TO THE EDITOR
Xeroderma pigmentosum (XP) is an inherited human disorder in which patients have greatly elevated incidence of solar-induced cancers caused by mutations in genes for nucleotide excision repair or polymerase η that are required for repair or replication of damaged DNA (Hoeijmakers, 2001; Wood et al., 2001; Cleaver and Mitchell, 2006). We report a cluster of XP-C patients within an isolated Guatemala community containing a new mutation. A recently diagnosed US XP-C patient was also analyzed for comparison and found to be a compound heterozygote for two novel mutations in the XPC gene.
A Guatemala village containing many XP patients was identified by missionaries from Good Samaritans International, in consultation with physicians in Guatemala City. Among a total of about 50 children, approximately 12 had evidence of photosensitivity, severe poikloderma, and multiple non-melanoma skin cancers. Their tumors were often advanced, showing necrosis and infection and the children suffered continual pain from sun exposure. There was no evidence of neurological symptoms seen in the XP patients identified, such as hearing loss and ataxia (Cleaver and Mitchell, 2006). However, the majority of the affected children do not live beyond their 10th birthday and later findings of neurological degeneration would not be identified. There were no reported skin cancers among parents. According to oral tradition, the village originated in a common family approximately 100 years ago. Intermarriage is high and we hypothesized that the XP patients would be homozygous for the same mutation in an XP gene (see Table 1).
Table 1.
Sensitivity to UV light and mutations identified in candidate XP cell lines
| Designation | Status/origin | Age (years) |
D501 (Jm−2) |
Allele 1 | Allele 2 |
|---|---|---|---|---|---|
| XP202BA | Proband (Guatemala) | 8 | 1.2 (n=4) | Exon 8, C940del-1, 969 stop | Same |
| XP211BA | Father of XP202BA | Adult | 6.6 (n=1) | Exon 8, C940del-1, 969 stop | Wild-type |
| XP212BA | Father of another XP child | Adult | 7.5 (n=1) | Exon 8, C940del-1, 969 stop | Wild-type |
| XP207SF (UDS 32.1%)2 | Proband (UCSF) | 2 | 0.65 (n=3) | Exon 3, A362del-1, 438 stop | Exon 4, GA444, 445 del-2, 498 stop |
| XP9BE3 (UDS 10–20%)2 | Coriell repository | 16 | 1.2 (n=1) | Exon 11, GTG2092ins3 | Same and Exon 16, A2815C |
| Normal (PPO34) | UCSF | Adult | 8.2 (n=4) | Wild-type | Wild-type |
UCSF, University of California San Francisco; UDS, unscheduled DNA synthesis; XP, xeroderma pigmentosum.
All nucleotide numbers refer to the coding DNA sequence (CDS) described in Genbank NM_004628 in which exons are numbered sequentially 1–16.
The UV survival curves from these lymphoid cells were exponential (linear on a semi-log plot). The UV sensitivity is therefore unambiguously defined by the slope, which is designated here by the dose for 50% survival. The survival curves are represented by the formula: “Relative survival=exp(−D/1.443D50)”, for the relative survival at dose D where D50 is the dose required for 50% survival in units of Jm−2. The value “n” represents the number of independent survival curves determined to obtain the D50 value.
Unscheduled DNA synthesis (Robbins et al., 1974). These values are typical of XP-C and we expect the Guatemalan cells to be similar (Cleaver and Mitchell, 2006).
This insertion was previously reported and confirmed by our sequencing. We also identified the non-conservative A2815C (lysine to glutamine) in exon 16 in one allele that had been previously reported in this cell line as A2920C in exon 15 using nomenclature in which our exons 5 and 6 is annotated as exons 5.1 and 5.2 (Khan et al., 2000, 2002).
