Abstract
Genetic material in the nucleus governs mechanisms related to cell proliferation, differentiation, and function. Thus, senescence and aging are directly tied to the change of nuclear function and structure. The most important mechanisms that affect cell senescence are: (i) telomere shortening; (ii) environmental stress-mediated accumulation of DNA mutations, and (iii) the intrinsically encoded biological clock that dictates lifespan events of any particular cell type. Overall, these changes lead to modification of the expression of genes that are responsible for: (i) organization of the nuclear structure; (ii) integrity of transcriptionally inactive heterochromatin, and (iii) epigenetic modification of chromosomes due to DNA methylation and/or histone modifications. These aging-related nuclear alterations do not only affect somatic cells. More importantly, they affect stem cells, which are responsible for proper tissue rejuvenation. In this review, we focus on epigenetic changes in the chromatin structure and their impact on the biology and function of adult cells as they age. We will also address aging-related changes in a compartment of the most primitive pluripotent stem cells that were recently identified by our team and named ‘very small embryonic/epiblast-like stem cells’.
Key Words: Aging, Telomeres, Heterochromatin, Very small embryonic/epiblast-like stem cells
Introduction
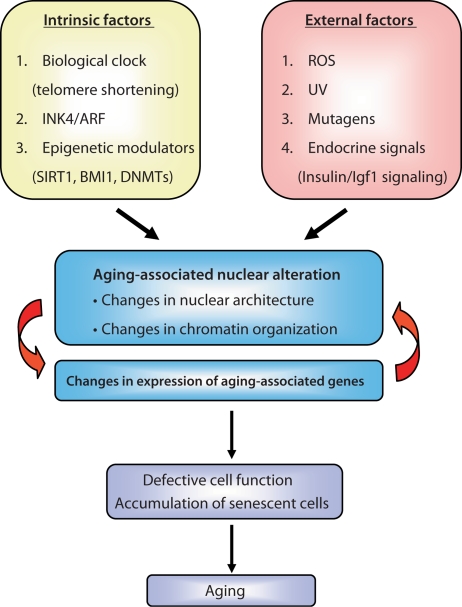
The aging process is an inevitable consequence of life. As a result of exposure to intrinsic- as well as extrinsic-aging factors, cellular aging is triggered by gradually accumulating DNA damage and epigenetic changes in DNA's structure [1,2,3]. Thus, aging could be envisioned at the cellular level as a result of altered cell function in response to changes in the DNA structure that directly affect proper gene expression (fig. 1).
Fig. 1.
Aging-associated nuclear alterations as a result of intrinsic and external factors. Both intrinsic factors (e.g. telomere shortening, epigenetic modulators, and an increase in INK4a/ARF expression) as well as external factors (e.g. free radicals (ROS), ultraviolet (UV) mutagens, and some endocrine signals) lead to changes in the nuclear architecture and chromatin organization. This leads to changes in expression of aging-associated genes that impact cell function and lead to accumulation of senescent cells in various tissues.
Telomere shortening also plays an important role [4] in aging. It is well known that DNA replication machinery is not able to copy the ends of chromosomes (telomeres). Thus, with every round of DNA replication during cell division, telomeres become shorter. The process of telomere shortening can be prevented by specific DNA polymerase, called telomerase [4]. However, most adult somatic cells do not show telomerase activity at the level that could compensate telomere shortening during consecutive cell divisions. The shortening of telomeres below a critical length could be falsely sensed by the cell as DNA double-strand breaks. In response to this, the cell triggers DNA damage response mechanisms such as activation of the p53 tumor suppressor protein, for example [4]. The telomere shortening creates an ‘intrinsic biological clock’ for cellular aging and explains the so-called Hayflick's limit of cell division at the molecular level [5]. In contrast to somatic cells, telomerase activity is expressed at a higher level in stem cells [4]. However, as demonstrated, even in this population of cells, telomeres shorten with time [4].
In addition to mechanisms triggered by telomere shortening, DNA damage is induced by intra- and extracellular processes that perturb DNA replication and function (e.g. release of free radicals from the mitochondria during cellular respiration and after γ-irradiation, ultraviolet (UV) exposure, or exposure to chemical mutagens; fig. 1). Overall, DNA damage due to oxidative stress could be repaired by specialized cellular defense mechanisms such as DNA repair machineries and enzymes that metabolize reactive oxygen species (ROS). However, the dysfunction of these protective mechanisms observed during aging leads to the accumulation of damaged DNA. Indeed, a significant increase in DNA damages such as DNA mutations, DNA oxidation, chromosomal aneuploidy, and translocations have been demonstrated during aging in all types of adult tissues [6,7,8].
