Abstract
Current models for cleavage plane determination propose that metaphase spindles are positioned and oriented by interactions of their astral microtubules with the cellular cortex, followed by cleavage in the plane of the metaphase plate [1, 2]. We show that in early frog and fish embryos, where cells are unusually large, astral microtubules in metaphase are too short to position and orient the spindle. Rather, the preceding interphase aster centers and orients a pair of centrosomes prior to nuclear envelope breakdown, and the spindle assembles between these prepositioned centrosomes. Interphase asters center and orient centrosomes using dynein-mediated pulling forces. These forces act before astral microtubules contact the cortex; thus, dynein must pull from sites in the cytoplasm, not the cell cortex as is usually proposed for smaller cells. Aster shape is determined by interactions of the expanding periphery with the cell cortex, or with an interaction zone that forms between sister-asters in telophase. We propose a model to explain cleavage plane geometry in which the length of astral microtubules is limited by interaction with these boundaries, causing length asymmetries. Dynein anchored in the cytoplasm then generates length–dependent pulling forces, which move and orient centrosomes.
Results and discussion
Cleavage furrows initiate at a position equidistant between two radial arrays of microtubules (asters), which grow out from sister centrosomes at the end of mitosis. The position of these asters depends on the prior position of the metaphase spindle. In typical animal cells, the mitotic spindle is positioned in the center of the cell, and oriented parallel to the cell's long axis, by forces acting on its astral microtubules during metaphase [1, 2]. In frog eggs, the sperm enters on the side; its centrosome nucleates a huge sperm aster. This aster captures the female pronucleus, and moves it, together with the male pronucleus and centrosomes, to a position near the cell center, where the first mitotic spindle assembles. The spindle is small compared to cell size, and its astral microtubules are too short to contact the cortex in metaphase [3]. The first cleavage plane tends to cut through the sperm entry point [4]. The reason for this is unclear. (For an overview of microtubule organization in early frog embryos see Fig. S4 A)
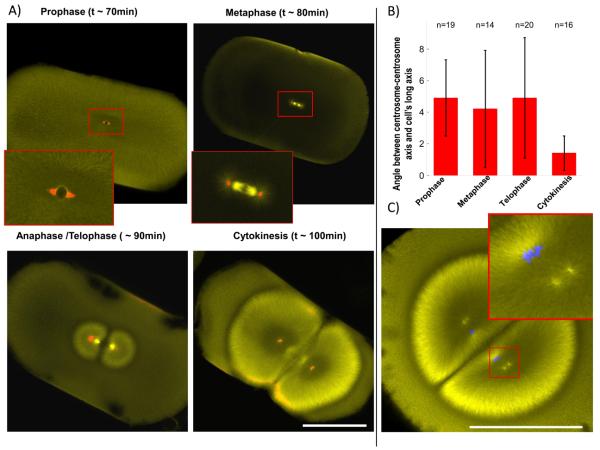
To determine when the orientation of the first cleavage plane is established, we repeated Hertwig's experiment of compressing the embryo, which imposes a cleavage plane normal to the long axis of the cell [5]. Shortly after fertilization embryos of the African clawed frog Xenopus laevis were compressed between glass slides. By fixing at different times we asked when the spindle axis is determined (Fig. 1 A, B). Immunofluorescence staining of α-tubulin and γ-tubulin was used to distinguish cell cycle stages, and to measure the angle between sister centrosomes and the imposed long axis of the cell. Already in prophase, before nuclear envelope breakdown, the intercentrosome axis was accurately positioned, differing from the long axis by only 4.9°± 2.4 (compare to random orientation of 45°). This orientation did not improve significantly in metaphase (4.2° ± 3.7, p=0.5, Student's t-test). Between anaphase and cytokinesis alignment improved significantly (1.4° ± 1.1, p=0.001), presumably because the expanding asters, which we will call telophase asters, begin to contact the cortex and thus sense cellular shape. However, the sister-asters are only able to fine-tune the angle of cleavage, and not completely reorient it (Fig. S1 A and previous work [6]). Cleavage planes were oriented with an average of 86.1° ± 2.8 relative to the artificial long axis, showing that cleavage planes accurately respect the centrosome orientation imposed before mitosis. To ask why cleavage planes tend to cut through the sperm entry point in unperturbed embryos [4], we performed immunofluorescence after fertilization. We observed that sperm aster expansion is limited by the nearest cortex (i.e. the cortex near where the sperm entered), resulting in an aster with an oblate ellipsoid shape, with its long axis parallel to a tangent to the cortex at the sperm entry point (Fig. S1 B). In favourable images, we could visualize paired centrosomes already oriented along this axis by ~35min post fertilization. We propose that the centrosomes preserve this orientation at the center, which serves to orient the 1st mitotic spindle, and in turn to orient the 1st cleavage plane. The 2nd cleavage plane is orthogonal to the first. When we visualized centrosomes in telophase of first mitosis in Xenopus, we found that sister centrosomes are already oriented orthogonal to the first mitotic spindle before astral microtubules have reached the cortex, which is long before nuclear envelope break down for 2nd mitosis (Fig. 1 C). In summary, the positions and orientations of both the 1st and 2nd mitotic spindles are determined by the position of centrosome pairs before mitosis onset, which in turn are determined by the behaviour of centrosomes inside interphase (or telophase) asters. (In early embryos, where there is no G1 or G2, interphase and telophase are equivalent)
Figure 1. Spindles are positioned and oriented by asters prior to mitosis onset.
