Abstract
Aberrant promoter DNA-hypermethylation and repressive chromatin constitutes a frequent mechanism of gene inactivation in cancer. There is great interest in dissecting the mechanisms underlying this abnormal silencing. Studies have shown changes in nuclear organization of chromatin in tumor cells as well as association of aberrant methylation with long range silencing of neighboring genes. Further, certain tumors show a high incidence of promoter methylation termed as the CpG island methylator phenotype (CIMP). Here we have analyzed the role of nuclear chromatin architecture for genes in hypermethylated inactive versus non-methylated active states and its relation with long range silencing and CIMP. Using combined immunostaining for active/repressive chromatin marks and FISH in colorectal cancer cell lines we show that aberrant silencing of these genes occurs without requirement for their being positioned at heterochromatic domains. Importantly, hypermethylation, even when associated with long-range epigenetic silencing of neighboring genes, occurs independent of their euchromatic or heterochromatic location. Together, these results indicate that, in cancer, extensive changes around promoter chromatin of individual genes, or gene clusters, can potentially occur locally without preference for nuclear position and/or causing repositioning. These findings have important implications for understanding relationships between nuclear organization and gene expression patterns in cancer.
Keywords: DNA hypermethylation, nuclear organization, long range silencing, heterochromatin, colon cancer
INTRODUCTION
Epigenetic abnormalities, especially aberrant DNA methylation of promoter CpG islands of cancer-related (CR) genes, are common and early events contributing to gene inactivation during tumorigenesis (1). In addition to DNA methylation, our studies analyzing selected hypermethylated genes as well as global analysis of hypermethylated genes in cultured colorectal cancer (CRC) cell lines, have shown that these repressed promoters are marked by tri-methylation of H3K27 residues (H3K27Me3) concomitant with decreased levels of the activating mark, di-methylation of H3K4 (H3K4Me2) (2–3).
The mechanisms underlying CpG hypermethylation in cancer are unknown. It has been shown that aberrantly silenced CR genes can be reactivated by the DNA-methyltransferase (DNMT) inhibitor, 5-aza-2′-deoxycytidine (5-aza-CdR). However, re-expression in response to 5-aza-CdR is transient and the genes get re-silenced on drug removal (4–5). Genetic knockout, as well as 5-aza-CdR mediated inhibition of the DNMT’s, result in loss of promoter CpG methylation and de-repression of the CR genes. However, in CRCs, the levels of the inactive H3K27Me3 mark increases and co-exists with increases in the active H3K4Me2 mark indicating that the promoters may still reside in a H3K27Me3-marked heterochromatic environment (2–3). Another interesting feature associated with aberrant hypermethylation is the long range epigenetic silencing (LRES) wherein a cluster of adjacent genes across a large chromosomal segment undergoes coordinated silencing and show synergistic reactivation by a combination treatment with 5-aza-CdR and the HDAC inhibitor, trichostatin A (TSA) (6–7). This suggests that the entire chromosomal segment is under a common control mechanism involving DNA methylation and heterochromatic histone modification.
A little explored facet of epigenetic regulation in cancer cells concerns the increasing evidence for the role of spatial arrangement of chromosomes and genes in transcriptional regulation (8–9). Gene position has been shown to vary during development and disease states wherein genes reposition to heterochromatic compartments (the nuclear periphery or centromeric heterochromatin) when inactivated and relocate to the interior of the nucleus when activated (9). Furthermore, genes artificially tethered to the heterochromatic environment in the inner nuclear membrane undergo variable degrees of silencing (10–12). Physical association with heterochromatin accompanied by DNA methylation has been observed in a transgene induced to undergo stable silencing by transient, corepressor-mediated targeting (13). Thus, the nucleus can be viewed to have domains of gene activity (euchromatin) and inactivity (heterochromatin) which are proposed to optimize and regulate gene expression(8, 14–16).
In cancer, changes in the spatial organization of chromosome territories, centromeres, telomeres and specific genes have been observed (17–20). The functional significance of these changes is not well understood. It has been proposed that changes in nuclear strucure in cancer cells can influence gene expression (21). It is possible that nuclear position of genes might play a role in aberrant hypermethylation in cancer cells, especially during LRES where repositioning to a heterochromatic domain might coordinately silence the entire chromosomal segment.
Herein, we address whether or not higher order nuclear positioning of genes has a role in aberrant methylation or if aberrant methylation is associated only with local promoter changes. We have analyzed the relationship between the position of CR gene loci that undergo hypermethylation individually or in the context of LRES, and their nuclear microenvironment (eu-/hetero-chromatin) by Immuno-FISH in CRC cell lines. We analyzed the position of the MLH1, SFRP4, SFRP5 and ICAM1 genes which are frequently DNA hypermethylated, and silenced, in CRC lines. We show that hypermethylation mediated aberrant silencing of individual genes or in the context of LRES can occur both in an euchromatic or heterochromatic environment. We observe that aberrant silencing involves local chromatin changes in the absence of a requirement for global positioning to a heterochromatic compartment. These studies have important implications on the understanding of aberrant CpG hypermethylation and the role of nuclear position in gene regulation.
MATERIALS AND METHODS
Combined immunostaining and FISH and Microscopy
Cell lines used in this study were obtained from ATCC and were authenticated on June 9, 2010 by short tandem repeat (STR) profiling and by matching with the profile published in ATCC. Cells grown on coverslips were processed for immuno-FISH using modifications of previously described protocols (22–23). Immunostained cells were fixed in 50 mM Ethylene glycolbis(succinimidylsuccinate) (EGS, Pierce Biochemicals) followed by FISH. See Supplementary Methods for protocol/microscopy details.
ChIP-chip
ChIP on chip analysis are from single, experiments and were performed on RKO and SW480 cells as previously described (3). Additionally, previously published ChIP-chip data from HCT116 and DKO were analyzed (3).
