us to capture far more genetic variation than has previously been captured by candidate gene studies. However, use of whole-genome data incurs a considerable cost, in the form of exacting a heavy penalty for multiple testing.
Here, we pursue an approach that combines the theory-driven candidate gene approach with the genotypic data available from genome-wide high-throughput genotyping technology. From all SNPs located in dopamine genes that were available from a dense-coverage commercially-available genotyping platform, we first selected those SNPs that were individually associated with sensation seeking. We then fit nested regression models, in which sensation seeking was predicted by 1) demographic covariates; or 2) variables from model 1 (covariates) and all SNPs identified as significant in the individual association tests. By comparing model fit statistics for these models, we addressed a specific research question. Does the aggregation of multiple SNPs within dopamine genes explain significant variance in sensation seeking, over and above that explained by demographic covariates (i.e. age, sex, and ancestry)?
Methods
Participants
Participants were 635 unrelated individuals who had participated in the Study of Addiction: Genetics and the Environment (SAGE; Bierut et al., under review; http://zork.wustl.edu/gei/). Participants in the current research were a subset of the SAGE sample, all of whom were drawn from a primary study of alcohol dependence (COGA; Reich et al., 1998) because these individuals had completed the Sensation Seeking Scale (Zuckerman, 1978; Zuckerman, 1996). Written informed consent was obtained from all participants following thorough description of the study. The average age of our sample was 45.3 years (with a range of 22 to 77), 55.1% were female, 65.2% met criteria for a lifetime DSM-IV alcohol dependence diagnosis, and 18.9% and 8.2% self-reported African and/or Hispanic ancestry, respectively.
Measure
Participants were administered Zuckerman’s Sensation Seeking Scale Form V (SSS-V; Zuckerman, 1978; Zuckerman, 1994). The SSS-V yields an overall Sensation Seeking score from four 10-item subscale scores. The subscales are Boredom Susceptibility (e.g. “When you can predict almost everything a person will do and say, he or she must be a bore.”), Disinhibition (e.g. “I like wild ‘uninhibited’ parties.”), Experience Seeking (e.g. “I often find beauty in the ‘clashing’ colors and irregular form of modern painting.”), and Thrill/Adventure Seeking (e.g. “I would like to try parachute jumping.”). The total sensation seeking score has good reliability in both American males (Cronbach’s α = 0.84) and females (α = 0.85; Zuckerman, 1979). Our analysis of the total sensation seeking score, rather than its subscales, provides generally greater measurement reliability, as well as a reduced number of statistical tests.
Genotyping
DNA was obtained from blood samples and genotyping was carried out at the Johns Hopkins University Center for Inherited Disease Research (CIDR) using the Illumina Human IM Bead Chip. The median missing call rate was less than 0.05%, with 95% of SNPs resulting in less than 1.4% missingness. Strict quality control procedures were implemented, including assessment of population structure, missing call rates, Mendelian errors, duplication errors, gender and chromosomal anomalies, hidden relatedness, batch effects, and Hardy-Weinberg disequilibrium. Duplicates, related subjects, and outliers were removed. A total of 948,142 SNPs passed this thorough cleaning procedure (Laurie et al., under review; http://www.genevastudy.org/).
Gene and SNP selection
Dopamine-related genes were identified from a search of the relevant candidate-gene literature. Only genes located on autosomal (i.e. non-sex) chromosomes that have definite and direct effects on the dopaminergic system were included. SNPs in each of these genes (identified from dbSNP; Database of Single Nucleotide Polymorphisms, build 129) that were available on the Illumina Human 1M Bead Chip were then selected. Functionality of each SNP was identified using dbSNP. A total of 273 SNPs were chosen for inclusion in our analyses. Table 1 reports the genes included in our analyses, the number of SNPs available in each gene from the SAGE genotype data, and the general function of each gene as it relates to dopamine. (Note. No SNPs from the DRD5 gene were included in our analysis, because the single SNP available in this gene on the Illumina 1M platform did not pass genotyping quality control procedures in the SAGE sample. In addition, our sample did not include genotyping of rs1800955, or a reasonable proxy, a SNP in DRD4 showing significant association with impulsivity-related personality traits in a recent meta-analysis by Munafò and colleagues, 2008.)
Table 1.
Genes from which SNPs were chosen for analyses.
| Gene | Location | SNPs | Role in Dopamine (DA) |
|---|---|---|---|
| DRD3 | 3q13.3 | 32 | codes D3 subtype of DA receptors |
| SLC6A3 (a.k.a. DAT1) | 5p15.3 | 35 | DA transporter that mediates reuptake of DA from the synapse |
| DRD1 | 5q35.1 | 9 | codes D1 subtype of DA receptors |
| DDC | 7p12.2 | 81 | codes a protein that converts L-DOPA to DA |
| DBH | 9q34 | 37 | converts DA to norepinephrine |
| DRD4 | 11p15.5 | 4 | codes D4 subtype of DA receptors |
| DRD2 | 11q23 | 40 | codes D2 subtype of DA receptors |
| COMT | 22q11.21 | 35 | affects degradation of catecholamines (including DA) |
Note. Location = chromosome location of the gene; SNPs = number of single nucleotide polymorphisms in that gene available in the SAGE data from the Illumina Human IM Bead Chip following quality control procedures; DRD3 = dopamine receptor D3; SLC6A3 = solute carrier family 6 (DAT1 = dopamine transporter 1); DRD1 = dopamine receptor D1; DDC = dopa decarboxylase; DBH = dopamine beta-hydroxylase; DRD4 = dopamine receptor D4; DRD2 = dopamine receptor D2; COMT = catechol-O-methyltransferase; L-DOPA = L-3,4-dihydroxyphenylalanine.