A) Frog embryos were artificially elongated by compression and fixed at different time points. Immunofluorescence against α- (yellow) and γ-tubulin (red) allows scoring of centrosome orientation and cell cycle stage. Note centrosomes are already aligned in prophase, before nuclear envelope breakdown. B) Quantification of average centrosome orientation relative to longest cell axis (Angle measured between 0° and 90°; random orientation would be 45°) Centrosomes are already well aligned (4.9 °± 2.4 SD) before nuclear envelope breaks down, as soon as they can be visualized with γ-tubulin staining. Alignment does not improve significantly in metaphase (4.2 °± 3.7) or anaphase/telophase (4.9 °± 3.8). Once the expanding telophase asters touch the cortex, just before cytokinesis, centrosome alignment is fine-tuned significantly (1.4 °± 1.1) C) α- tubulin (yellow) immunofluorescence in a frog embryo at anaphase-telophase of first mitosis, before cytokinesis. Duplicated centrosomes are already aligned parallel to the longest axes of the daughter cells. DNA (blue) follows centrosomes. Bars = 500 μm. See also Figure S1.
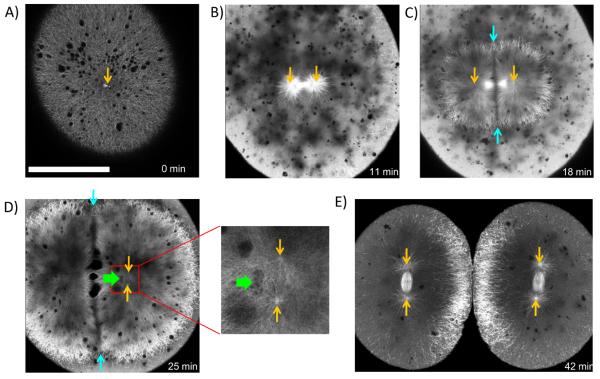
Internal imaging of amphibian embryos requires fixation and clearing, because yolk with high refractive index is distributed throughout the cells. To image microtubules in living embryos with large cells we generated a zebrafish line stably expressing the microtubule binding domain of ensconsin fused to three GFPs (EMTB-3GFP) [7, 8]. Zebrafish sperm enters the egg near the animal pole, nucleating a sperm aster that spans the whole cell (excluding the lower, yolk-filled part of the egg) (Fig. 2 A), and captures the female pronucleus. This aster breaks down at the onset of mitosis, and the 1st mitotic spindle forms. As in the frog, astral microtubules at metaphase are too short to contact the cortex, and the spindle forms where centrosomes and DNA were deposited by the sperm aster (Fig. 2 B, movie S1). After anaphase onset, telophase asters expand dramatically from the separating sister centrosomes (Fig. 2 C). At the plane where the sister telophase asters overlap, a zone of reduced microtubule density emerges (Fig. 2 C, D). We will call this region, which we also observed in frog embryos (Fig. 1 A, C), the aster-aster interaction zone. It seems to form because interpenetration of microtubules from the two asters is blocked, by unknown mechanisms.
Figure 2. EMTB-3GFP transgenic zebrafish embryos allow live imaging of microtubule organization in large cells.