RT-PCR
RT-PCR were done as previously described (2).
Gene Density calculations
HGNC genes in 2 Mb and 6 Mb regions around the genes of interest was obtained from BioMart (Ensembl) and gene densities were calculated as number of genes per Mb.
Analysis of Neighborhood Expression
Agilent microarray data was used to analyze neighborhood expression. Details of the procedure are given in Supplementary Methods. The data discussed in this publication is accessible through NCBI’s Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE23693).
Infiinium Methylation Array
DNA methylation was assessed using the Illumina Infinium platform as previously described (24, 25).
RESULTS
DNA methylation is independent of gene association with chromatin domains
The relationship between the nuclear positions of aberrantly methylated CR genes relative to the chromatin environment was explored by immunostaining for H3K4Me2 or H3K27Me3 domains and DNA-FISH in RKO, SW480 and HCT116 cells. H3K4Me2 and H3K27Me3 respectively mark active euchromatin and facultative heterochromatin, which are visible as distinct subnuclear domains (26–28). Technical artifacts could arise during the FISH procedure compromising the distribution of chromatin domains; interestingly the H3K27Me3 mark in SW480 nuclei was especially sensitive to FISH whereas the H3K4Me2 mark was resilient to immuno-FISH (Supplementary Fig. S1). To overcome this, we evaluated various FISH protocols and employed a modified protocol that preserves the chromatin pattern after FISH (see Supplementary Methods; Supplementary Fig. S2A). To demonstrate that the H3K27Me3 patterns are maintained before and after FISH, cells were fixed and immunostained and the same nuclei were imaged before and after FISH. Supplementary Fig. S2B is a typical image of a SW480 nucleus stained for H3K27Me3 before and after FISH. Although there is a 15 to 20% reduction in the immunostaining signal following FISH, the different z-stacks analyzed show that the histone staining pattern is robustly maintained.
Previous studies have shown that euchromatin and heterochromatin are marked by low and high DNA staining respectively (29). Consistent with this, H3K4Me2 and H3K27Me3 domains showed weak and dense DNA staining, respectively, indicating that these marks differentiate euchromatin from heterochromatin (Fig. 1A–B). As a control, we first studied the position of the ubiquitously active housekeeping gene, ACTB, with respect to eu-/heterochromatin. In SW480 and RKO cells ACTB associated with H3K4Me2-marked euchromatin (Supplementary Fig. S3: A–B, G–H). Similarly, we used the β-globin gene (HBB), which is not expressed in the CRC lines, as a control for an inactive gene. Previous studies have shown that HBB localization is developmentally regulated and that it is positioned close to heterochromatin in lineages where this gene is not expressed (30–31). In both SW480 and RKO cells, HBB associated with H3K27Me3 domains or conversely is excluded from H3K4Me2 domains (Supplementary Fig. S3: E–F, K–L).
Figure 1. Association of MLH1 with the H3K27Me3 domains.
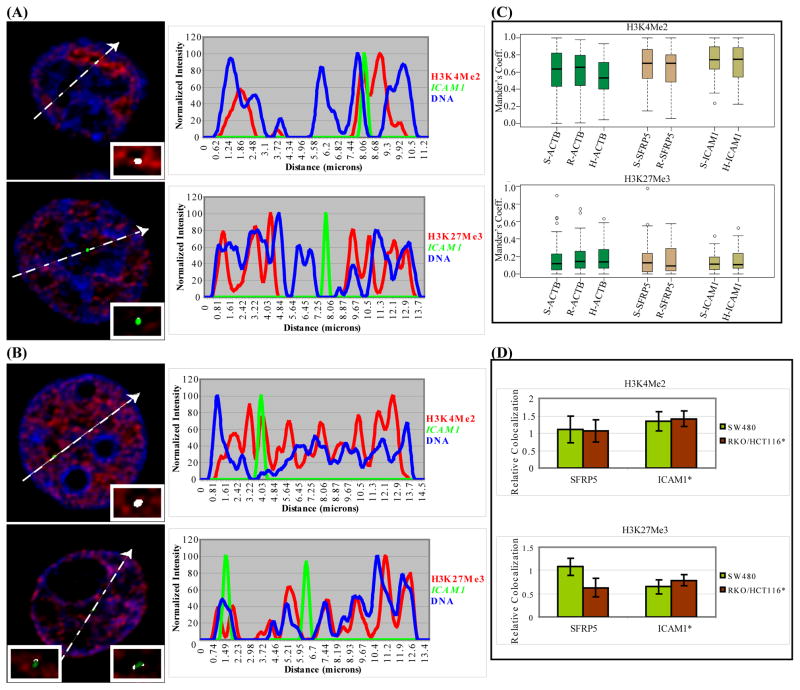
RKO (A) and SW480 cells (B) immunostained for H3K4Me2 (top) or H3K27Me3 (bottom) and MLH1 and ACTB (shown in Supplementary Fig. S3 A-B, G-H) by DNA-FISH. Line scan plots the intensities of the modified histone (red), gene signal (green) and DNA (blue) along the line. Inset shows the degree of colocalization of the FISH and modified histone signal as white pixels. Only allele(s) in a single z-slice are shown here (all alleles are shown in Supplementary Fig. S3). (C) Boxplots show colocalization (Manders’ coefficient) between the FISH and modified histone signal from a single experiment (n=25 nuclei). X-axis labels shows the genes analyzed in SW480 (prefix S) and RKO (prefix R). (D) Colocalization values normalized to ACTB. The median Manders’ coefficients of MLH1, SFRP4 and HBB colocalization with the modified histones from three experiments (n=25, 10, 10 nuclei) were normalized to that of ACTB and plotted as relative colocalization (y-axis). Error bars indicate standard deviation.