Analyses
To characterize ethnic heterogeneity in our sample, principal components were estimated based on the SAGE genome-wide data, using the procedure described by Price and colleagues (2006). Two major principal components emerged, corresponding to European vs. African ancestry (PC1) and Hispanic vs. non-Hispanic ancestry (PC2). Treating ancestry as a covariate assumes that while minor (i.e. less common) allele frequencies may differ between races, the biological impact of SNPs does not (Ioannidis, Ntzani, & Trikalinos, 2004). While ideally we would model ethnicities separately to explore this, our sample size prevented this approach. PC1 and PC2 were used as covariates in our analyses, along with age (coded in quartiles as three dummy codes, as has been done in previous SAGE analyses, corresponding to ≤34, 35–39, and 40–44, with 45+ as the reference group), and sex (1 for males and 2 for females). SNPs were coded as 0/1/2 to indicate the number of minor alleles present for a given individual. We did not control for alcohol dependence case status for our main analyses, because sensation seeking is substantially related to alcohol use behaviors (Zuckerman, 1994), and including it as a covariate may eliminate meaningful variance (Meehl, 1971). Nevertheless, post-hoc analyses revealed that our pattern of results and conclusions did not change if alcohol dependence was included as a covariate in the aggregated SNP tests (i.e. the p-value reported in Table 3 remained significant at p < 5 × 10−12).
Table 3.
Comparisons between models predicting sensation seeking from 1) only covariates; and 2) covariates and all SNPs meeting p < 0.05 and FDR < 0.10 in the association tests.
| Model | SNPs | R2 | ΔR2 | −2LL | df | p | AIC | BIC |
|---|---|---|---|---|---|---|---|---|
| 1. Covariates only | 0 | 27.9% | 4113.0 | 8 | 4129.0 | 4164.7 | ||
| 2. All significant SNPs | 12 | 31.7% | 3.9% | 4027.5 | 20 | < 4 × 10−13 | 4067.5 | 4156.5 |
Note. SNPs = number of single nucleotide polymorphisms included in the model; R2 = proportion of variance in the dependent variable explained by the covariates and any SNPs included in that regression model; ΔR2 = additional variance explained by adding SNPs (model 2) to the covariate model (model 1); −2LL = −2 times the log-likelihood of the regression model; df = degrees of freedom in the regression model; p = the p-value when comparing model 2 to model 1, estimated by the change in −2LL on a chi-square distribution with df equal to the difference in dfs between the models; AIC = Akaike’s Information Criterion, where lower values indicate better model fit to the data; BIC = Bayesian Information Criterion, where lower values indicate better model fit to the data.
Association tests were initially run on each individual SNP (coded as 0, 1, or 2, representing the number of minor alleles), performing a linear regression in R (an open-source statistical program; R Development Core Team, 2009) of the sensation seeking score on that SNP and all covariates (i.e. PC1, PC2, age, and sex). We implemented two additional methods to ensure that any significant results were greater than chance. First, in addition to p-values, we calculated the false discovery rate (FDR) for each regression-weight p-value reaching significance. FDR controls the proportion of false-positive results expected from all those tests declared significant and is calculated as:
| (1) |
Where i is the rank order of the test (ranked in terms of ascending p-values), m is the total number of independent tests, α is the p-value cut-off for significance, and Pi is the p-value for test i (Benjamini & Hochberg, 1995). We set our maximum FDR at 0.10, interpreted as no more than 10% of the SNPs declared significant based on p < 0.05 would be false positives. For our purposes, we set the value of m to 8, the number of genes included in our analyses.
To account for linkage disequilibrium (LD; the correlation between SNPs) in our heterogeneous sample, our second method for estimating false positives was to calculate the number of statistically significant associations observed when genotypes were randomly assigned to individuals (i.e., a permutation analysis). By randomly assigning intact genotypes to individuals (and keeping each individual’s sensation seeking and covariate scores the same as in the real association tests), we were able to observe the number of false-positive results obtained when keeping allelic distributions and LD patterns in our sample intact.