Orange arrows indicate positions of centrosomes. A) Shortly after fertilization sperm aster expands throughout the cell. B) Metaphase: Sperm aster breaks down and first mitotic spindle forms. C) During anaphase-telophase, astral microtubules grow out, and centrosomes move apart. An interaction zone forms in the plane where sister asters contact each other (between blue arrows). D) Centrosomes separated and align in the direction of the future spindle during late telophase (see enlargement). The centrosomes in the left aster are out of focus. Nuclei (green arrow) follow centrosomes, lagging behind. E) Second mitotic spindles assemble after cytokinesis (E is taken from different embryo). Bar = 200 μm. (See also Fig. S2, movies S1 and S2)).
The interaction zone limits the length of microtubules growing towards the sister aster, creating a length asymmetry in the left-right direction in figure 2 (For quantification of asymmetry see Fig. S2). As the telophase asters expand, and before astral microtubules reach the cortex, the centrosomes at their centers start to move apart, towards a point mid-way between the interaction zone and the far cortex (Fig. 2 B–E, movie S1, S2). This corresponds to the presumptive center of the daughter cell following cytokinesis, where the second mitotic spindle must assemble. In movie S1 the aster expands at approximately 15 μm per minute, while centrosomes move away from the mid-plane at approximately 3 μm per minute. During their outwards movement the centrosomes duplicate and separate, and each pair orients parallel to the interaction zone, which are the planes containing the longest axes of the telophase asters (Fig. 2 D). Interphase nuclei follow centrosomes (Fig. 1 C, 2 D), presumably by recruiting dynein to their surfaces [9]. The spindles of 2nd mitosis assemble between the separated centrosomes, shortly after cytokinesis (Fig. 2 D, E). Again, their astral microtubules at metaphase are too short to reach the cortex. We conclude the position (at the cell center) and orientation (orthogonal to the first mitotic spindle) of second mitotic spindles in zebrafish are determined by the prior position of centrosome pairs in prophase, which was in turn determined in some way by the geometry of the telophase asters they nucleated in the preceding interphase. Because microtubule organization in fish and frog embryos were similar, we combined their technical advantages to probe the mechanisms that locate and orient centrosomes in interphase, and thus determine cleavage plane geometry.
To determine whether centrosomes are moved by pushing or pulling forces, we depolymerised microtubules in selected regions of zebrafish embryos by uncaging a photo-activatable derivative of the microtubule depolymerising drug Combretastatin 4A (Fig. 3A). When the caged drug was activated with UV-light in defined regions close to asters (red area in Fig. 3B), the asters started to disassemble preferentially on the irradiated side. As soon as strong asymmetry in microtubule distribution was observed, centrosomes moved away from the depolymerisation zone (Fig. 3B, movie S3). The exact geometry and degree of depolymerization were difficult to control, and the speed of aster movement initiated by localized microtubules depolymerisation varied significantly. However, asters reproducibly moved away from the region of photo-activation (12/13 experiments, in 1/13 no movement was seen; Fig. S3 A). This observation argues strongly for movement by pulling forces. At the concentrations used (10μM) the caged drug did not significantly interfere with microtubule dynamics, and similar irradiation of embryos without drug had no effect (not shown). Movement induced by local microtubule depolymerisation occurred before the remaining part of the aster touched the cortex, arguing against a primary role for cortex-attached motors. We conclude that astral microtubules in zebrafish embryos exert pulling forces that are independent of aster-cortex contact. Our approach mimics Hiramoto's classic local photo-inactivation of Colcemid in marine eggs, which led to similar conclusions [10].
Figure 3. Aster movement depends on dynein dependent pulling forces.