We then tested whether CR genes are subject to changes in their association with heterochromatic/euchromatic domains in response to hypermethylation. We first studied MLH1 and SFRP4, which are both active and non-DNA methylated in SW480 cells (Supplementary Fig. S4), and their promoters are enriched for the H3K4Me2 mark and have reduced H3K27Me3 upstream of the transcription start site (TSS) (2–3, 32) (Supplementary Fig. S5A–B). Both genes are DNA methylated, silenced and have reduced H3K4Me2 in RKO cells (2–3, 32) (Supplementary Fig. S4 and S5A–B). Although in RKO cells H3K27Me3 showed increased enrichment at the SFRP4 promoter, MLH1 showed only a moderate enrichment of H3K27Me3 upstream of the TSS (Supplementary Fig. S5A). ChIP-PCR analysis has shown that the MLH1 promoter in RKO cells is enriched for H3K27Me3 (2; data not shown). In both cell types, MLH1 and SFRP4 showed an increased association with H3K27Me3 staining similar to HBB and in contrast to ACTB (MLH1 shown in Fig. 1A–B; all alleles of MLH1 and SFRP4 in both cell types shown in Supplementary Fig. S3A–D and G–J). Quantitation of colocalization between the gene signal and the modified histone signal reveal that most alleles of MLH1 and SFRP4 show high association with H3K27Me3 domains in both cell lines (Fig. 1C), with no significant differences between the two cell lines (P-value>0.1). To enable direct comparison of the colocalization values across cell lines, multi-colored FISH was performed for the genes of interest and ACTB and the median colocalization was normalized to this latter gene. This normalization, in independent experiments, verified that most alleles of MLH1 and SFRP4 associate with the H3K27Me3 mark and less with the H3K4Me2 mark in both cell lines (Fig. 1D).
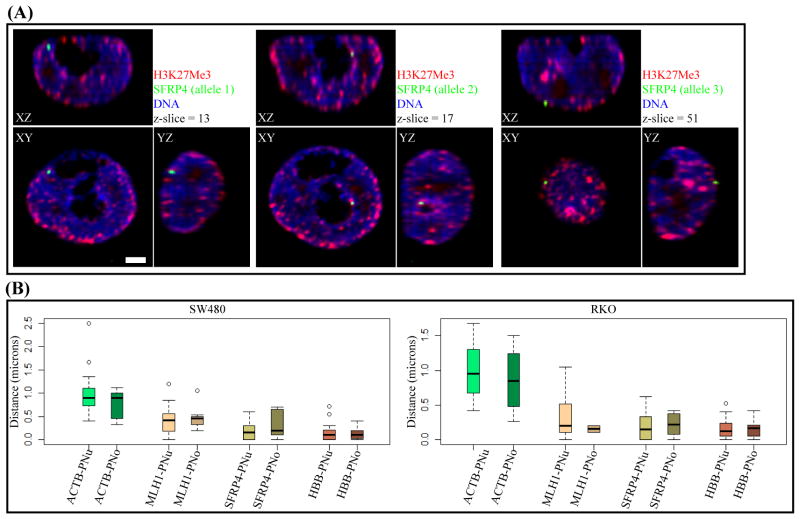
Previous studies have shown that H3K27Me3 domains are enriched at the perinuclear and perinucleolar regions (29). In concordance with above results showing a high degree of association with the H3K27Me3 domains, MLH1, SFRP4 and HBB alleles are preferentially located at the perinuclear or perinucleolar regions (Fig. 2A–B; Supplementary Fig. S6 A–C), with a median distance from these regions of <0.5μm. There are three aneuploid alleles of the SFRP4 and HBB loci in SW480 cells, and interestingly, like the diploid alleles of RKO (data not shown), these are all positioned either at the perinuclear or perinucleolar regions indicating that extra gene copies consistently tend to associate with the same chromatin domains.
Figure 2. Position of MLH1, SFRP4, HBB, ACTB relative to the perinuclear and perinucleolar domains.
(A) Orthogonal sections passing through the three aneuploid alleles of SFRP4 locus (green) in SW480 cells showing proximity to the perinuclear or perinucleolar regions, which are stained with H3K27Me3 (red). Nucleoli are devoid of DNA staining (blue). Scale-bar is 2 μm. (B) Quantitation of gene position relative to perinuclear (PNu) or perinucleolar (PNo) regions in SW480 and RKO cells. The nearest distance to the perinuclear or perinucleolus for 30–45 alleles are plotted (y-axis).
The above analyses show that MLH1 and SFRP4 are positioned close to heterochromatin independent of their silencing status. However, such positioning could predispose the genes to permanent silencing by DNA methylation. To test if abnormally silenced CR genes generally tend to position close to heterochromatin, two additional genes silenced in CRC lines were analyzed. SFRP5 was analyzed in RKO cells where it is DNA hypermethylated and silenced versus in SW480 cells where it is unmethylated and active (33; Supplementary Fig. S4). To place these studies in perspective, we first analyzed the local promoter marks from the ChIP-chip data which showed that SFRP5 is enriched for H3K4Me2 in SW480 while it lacks this mark in RKO (Supplementary Fig. S5C). Interestingly, the silenced SFRP5 promoter did not show any enrichment of H3K27Me3. The other gene, ICAM1 is unmethylated and active in both RKO and SW480 cells but in HCT116 cells, it is DNA hypermethylated and silenced (33; Supplementary Fig. S4). In both SW480 and RKO cells, ICAM1 is enriched for H3K4Me2 around the TSS consistent with its active state. Using previous data (3), we compared the ICAM1 promoter between HCT116 and its isogenic partner, DKO cells, which has genetic disruption of the major DNA methyltransferases-DNMT1 and DNMT3B (34). In HCT116, the silenced ICAM1 promoter showed a modest decrease in H3K4Me2 along with slight enrichment of H3K27Me3 compared to the reactivated promoter in DKO cells (Supplementary Fig. S5 D–E).