Epigenetic Regulation of Chromosome Structure
In nucleated cells (eukaryotes), DNA exists as a highly folded and compacted structure known as chromatin, in which DNA double helix is wrapped over core histone protein octamers to form nucleosomes, the fundamental units of chromatin [2]. The histone core proteins consist of two subunits of histones H2A, H2B, H3, and H4. Histone H1 functions as a linker histone to integrate other neighboring nucleosomes. Depending on the level of DNA compaction, we distinguish two types of chromatin, i.e. euchromatin and heterochromatin. Euchromatin is transcriptionally active and thus less organized. In contrast, heterochromatin is transcriptionally inactive, contains tightly compacted hypermethylated DNA, and includes several genetically inactive regions (e.g. satellite sequences, centromeres, telomeres, and the Barr body of the inactivated X chromosome in females). The heterochromatin is usually localized to the periphery of the nucleus. Due to the dense arrangement of DNA, heterochromatin is responsible for gene silencing and genomic stability.
Overall, the chromatin structure is regulated through the chemical modification of histone proteins and DNA itself. The compact nature of chromatin, where histones are wrapped by DNA, makes them marginally accessible to nuclear enzymes. However, both N- and C-terminal ends of histone proteins protrude from the chromatin, making them susceptible to a number of enzymatic post-translational modifications including phosphorylation, methylation, acetylation, ubiqutination, and mono- or poly(ADP-ribosyl)ation [9]. All these chemical modifications of histones are reversible and affect their secondary structure and interaction with DNA. Histone modification has a direct impact on the dynamic status of chromatin and gene expression.
In addition to histone modification, another important mechanism that plays a crucial role in chromatin organization is DNA methylation. DNA methylation results from a presence of cytosine nucleotides, which could be methylated at the C-5 position by DNA methyltransferases (DNMTs). Based on the substrate preference, we can distinguish two major classes of DNMTs in mammalian cells [10]. Accordingly, the first so-called de novo DNMTs (DNMT3a and DNMT3b) catalyze the methylation on previously unmethylated cytosine residues. This established DNA methylation pattern is subsequently maintained during DNA replication by DNMT1. It was also identified that the DMNT3-like (DNMT3L) protein shows a high homology to de novo DNMTs, but lacks enzymatic activity. However, DNMT3L modulates DNA methylation patterns as a regulatory factor for interaction with other DNMTs [11]. DNA methylation is involved in several biological phenomena including: (i) heterochromatin formation; (ii) X chromosome inactivation; (iii) genomic imprinting, and (iv) inhibition of gene transcription through recruitment of transcriptional co-repressors and chromatin-modifying enzymes [10].
All these epigenetic modifications of chromatin (e.g. histone modifications and DNA methylation) play a crucial role in maintaining genomic integrity, thus assuring proper gene transcription and DNA repair [9,10,12]. This explains why epigenetic modification of chromatin's structure was proposed to be a crucial factor in cellular aging in budding yeast, i.e. Saccharomyces cerevisiae[2]. Recently, evidence emerged to support that similar processes mediate aging in cells of organisms at a higher evolutionary level [3].
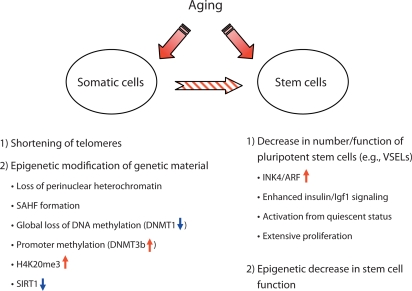
Therefore, in this review, we focus on age-related alteration in chromatin organization and discuss the epigenetic features that can influence the aging of adult somatic cells and stem cells (fig. 2).
Fig. 2.
Aging affects pools of somatic and stem cells. Somatic cells undergo an agerelated process of telomere shortening and epigenetic modification of their genetic material. The stem cell compartment is affected by decreases in the number and function of pluripotent stem cell populations (e.g. VSELs) as well as by epigenetic changes in a pool of tissue-committed stem cells. Overall, the age-related nuclear alterations in the stem cell compartment are more relevant for those somatic cells that show a short half-life duration (e.g. hematopoietic cells, intestinal epithelium, and keratinocytes). However, such a nuclear alteration also affects tissues and organs that show even relatively low cell turnover (e.g. heart, brain, skeletal muscles).
Age-Related Changes of Nuclear and Chromatin Organization
The significance of nuclear reorganization in aging has been described in several human progeroid syndromes such as Hutchinson-Gilford progeria syndrome (HGPS), Werner syndrome (WS), and ataxia telangiectasia (AT). It has been reported that defective mutations in genes encoding nuclear intermediate filaments, DNA helices, and DNA damage sensors cause genomic instability in HGPS, WS, and AT patients, respectively, which leads to accelerated aging in these patients. The molecular consequences of these mutations are discussed below.