A) Structure of caged-Combretastatin 4A. B) EMTB-3GFP zebrafish embryos were incubated in caged-Combretastatin and subjected to UV-irradiation in defined region (marked in red). Within seconds, microtubules depolymerised selectively on the irradiated side. The remaining aster moved away from the irradiated region, arguing for pulling forces on asters. Full arrows mark the positions of centrosomes, hollow arrows on right mark their original positions. C) Injection of p150-CC1 blocks aster movement. Asters still grew out and broke down under cell cycle control, but lost their ability to move or orient centrosomes. (See also Fig. S3, movies S3 and S4)
Dynein is implicated in centrosome movements in other systems [11]. To test its role in zebrafish, we injected a dominant-negative fragment of the dynactin complex (p150-CC1) into embryos shortly after fertilization. This fragment binds to dynein, blocks its interaction with dynactin, and inhibits most dynein functions [12]. Expansion of sperm and telophase asters, and aster breakdown in mitosis, were unperturbed (Fig. 3 C, movie S4). However, centrosome movement, and orientation of paired centrosomes, in telophase were inhibited. As a result, un-oriented centrosome pairs accumulated, and the asters they nucleated appeared to fuse. To test for a role of dynein in sperm centrosome centering in frog embryos, we injected eggs with p150-CC1 shortly after fertilization, and fixed them at several time points. By immunofluorescence, aster centering was severely inhibited (Fig. S3 B-D). We conclude that the forces that center and orient centrosomes in frog and fish embryos depend on dynein-dynactin interaction, and thus that pulling force is most likely generated by dynein. To explain how asters move long before their microtubules contact the cortex toward which they are moving (Fig. 2, see also discussion in [13]), we propose that dynein pulls from sites dispersed throughout the cytoplasm. Similar mechanisms have been proposed for pronuclear migration in sand dollar and C. elegans [10, 14, 15].
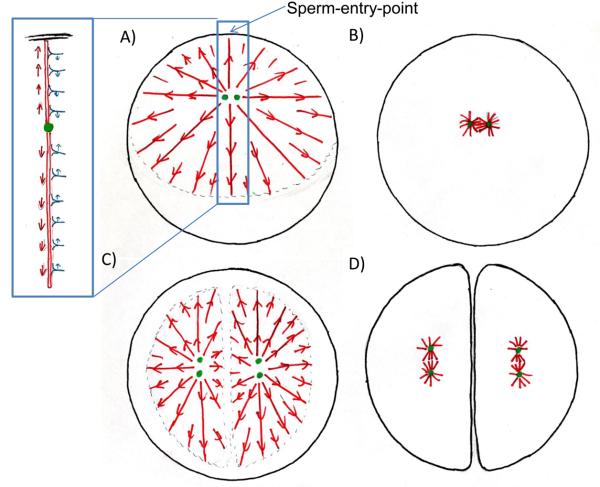
In light of these findings, we propose a simple model for centrosome positioning, with three essential components (Fig. 4): i) During interphase/telophase a pair of centrosomes center and orient with the longest axis of the aster determining the orientation and position of the following mitotic spindle (observation). ii) force on centrosomes is generated by a tug-of-war mechanism, where dynein anchored throughout the cytoplasm engages astral microtubules, resulting in length dependent pulling forces on centrosomes (hypothesis), and (iii) microtubule length is determined by interaction of the periphery of the interphase aster with cortex and interaction zone that limit its expansion (observation, mechanism unknown). These boundaries shape the aster, giving rise to microtubule length asymmetries that the dynein tug-of-war operates on to move and orient centrosomes.
Figure 4. Model for cleavage plane determination in large cells.
Astral microtubules (red) pull with dynein (blue) on cytoplasm to determine center and longest axis for cell division. A) Sperm enters at periphery. Cellular boundary causes asymmetry in sperm aster. Numbers of dynein bound is proportional to microtubule length resulting in net force on centrosome towards cell's center, but strongest stress on duplicated centrosomes (green) is perpendicular to movement. B) Sperm aster breaks down, small first mitotic spindle forms. C) At onset of anaphase asters expand but do not grow into each other. The microtubules free interaction zone between them generates the asymmetry in the aster leading to a net force on the pair of centrosomes towards the future centers of the daughter cells. The forces on the individual centrosomes cause the largest stress perpendicular to this movement resulting in the alignment of the linked centrosomes with the aster's longest axis. D) The cytokinetic furrow divided the cell into two, where the telophase asters overlapped, cutting through the sperm-entry -point. Asters break down; small mitotic spindles form at center and along longest axis of daughter cells. (See also Fig. S4)
The sperm aster in frog is shaped by the fact that microtubules on the cortex side near the sperm entrance point are length-limited by the cortex (Fig. 4A). Microtubules pointing in any other direction can grow, so our proposed length-dependent dynein force pulls the centrosome towards the cell center and also orient sister centrosomes (Fig. S1 B). The first mitotic spindle forms centered and oriented parallel to the tangent of the sperm entry point (Fig. 4B). Egg compression in frog limits aster growth in the short axis of the cell (Fig. 1A, B). This generates a length asymmetry that is converted into centrosome orientation along the long axis by Dynein pulling. In telophase of first mitosis in both frog and fish, the interaction zone between telophase asters limits aster expansion towards the center of the cell (Fig. 4C). The asters become dome-shaped, with microtubules longer in directions pointing away from the mid-line (Fig. S2). This length imbalance leads to dynein-mediated pulling of centrosomes towards a position equidistant between the interaction zone and the cortex, which segregates sister nuclei, and positions them appropriately for the next mitosis. At the same time, sister centrosome pairs orient parallel to the interaction zone as the sisters separate in telophase (Fig. 4C). Cytokinesis furrows ingress where the interaction zones between telophase asters touch the cortex [16] using partially established mechanisms [17, 18]. Shortly thereafter comparatively small mitotic spindles form centred and aligned with the long axes of the daughter cells (Fig. 4D).