In all the cell lines, regardless of the above methylation and expression status, most alleles of ICAM1 and SFRP5, like ACTB, in contrast to MLH1 and SFRP4, exhibit a preference to be in the H3K4Me2-labeled euchromatin and are excluded from the H3K27Me3-marked heterochromatin (Fig. 3A–B, Supplementary Fig. S7). Colocalization analysis showed that the majority of ICAM1 and SFRP5 alleles associate with the euchromatic mark with little difference between their active and inactive states in SW480 and HCT116/RKO cells (P-value>0.1) (Fig. 3C–D). Thus, this data indicate that there is no general requirement for aberrantly DNA methylated and silenced genes to be positioned close to heterochromatin.
Figure 3. Association of SFRP5 and ICAM1 with the H3K4Me2 domains.
HCT116 (A) and SW480 cells (B) immunostained for H3K4Me2 (top) or H3K27Me3 (bottom) and ICAM1 and ACTB (not shown) loci by DNA-FISH. Figure details same as Fig. 1A-B. All alleles are shown in Supplementary Fig. S6. (C) Quantitation of colocalization from a single experiment (n=20 nuclei) as in Fig. 1C. SW480, RKO and HCT116 are labeled with prefix S, R and H respectively. (D) Colocalization values of SFRP5 and ICAM1 normalized to ACTB as in Fig. 1D (n=20, 10, 10 nuclei) plotted as relative colocalization (y-axis). Asterisk indicates that ICAM1 localization was compared between SW480 and HCT116 cells.
The data here also show that a gene can be active in a heterochromatic environment and still be enriched for euchromatic marks locally at the promoter, as is the case with MLH1 and SFRP4 in SW480 cells. These results, in the aggregate, again emphasize that the position of CR genes relative to eu-/hetero-chromatin in CRC lines is independent of their promoter CpG island methylation status, and local epigenetic alterations can exist in the absence of global changes in positioning.
Relation between nuclear position and LRES
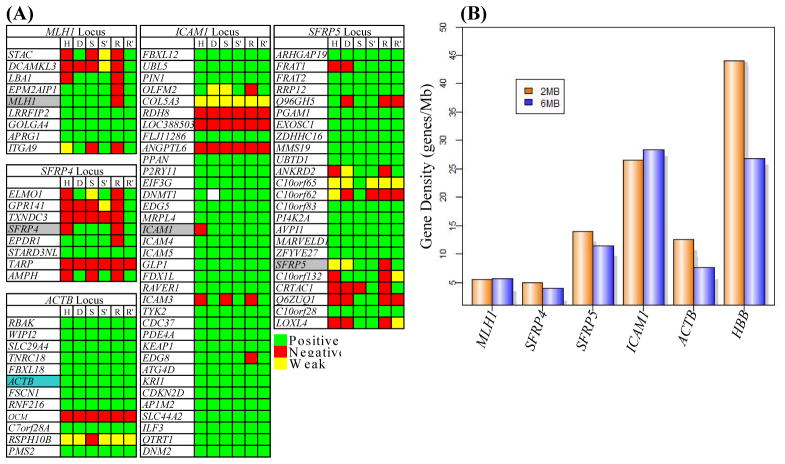
The data above reveal that MLH1, SFRP4 and HBB show association with heterochromatin while ICAM1, SFRP5 and ACTB reside in euchromatin. Among the factors that could dictate nuclear positions of these gene loci include their relationships to the gene density of the regions in which they reside or the activity of neighboring genes. Previous studies have shown that gene rich loci reside in euchromatic domains (27). Further it has been shown in colon cancers that aberrant methylation of MLH1 is accompanied by LRES of a cluster of neighboring genes in the locus (7). Since the data above showed that MLH1 associates with heterochromatic compartments, we explored the relationships between nuclear position and LRES. We first analyzed the expression pattern of neighboring genes in a 1 Mb domain around the genes of interest (Fig. 4A). To determine the dependency on DNA methylation mediated silencing of neighboring genes, RT-PCR was performed on RNA from SW480 and RKO cells that were mock treated or 5-aza-CdR treated. In the case of HCT116, its isogenic line, DKO, was used for RT-PCR analysis. Fig. 4 shows that MLH1, SFRP4 and SFRP5 reside in areas of local spreading of silencing to neighboring genes in a cell-type specific manner RKO showing the highest degree of spreading of silencing, HCT116 an intermediate degree, and SW480 the least. ICAM1 and ACTB loci did not show any spreading of silencing to neighboring genes. Thus, the activity of neighboring genes in these analyzed loci does not always render genes predisposed to abnormal silencing and LRES is independent of the nuclear position of involved genes because: (a) ICAM1 can be silenced even though it resides in a region where neighboring genes are always active; (b) although MLH1 and SFRP4 are positioned in heterochromatin in both RKO and SW480, these loci show differential spreading of silencing in different cell lines; (c) even though SFRP5 is positioned in euchromatin, it is involved in differential, local spreading of silencing to neighboring genes in different cell lines.
Figure 4. Relationship between nuclear position, LRES and gene density.
(A) Expression of genes in a 1Mb domain around the genes studied here in HCT116 (H), DKO (D), SW480 (S), SW480 + 5-aza-CdR (S′), RKO (R) and RKO + 5-aza-CdR (R′). The CR genes are highlighted in grey and ACTB control is highlighted in blue; genes are listed top to bottom in the order of their 5′ to 3′ position in the genome (UCSC). Positive expression (green), no expression (red) and weak expression (yellow) are shown. White box in DKO in the ICAM1 locus indicates the absence of DNMT1 gene due to genetic knockout. MLH1, SFRP4 and SFRP5 loci tend to show variable degree of silencing of adjacent genes in RKO, HCT116 and SW480, in that order. (B) Gene densities (y-axis) in a 2 and 6 Mb window centered on the listed genes plotted as genes/Mb.