Accordingly, the HGPS patients mentioned above have been identified with a defective mutation of the LMNA gene, which encodes lamin A, a crucial component of nuclear lamina that forms a perinuclear meshwork of nuclear intermediate filaments located on the inside of the nuclear membrane [13]. The mutant LMNA gene generates the truncated splice variant that perturbs normal nuclear organization. Consequently, HPGS patient cells show: (i) an aberrant nuclear shape; (ii) loss of heterochromatin-associated protein (e.g. heterochromatin protein-1 (HP1)) leading to decreased heterochromatin content, and (iii) altered patterns of histone modification [13,14]. The WS patients display an inherited mutation in the WRN gene, which encodes a member of the RecQ DNA helicase involved in DNA repair and recombination [15]. As result of this particular mutation, WS patient cells show high genomic instability, especially in repetitive loci. Finally, AT patients show an inherited defect in the ATM gene, which encodes a protein kinase that functions as a sensor for DNA damage and initiates the DNA damage cascade response when required [16]. In sum, changes in HGPS, WS, and AT progeroid syndrome patients are characterized by genomic instability, disorganization of the nuclear structure, progressive neurodegeneration, growth retardation, and premature aging. This shows that the genomic instability resulting from the disturbed nuclear architecture and heterochromatin structure may accelerate the aging process. In fact, to support this notion, some similar changes in nuclear reorganization have been reported in normal cells during their senescence [2].
As mentioned above, heterochromatin governs gene silencing and genomic stability. To explain this in the human genome, numerous repetitive elements could be susceptible to recombination due to their repetitive nature. Therefore, the confining of the repetitive elements into heterochromatin ensures the genomic stability by preventing improper recombination that, in consequence, could lead to chromosome breakage and fusion [2]. The disorganization of the nuclear architecture with concomitant loss of perinuclear heterochromatin is a common finding in senescent cells [2,13,14]. This aging-associated heterochromatin reorganization can influence both the genomic integrity and transcription of some aging-associated genes [2]. Indeed, it has been reported that in aged cardiomyoctes [17], similarly as in fibroblasts from HGPS patients [14], the reduction in repetitive sequence-associated heterochromatin structure may lead to increased transcription of the major satellite repeat that forms heterochromatin around chromosome centromeres. Moreover, the aging-associated changes in inactivation of the X chromosome and genomic imprinting result in the loss of allelic repression of selected genes (e.g. X-linked ATPase copper-transporting type 7a and one paternally imprinted insulin-like growth factor 2 (Igf2)) [18]. Thus, loss of epigenetic silencing of heterochromatin-related genes in aged tissue contributes to age-associated aberrant gene expression.
As another example, the existence of a link between perinuclear heterochromatin and transcription explains increased expression of collagenase in cells isolated from aged individuals as well as from WS patients [19]. To explain this phenomenon at the molecular level, the expression of the collagenase gene is repressed in an age-dependent manner by the Oct1 gene, a member of the POU domain family [20]. Accordingly, in cells from young individuals, Oct1 protein is located in the heterochromatin-enriched periphery of the nucleus, where it co-localizes with laminin B, a component of nuclear lamina. In senescent cells, by contrast, loss of heterochromatin content induces the release of Oct1 protein from the nuclear periphery, which results in a loss of Oct1-mediated repression of the collagenase gene. Thus, this age-related reorganization of heterochromatin may directly affect aging by modulating the transcription of aging-associated genes.
Another example of age-related changes in chromatin's structure is the formation of so-called senescence-associated heterochromatin foci (SAHF) [21]. The SAHF is a transcriptionally repressive heterochromatin structure, which is highly enriched in heterochromatin-associated modified histone (trimethylated lysine 9 (K9) of H3) and HP1. SAHF formation around E2F-responsive promoters induces the stable repression of E2F target genes through the recruitment of the retinoblastoma tumor suppressor. Thus, SAHF is one of the aging-associated chromatin structures that repress some of the growth-promoting genes in senescent cells.
Overall, the dynamic structure of chromatin is orchestrated by chemical modification of DNA and histones. Based on this, aging-associated chromatin reorganization (e.g., loss of heterochromatin and SAHF) could be explained by concomitant changes in DNA methylation and histone modifications. To support this notion, it has been shown that the amount of the trimethylation of H4-K20 (H4K20me3) that is highly enriched in pericentric heterochromatin regions increases in senescent cells [3] as well as in fibroblasts derived from HGPS patients [14]. Moreover, HGPS patients’ cells display a loss of the trimethylated H3K27 on the inactivated X chromosome.