While it is clear, in outline, how length asymmetry can center an aster in the cell by our model, it is less clear how aster shape determines the orientation of the centrosome pair with the aster, parallel to its longest axis . We propose a simple model in figure S4 B where we assume i) radial organization of microtubules, ii) length dependent pulling force on a given microtubule is exerted mostly on the centrosome closest to the minus end of that microtubules and iii) sister centrosmes are connected, and can exert force on each other. This should result in a net force on the centrosome pair that orients it along the longest axis of the aster (Fig. S4 B).
Cleavage planes are also oriented in z, the animal-vegetal axis (axes are defined in Fig. S4 A). In both frog and fish, yolk is more concentrated towards the vegetal side of the embryo, which breaks spherical symmetry in this direction. In fish, where yolk and cytoplasm seem mostly separated, yolk seems to provide a physical barrier to microtubule growth. Thus yolk may simply inhibit aster expansion downwards, towards the vegetal pole, controlling aster shape in the z-axis. This orients spindles in the x-y plane for many divisions (Movie S2), perhaps by the mechanism proposed in figure S4B. In frog yolk is dispersed but more concentrated on the vegetal side of the embryo. The position of first three mitotic spindles is shifted towards the animal pole [3]. We can envision two mechanisms that might explain z-asymmetry: 1) Yolk could act as a boundary and stop or slow down aster growth resulting in length asymmetry as in fish. 2) Less force per unit microtubules length is exterted in more yolky cytoplasm.
Hertwig first proposed that mitotic spindles are oriented by cell shape, so that cleavage planes bisect the cell's long axis [5], to explain the normally orthogonal succession of the first three cleavage planes, and the artificial orientation imposed by egg compression. We have updated his theory by showing that cell shape in fact orients centrosomes within interphase asters in early frog and fish embryos, and we propose a unified model that accounts for these observations using plausible molecular mechanisms. Many details remain to be elucidated, but our model can explain, in outline, how pronuclei center in large vertebrate embryos, how sister nuclei segregate when the spindle is small compared to the cell, and how successive cleavage planes orient.
It takes until approximately cell cycle 5 in fish, and 8 in frog, for cells to become small enough that metaphase asters can touch the cortex (Movie S2 and previous work [3]). It is possible that in more normal sized cells forces from cortical dynein dominate. However, support for cortical pulling forces comes mostly from systems which position spindles asymmetrically [19, 20]. We suggest that dynein anchored in bulk cytoplasm might be generally used to promote aster centering by a tug-of-war mechanism, whereas dynein localized at specialized cortical sites might promote off-centering during asymmetric divisions and cell polarization.
Experimental procedures
Imaging zebrafish embryos
The EMTB-3GFP transgenic zebrafish line was generated by use of the Tol2kit [21-23]. Shortly after fertilization, transgenic zebrafish embryos were mounted by use of an agarose mould in 0.1 × MMR (10mM NaCl; 0.2 mM KCl; 0.1 mM MgCl2; 0.2 mM CaCl2; 0.5 mM HEPES, pH 7.5). Embryos were imaged with an upright or inverted Zeiss LSM 710 equipped with 20x lenses. Centrosome position could be derived by microtubules morphology. For figure 2, picture of D is taken from different embryo than A-C, as regions of interest drifted out of focus. Time of D was estimated based on similar cell cycle stage in embryo used for A-C.