Following the identification of all statistically significant SNPs (i.e. those that were significant at the two-tailed p < 0.05 level in the individual SNP association tests, and that met FDR < 0.10), we compared two models of sensation seeking. The baseline model regressed sensation seeking on the covariates included in the initial association tests. The second model regressed sensation seeking on covariates and those significant SNPs identified from our initial association tests. We evaluated the relative goodness-of-fit of these models to the raw data by comparing a) the total proportion of variance in sensation seeking explained, b) the model likelihoods, and c) the Akaike’s Information Criterion (AIC) and Bayesian Information Criterion (BIC) from each model. AIC and BIC are information theoretic measures of goodness-of-model fit, and account for model parsimony in evaluating fit. Relative to a comparison of model likelihoods, AIC and BIC are more conservative, requiring greater evidence of the predictive utility of additional predictor variables to show improved fit. Lower values of AIC and BIC indicate a relatively better fit to the data (Akaike, 1974; Schwarz, 1978).
Results
Tests for association between individual SNPs and sensation seeking
A total of 273 SNPs from eight dopamine-related genes were individually tested for association with sensation seeking, controlling for demographic characteristics. Results for the individual-SNP association tests (for those SNPs whose regression weights met p<0.05) are reported in Table 2. Table S1 in the supporting information available on-line presents results from all 273 association tests. Twelve SNPs met significance criteria for association with sensation seeking, as defined by p < 0.05 and FDR < 0.10. By comparison, only three SNPs in the randomized genotype condition (rs2042449 and rs9312866 in SLC6A3, and rs1611114 in DBH) were significant by chance (at p < 0.05), and none of these passed FDR criteria (i.e. the p-value of the top-ranked SNP was greater than its FDR value). Because the number of SNPs meeting p < 0.05 was substantially greater in the correct genotype tests than in the tests using randomly assigned genotypes, and because all of the SNPs in the correct genotype condition met FDR criteria, compared to none of the SNPs in the random genotype condition, we concluded that the implicated SNPs are likely true associations with sensation seeking, at least in the current data set.
Table 2.
SNPs significantly associated with sensation seeking (p < 0.05 and FDR < 0.10) from individual SNP tests.
| SNP | Gene | Chr | Pos (bp) | Function | Allele | MAF | B | Z | p | FDR |
|---|---|---|---|---|---|---|---|---|---|---|
| rs11575551 | DDC | 7 | 50,493,757 | UTR-3’ | C | 0.03 | −3.266 | −2.756 | 0.006 | 0.006 |
| rs11575522 | DDC | 7 | 50,502,889 | Intron | A | 0.05 | −2.185 | −2.581 | 0.010 | 0.013 |
| rs11575542 | DDC | 7 | 50,498,481 | Missense | A | 0.05 | −2.125 | −2.512 | 0.012 | 0.019 |
| rs11575543 | DDC | 7 | 50,498,363 | Intron | T | 0.05 | −2.170 | −2.506 | 0.012 | 0.025 |
| rs3829897 | DDC | 7 | 50,597,258 | Intron | T | 0.42 | −0.896 | −2.456 | 0.014 | 0.031 |
| rs7876027 | DBH | 9 | 135,504,360 | Intron | G | 0.07 | 1.688 | 2.335 | 0.020 | 0.038 |
| rs174699 | COMT | 22 | 18,334,458 | Intron | C | 0.06 | −1.748 | −2.290 | 0.022 | 0.044 |
| rs10278338 | DDC | 7 | 50,597,764 | Intron | T | 0.34 | −0.842 | −2.277 | 0.023 | 0.050 |
| rs11575552 | DDC | 7 | 50,493,740 | UTR-3’ | C | 0.02 | −3.118 | −2.250 | 0.024 | 0.056 |
| rs933271 | COMT | 22 | 18,311,407 | Intron | C | 0.32 | −0.827 | −2.127 | 0.033 | 0.063 |
| rs12669770 | DDC | 7 | 50,597,382 | Intron | A | 0.33 | −0.732 | −1.984 | 0.047 | 0.069 |
| rs2975284 | SLC6A3 | 5 | 1,485,552 | Intron | T | 0.01 | −2.902 | −1.970 | 0.049 | 0.075 |
Note. SNP = single nucleotide polymorphism in the regression equation; Gene = gene location of the SNP (see Table 1 for gene function descriptions); Chr = chromosome location of the SNP; Pos (bp) = position on the chromosome (in base pairs) of the SNP; Function = SNP function; UTR-3’ = SNP located in an untranslated region (UTR) at the 3’ end of the gene that may affect the stability, localization, and/or efficiency of messenger RNA (mRNA) translation of the gene; Intron = non-coding SNP; Missense = missense mutation, in which each allele produces a different amino acid; Allele = minor allele (i.e. less frequent nucleotide) for which the regression effect, B, is reported; MAF = minor allele frequency; B = regression coefficient for the effect of the SNP; Z = z-score for the regression coefficient (calculated as B / std.error(B)); p = two-tailed p-value for B; FDR = False Discovery Rate (see Equation 1 in text).