Blood samples were obtained from a male patient (XP202BA), his father (XP211BA), and one other father (XP212BA) of an XP patient who was not directly related. Blood samples were also obtained from a newly diagnosed US patient seen at the University of California San Francisco (UCSF) (XP207SF). This patient was a 2-year-old female born of non-consanguineous parents who had extensive facial freckling and a small squamous cell carcinoma on the bridge of the nose that was biopsied for diagnosis. The purpose of the sample collection was carefully explained to donors and parents under the terms of a current UCSF Committee on Human Research permit and the Declaration of Helsinki Principles. Parents of subjects consented to blood draws in Guatemala via translators in the presence of the local health official and documentation was retained by their physicians. Lymphocytes were transformed by Epstein–Barr virus infection and permanent lymphoblastoid cell lines were established and given UCSF designations. A previously classified XP-C lymphoid cell line (GM02498) was obtained from the Coriell Cell Repository (Camden, NJ) and a normal cell line (PPO34) from a UCSF bank. Fibroblast cultures were obtained from the UCSF patient and for GM02498 (GM00676 or XP9BE) (Robbins et al., 1974), but could not be obtained for the Guatemalan patients. UV sensitivity was determined by irradiation with increasing doses of UV light (254 nm, 1.3 Jm−2 second), growth for 7 days, followed by counting the total surviving cell number, a method previously shown equivalent to colony formation (Cleaver and Thomas, 1988). From the survival curve, the values of the dose for 50% survival was determined (Table 1). The UV sensitivities of the XP202BA and XP207SF cell lines were close to that of the standard XP-C cell line (GM02498) (see D50 values in Table 1). These sensitivities combined with the absence of neurological symptoms were suggestive of XP group C.
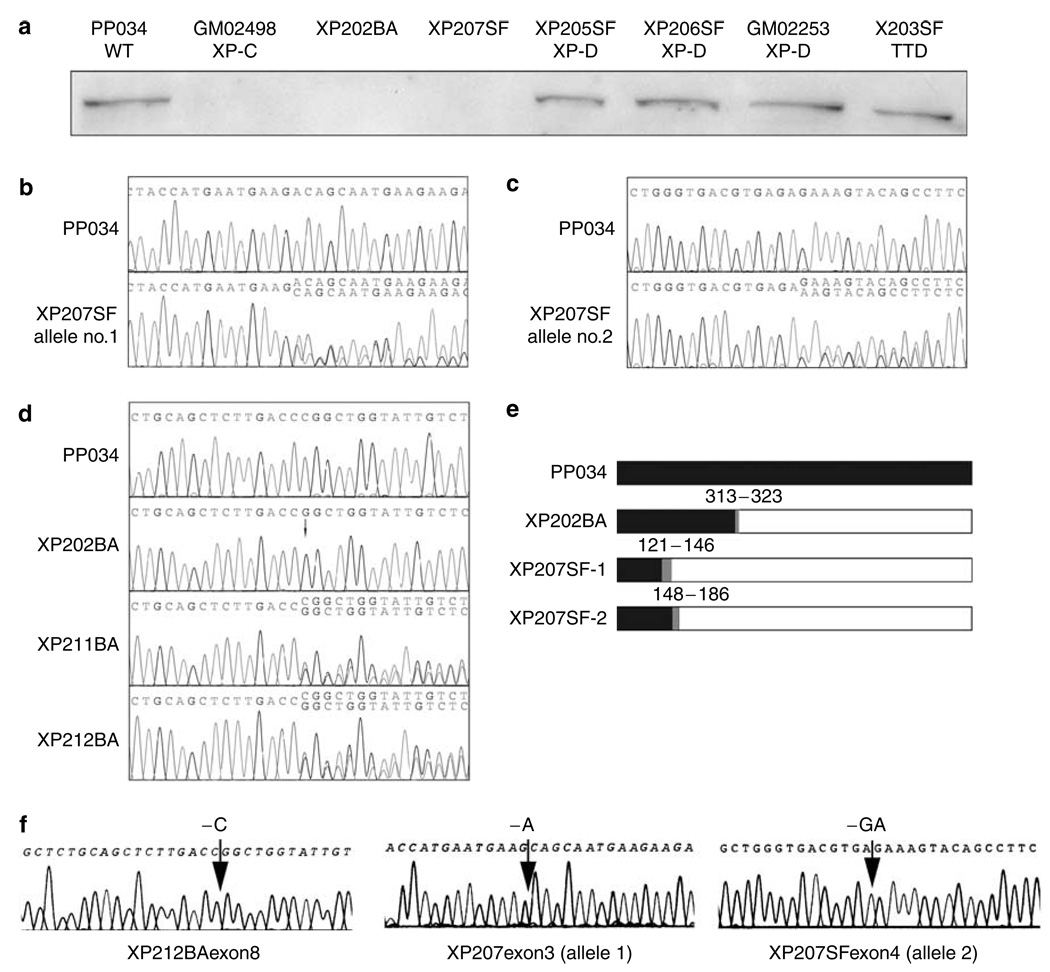
Western blot analysis was carried out using an antibody to XPC (Abcam ab6264, Abcam, Cambridge, MA) followed by anti-mouse antibody conjugated to horseradish peroxidase. XP202BA, XP207SF, and GM02498 lacked detectable full-length protein, whereas similar protein levels were seen in normal and XP-D cell lines (Figure 1a). Immunohistochemical analysis of a fixed tumor from another Guatemalan patient was also negative for XPC (to be reported later).
Figure 1. Protein expression and sequence analysis of XPC.