Caged Combretastatin
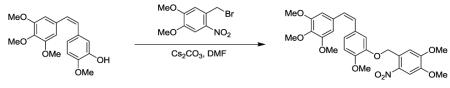 O-[4,5-dimethoxy-2-nitrobenzyl]-2-methoxy-5-[(Z)-2-(3,4,5 trimethoxyphenyl)ethenyl]phenol. To a solution of Combretastatin A4 (4 mg, 0.013 mmol) in DMF (1.5 ml) was added 4,5-(MeO)2-2-NO2-benzyl bromide (10.5 mg, 0.038 mmol), and Cs2CO3 (12.4 mg, 0.038 mmol). After stirring at room temperature for 22 h, the reaction mixture was diluted with dichloromethane. The organic layer was washed with water and brine, dried over MgSO4, filtered, and concentrated under reduced pressure. The crude mixture was purified via flash SiO2 column chromatography [EtOAc/hexanes (20%/80% to 25%/75%)] to give the desired compound with some impurities as a yellow solid (9.9mg, 96% yield). 1H NMR (400 MHz, chloroform-d) δ 7.47 (s, 1H), 7.18 (s, 1H), 7.05 (s, 1H), 6.94 (dd, J=2.0 and 8.2 Hz, 1H), 6.92 (d, J=1.6 Hz, 1H), 6.82 (d, J=8.6 Hz, 1H), 6.48 (s, 2H), 6.45 (d, J=2.7 Hz, 1H), 4.01 (s, 3H), 3.97 (s, 3H), 3.88 (s, 3H), 3.82 (s, 3H), 3.68 (s, 6H). Zebrafish embryos were mounted in 0.1 MMR containing 10 μM Caged Combretastatin. The drug was activated with a 405nm laser (30mW) with 13μsec pixel dwell time on a LSM 710.
O-[4,5-dimethoxy-2-nitrobenzyl]-2-methoxy-5-[(Z)-2-(3,4,5 trimethoxyphenyl)ethenyl]phenol. To a solution of Combretastatin A4 (4 mg, 0.013 mmol) in DMF (1.5 ml) was added 4,5-(MeO)2-2-NO2-benzyl bromide (10.5 mg, 0.038 mmol), and Cs2CO3 (12.4 mg, 0.038 mmol). After stirring at room temperature for 22 h, the reaction mixture was diluted with dichloromethane. The organic layer was washed with water and brine, dried over MgSO4, filtered, and concentrated under reduced pressure. The crude mixture was purified via flash SiO2 column chromatography [EtOAc/hexanes (20%/80% to 25%/75%)] to give the desired compound with some impurities as a yellow solid (9.9mg, 96% yield). 1H NMR (400 MHz, chloroform-d) δ 7.47 (s, 1H), 7.18 (s, 1H), 7.05 (s, 1H), 6.94 (dd, J=2.0 and 8.2 Hz, 1H), 6.92 (d, J=1.6 Hz, 1H), 6.82 (d, J=8.6 Hz, 1H), 6.48 (s, 2H), 6.45 (d, J=2.7 Hz, 1H), 4.01 (s, 3H), 3.97 (s, 3H), 3.88 (s, 3H), 3.82 (s, 3H), 3.68 (s, 6H). Zebrafish embryos were mounted in 0.1 MMR containing 10 μM Caged Combretastatin. The drug was activated with a 405nm laser (30mW) with 13μsec pixel dwell time on a LSM 710.
Dynein inhibition
P150-CC1 was expressed and purified as described previously [24] and dialyzed in XB +150mM sucrose (10 mM HEPES (pH 7.7), 1 mM MgCl2 0.1 mM CaCl2 100 mM KCl, 200mM sucrose) and flash frozen (25mg/ml). Zebrafish were injected shortly after fertilization with ~4 nL of protein solution and prepared for imaging as described above. For movie S4, maximum intensity of z-stacks (5 planes 9μm apart) were projected. Xenopus laevis embryos were fertilized at ~16°C. Shortly thereafter, embryos were injected with ~25 nL of p150-CC1 solution. Embryos were fixed 60 and 90 min in low FG-buffer [25] (0.3% formaldehyde, 0.1% glutaraldehyde, 80 mM K Pipes, pH 6.8,1 mM MgCl2, 5 mM EGTA, 0.2% Triton X-100) and after 25 minutes post-fixed in 90% MeOH, 10% (0.5 M EGTA (pH=7.8) in H2O). Embryos were cut perpendicular to the animal-vegetal-axis, and prepared for immunofluorescence against α-tubulin and γ-tubulin as described [3] (Antibodies B-5-1-2 and GTU-88 were purchased from Sigma). Because the number of embryos that can be injected after fertilization is limited, uninjected embryos served as control for fixation and immunostaining. Later control embryos from same parents were injected with ~25nl of p150-CC1 dialysis buffer, these embryos showed a normal cleavage pattern (not shown).