Table 2 shows that eight of our twelve significant SNPs were located in DDC, and two were located in COMT. To examine whether these SNPs could be expected to explain unique variance in sensation seeking, we estimated the intercorrelations among all significant SNPs located within a single gene. In our sample, the two COMT SNPs (rs174699 and rs933271) had an r-squared value of 0.03, suggesting that linkage disequilibrium (LD, i.e., correlation between SNPs) was not responsible for the significant association of both with sensation seeking. Among the eight DDC SNPs significantly associated with sensation seeking, three (rs11575522, rs11575542, and rs11575543) were highly intercorrelated (r2 = 0.95–0.98). Nevertheless, even including these highly intercorrelated SNPs, the median r-squared value among the significant DDC SNPs was 0.04, indicating that linkage disequilibrium was likely not driving the inclusion of a relatively large number of DDC SNPs in our aggregate score.
Predictive utility of aggregate SNPs
We examined the utility of including all SNPs that were significant in the initial association tests when predicting sensation seeking. Results from models comparing the utility of covariates to covariates plus all significant SNPs are presented in Table 3. The model that included dopamine-related SNPs fit significantly better than the covariates-only model (as indicated in Table 3 by p < 4 × 10−13 and the lower AIC and BIC of model 2 compared to model 1). Including the 12 significant SNPs explained 3.9% more variance (an average of 0.35% per exonic SNP, 0.31% per intronic SNP) in sensation seeking than the covariates-only models.
To illustrate the influence of all significantly associated SNPs on the total sensation seeking score, we calculated a “genetic risk score” (Evans, Visscher, & Wray, in press; Purcell et al., 2009; Wray, Goddard, & Visscher, 2007), defined for each individual as the sum of the number of minor alleles at each associated SNP multiplied by that SNPs regression weight from the aggregate SNP model. That is:
| (2) |
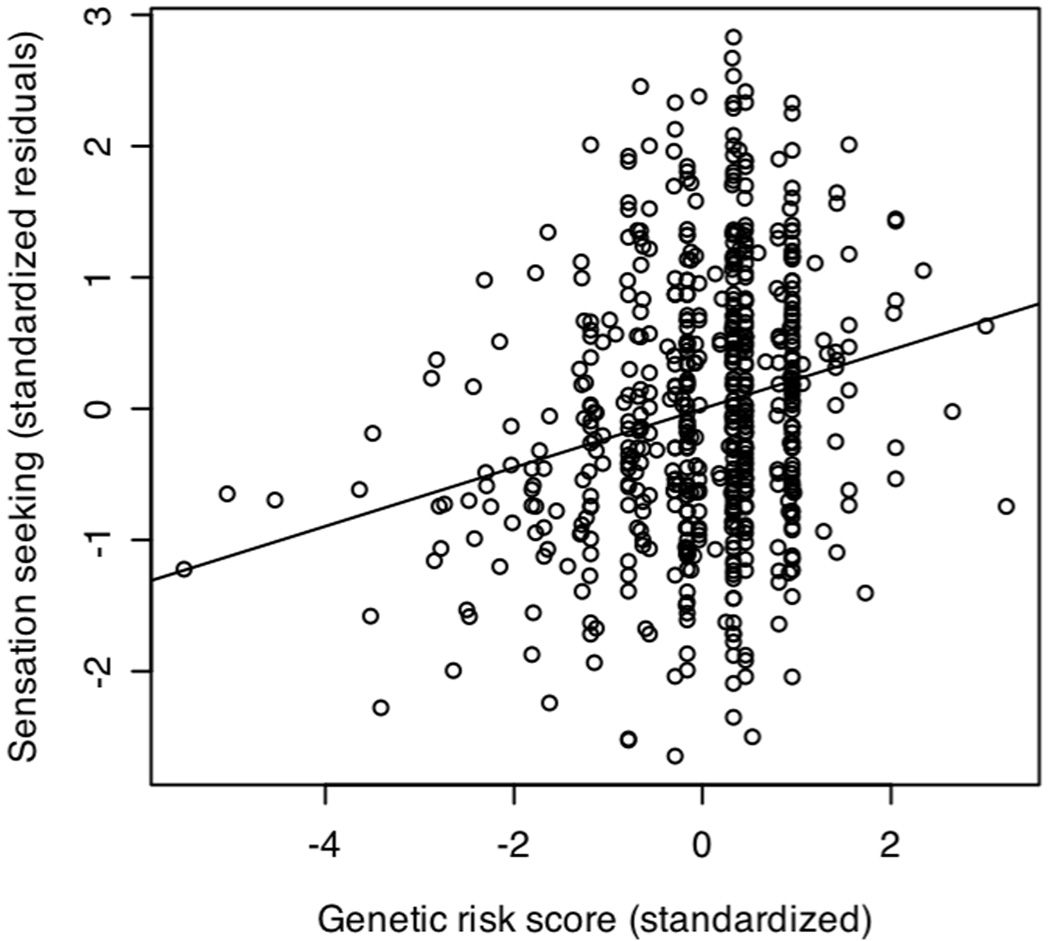
Figure 1 illustrates the correlation (r = 0.20; p < 2 × 10−8) between this genetic risk score and the residualized total sensation seeking score1 (after accounting for age, sex, and ancestry; both the genetic risk score and the residual sensation seeking score were standardized to a mean of zero and a standard deviation of one to increase interpretability). With the caveat that calculating the genetic risk score in the same sample used to identify significant SNPs may represent an optimistic population effect size estimate, this correlation represents a non-trivial effect in the behavioral sciences (e.g. Cohen, 1992) and is notable in the context of the effect sizes of accepted physical and mental health associations (e.g. aspirin and heart attack survival; chemotherapy and breast cancer survival; lead exposure and childhood IQ; nicotine patch use and smoking cessation; Meyer et al., 2001). This is also a non-trivial effect in the context of the candidate gene and genome-wide association literatures, where effect sizes for single genetic polymorphisms are typically small (Maher, 2008).