(a) Western analysis of cell extracts probed by anti-XPC mAb (Abcam ab6264). Cell line complementation groups designated where known. (b) DNA sequence within the regions of XPC exon 3 for wild-type PP034 and XP207SF showing Adel and wild-type sequence overlapping downstream of the deletion (accession number DQ076467). (c) DNA sequence within the regions of XPC exon 4 for wild-type PP034 and XP207SF showing GAdel and wild-type sequence overlapping downstream of the deletion (accession number DQ076476). (d) DNA sequence within the regions of XPC exon 8 showing C940del. Top to bottom: wild-type, XP202BA, XP211BA, XP212BA. The presence of two sequences can be seen as overlapping sequences downstream from the deletion in both of the heterozygous samples (accession number DQ076464). (e) Predicted translation products in XPC showing normal sequences (black), frameshift (gray), and amino-acid sites of mutations and downstream termination for XP202BA and XP207SF. (f) Subcloned sequences showing mutations from XP211BA exon 8, XP207SF exon 3, and XP207SF exon 4. Subcloned exon sequences identified mutations in five of nine clones for XP207SF exon 3; three of six clones in XP207SF exon 4; one of six in XP211BA; and two of six in XP211BA.
The XPC gene was sequenced using the PCR-ready primer pairs (Variant-SEQr) from Applied Biosystems (AB, Foster City, CA) that amplified each exon and flanking regions from genomic DNA and data were analyzed using their custom software (Table 1). Direct sequencing of the PCR products from the UCSF patient, XP207SF, detected a complex heterozygote with two different mutations: one was in exon 3 (a single base pair deletion) (Figure 1b) and the other in exon 4 (a two base pair deletion) (Figure 1c), resulting in overlapping sequences downstream of the mutations. These both resulted in chain terminations and predicted truncated protein products (Figure 1e). No other mutations were detected, so we assume that these two mutations occurred in the two different alleles. Direct sequencing of XP202BA identified a deletion of a single base pair in exon 8, with no downstream overlapping sequences, that caused a frameshift and premature termination (Figure 1d and e).
The presence of a single mutant sequence in DNA from the proband could indicate homozygosity or hemizygosity for the -c deletion. We resolved these alternatives by determining the copy numbers of the wild-type and deletion sequences using real-time PCR (Taqman analysis). Probes were designed to detect the wild-type (-ccc-) and deleted (-cc-) sequences in exon 8, and copy numbers determined against a pool of ovarian DNA (Table 2). As the target sequences differed by only one base, discrimination of wild-type and mutant sequences was not complete, but adequate to distinguish the two alternatives. The normal control, PPO34, and XP207SF both showed high relative copy numbers for the normal sequence, as expected for diploid DNA content. The heterozygote XP211BA showed a similar copy number for mutant and wild-type sequences. XP202BA showed a signal for mutant sequence approximately double that of the heterozygote, and low signal for wild-type sequence. This result is consistent with the proband containing two copies of the mutant sequence, and not with a complete deletion of one allele. The proband is therefore homozygous for the -c deletion in exon 3.