In addition to the histone modifications, genomic global DNA methylation declines with age in cells located in several tissues [22]. This loss of global DNA methylation could be explained by the progressive loss of DNMT1 activity that results in the passive demethylation of heterochromatic DNA [23]. The precise role of this aging- associated phenomenon remains somewhat unclear. However, based on DNA methylation maintaining heterochromatin's integrity, this may suggest that the global change of DNA methylation can be associated with the reorganization of the heterochromatin structure during aging. Interestingly, in response to the global decrease of DNA methylation, senescent cells upregulate expression of de novo DNMT3b as a compensatory mechanism [23]. The overexpressed DNMT3b leads to the hypermethylation of the CpG islands in promoters of selected genes. To support this, senescent cells are known to exhibit the hypermethylated promoters in genes associated with aging, such as: (i) estrogen receptor; (ii) E-cadherin; (iii) collagen α1(I); (iv) c-fos; (v) Forkhead box O transcription factors (FoxOs); (vi) Igf2, and (vii) tumor suppressor candidate 33 (N33) [3]. This leads to the aging-associated changes in expression of these genes. In conclusion, the cellular aging and senescence could be result of reorganization of the nuclear architecture, changes in heterochromatin's structure, epigenetic modification of histones, and changes in DNA methylation.
Changes in the Expression and Function of Chromatin-Remodeling Factors during Aging
The aging-associated reorganization of chromatin's structure supports the idea that chromatin-remodeling factors play an important role in the aging process. As mentioned above, the alternation in DNMTs expression affects the transcription of aging-associated genes at the level of global DNA and promoter methylation [3]. On other hand, histone-modifying enzymes are also important contributors in the aging process [2,3]. It is also well known that Sirtuins, nicotinamide adenine dinucleotide-dependent histone deacetylases, are important players in the aging process.
Accordingly, the silent information regulator 2 (Sir2), the yeast ortholog of mammalian Sirtuins, delays the aging process of budding yeastby promoting heterochromatin formation through histone deacetylation in the repetitive genomic regions that are present in mating genes, telomeres, and ribosomal (r)DNA. The genetic deletion of Sir2 accelerates the aging process and, in contrast, overexpression of this gene leads to increased lifespan [24]. To support this further, calorie restriction (CR), which extends the lifespan of numerous organisms, was shown to increase the expression of Sir2 in budding yeast [24].
In the mammalian genome, seven Sirtuins (SIRT1–7) genes were found [25] and SIRT1, a close homolog of Sir2, has been extensively scrutinized in aging studies. The expression of SIRT1 is age-dependent [25] and induced by CR both in rodent and human cells [26]. In addition, SIRT1 is activated by resveratrol, which is known to increase the lifespan in lower organisms [24] and reduce the death rate in a high-fat diet rodent model [27]. SIRT1 removes the acetyl groups predominantly on histones H4-K16 and H3-K9 and promotes formation of heterochromatin [25]. However, it remains to be determined whether heterochromatin formations through SIRT1-mediated histone modifications found similarly in yeast are also directly involved in preventing aging of mammalian cells. In addition, SIRT1 also affects the function of several other non-histone proteins including p53, FoxO transcription factors (FoxOs), p300 histone acetyltransferase, tumor protein p73, E2F1, DNA repair factor Ku antigen 70-kDa subunit, nuclear factor κB, and the androgen receptor [24,25]. Therefore, due to its pleiotropic effects, SIRT1 could be involved in modulating aging at multiple levels, including: (i) gene transcription; (ii) cell cycle; (iii) signal transduction; (iv) stress response; (v) chromatin structure; (vi) DNA repair, and (vii) genomic integrity.
One group of interesting targets for SIRT1 is FoxOs that positively regulate several anti-aging-related genes (e.g. those involved in increasing levels of antioxidants, chaperones, and metabolism-related genes) and, at the same time, negatively regulate expression of putative pro-aging genes [28]. It is believed that the final biological outcome of SIRT1-mediated FoxOs deacetylation enhances expression of anti-aging genes.
Conversely, the activity of FoxOs is regulated by phosphorylation, which sequestrates phosphorylated FoxOs in the cytoplasm and, as result of this, inhibits FoxOs-mediated transcription [28]. Interestingly, the insulin/insulin-like growth factor 1 (Igf1), which regulates the lifespan in worms, flies, and mammals, phosphorylates FoxOs in a serine/threonine kinase AKT/protein kinase B (PKB) manner [29]. Thus, the increase of lifespan induced by the inhibition of the insulin/Igf1 signaling cascade is partially a result of the blockage of AKT/PKB- mediated FoxO phosphorylation. Because SIRT1 inhibits the insulin-stimulated phosphorylation of FoxOs [28], it negatively regulates the insulin/Igf1 signaling by the post-translational modification of these transcription factors. In addition, SIRT1 is involved in insulin secretion and expression of IGF-binding protein-1 [29]. Thus, SIRT1 could be envisioned as a central mediator for cellular aging that orchestrates cell-intrinsic (nuclear reorganization-dependent) and cell-extrinsic (oxidative stress- and endocrine signal-mediated) processes.