Figure 1.
Scatterplot and least-squares regression line for additive effects of twelve SNPs from four dopamine-related genes predicting sensation seeking (r = 0.20; p < 2 × 10−8), after accounting for demographic covariates.
Discussion
We implemented a multivariate approach to investigating the effects of SNPs in dopamine genes, a theoretically implicated neurobiological system, on sensation seeking, a personality trait associated with costly outcomes, such as substance use disorders. Working with data from 635 individuals, we selected 273 SNPs covering eight dopamine genes, and conducted initial association analyses to identify individual SNPs significantly associated (at p<0.05, FDR < 0.10) with sensation seeking. We then estimated the variance in sensation seeking explained by 1) demographic covariates; and 2) all significantly associated SNPs. Increased variance explained and improved model fit statistics (see Table 3) indicated that aggregated SNPs from dopamine genes explained significant variation between individuals in sensation seeking, even when controlling for demographic characteristics. Despite our relatively dense coverage of these selected genes, not all possible SNPs (or other genetic variants) were included on our genotyping platform. To the extent that the genotyped SNPs are not themselves functional, but are instead in linkage disequilibrium (i.e. correlated) with ungenotyped functional variants, these proportions of variance may be underestimates compared to the true variance in sensation seeking explainable by these dopamine genes.
Strengths and weaknesses
The primary weakness of this study was its modest sample size, and our lack of a sample in which to examine replication of our findings. However, our sample was demographically diverse, with an overrepresentation of individuals meeting criteria for alcohol dependence, a disorder where sensation seeking may be a particularly relevant risk factor. Our genotypic data provided more thorough coverage of genetic variation in the candidate genes examined here than in previous association studies related to sensation seeking (Beaver, 2009; Ebstein, 2006; Heck et al., 2009), and we made use of our additional genomic data by taking a systems approach and considering the aggregate effect of numerous SNPs in multiple genes associated with dopamine. This approach resulted in significant improvement in our ability to predict sensation seeking scores, beyond the prediction afforded by covariates, with an overall non-trivial aggregate effect (Cohen, 1992) of SNPs on sensation seeking.
Conclusions
Our results indicated that dopamine genes are associated with sensation seeking at the system level, that is, at the level of multiple SNPs in multiple dopamine genes. This systems approach had the ability to account for nontrivial variance; 3.9% (corresponding to a correlation of 0.20) of the variance in sensation seeking was explained using only 12 SNPs. Given a heritability of sensation seeking of 58.3% (Fulker & Eysenck, 1980), we were able to account for 6.6% of the heritable variance in sensation seeking. However, our sample was not of sufficient size to allow for cross-validation, and so this effect size estimate may be reduced when replication is attempted. Nevertheless, model fit indices demonstrated that significant independent variance was accounted for by the inclusion of multiple SNPs.
The lack of evidence for linkage disequilibrium accounting for our detection of multiple SNPs within both the COMT and DDC genes associated with sensation seeking suggests that dopamine-related candidate genes likely contain multiple markers that affect sensation seeking, rather than a single SNP (or other individual unit of genetic variation) of relatively large effect which is simply being “tagged” (due to inter-SNP correlations, i.e., linkage disequilibrium) by surrounding SNPs. The apparent overrepresentation of significant SNPs located in the gene DDC in the individual association tests might imply the relevance of production over receptor characteristics in sensation seeking. While our current results suggest this conclusion, it would require replication in future research before being considered a reliable finding. Future research should also include genes corresponding with other candidate systems, such as serotonin-related genes (e.g. Heck et al., 2009), that have also been implicated in the etiology of sensation seeking and associated traits.
Our model of aggregating multiple SNP effects from across genes within a single system is consistent with an additive model of genetic influence (the model of genetic influence employed for heritability estimates of sensation seeking; Fisher, 1918). The aggregation of multiple SNP effects into a genetic risk score is also well-aligned with current thinking on the nature of genetic influence on complex continuous traits. It is likely that numerous (e.g., thousands of) genetic polymorphisms, each with a small effect, contribute to the wide variation in observable human traits (e.g. Maher, 2008). The construction of theory-driven genetic risk scores (as in the candidate system approach demonstrated here) provides a promising direction for predicting phenotypic variation. Future work should focus on refining the genetic risk score, by using larger samples that would allow for greater accuracy in SNP selection and cross-validation.
