Abstract
Lipoproteins are a largely uncharacterized class of proteins in bacteria. In this study, metabolic labeling of bacteria with fatty acid chemical reporters allowed rapid profiling of lipid-modified proteins. We identified many candidate lipoproteins in Escherichia coli and detected a novel modification on YjgF. This chemical approach should facilitate future characterization of lipoproteins.
Bacterial lipoproteins (LPPs) are a functionally diverse subset of membrane-associated proteins that are involved in various cellular processes including membrane maintenance, transport, and signal transduction1. A number of LPPs have also been implicated in the virulence mechanisms of bacterial pathogens and are recognized by pattern recognition receptors of host immune systems2, 3, making them attractive targets for antibiotic4 and vaccine development. Despite their clinical relevance and role in numerous membrane processes, many LPPs remain uncharacterized. This is due, in part, to a lack of robust and convenient tools for the detection and large-scale analysis of LPPs in bacteria.
Existing methods available for identification and analysis of bacterial LPPs include radiolabeling and bioinformatic approaches, both of which present limitations. Labeling with radioactive fatty acids has low sensitivity and is not amenable to large-scale proteomic studies. Bioinformatic predictions are limited to lipoproteins containing a recognizable N-terminal lipobox signal motif5. Although this motif directs the canonical form of protein lipidation in Gram-negative bacteria (Figure 1A), alternative mechanisms of lipidation have been reported6. The prevalence of such non-canonical mechanisms of lipidation is not known, making proteomic analysis essential for exploring the diversity of lipid-modified proteins in bacteria. Herein, we demonstrate that metabolic labeling of bacteria with fatty acid reporters provides an efficient and robust chemical approach for the detection and discovery of bacterial lipoproteins.
Figure 1.
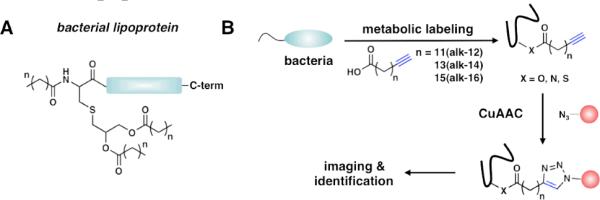
A) Structure of a bacterial lipoprotein. (n = 12 – 20). B) Two-step bioorthogonal labeling strategy using alkynyl-fatty acids.
To explore the utility of fatty acid chemical reporters in bacteria, we metabolically labeled E. coli K12 with alkynyl-fatty acid (alk-FA) reporters of varying chain lengths (alk-12, alk-14, and alk-16)7 and reacted bacterial cell lysates with azido-rhodamine (az-Rho) (Figure S1) via Cu(I)-catalyzed azide-alkyne cycloaddition (CuAAC)8 for in-gel fluorescence profiling (Figure 1B). While this approach has been utilized for the visualization and enrichment of lipidated proteins in mammalian cells9 as well as the functionalization of an overexpressed eukaryotic N-myristoylated protein in bacteria10, fatty acid reporters have not been used to explore the diversity of endogenous bacterial LPPs. Because the enzymatic machinery required for protein lipidation is different in bacteria from eukaryotes1, 9, we evaluated the utility of alk-FA reporters for the rapid detection and profiling of bacterial LPPs. Fluorescence visualization of alk-FA-modified proteins revealed selective- and chain-length-dependent labeling of proteins (Figure 2A). Since alk-14 afforded the most efficient labeling of E. coli proteins, we focused on characterizing this fatty acid reporter for subsequent experiments. Protein labeling with alk-14 was rapid, dose-dependent, and not toxic to bacteria (Figure S2). Importantly, alk-14 specifically labels a known bacterial LPP, NlpE11, as judged by in-gel fluorescence visualization of a purified C-terminal His6-tagged construct (NlpE-His6) (Figure 2B).
Figure 2.
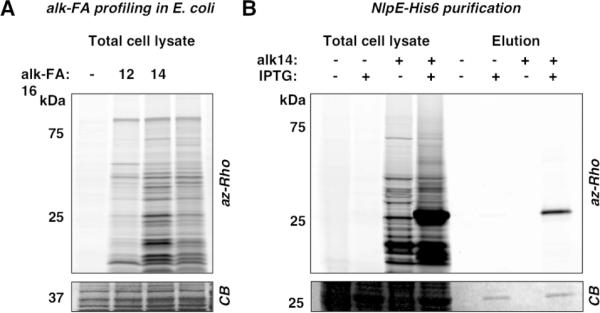
A) Profiling of E. coli lipoproteins with alk-FA reporters. B) Specific labeling of NlpE-His6 with alk-14. Coomassie blue staining (CB) indicates level of protein loading.