It can also be noted, from Fig. 4A, that the gene densities in these loci differ widely. Previous studies have shown that gene density may be an integral determinant and/or marker of the radial positioning of genomic elements in the nucleus (35–36) and that the gene density in a window of 2Mb is a good predictor of radial gene position (37). We compared the gene densities of our genes of interest in a window of 2 and 6 Mb (Fig. 4B) which revealed that the MLH1 and SFRP4 loci, which are positioned in heterochromatin, are in gene poor regions. In contrast, SFRP5, ICAM1 and ACTB loci, which are positioned in euchromatin, are gene rich. These data agree with a previous study showing correlation between high gene density (>16 genes/Mb) and H3K4Me3 domains (27). Thus, we observe a correlation between high gene density and positioning at euchromatic domains, and low gene density and heterochromatic domains.
One exception to the relationships between gene density and gene position is the HBB gene which has a very high gene density but was observed to associate with H3K27Me3-marked heterochromatin. This could be because the majority of genes present in the HBB locus belong to the Olfactory Receptor family genes. Analysis of whole transcriptome expression arrays (Agilent) of all the cell lines in this study show that the olfactory genes and other genes in a >1Mb domain around HBB are not expressed in any of the CRC lines (data not shown). Further, analysis of ChIP-chip data from all three cell lines revealed H3K27Me3 enrichment at the promoters of 27 of 30 genes in the 2Mb domain around HBB (data not shown) confirming the silent status of genes in this neighborhood. Thus, the fact that HBB locus is an exception to the relationship between gene density and chromatin domains may reflect the unusual situation wherein a high proportion of genes in a locus are under regional control in a tissue-specific manner and indicates that other parameters might play an important role in nuclear organization of gene loci.
Relation between DNA methylation, activity of neighboring genes and CIMP
Fig. 4A indicates that MLH1, SFRP4 and SFRP5 loci in RKO and HCT116 show silencing of more neighboring genes compared to SW480, especially in the SFRP5 loci. An important feature of these cells is that RKO has typical features of the CIMP+ phenotype including having a BRAF mutation, while SW480 is CIMP− and has wild type BRAF. Analysis of methylation of a set of CIMP-markers shows that majority of these markers are methylated in RKO compared to SW480 indicating the differential CIMP status of SW480 and RKO (Supplementary Fig. S8). A previous study has suggested a link between LRES and CIMP in colon cancer (38). To test if CIMP-dependent long range silencing at methylated gene loci is a widespread phenomenon, the activity of genes residing near methylated genes was analyzed on a genome wide scale in RKO and SW480. Agilent Whole Genome expression data was processed to determine the median expression level of probes in a window of seven genes centered around every gene, hereafter referred to as the Neighborhood Expression Score (NES; see methods). In essence, a low NES score for a gene indicates that the gene is present in a genomic neighborhood with low gene expression while a high score means that the gene resides in a region with high gene expression. Methylation status of genes in SW480 and RKO was determined using the Infinium methylation array (24).
Previous studies have shown that the genome tends to be organized into regions of high and low gene expression (39). To test if the NES captures the activity dependent organization of the genome, the distribution of the NES values of low and high expressing genes was compared with that of all genes. Supplementary Fig. S9A–B show that low and high expressing genes tend to have low and high NES values respectively (P-values<10−16) indicating that the NES is a good measure of the neighborhood expression status. This is further supported by the observation that randomization of the genomic positions of the probes abolishes this gene-activity dependent NES values (Supplementary Fig. S9C–D).
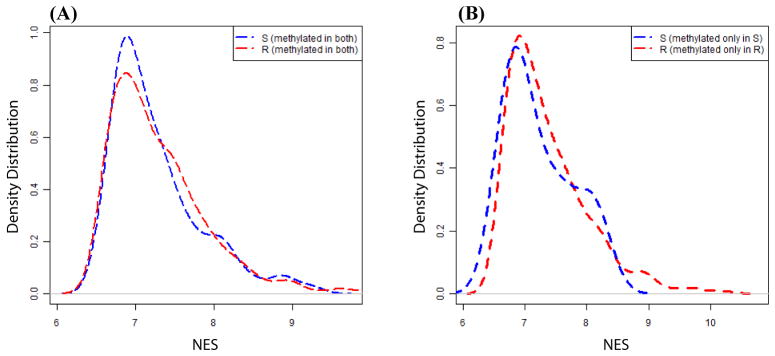
To test if long range silencing at methylated gene loci is a general phenomenon in CIMP+ cells, the NES score distributions of genes methylated in both SW480 and RKO (285 genes) were analyzed (Fig. 5A). In both SW480 and RKO, methylated genes reside in a continuum of low to high expressing regions. However, there are no significant differences in the distribution of NES values between the two cell-types (P-value>0.2). Similarly, genes methylated either only in SW480 (27 genes) or only in RKO (438 genes) show similar NES values in both cell types (Fig. 5B). Further, based on the Infinium array data, SW480 cells have 312 methylated genes of which only two genes (NMBR, RASGRF1) have 2-fold lower NES values in SW480 compared to RKO. RKO has 723 methylated genes of which only ten genes (SFRP5, ALK, CYP1B1, GBX2, CLSTN2, SHOX2, ADAMTS14, FGF4, BARX2, CHODL) have 2-fold lower NES values in RKO compared to SW480. Thus, except for a few loci, the excess methylated genes in CIMP+ RKO do not show a general tendency to be in loci with low neighborhood gene expression.
Figure 5. Relationship between LRES and CIMP.