Effect of Aging on the Stem Cell Compartment
Aging results from a decrease in the rejuvenation and regeneration potential of several vital organs and tissues. Because tissue regeneration depends on a proper function of stem cells, multipotent stem/primitive cells become a major focus of interest in regenerative medicine. The previously discussed epigenetic mechanisms above may directly affect the stem cell compartment. Recent data (fig. 2) also suggest that aging could be explained partially by the decline in the regenerative potential of adult stem cells [30]. This could be explained by: (i) age-related dysfunction; (ii) exhaustion of the stem cell pool due to tissue regeneration; (iii) damage by oxidative or environmental cues, and (iv) aging-associated epigenetic changes [31]. What is important to mention is that any stable changes in genetic material within stem cells are passed to the daughter stem cells and are maintained in the stem cell compartment. Below, we briefly discuss the contribution of nuclear and chromatin alterations in stem cells to aging.
Pluripotent ‘Very Small Embryonic/Epiblast-Like Stem Cells’ and Aging
A population of the most primitive pluripotent stem cells in adult tissues was recently identified by our team. Accordingly, we isolated rare stem cells from murine bone marrow (BM) by multiparameter fluorescence-activated cell sorter (FACS) as a population of Sca-1+lin−CD45−s. We named these cells ‘very small embryonic/epiblast-like stem cells’ (VSELs) as they: (i) are very rare (∼0.01% of nucleated cells); (ii) are small in size (∼3–6 μm); (iii) express pluripotent markers such as Oct-4, Nanog, Rex-1, and SSEA-1; (iv) possess large nuclei containing unorganized chromatin (euchromatin), and (v) are capable of differentiation (Oct-4+) in vitro into cells from all three germ lineages [32]. In ImageStream system analysis, which is flow cytometry combined with high-resolution fluorescence microscopy, we found that VSELs exhibit a significantly higher nuclear/cytoplasm ratio and a lower cytoplasmic area as compared with hematopoietic stem cells [33].
The rare population of similar Oct-4+ VSEL-like cells is also present in multiple murine organs [33] and human umbilical cord blood, BM, and peripheral blood [34]. When purified VSELs are plated over a C2C12 myoblast feeder layer, they form spheres that resemble embryoid bodies. The VSEL-derived spheres (VSEL-DSs) contain primitive stem cells that, after replating into media, promote tissue differentiation and are able to differentiate into cells from all three germ layers. Furthermore, we reported that VSELs are mobilized into peripheral blood during organ injuries (e.g., heart infarct, stroke), which suggests that these cells could participate in the regeneration of damaged tissues [35,36].
The number of VSELs in murine BM gradually declines with age, ranging from 0.052 ± 0.018% to 0.003 ± 0.002% between ages of 2 months and 3 years, respectively [31]. More importantly, the ability of VSEL-DS formation decreases with age, thus no VSEL-DSs were observed in cells isolated from older mice (>2 years) [40]. The aging-dependent decrease of the pool and function of VSELs in BM may explain the decline of the regeneration potential during aging. This hypothesis has been further confirmed by looking for differences in the content of these cells among BM mononuclear cells (BMMNCs) in long- and short-lived mouse strains. The concentration of VSELs was much higher in the BM of long-lived (e.g., C57B6) as compared to short-lived (DBA/2J) mice [32]. Because we reported that the Oct-4 promoter is hypomethylated and displays an open chromatin structure in BM-derived VSELs from 2-month-old mice [37], it would be important to see whether this changes with the age.
Epigenetic Changes of Imprinted Genes Regulate VSELs Pluripotency
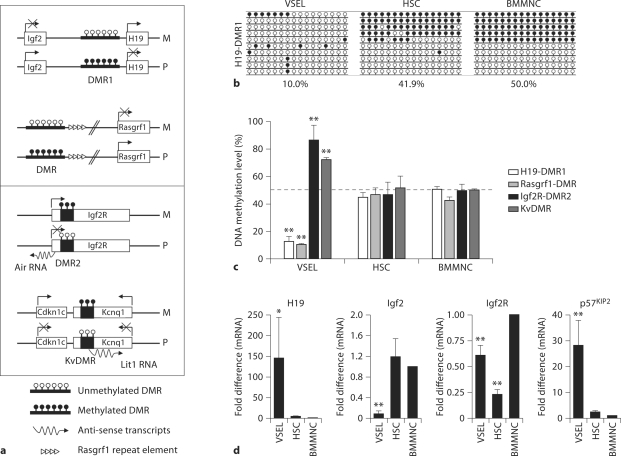
We noticed that Oct-4+ VSELs do not proliferate in vitro if cultured alone and that the quiescence of these cells is epigenetically regulated by DNA methylation of genomic imprinting, which is an epigenetic program that ensures the parent-of-specific monoallelic transcription of imprinted genes [37]. The imprinted genes play a crucial role in embryogenesis, fetal growth, totipotential status of the zygote, and pluripotency of developmentally early stem cells [38]. The expression of imprinted genes is regulated by DNA methylation on differential methylated regions (DMRs), which are CpG-rich cis-elements in their loci [38]. We noticed that VSELs freshly isolated from murine BM erase the paternally methylated imprints (e.g. Igf2-H19, Rasgrf1 loci), however they hypermethylate the maternally methylated ones (e.g. Igf2 receptor (Igf2R), Kcnq1-p57KIP2, Peg1 loci) (fig. 3b, c).