Supplementary Material
Acknowledgments
Study of Addiction: Genetics and Environment (SAGE). Funding support for the Study of Addiction: Genetics and Environment (SAGE) was provided through the NIH Genes, Environment and Health Initiative [GEI] (U01 HG004422). SAGE is one of the genome-wide association studies funded as part of the Gene Environment Association Studies (GENEVA) under GEI. Assistance with phenotype harmonization and genotype cleaning, as well as with general study coordination, was provided by the GENEVA Coordinating Center (U01 HG004446). Assistance with data cleaning was provided by the National Center for Biotechnology Information. Support for collection of datasets and samples was provided by the Collaborative Study on the Genetics of Alcoholism (COGA; U10 AA008401), the Collaborative Genetic Study of Nicotine Dependence (COGEND; P01 CA089392), and the Family Study of Cocaine Dependence (FSCD; R01 DA013423). Funding support for genotyping, which was performed at the Johns Hopkins University Center for Inherited Disease Research, was provided by the NIH GEI (U01HG004438), the National Institute on Alcohol Abuse and Alcoholism, the National Institute on Drug Abuse, and the NIH contract "High throughput genotyping for studying the genetic contributions to human disease"(HHSN268200782096C).
Collaborative Study on the Genetics of Alcoholism (COGA). The Collaborative Study on the Genetics of Alcoholism (COGA), Co-Principal Investigators B. Porjesz, V. Hesselbrock, H. Edenberg, L. Bierut, includes nine different centers where data collection, analysis, and storage take place. The nine sites and Principal Investigators and Co-Investigators are: University of Connecticut (V. Hesselbrock); Indiana University (H.J. Edenberg, J. Nurnberger Jr., T. Foroud); University of Iowa (S. Kuperman); SUNY Downstate (B. Porjesz); Washington University in St. Louis (L. Bierut, A. Goate, J. Rice); University of California at San Diego (M. Schuckit); Howard University (R. Taylor); Rutgers University (J. Tischfield); Southwest Foundation (L. Almasy). Q. Max Guo is the NIAAA Staff Collaborator. This national collaborative study was supported by the NIH Grant U10AA008401 from the National Institute on Alcohol Abuse and Alcoholism (NIAAA) and the National Institute on Drug Abuse (NIDA). In memory of Henri Begleiter and Theodore Reich, Principal and Co-Principal Investigators of COGA since its inception; we are indebted to their leadership in the establishment and nurturing of COGA, and acknowledge with great admiration their seminal scientific contributions to the field.
Dr. S Saccone was supported by NIH Grant DA024722.
Footnotes
Financial Disclosures. Drs. LJ Bierut and S Saccone are listed as inventors on the patent "Markers for Addiction" (US 20070258898) covering the use of certain SNPs in determining the diagnosis, prognosis, and treatment of addiction. Dr. Bierut has acted as a consultant for Pfizer, Inc. in 2008. All other authors report no competing interests.
We conducted post hoc analyses to examine the generality of the genetic risk score across sensation seeking subscales. Each subscale was residualized over all covariates and then correlated with the overall genetic risk score. Pearson correlations were 0.19, 0.18, 0.14, and 0.12 for disinhibition, boredom susceptibility, thrill and adventure seeking, and experience seeking, with p-values of 8 × 10−7, 7 × 10−6, 4 × 10−4, and 2 × 10−3, respectively. These correlations were within the range of those reported for the total sensation seeking score (i.e., r = 0.20). A reasonable conclusion is that the dopaminergic genetic risk score explained variance in general sensation seeking, rather than variance in only a specific subscale.
Contributor Information
Jaime Derringer, Washington University in St. Louis.
Robert F Krueger, Washington University in St. Louis.
Danielle M Dick, Virginia Institute for Psychiatric and Behavioral Genetics.
Scott Saccone, Washington University in St. Louis.
Richard A Grucza, Washington University in St. Louis.
Arpana Agrawal, Washington University in St. Louis.
Peng Lin, Washington University in St. Louis.
Laura Almasy, Southwest Foundation.
Howard J Edenberg, Indiana University.
Tatiana Foroud, Indiana University.
John I Nurnberger, Jr, Indiana University.
Victor M Hesselbrock, University of Connecticut.
John R Kramer, University of Iowa.
Samuel Kuperman, University of Iowa.
Bernice Porjesz, SUNY Downstate.
Marc A Schuckit, University of California at San Diego.
Laura J Bierut, Washington University in St. Louis.