(A and B) Neighborhood expression score (NES) for every gene was calculated as the median expression values of three upstream and three downstream genes. The NES was used as a measure of LRES. A and B shows distribution of the NES values for genes methylated in both SW480 and RKO (A) and genes methylated only in SW480 or only in RKO (B). S and R in legend denote SW480 and RKO.
DISCUSSION
Our data indicate that stable silencing by aberrant DNA methylation of the CR genes analyzed is independent of their position within the nuclear microenvironment or nuclear sub-compartments, viz. the perinuclear or perinucleolar domains. These results are, perhaps, contrary to what might have been expected based on work from others, using development/differentiation systems. These previous studies have suggested a general model wherein genes reposition away from the heterochromatin (perinuclear or pericentromeric) when activated and gravitate to heterochromatin when silenced (40–41). In these models, the perinuclear and pericentric heterochromatin is purported to play a role in establishing/maintaining domains of transcriptionally inactive regions wherein genes get recruited for stable silencing during differentiation (42).
Despite the above general models of development, other recent studies may help explain the lack of an obligatory requirement for hypermethylated CR genes to be positioned in heterochromatic domains to maintain stability of their expression patterns. It has been observed that active genes dynamically shuttle in and out of transcription hubs wherein the active phase of the gene is characterized by association with the transcription hub (16). Similarly, the active genes observed to be present in heterochromatic domains in a majority of the cells in this study might be expressed by dynamic and temporally short association with neighboring euchromatic domains. Further, recent reports analyzing the influence of the heterochromatic environment at the nuclear periphery on gene expression have shown that although artificial tethering of a gene to the periphery can downregulate expression of some genes, other neighboring genes relocated to this new environment remain transcriptionally active (10–12). Thus, the response elicited from a gene by the environment is gene-specific and our results support the idea that genes could reside in euchromatin and be silenced and vice versa. This is highlighted in the observation that the active MLH1 and SFRP4 genes in SW480 cells are enriched for H3K4Me2 locally around the TSS but a majority of the alleles are in H3K27Me3-labeled microenvironment.
In the last few years LRES has been reported in colon, bladder and lung cancers (6–7, 43–44). The mechanism underlying LRES is not known and could possibly involve long-range interactions between genomic elements and chromatin remodeling (45). If so, our data indicate that the mechanisms responsible for LRES can encompass loci that reside in either eu-/heterochromatic domains and excludes a major role for gene position with respect to chromatin environment in this process. Our data in this regard, again, support the notion that aberrant promoter CpG hypermethylation and its association with CR gene silencing is independent of nuclear position of the affected genes.
Tumors differ greatly in the incidence of gene methylation resulting in the CIMP+ and CIMP− phenotype. In a recent study, Karpinski et al observed that LRES at the 2q14.2 loci correlated with the CIMP phenotype in a panel of colon tumors samples (38). In the current study, gene expression analysis by PCR showed that MLH1, SFRP4 and SFRP5 reside in genomic region that shows long range silencing of neighboring genes in a CIMP+ cell type. However, our global analyses of the direct relationship between gene methylation and long range silencing as a function of CIMP show that, except for a few loci, the majority of methylated gene loci in SW480 and RKO display similar levels of neighborhood gene expression. Thus, it appears that CIMP-dependent long range silencing of methylated genes seems to occur only at a few loci and that the majority of methylated genes do not show CIMP-dependent long range silencing. A caveat in the current analysis of CIMP-dependent long range silencing is that cancer cell lines were compared. Further understanding of the relation between CIMP and long range silencing will require direct comparison of matched tumor and normal colonic epithelium.
To our knowledge, our data here is the first study analyzing the relationships among nuclear position of genes under epigenetic regulation individually or in clusters (LRES), chromatin domains, and nuclear compartments (periphery and perinucleolar) in cancer cell model. It is clearly established that the organization of genes and chromosomes are very different in tumor cells compared to normal cells (17, 20). Based on these reports, it is plausible that the position of the CR genes analyzed here may differ from the normal colonic epithelia. Also it is possible that large scale changes in nuclear organization might be an early event in tumorigenesis and might play a role in the initial establishment of methylation patterns. However, our data here establish that silencing of CR genes by aberrant CpG methylation does not require positioning of the genes at heterochromatic compartments. A previous study in a breast cancer model system showed that the changes in position of a panel of gene loci is independent of gene expression changes (20). It is not clear what causes the changes in nuclear organization in cancer cells and its effect on cancer progression. In future work it will be interesting to understand the significance of the nuclear reorganization that accompanies tumorigenesis.
Supplementary Material
Acknowledgments
Grant Support: This work was funded by National Institute of Environmental Health Sciences grant ES011858; HE was supported by the Hodson Foundation postdoctoral fellowship.
We thank Eriko Greene, Drs. Joo Mi Y, Mashaal Dhir, Nita Ahuja and Emi Ota-Machida for reagents; Drs. Peter Fraser and Lyubomira Chakalova for FISH protocols; Dr. Mark Groudine for HBB plasmid; Dr. Thomas Jenuwein for anti-H3K27Me3 antibodies; Dr. Kundan Sengupta for invaluable suggestions; Tina Largent and Kathy Bender for technical support.