Table 2.
Relative copy number of exon 3 sequences normalized to an ovarian DNA pool
| Mutant sequence -cc- | Wild-type sequence -ccc- | |||
|---|---|---|---|---|
| Cell line | Observed | Ratios1 | Observed | Ratios1 |
| Normal (PPO34) | 0.11 | 0.35 | 0.76 | 1.9 |
| XP207SF | 0.12 | 0.39 | 0.78 | 2.0 |
| XP211BA | 0.31 | 1 | 0.4 | 1 |
| XP202BA | 0.59 | 1.9 | 0.07 | 0.18 |
Normalized to heterozygote values of 1.0 for each allele.
The deletion was present in the heterozygous form in both fathers of XP children resulting in overlapping sequences downstream of the deletion (Figure 1d). As the mutations could not be unambiguously identified from the sequence of the PCR products when both a wild-type and mutant sequence were present, we subcloned the PCR products via TA-TOPO cloning (Invitrogen, Carlsbad, CA) and resequenced to confirm the mutations (Figure 1f). The mutations in XP202BA and XP207SF have not been reported before (Chavanne et al., 2000; Khan et al., 2002, 2004, 2006), and each cause loss of protein and consequent failure in nucleotide excision repair. The novelty of the Guatemalan mutation is, however, insufficient to decide whether the XPC mutation is newly derived or of immigrant origin.
The high incidence of XP in this village is most likely due to the immigration of a single individual carrying the –c deletion that then spread through the population owing to intermarriage. A similar “founder effect” has been reported for XP elsewhere. XP patients in Tunisia show homozygosity for an exon 6 mutation, R228ter, in XPA (Nishigori et al., 1993). Many XP patients in Japan have the same splice site mutation in XPA (Satokata et al., 1992). Two families in Italy and Turkey were found to have common descent with the same mutation in XPC (Gozukara et al., 2001).
This disease presents a major health crisis to an extremely isolated community. They are subsistence coffee farmers, very distant from health services, and bringing care to the patients and families presents major difficulties. The clinical conditions are severe, with life-threatening cancers and associated infectious disease, such that patients die in their early teenage years. This is in contrast to XP-C patients in the USA who can modify their life-styles to avoid sun exposure and achieve a near normal lifespan.
ACKNOWLEDGMENTS
The work was supported by the National Institutes of Environmental Health Sciences Grant 1 RO1 ES 8061. We thank the XP Society, Poughkeepsie, NY, for their continued support (JEC). We also thank Dr R. Skurat for initial sequencing, Dr A.R. Lehmann for carrying out the unscheduled DNA synthesis measurements on XP207SF who graciously allowed us to cite his data. We thank Drs David Ginzinger, John Weger, and Kirsten Copren, Molecular Diagnostics, UCSF Cancer Center for assistance in DNA sequencing and Taqman analysis. The NCBI accession numbers are DQ076457 through DQ076480.
Abbreviations
- CSF
University of California San Franciso
- XP
xeroderma pigmentosum.
Footnotes
CONFLICT OF INTEREST
The authors state no conflict of interest.