Fig. 3.
The unique DNA methylation pattern and expression of imprinted genes in VSELs. a Schematic diagram of paternally (Igf2-H19 and Rasgrf1, upper panel) and maternally (Igf2R and Kcnq1) methylated loci. Arrow indicates the active transcription from indicated genes. DMR = Differential methylated regions, M = maternal chromosome, P = paternal chromosome, KvDMR = DMR for Kcnq1 locus. b Bisulfite sequencing results of DNA methylation of DMR1 for Igf2-H19 locus in the indicated cells (VSELs = very small embryonic like stem cells, HSC = hematopoietic stem cells, BMMNC = bone marrow mononuclear cells). Methylated and unmethylated CpG sites are shown in filled and open circles, respectively. The numbers under the bisulfite sequencing results indicate the percentage of methylated CpG sites. c The DNA methylation level of DMRs for imprinted genes. Mean values for percentage of methylated CpG sites are shown as the mean ± SD from at least three independent experiments. ∗ p < 0.05, ∗∗ p < 0.01 compared to BMMNCs. d RQ-PCR analysis of H19, Igf2, Igf2R, and p57KIP2 in the indicated cells. Relative quantification of the expression level of the imprinted genes was performed with the 2−ΔΔCt method, using the mRNA level of 2-microglobulin as an endogenous control gene and that of BMMNCs as a calibrator. The relative expression level is represented as the fold difference to the value of BMMNCs and shown as the mean ± SD from at least four independent experiments on different samples of FACS sorted VSELs, HSCs, and BMMNCs. ∗ p < 0.05, ∗∗ p < 0.01 compared to BMMNCs.
Because paternally expressed imprinted-genes (Igf2, Rasgrf1) enhance the embryo growth and maternally expressed genes (H19, p57KIP2, Igf2R) inhibit cell proliferation [38], the unique genomic imprinting pattern observed on VSELs demonstrates growth-repressive imprints in these cells. As supported, VSELs highly express growth-repressive genes (H19, p57KIP2, Igf2R) and downregulate growth-promoting genes (Igf2, Rasgrf1), which explains the quiescent status of VSELs (fig. 3d). Importantly, the quiescent pattern of genomic imprinting was progressively recovered during the formation of VSEL-DSs, in which stem cells proliferate and differentiate. These results suggest that epigenetic reprogramming of genomic imprinting should maintain the quiescence of the most primitive pluripotent adult stem cells (e.g. Oct-4+ VSELs) deposited in the adult body and protect them from premature aging and tumor formation. Therefore, it will be important to investigate whether this genomic imprinting pattern differs between VSELs isolated from young versus old mice and whether these potential epigenetic changes could contribute to the previously mentioned decrease in the pool and function of VSELs during aging. Furthermore, because the pluripotency of VSELs is regulated by the DNA methylation of the Oct-4 promoter [37], potential changes of Oct-4 promoter methylation could contribute to the VSELs aging and decrease the number of these cells with age.
Quiescence of Pluripotent VSELs and Its Potential Link to Insulin/Igf1 Signaling Pathways
As a result of the unique DNA methylation pattern of imprinted genes, VSELs express lower amounts of Igf2, but highly express the non-signaling receptor Igf2R, which functions at the cell surface as a decoy receptor for Igf2 and prevents its binding to tyrosine kinase receptor, i.e. Igf1R [38] (fig. 3d). Furthermore, we noticed that another paternally imprinted gene, Rasgrf1, which is one of the Ras-specific guanine nucleotide exchange factors and activates the Ras protein, is expressed at very low levels in VSELs [37]. This gene could be also linked to insulin/Igf1 signaling. Accordingly, Rasgrf1-deficient mice show reduced levels of Igf1 (∼32–40%) and growth hormone (∼42–35%) relative to wild-type mice [39]. Moreover, it has been reported that Igf1-mediated signaling is impaired in Rasgrf1-deficient pancreatic β cells [40]. Therefore, these results suggest that the unique expression of imprinted genes in VSELs could negatively regulate the insulin/Igf1 signaling in these pluripotent stem cells and maintain their quiescence. As such, it is important to identify potential mechanisms responsible for regulating insulin/Igf1 signaling in VSELs together with potential involvement of SIRT1 and FoxOs, which, as mentioned above, are involved in the aging process.