Compression of Xenopus embryos
Embryos were fertilized at room temperature, dejellied, and squeezed between two glass plates. The plates were pressed together with a spring, and the compression distance controlled by short pieces of wire (0.8 mm diameter). Embryos were fixed while compressed by immersion in 90% MeOH, 10% (0.5 M EGTA (pH 7.8) in H2O). Immunofluorescence was performed as described [3].
Highlights.
Orientation and location of mitotic spindle is determined by preceding interphase aster.
Aster-aster interactions govern aster geometry in telophase.
Aster movement depends on dynein dependent pulling forces.
Proposed model explains how large cells divide symmetrically, perpendicular to their long axis.
Supplementary Material
Acknowledgment
The authors would like to thank Chi-Bin Chien for the Tol2-kit plasmids, Chloe Bulinski for the EMTB-3GFP plasmid and Trina Schroer for the p150-CC1 plasmid. Nick Obholzer, Ramil Noche, and members of the Mitchison lab provided helpful suggestions and discussion. We gratefully acknowledge Will Ludington for help developing the embryo compression assay, Sean Megason and Angela De Pace for allowing us to use their microscopes, and Jonathan Wong for assistance microinjecting zebrafish embryos. We appreciate Rebecca Ward and Akatsuki Kimura for reading the manuscript. E.S.T. was supported by the American Cancer Society Fellowship (PF-09-024-01-CCG). H.W.D. and S.K.P were supported by NSF grant ANT-0635470. This work was supported by the National Institutes of Health (NIH) grant GM39565.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Dogterom M, Kerssemakers JW, Romet-Lemonne G, Janson ME. Force generation by dynamic microtubules. Curr Opin Cell Biol. 2005;17:67–74. doi: 10.1016/j.ceb.2004.12.011. [DOI] [PubMed] [Google Scholar]
- 2.Grill SW, Hyman AA. Spindle positioning by cortical pulling forces. Dev Cell. 2005;8:461–465. doi: 10.1016/j.devcel.2005.03.014. [DOI] [PubMed] [Google Scholar]
- 3.Wühr M, Chen Y, Dumont S, Groen AC, Needleman DJ, Salic A, Mitchison TJ. Evidence for an upper limit to mitotic spindle length. Curr Biol. 2008;18:1256–1261. doi: 10.1016/j.cub.2008.07.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roux W. Ueber die Ursachen der Bestimmung der Hauptrichtungen des Embryo im Froschei. Anat. Anz. 1903;23 [Google Scholar]
- 5.Hertwig O. Ueber den Werth der ersten Furchungszellen fuer die Organbildung des Embryo. Experimentelle Studien am Frosch- und Tritonei. Arch. mikr. Anat. 1893;xlii:662–807. [Google Scholar]
- 6.Black SD, Vincent JP. The first cleavage plane and the embryonic axis are determined by separate mechanisms in Xenopus laevis. II. Experimental dissociation by lateral compression of the egg. Dev Biol. 1988;128:65–71. doi: 10.1016/0012-1606(88)90267-9. [DOI] [PubMed] [Google Scholar]
- 7.Faire K, Waterman-Storer CM, Gruber D, Masson D, Salmon ED, Bulinski JC. E-MAP-115 (ensconsin) associates dynamically with microtubules in vivo and is not a physiological modulator of microtubule dynamics. J Cell Sci. 1999;112(Pt 23):4243–4255. doi: 10.1242/jcs.112.23.4243. [DOI] [PubMed] [Google Scholar]
- 8.von Dassow G, Verbrugghe KJ, Miller AL, Sider JR, Bement WM. Action at a distance during cytokinesis. J Cell Biol. 2009;187:831–845. doi: 10.1083/jcb.200907090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reinsch S, Karsenti E. Movement of nuclei along microtubules in Xenopus egg extracts. Curr Biol. 1997;7:211–214. doi: 10.1016/s0960-9822(97)70092-7. [DOI] [PubMed] [Google Scholar]
- 10.Hamaguchi MS, Hiramoto Y. Analysis of the role of astral rays in pronuclear migration in sand dollar eggs by the colcemid-UV method. Development Growth & Differentiation. 1986;28:143–156. doi: 10.1111/j.1440-169X.1986.00143.x. [DOI] [PubMed] [Google Scholar]