REFERENCES
- Chavanne F, Broughton BC, Pietra D, Nardo T, Browitt A, Lehmann AR, et al. Mutations in the XPC gene in families with xeroderma pigmentosum and consequences at the cell, protein, and transcript levels. Cancer Res. 2000;60:1974–1982. [PubMed] [Google Scholar]
- Cleaver JE, Mitchell DL. Ultraviolet radiation carcinogenesis. In: Kufe DW, Bast RC Jr, Hait WN, Kong WK, Pollock RE, Weichselbaum RR, Holland JF, Frei E III, editors. Cancer Medicine. 7th edn. Hamilton-London: BC Decker, Inc.; 2006. pp. 283–291. [Google Scholar]
- Cleaver JE, Thomas GH. Rapid diagnosis of sensitivity to ultraviolet light in fibroblasts from dermatological disorders with particular reference to xeroderma pigmentosum. J Invest Dermatol. 1988;90:467–471. doi: 10.1111/1523-1747.ep12460917. [DOI] [PubMed] [Google Scholar]
- Gozukara EM, Khan SG, Metin A, Emmert S, Busch DB, Shahlavi T, et al. A stop codon in xeroderma pigmentosum group C families in Turkey and Italy: molecular genetic evidence for a common ancestor. J Invest Dermatol. 2001;117:197–204. doi: 10.1046/j.1523-1747.2001.01424.x. [DOI] [PubMed] [Google Scholar]
- Hoeijmakers JH. Genome maintenance mechanisms for preventing cancer. Nature. 2001;411:366–374. doi: 10.1038/35077232. [DOI] [PubMed] [Google Scholar]
- Khan SG, Metin A, Gozukara E, Inui H, Shahlavi T, Muniz-Medina V, et al. Two essential splice lariat branchpoint sequences in one intron in a xeroderma pigmentosum DNA repair gene: mutations result in reduced XPC mRNA levels that correlate with cancer risk. Hum Mol Genet. 2004;13:343–352. doi: 10.1093/hmg/ddh026. [DOI] [PubMed] [Google Scholar]
- Khan SG, Metter EJ, Tarone RE, Bohr VA, Grossman L, Hedayati M, et al. A new xeroderma pigmentosum group C poly(AT) insertion/deletion polymorphism. Carcinogen. 2000;21:1821–1825. doi: 10.1093/carcin/21.10.1821. [DOI] [PubMed] [Google Scholar]
- Khan SG, Muniz-Medina V, Shahlavi T, Baker CC, Inui H, Ueda T, et al. The human XPC DNA repair gene: arrangement, splice site information content and influence of a single nucleotide polymorphism in a splice acceptor site on alternative splicing and function. Nucleic Acids Res. 2002;30:3624–3631. doi: 10.1093/nar/gkf469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan SG, Oh KS, Shahlavi T, Ueda T, Busch DB, Inui H, et al. Reduced XPC DNA repair gene mRNA levels in clinically normal parents of xeroderma pigmentosum patients. Carcinogen. 2006;27:84–94. doi: 10.1093/carcin/bgi204. [DOI] [PubMed] [Google Scholar]
- Nishigori C, Zghal M, Yagi T, Imamura S, Komoun MR, Takebe H. High prevalence of point mutations in exon 6 of xeroderma pigmentosum group A-complementing (XPAC) gene in xeroderma pigmentosum group A patients in Tunisia. Am J Hum Genetics. 1993;53:1001–1006. [PMC free article] [PubMed] [Google Scholar]
- Robbins JH, Kraemer KH, Lutzner MA, Festoff BW, Coon HG. Xeroderma pigmentosum: an inherited disease with sun sensitivity, multiple cutaneous neoplasms, and abnormal DNA repair. Ann Inter Med. 1974;80:221–248. doi: 10.7326/0003-4819-80-2-221. [DOI] [PubMed] [Google Scholar]
- Satokata I, Tanaka K, Yuba S, Okada Y. Identification of splicing mutations of the last nucleotide of exons, a nonsense mutation, and a missense mutation of the XPAC gene as causes of group A xeroderma pigmentosum. Mutat Res. 1992;273:203–212. doi: 10.1016/0921-8777(92)90081-d. [DOI] [PubMed] [Google Scholar]
- Wood RD, Mitchell M, Sgouros J, Lindahl T. Human DNA repair genes. Science. 2001;291:1284–1289. doi: 10.1126/science.1056154. [DOI] [PubMed] [Google Scholar]