For the large-scale retrieval and identification of fatty acid modified proteins, alk-14-labeled E. coli cell lysates were reacted with a cleavable azido-diazo-biotin affinity tag12 (Figure S1). Biotinylated proteins were then enriched with streptavidin beads, selectively eluted with sodium dithionite (Na2S2O4), and separated by SDS-PAGE for gel-based proteomics. As a positive control for LPP recovery, we labeled the E. coli strain expressing NlpE-His6 (Figure S4). Visualization of the Na2S2O4-eluted proteins showed that NlpE-His6 was selectively recovered in the IPTG-induced samples (Figure S4). Analysis of three independent runs revealed ~44 high confidence (≥10 peptide spectral counts and 10-fold enrichment above the unlabeled control) LPP candidates, of which 70% are known or predicted LPPs, based on annotation in DOLOP5 (Figure 3A, Table S1). An additional ~42 candidates were identified with medium confidence (≥5 peptide spectral counts and 5-fold enrichment above the unlabeled control) (Table S1). Functional annotation of the high confidence candidates revealed that 96% of the unannotated proteins were LPPs (Figure 3A). As expected, NlpE was substantially enriched in the IPTG-induced samples (Table S1). Other known fatty acid modified proteins such as acyl-carrier protein (AcpP) were also selectively recovered (Table S1). An alkynyl-acetic acid analog, alk-2, did not efficiently label bacterial proteins by fluorescence profiling or gel-based proteomics, demonstrating LPP labeling is specific to alk-14 and not due to general labeling of proteins by alkynyl-functionalized substrates (Figure S4, Table S1).
Figure 3.
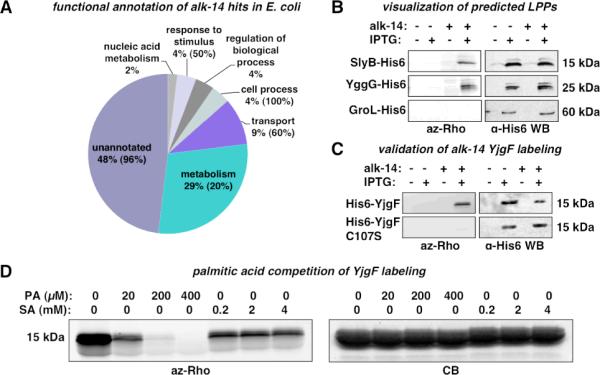
Validation of alk-FA labeled proteins in bacteria. A) GO annotation of high confidence candidates identified from E. coli. Percent LPPs in each category is indicated in parenthesis. B. alk-14 labeling of SlyB and YggG. C) alk-14 labeling of YjgF and YjgFC107S. D) Competition of alk-14 labeling of YjgF with palmitic acid (PA) and sodium acetate (SA).
We then validated alk-14 labeling of proteins that were selectively recovered from E. coli. Labeling of SlyB, a known LPP, and YggG, a predicted but unvalidated LPP, was confirmed by fluorescence detection of C-terminal His6-tagged constructs after affinity purification (Figure 3B). Non-specific labeling due to over-expression is unlikely since no labeling was observed for the abundant and non-lipidated protein, GroL-His6 (Figure 3B). Interestingly, a 14 kDa bacterial protein that does not carry a lipobox motif, YjgF13, was considerably enriched in alk-14-labeled samples (Table S1). To validate the alk-14 labeling of YjgF, we generated an N-terminal His6-tagged YjgF construct (His6-YjgF) and confirmed covalent modification with alk-14 by in-gel fluorescence detection (Figure 3C). YjgF contains one Cys residue at position 107 as a potential site of modification that is conserved in most bacterial homologs of YjgF. Analysis of a Cys107 to Ser construct (His6-YjgF-C107S) revealed that alk-14 labeling is abrogated in this mutant, suggesting that YjgF is modified at Cys107 (Figure 3C).
To determine whether the alk-14 labeling we observe on YjgF is reflective of fatty acid modification, we performed competition experiments with palmitic acid or sodium acetate (Figure 3D). In-gel visualization of affinity purified His6-YjgF reveals that alk-14 labeling is effectively competed away with increasing amounts of palmitic acid, but not with sodium acetate (Figure 3D, Figure S5). Analysis of YjgF and YjgFC107S mutant did not reveal a difference in the partitioning between membrane and cytosolic fractions, suggesting that Cys107 does not influence the membrane association of YjgF (Figure S6). These results suggest that Cys107 of YjgF is covalently modified with a fatty acid metabolite, but does not play a role in subcellular distribution of YjgF (Figure S6).
The precise function of YjgF and its homologs is unknown, but they have been implicated in the regulation of isoleucine biosynthesis14. Structural and biochemical studies have demonstrated that YjgF13 and its homologs form trimers and bind metabolites associated with isoleucine biosynthesis15, 16. X-ray crystallography of YjgF has suggested that Cys107 is modified by a phosphate group based on electron density13, but this modification has not been validated biochemically or genetically, and has not been recapitulated with other homologs15, 16. These biochemical and genetic studies suggest that the YjgF protein family is involved in amino acid metabolism. We discovered that YjgF is covalently labeled with fatty acid metabolites using alk-14, which may reflect a coupling of fatty acid metabolism to amino acid synthesis that warrants future investigation.