Conclusion
The aging-associated changes in the nuclear architecture, chromatin structure, altered expression and activity of chromatin remodeling factors, and change in pattern of epigenetic marks (DNA methylation and histone modification) affect all cells in the adult body including the population of stem cells responsible for proper tissue rejuvenation. Therefore, the elucidation of these precise mechanisms will help to develop more efficient anti-aging strategies as well as facilitate a better understanding of age-related risks for cancer genesis.
Acknowledgements
This work was supported by NIH R01 CA106281-01, NIH R01 DK074720, and Stella and Henry Endowment and European Union structural funds, Innovative Economy Operational Program POIG 01.02-00-109/09 to M.Z.R., and by NIH P20RR018733 from the National Center for Research Resources to M.K.
References
- 1.Bergmann O, Bhardwaj RD, Bernard S, Zdunek S, Barnabé-Heider F, Walsh S, Zupicich J, Alkass K, Buchholz BA, Druid H, Jovinge S, Frisén J. Evidence for cardiomocyte renewal in humans. Science. 2009;324:98–102. doi: 10.1126/science.1164680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Oberdoerffer P, Sinclair DA. The role of nuclear architecture in genomic instability and aging. Nat Rev Mol Cell Biol. 2007;8:692–702. doi: 10.1038/nrm2238. [DOI] [PubMed] [Google Scholar]
- 3.Fraga MF, Esteller M. Epigenetics and aging: the targets and the marks. Trends Genet. 2007;23:413–418. doi: 10.1016/j.tig.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 4.Blasco MA. Telomere length, stem cells and aging. Nat Chem Biol. 2007;3:640–649. doi: 10.1038/nchembio.2007.38. [DOI] [PubMed] [Google Scholar]
- 5.Collado M, Blasco MA, Serrano M. Cellular senescence in cancer and aging. Cell. 2007;130:223–233. doi: 10.1016/j.cell.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 6.Vijg J. Somatic mutations and aging: a re-evaluation. Mutat Res. 2000;447:117–135. doi: 10.1016/s0027-5107(99)00202-x. [DOI] [PubMed] [Google Scholar]
- 7.Hamilton ML, Van Remmen H, Drake JA, Yang H, Guo ZM, Kewitt K, Walter CA, Richardson A. Does oxidative damage to DNA increase with age? Proc Natl Acad Sci USA. 2001;98:10469–10474. doi: 10.1073/pnas.171202698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rehen SK, Yung YC, McCreight MP, Kaushal D, Yang AH, Almeida BSV, Kingsbury MA, Cabral KMS, McConnell MJ, Anliker B, Fontanoz M, Chun J. Constitutional aneuploidy in the normal human brain. J Neurosci. 2005;25:2176–2180. doi: 10.1523/JNEUROSCI.4560-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Margueron R, Trojer P, Reinberg D. The key to development: Interpreting the histone code? Curr Opin Genet Dev. 2005;15:163–176. doi: 10.1016/j.gde.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 10.Klose RJ, Bird AP. Genomic DNA methylation: the mark and its mediators. Trends Biochem Sci. 2006;31:89–97. doi: 10.1016/j.tibs.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 11.Hata K, Okano M, Lei H, Li E. Dnmt3L cooperates with the Dnmt3 family of de novo DNA methyltransferases to establish maternal imprints in mice. Development. 2002;129:1983–1993. doi: 10.1242/dev.129.8.1983. [DOI] [PubMed] [Google Scholar]
- 12.Van Attikum H, Gasser SM. The histone code at DNA breaks: a guide to repair? Nat Rev Mol Cell Biol. 2005;6:757–765. doi: 10.1038/nrm1737. [DOI] [PubMed] [Google Scholar]
- 13.Hennekam RC. Hutchinson-Gilford progeria syndrome: review of the phenotype. Am J Med Genet A. 2006;140A:2603–2624. doi: 10.1002/ajmg.a.31346. [DOI] [PubMed] [Google Scholar]