ABBREVIATIONS LIST
- 5-aza-CdR
5-aza-2′-deoxycytidine
- LRES
Long-range epigenetic silencing
- FISH
Fluorescent in situ hybridization
- ChIP
Chromatin Immuno-precipitation
References
- 1.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–92. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McGarvey KM, Fahrner JA, Greene E, Martens J, Jenuwein T, Baylin SB. Silenced tumor suppressor genes reactivated by DNA demethylation do not return to a fully euchromatic chromatin state. Cancer Res. 2006;66:3541–9. doi: 10.1158/0008-5472.CAN-05-2481. [DOI] [PubMed] [Google Scholar]
- 3.McGarvey KM, Van Neste L, Cope L, et al. Defining a chromatin pattern that characterizes DNA-hypermethylated genes in colon cancer cells. Cancer Res. 2008;68:5753–9. doi: 10.1158/0008-5472.CAN-08-0700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bender CM, Pao MM, Jones PA. Inhibition of DNA methylation by 5-aza-2′-deoxycytidine suppresses the growth of human tumor cell lines. Cancer Res. 1998;58:95–101. [PubMed] [Google Scholar]
- 5.Egger G, Aparicio AM, Escobar SG, Jones PA. Inhibition of histone deacetylation does not block resilencing of p16 after 5-aza-2′-deoxycytidine treatment. Cancer Res. 2007;67:346–53. doi: 10.1158/0008-5472.CAN-06-2845. [DOI] [PubMed] [Google Scholar]
- 6.Frigola J, Song J, Stirzaker C, Hinshelwood RA, Peinado MA, Clark SJ. Epigenetic remodeling in colorectal cancer results in coordinate gene suppression across an entire chromosome band. Nat Genet. 2006;38:540–9. doi: 10.1038/ng1781. [DOI] [PubMed] [Google Scholar]
- 7.Hitchins MP, Lin VA, Buckle A, et al. Epigenetic inactivation of a cluster of genes flanking MLH1 in microsatellite-unstable colorectal cancer. Cancer Res. 2007;67:9107–16. doi: 10.1158/0008-5472.CAN-07-0869. [DOI] [PubMed] [Google Scholar]
- 8.Lanctot C, Cheutin T, Cremer M, Cavalli G, Cremer T. Dynamic genome architecture in the nuclear space: regulation of gene expression in three dimensions. Nat Rev Genet. 2007;8:104–15. doi: 10.1038/nrg2041. [DOI] [PubMed] [Google Scholar]
- 9.Misteli T. Beyond the sequence: cellular organization of genome function. Cell. 2007;128:787–800. doi: 10.1016/j.cell.2007.01.028. [DOI] [PubMed] [Google Scholar]
- 10.Finlan LE, Sproul D, Thomson I, et al. Recruitment to the nuclear periphery can alter expression of genes in human cells. PLoS Genet. 2008;4:e1000039. doi: 10.1371/journal.pgen.1000039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reddy KL, Zullo JM, Bertolino E, Singh H. Transcriptional repression mediated by repositioning of genes to the nuclear lamina. Nature. 2008;452:243–7. doi: 10.1038/nature06727. [DOI] [PubMed] [Google Scholar]
- 12.Kumaran RI, Spector DL. A genetic locus targeted to the nuclear periphery in living cells maintains its transcriptional competence. J Cell Biol. 2008;180:51–65. doi: 10.1083/jcb.200706060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ayyanathan K, Lechner MS, Bell P, et al. Regulated recruitment of HP1 to a euchromatic gene induces mitotically heritable, epigenetic gene silencing: a mammalian cell culture model of gene variegation. Genes Dev. 2003;17:1855–69. doi: 10.1101/gad.1102803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shopland LS, Johnson CV, Byron M, McNeil J, Lawrence JB. Clustering of multiple specific genes and gene-rich R-bands around SC-35 domains: evidence for local euchromatic neighborhoods. J Cell Biol. 2003;162:981–90. doi: 10.1083/jcb.200303131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moen PT, Jr, Johnson CV, Byron M, et al. Repositioning of muscle-specific genes relative to the periphery of SC-35 domains during skeletal myogenesis. Mol Biol Cell. 2004;15:197–206. doi: 10.1091/mbc.E03-06-0388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Osborne CS, Chakalova L, Brown KE, et al. Active genes dynamically colocalize to shared sites of ongoing transcription. Nat Genet. 2004;36:1065–71. doi: 10.1038/ng1423. [DOI] [PubMed] [Google Scholar]
- 17.Cremer M, Kupper K, Wagler B, et al. Inheritance of gene density-related higher order chromatin arrangements in normal and tumor cell nuclei. J Cell Biol. 2003;162:809–20. doi: 10.1083/jcb.200304096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sarkar R, Guffei A, Vermolen BJ, Garini Y, Mai S. Alterations of centromere positions in nuclei of immortalized and malignant mouse lymphocytes. Cytometry A. 2007;71:386–92. doi: 10.1002/cyto.a.20395. [DOI] [PubMed] [Google Scholar]
- 19.Chuang TC, Moshir S, Garini Y, et al. The three-dimensional organization of telomeres in the nucleus of mammalian cells. BMC Biol. 2004;2:12. doi: 10.1186/1741-7007-2-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meaburn KJ, Misteli T. Locus-specific and activity-independent gene repositioning during early tumorigenesis. J Cell Biol. 2008;180:39–50. doi: 10.1083/jcb.200708204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stein GS, Montecino M, van Wijnen AJ, Stein JL, Lian JB. Nuclear structure-gene expression interrelationships: implications for aberrant gene expression in cancer. Cancer Res. 2000;60:2067–76. [PubMed] [Google Scholar]
- 22.Tam R, Smith KP, Lawrence JB. The 4q subtelomere harboring the FSHD locus is specifically anchored with peripheral heterochromatin unlike most human telomeres. J Cell Biol. 2004;167:269–79. doi: 10.1083/jcb.200403128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson CV, Singer RH, Lawrence JB. Fluorescent detection of nuclear RNA and DNA: implications for genome organization. Methods Cell Biol. 1991;35:73–99. [PubMed] [Google Scholar]