References
- Akaike H. A new look at the statistical model identification. IEEE Transactions on Automatic Control. 1974;19:716–723. [Google Scholar]
- Ball SA, Carroll KM, Rounsaville BJ. Sensation seeking, substance abuse, and psychopathology in treatment-seeking and community cocaine abusers. Journal of Consulting and Clinical Psychology. 1994;62:1053–1057. doi: 10.1037//0022-006x.62.5.1053. [DOI] [PubMed] [Google Scholar]
- Beaver KM. The interaction between genetic risk and childhood sexual abuse in the prediction of adolescent violent behavior. Sexual Abuse: A Journal of Research and Treatment. 2009;20:426–443. doi: 10.1177/1079063208325204. [DOI] [PubMed] [Google Scholar]
- Benjamin J, Ebstein RP, Belmaker RH. Genes for human personality traits: “Endophenotypes” of psychiatric disorders? World Journal of Biological Psychology. 2001;2:54–57. doi: 10.3109/15622970109027494. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. Journal of the Royal Statistical Society. Series B, Statistical Methodology. 1995;57:289–300. [Google Scholar]
- Bierut LJ, Agrawal A, Bucholz KK, Doheny KF, Laurie C, Pugh E GENEVA Consortium. A genome wide association study of alcohol dependence. Proceedings of the National Academy of Sciences. doi: 10.1073/pnas.0911109107. (under review) http://zork.wustl.edu/gei/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen J. A power primer. Psychological Bulletin. 1992;112:155–159. doi: 10.1037//0033-2909.112.1.155. [DOI] [PubMed] [Google Scholar]
- Dalley JW, Fryer TD, Brichard L, Robinson ES, Theobald DE, Lääne, Robbins TW. Nucleus accumbens D2/3 receptors predict trait impulsivity and cocaine reinforcement. Science. 2007;315:1267–1270. doi: 10.1126/science.1137073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Database of Single Nucleotide Polymorphisms (dbSNP) Bethesda, MD: National Center for Biotechnology Information, National Library of Medicine; (dbSNP Build ID: 129). Available from: http://www.ncbi.nlm.nih.gov/SNP/
- Ebstein RP. The molecular genetic architecture of human personality: beyond self-report questionnaires. Molecular Psychiatry. 2006;11:427–445. doi: 10.1038/sj.mp.4001814. [DOI] [PubMed] [Google Scholar]
- Eisenberg DTA, Campbell B, MacKillop J, Lum JK, Wilson DS. Season of birth and dopamine receptor gene associations with impulsivity, sensation seeking and reproductive behaviors. PLoS One. 2007;2:e1216. doi: 10.1371/journal.pone.0001216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans DM, Visscher PM, Wray N. Harnessing the information contained within genome-wide association studies to improve individual prediction of complex disease risk. Human Molecular Genetics. doi: 10.1093/hmg/ddp295. (in press) [DOI] [PubMed] [Google Scholar]
- Eysenck HJ. A biometrical-genetical analysis of impulsive and sensation seeking behavior. In: Zuckerman M, editor. Biological Bases of Sensation Seeking. Hillsdale NJ: Erlbaum; 1983. pp. 1–27. [Google Scholar]
- Fisher RA. The correlation between relatives on the supposition of Mendelian inheritance. Philosophical Transactions of the Royal Society of Edinburgh. 1918;52:399–433. [Google Scholar]
- Fulker DW, Eysenck SB, Zuckerman M. A genetic and environmental analysis of sensation seeking. Journal of Research in Personality. 1980;14:261–281. [Google Scholar]
- Gottesman II, Gould TD. The endophenotype concept in psychiatry: Etymology and strategic intentions. American Journal of Psychiatry. 2003;160:636–645. doi: 10.1176/appi.ajp.160.4.636. [DOI] [PubMed] [Google Scholar]
- Heck A, Lieb R, Ellgas A, Pfister H, Lucae S, Roeske D, Ising M. Investigation of 17 candidate genes for personality traits confirms effects of the HTR2A gene on novelty seeking. Genes, Brain, and Behavior. 2009;8:464–472. doi: 10.1111/j.1601-183X.2009.00494.x. [DOI] [PubMed] [Google Scholar]
- Hur YM, Bouchard TJ. The genetic correlation between impulsivity and sensation seeking traits. Behavior Genetics. 1997;27:455–463. doi: 10.1023/a:1025674417078. [DOI] [PubMed] [Google Scholar]
- Ioannidis JP, Ntzani EE, Trikalinos TA. ‘Racial’ differences in genetic effects for complex diseases. Nature Genetics. 2004;36:1312–1318. doi: 10.1038/ng1474. [DOI] [PubMed] [Google Scholar]
- Joseph JE, Liu X, Jiang Y, Lynam D, Kelly TH. Neural correlates of emotional reactivity in sensation seeking. Psychological Science. 2009;20:215–223. doi: 10.1111/j.1467-9280.2009.02283.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koopmans JR, Boomsma DI, Heath AC, van Doornen LJP. A multivariate genetic analysis of sensation seeking. Behavior Genetics. 1995;25:349–356. doi: 10.1007/BF02197284. [DOI] [PubMed] [Google Scholar]