- 11.Gonczy P, Pichler S, Kirkham M, Hyman AA. Cytoplasmic dynein is required for distinct aspects of MTOC positioning, including centrosome separation, in the one cell stage Caenorhabditis elegans embryo. J Cell Biol. 1999;147:135–150. doi: 10.1083/jcb.147.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Quintyne NJ, Gill SR, Eckley DM, Crego CL, Compton DA, Schroer TA. Dynactin is required for microtubule anchoring at centrosomes. J Cell Biol. 1999;147:321–334. doi: 10.1083/jcb.147.2.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wühr M, Dumont S, Groen AC, Needleman DJ, Mitchison TJ. How does a millimeter-sized cell find its center? Cell Cycle. 2009;8:1115–1121. doi: 10.4161/cc.8.8.8150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kimura A, Onami S. Local cortical pulling-force repression switches centrosomal centration and posterior displacement in C. elegans. J Cell Biol. 2007;179:1347–1354. doi: 10.1083/jcb.200706005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kimura A, Onami S. Computer simulations and image processing reveal length-dependent pulling force as the primary mechanism for C. elegans male pronuclear migration. Dev Cell. 2005;8:765–775. doi: 10.1016/j.devcel.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 16.Rappaport R. Experiments concerning the cleavage stimulus in sand dollar eggs. J Exp Zool. 1961;148:81–89. doi: 10.1002/jez.1401480107. [DOI] [PubMed] [Google Scholar]
- 17.Eggert US, Mitchison TJ, Field CM. Animal cytokinesis: from parts list to mechanisms. Annu Rev Biochem. 2006;75:543–566. doi: 10.1146/annurev.biochem.74.082803.133425. [DOI] [PubMed] [Google Scholar]
- 18.von Dassow G. Concurrent cues for cytokinetic furrow induction in animal cells. Trends Cell Biol. 2009;19:165–173. doi: 10.1016/j.tcb.2009.01.008. [DOI] [PubMed] [Google Scholar]
- 19.Grill SW, Howard J, Schaffer E, Stelzer EH, Hyman AA. The distribution of active force generators controls mitotic spindle position. Science. 2003;301:518–521. doi: 10.1126/science.1086560. [DOI] [PubMed] [Google Scholar]
- 20.Adames NR, Cooper JA. Microtubule interactions with the cell cortex causing nuclear movements in Saccharomyces cerevisiae. J Cell Biol. 2000;149:863–874. doi: 10.1083/jcb.149.4.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kawakami K, Takeda H, Kawakami N, Kobayashi M, Matsuda N, Mishina M. A transposon-mediated gene trap approach identifies developmentally regulated genes in zebrafish. Dev Cell. 2004;7:133–144. doi: 10.1016/j.devcel.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 22.Urasaki A, Morvan G, Kawakami K. Functional dissection of the Tol2 transposable element identified the minimal cis-sequence and a highly repetitive sequence in the subterminal region essential for transposition. Genetics. 2006;174:639–649. doi: 10.1534/genetics.106.060244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kwan KM, Fujimoto E, Grabher C, Mangum BD, Hardy ME, Campbell DS, Parant JM, Yost HJ, Kanki JP, Chien CB. The Tol2kit: a multisite gateway-based construction kit for Tol2 transposon transgenesis constructs. Dev Dyn. 2007;236:3088–3099. doi: 10.1002/dvdy.21343. [DOI] [PubMed] [Google Scholar]
- 24.King SJ, Brown CL, Maier KC, Quintyne NJ, Schroer TA. Analysis of the dynein-dynactin interaction in vitro and in vivo. Mol Biol Cell. 2003;14:5089–5097. doi: 10.1091/mbc.E03-01-0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Becker BE, Gard DL. Visualization of the cytoskeleton in Xenopus oocytes and eggs by confocal immunofluorescence microscopy. Methods Mol Biol. 2006;322:69–86. doi: 10.1007/978-1-59745-000-3_6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.