In this study, we demonstrate that fatty acid chemical reporters allow sensitive visualization and large-scale identification of LPPs in Gram-negative bacteria. This approach is quite general and can be applied to a number of different bacterial species, including Gram-positive bacteria and mycobacteria (Figure S7). Alkynyl-fatty acid reporters therefore represent robust chemical tools for the characterization and exploration of LPPs and non-canonical fatty acid modification of proteins in bacteria.
Acknowledgement
We thank The Rockefeller University Proteomics Resource Center for mass spectrometry analysis. Y-Y.Y. was supported in part by The Anderson Cancer Center postdoctoral fellowship. G.C. acknowledges the Weill-Cornell/Sloan-Kettering/Rockefeller Tri-institutional Program in Chemical Biology. H.C.H. acknowledges support from The Rockefeller University, Irma T. Hirschl/Monique Weill-Caulier Trust and Ellison Medical Foundation.
Footnotes
Supporting Information Available: Experimental Methods, Figures S1–S7, Table S1. This material is available free of charge via the Internet at http://pubs.acs.org
References
- 1.Tokuda H. Biosci. Biotechnol. Biochem. 2009;73(3):465–473. doi: 10.1271/bbb.80778. [DOI] [PubMed] [Google Scholar]
- 2.Moody DB, Young DC, Cheng TY, Rosat JP, Roura-Mir C, O'Connor PB, Zajonc DM, Walz A, Miller MJ, Levery SB, Wilson IA, Costello CE, Brenner MB. Science. 2004;303(5657):527–31. doi: 10.1126/science.1089353. [DOI] [PubMed] [Google Scholar]
- 3.Takeuchi O, Akira S. Microbes Infect. 2002;4(9):887–95. doi: 10.1016/s1286-4579(02)01615-5. [DOI] [PubMed] [Google Scholar]
- 4.Pathania R, Zlitni S, Barker C, Das R, Gerritsma DA, Lebert J, Awuah E, Melacini G, Capretta FA, Brown ED. Nat Chem Biol. 2009;5(11):849–56. doi: 10.1038/nchembio.221. [DOI] [PubMed] [Google Scholar]
- 5.Babu MM, Priya ML, Selvan AT, Madera M, Gough J, Aravind L, Sankaran K. Journal of Bacteriology. 2006;188(8):2761–73. doi: 10.1128/JB.188.8.2761-2773.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Issartel JP, Koronakis V, Hughes C. Nature. 1991;351(6329):759–61. doi: 10.1038/351759a0. [DOI] [PubMed] [Google Scholar]
- 7.Charron G, Zhang MM, Yount JS, Wilson J, Raghavan AS, Shamir E, Hang HC. J Am Chem Soc. 2009;131(13):4967–75. doi: 10.1021/ja810122f. [DOI] [PubMed] [Google Scholar]
- 8.Meldal M, Tornøe CW. Chem Rev. 2008;108(8):2952–3015. doi: 10.1021/cr0783479. [DOI] [PubMed] [Google Scholar]
- 9.Charron G, Wilson J, Hang HC. Curr Opin Chem Biol. 2009 doi: 10.1016/j.cbpa.2009.07.010. [DOI] [PubMed] [Google Scholar]
- 10.Heal WP, Wickramasinghe SR, Bowyer PW, Holder AA, Smith DF, Leatherbarrow RJ, Tate E. Chem Commun (Camb) 2008;(4):480–2. doi: 10.1039/b716115h. [DOI] [PubMed] [Google Scholar]
- 11.Hirano Y, Hossain MM, Takeda K, Tokuda H, Miki K. Structure. 2007;15(8):963–76. doi: 10.1016/j.str.2007.06.014. [DOI] [PubMed] [Google Scholar]
- 12.Yang Y-Y, Ascano JM, Hang HC. J. Am. Chem. Soc. 2010 doi: 10.1021/ja908871t. ASAP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Volz K. Protein Sci. 1999;8(11):2428–37. doi: 10.1110/ps.8.11.2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Christopherson MR, Schmitz GE, Downs DM. Journal of Bacteriology. 2008;190(8):3057–62. doi: 10.1128/JB.01700-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Parsons L, Bonander N, Eisenstein E, Gilson M, Kairys V, Orban J. Biochemistry. 2003;42(1):80–9. doi: 10.1021/bi020541w. [DOI] [PubMed] [Google Scholar]
- 16.Burman JD, Stevenson CE, Sawers RG, Lawson DM. BMC Struct Biol. 2007;7:30. doi: 10.1186/1472-6807-7-30. [DOI] [PMC free article] [PubMed] [Google Scholar]