- Krueger RF, Markon KE, Patrick CJ, Benning SD, Kramer M. Linking antisocial behavior, substance use, and personality: An integrative quantitative model of the adult externalizing spectrum. Journal of Abnormal Psychology. 2007;116:645–666. doi: 10.1037/0021-843X.116.4.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang UE, Bajbouj M, Sander R, Gallinat J. Gender-dependent association of the functional catechol-O-methyltransferase Val158Met genotype with sensation seeking personality trait. Neuropsychopharmacology. 2007;32:1950–1955. doi: 10.1038/sj.npp.1301335. [DOI] [PubMed] [Google Scholar]
- Laucht M, Becker K, Schmidt MH. Visual exploratory behavior in infancy and novelty seeking in adolescence: Two developmentally specific phenotypes of DRD4? Journal of Child Psychology and Psychiatry. 2006;47:1143–1151. doi: 10.1111/j.1469-7610.2006.01627.x. [DOI] [PubMed] [Google Scholar]
- Laurie CC, Doheny KF, Mirel DB, Pugh EW, Bierut L, Bhangale T GENEVA consortium. Quality control and quality assurance in genotypic data for genome-wide association studies. doi: 10.1002/gepi.20516. (under review) http://www.genevastudy.org/ [DOI] [PMC free article] [PubMed]
- Maher B. Personal genomes: The case of the missing heritability. Nature. 2008;456:18–21. doi: 10.1038/456018a. [DOI] [PubMed] [Google Scholar]
- Meehl PE. High school yearbooks: A reply to Schwarz. Journal of Abnormal Psychology. 1971;77:143–148. doi: 10.1037/h0031999. [DOI] [PubMed] [Google Scholar]
- Meyer GJ, Finn SE, Eyde SE, Kay GG, Moreland KL, Dies RR, Reed GM. Psychological testing and psychological assessment: A review of evidence and issues. The American Psychologist. 2001;56:128–165. [PubMed] [Google Scholar]
- Munafò MR, Yalcin B, Willis-Owen SA, Flint J. Association of the dopamine D4 receptor (DRD4) gene and approach-related personality traits: Meta-analysis and new data. Biological Psychiatry. 2008;63:197–206. doi: 10.1016/j.biopsych.2007.04.006. [DOI] [PubMed] [Google Scholar]
- Netter P, Hennig J, Roed LS. Serotonin and dopamine as mediators of sensation seeking behavior. Neuropsychobiology. 1996;34:155–165. doi: 10.1159/000119318. [DOI] [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nature Genetics. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- Purcell SM, Wray NR, Stone JL, Visscher PM, O'Donovan MC, Sullivan PF, Sklar P. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–752. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2009. http://www.R-project.org. [Google Scholar]
- Reich T, Edenberg HJ, Goate A, Williams JT, Rice JP, Van Eerdewegh P, Begleiter H. Genome-wide search for genes affecting the risk for alcohol dependence. American Journal of Medical Genetics. Part B, Neuropsychiatric Genetics. 1998;81:207–215. [PubMed] [Google Scholar]
- Schwarz G. Estimating the dimensions of a model. Annals of Statistics. 1978;6:461–464. [Google Scholar]
- Slutske WS, Heath AC, Madden PA, Bucholz KK, Statham DJ, Martin NG. Personality and the genetic risk for alcohol dependence. Journal of Abnormal Psychology. 2002;111:124–133. [PubMed] [Google Scholar]
- Staiger PK, Kambouropoulos N, Dawe S. Should personality traits be considered when refining substance misuse treatment programs? Drug and Alcohol Review. 2007;26:17–23. doi: 10.1080/09595230601036952. [DOI] [PubMed] [Google Scholar]
- Wray NR, Goddard ME, Visscher PM. Prediction of individual genetic risk to disease from genome-wide association studies. Genome Research. 2007;17:1520–1528. doi: 10.1101/gr.6665407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zald DH, Cowan RL, Riccardi P, Baldwin RM, Ansari MS, Li R, Shelby ES, Kessler RM. Midbrain dopamine receptor availability is inversely associated with novelty-seeking traits in humans. The Journal of Neuroscience. 2008;28:14372–14378. doi: 10.1523/JNEUROSCI.2423-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuckerman M, Neeb M. Sensation seeking and psychopathology. Psychiatry Research. 1979;1:255–264. doi: 10.1016/0165-1781(79)90007-6. [DOI] [PubMed] [Google Scholar]
- Zuckerman M. Sensation seeking: A comparative approach to a human trait. The Behavioral and Brain Sciences. 1984;7:413–471. [Google Scholar]
- Zuckerman M. Behavioral Expressions and Biosocial Bases of Sensation Seeking. Cambridge UK: Cambridge University Press; 1994. Smoking, drinking, drugs, and eating; pp. 225–257. [Google Scholar]
- Zuckerman M, Eysenck S, Eysenck HJ. Sensation seeking in England and America: Cross-cultural, age, and sex comparisons. Journal of Consulting and Clinical Psychology. 1984;7:413–471. doi: 10.1037//0022-006x.46.1.139. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.