- 24.Bibikova M, Fan JB. GoldenGate assay for DNA methylation profiling. Methods Mol Biol. 2009;507:149–63. doi: 10.1007/978-1-59745-522-0_12. [DOI] [PubMed] [Google Scholar]
- 25.Mohammad HP, Cai Y, McGarvey KM, et al. Polycomb CBX7 promotes initiation of heritable repression of genes frequently silenced with cancer-specific DNA hypermethylation. Cancer Res. 2009;69:6322–30. doi: 10.1158/0008-5472.CAN-09-0065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lachner M, O’Sullivan RJ, Jenuwein T. An epigenetic road map for histone lysine methylation. J Cell Sci. 2003;116:2117–24. doi: 10.1242/jcs.00493. [DOI] [PubMed] [Google Scholar]
- 27.Zinner R, Teller K, Versteeg R, Cremer T, Cremer M. Biochemistry meets nuclear architecture: Multicolor immuno-FISH for co-localization analysis of chromosome segments and differentially expressed gene loci with various histone methylations. Adv Enzyme Regul. 2007 doi: 10.1016/j.advenzreg.2007.01.005. [DOI] [PubMed] [Google Scholar]
- 28.Bartova E, Pachernik J, Harnicarova A, et al. Nuclear levels and patterns of histone H3 modification and HP1 proteins after inhibition of histone deacetylases. J Cell Sci. 2005;118:5035–46. doi: 10.1242/jcs.02621. [DOI] [PubMed] [Google Scholar]
- 29.Zinner R, Albiez H, Walter J, Peters AH, Cremer T, Cremer M. Histone lysine methylation patterns in human cell types are arranged in distinct three-dimensional nuclear zones. Histochem Cell Biol. 2006;125:3–19. doi: 10.1007/s00418-005-0049-1. [DOI] [PubMed] [Google Scholar]
- 30.Brown KE, Amoils S, Horn JM, et al. Expression of alpha- and beta-globin genes occurs within different nuclear domains in haemopoietic cells. Nat Cell Biol. 2001;3:602–6. doi: 10.1038/35078577. [DOI] [PubMed] [Google Scholar]
- 31.Schubeler D, Francastel C, Cimbora DM, Reik A, Martin DI, Groudine M. Nuclear localization and histone acetylation: a pathway for chromatin opening and transcriptional activation of the human beta-globin locus. Genes Dev. 2000;14:940–50. [PMC free article] [PubMed] [Google Scholar]
- 32.Suzuki H, Gabrielson E, Chen W, et al. A genomic screen for genes upregulated by demethylation and histone deacetylase inhibition in human colorectal cancer. Nat Genet. 2002;31:141–9. doi: 10.1038/ng892. [DOI] [PubMed] [Google Scholar]
- 33.Schuebel KE, Chen W, Cope L, et al. Comparing the DNA hypermethylome with gene mutations in human colorectal cancer. PLoS Genet. 2007;3:1709–23. doi: 10.1371/journal.pgen.0030157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rhee I, Bachman KE, Park BH, et al. DNMT1 and DNMT3b cooperate to silence genes in human cancer cells. Nature. 2002;416:552–6. doi: 10.1038/416552a. [DOI] [PubMed] [Google Scholar]
- 35.Croft JA, Bridger JM, Boyle S, Perry P, Teague P, Bickmore WA. Differences in the localization and morphology of chromosomes in the human nucleus. J Cell Biol. 1999;145:1119–31. doi: 10.1083/jcb.145.6.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kupper K, Kolbl A, Biener D, et al. Radial chromatin positioning is shaped by local gene density, not by gene expression. Chromosoma. 2007;116:285–306. doi: 10.1007/s00412-007-0098-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Murmann AE, Gao J, Encinosa M, et al. Local gene density predicts the spatial position of genetic loci in the interphase nucleus. Exp Cell Res. 2005;311:14–26. doi: 10.1016/j.yexcr.2005.07.020. [DOI] [PubMed] [Google Scholar]
- 38.Karpinski P, Ramsey D, Grzebieniak Z, Sasiadek MM, Blin N. The CpG island methylator phenotype correlates with long-range epigenetic silencing in colorectal cancer. Mol Cancer Res. 2008;6:585–91. doi: 10.1158/1541-7786.MCR-07-2158. [DOI] [PubMed] [Google Scholar]
- 39.Caron H, van Schaik B, van der Mee M, et al. The human transcriptome map: clustering of highly expressed genes in chromosomal domains. Science. 2001;291:1289–92. doi: 10.1126/science.1056794. [DOI] [PubMed] [Google Scholar]
- 40.Takizawa T, Meaburn KJ, Misteli T. The meaning of gene positioning. Cell. 2008;135:9–13. doi: 10.1016/j.cell.2008.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Deniaud E, Bickmore WA. Transcription and the nuclear periphery: edge of darkness? Curr Opin Genet Dev. 2009;19:187–91. doi: 10.1016/j.gde.2009.01.005. [DOI] [PubMed] [Google Scholar]
- 42.Fisher AG, Merkenschlager M. Gene silencing, cell fate and nuclear organisation. Curr Opin Genet Dev. 2002;12:193–7. doi: 10.1016/s0959-437x(02)00286-1. [DOI] [PubMed] [Google Scholar]
- 43.Stransky N, Vallot C, Reyal F, et al. Regional copy number-independent deregulation of transcription in cancer. Nat Genet. 2006;38:1386–96. doi: 10.1038/ng1923. [DOI] [PubMed] [Google Scholar]
- 44.Seng TJ, Currey N, Cooper WA, et al. DLEC1 and MLH1 promoter methylation are associated with poor prognosis in non-small cell lung carcinoma. Br J Cancer. 2008;99:375–82. doi: 10.1038/sj.bjc.6604452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Clark SJ. Action at a distance: epigenetic silencing of large chromosomal regions in carcinogenesis. Hum Mol Genet. 2007;16(1):R88–95. doi: 10.1093/hmg/ddm051. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.