- 14.Shumaker DK, Dechat T, Kohlmaier A, Adam SA, Bozovsky MR, Erdos MR, Eriksson M, Goldman AE, Khuon S, Collins FS, Jenuwein T, Goldman RD. Mutant nuclear lamin a leads to progressive alterations of epigenetic control in premature aging. Proc Natl Acad Sci USA. 2006;103:8703–8708. doi: 10.1073/pnas.0602569103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu C-E, Oshima J, Fu Y-H, Wijsman EM, Hisama F, Alisch R, Matthews S, Nakura J, Miki T, Ouais S, Martin GM, Mulligan J, Schellenberg GD. Positional cloning of the Werner's syndrome gene. Science. 1996;272:258–262. doi: 10.1126/science.272.5259.258. [DOI] [PubMed] [Google Scholar]
- 16.Shiloh Y. Ataxia telangiectasia: closer to unraveling the mystery. Eur J Hum Genet. 1995;3:116–138. doi: 10.1159/000472285. [DOI] [PubMed] [Google Scholar]
- 17.Gaubatz JW, Cutler RG. Mouse satellite DNA is transcribed in senescent cardiac muscle. J Biol Chem. 1990;265:17753–17758. [PubMed] [Google Scholar]
- 18.Bennett-Baker PE, Wilkowski J, Burke DT. Age-associated activation of epigenetically repressed genes in the mouse. Genetics. 2003;165:2055–2062. doi: 10.1093/genetics/165.4.2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Millis AJ, Hoyle M, McCue HM, Martini H. Differential expression of metalloproteinase and tissue inhibitor of metalloproteinase genes in aged human fibroblasts. Exp Cell Res. 1992;201:373–379. doi: 10.1016/0014-4827(92)90286-h. [DOI] [PubMed] [Google Scholar]
- 20.Imai S, Kitano H. Heterochromatin islands and their dynamic reorganization: a hypothesis for three distinctive features of cellular aging. Exp Gerontol. 1998;33:555–570. doi: 10.1016/s0531-5565(98)00037-0. [DOI] [PubMed] [Google Scholar]
- 21.Narita M, Nunez S, Heard E, Narita M, Lin AW, Hearn SA, Spector DL, Hannon GJ, Lowe SW. Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell. 2003;113:703–716. doi: 10.1016/s0092-8674(03)00401-x. [DOI] [PubMed] [Google Scholar]
- 22.Wilson VL, Jones PA. DNA methylation decreases in aging but not in immortal cells. Science. 1983;220:1055–1057. doi: 10.1126/science.6844925. [DOI] [PubMed] [Google Scholar]
- 23.Casillas MA, Lopatina N, Andrews LG, Tollefsbol TO. Transcriptional control of the DNA methyltransferases is altered in aging and neoplastically-transformed human fibroblasts. Mol Cell Biochem. 2003;252:33–43. doi: 10.1023/a:1025548623524. [DOI] [PubMed] [Google Scholar]
- 24.Guarente L, Picard FI. Calorie restriction – the SIR2 connection. Cell. 2005;120:473–482. doi: 10.1016/j.cell.2005.01.029. [DOI] [PubMed] [Google Scholar]
- 25.Vaquero A, Sternglanz R, Reinberg D. NAD+-dependent deacetylation of H4 lysine 16 by class III HDACs. Oncogene. 2007;26:5505–5520. doi: 10.1038/sj.onc.1210617. [DOI] [PubMed] [Google Scholar]
- 26.Cohen HY, Miller C, Bitterman KJ, Wall NR, Hekking B, Kessler B, Howitz KT, Gorospe M, de Cabo R, Sinclair DA. Calorie restriction promotes mammalian cell survival by inducing the sirt1 deacetylase. Science. 2004;305:390–392. doi: 10.1126/science.1099196. [DOI] [PubMed] [Google Scholar]
- 27.Lagouge M, Argmann C, Gerhart-Hines Z, Meziane H, Lerin C, Daussin F, Messadeq N, Milne J, Lambert P, Elliott P, Geny B, Laakso M, Puigserver P, Auwerx J. Resveratrol improves mitochondrial function and protects against metabolic disease by activating SIRT1 and PGC-1i. Cell. 2006;127:1109–1122. doi: 10.1016/j.cell.2006.11.013. [DOI] [PubMed] [Google Scholar]
- 28.Van der Horst A, Burgering BMT. Stressing the role of FoxO proteins in lifespan and disease. Nat Rev Mol Cell Biol. 2007;8:440–450. doi: 10.1038/nrm2190. [DOI] [PubMed] [Google Scholar]
- 29.Russell SJ, Kahn CR. Endocrine regulation of aging. Nat Rev Mol Cell Biol. 2007;8:681–691. doi: 10.1038/nrm2234. [DOI] [PubMed] [Google Scholar]
- 30.Sharpless NE, DePinho RA. How stem cells age and why this makes us grow old. Nat Rev Mol Cell Biol. 2007;8:703–713. doi: 10.1038/nrm2241. [DOI] [PubMed] [Google Scholar]
- 31.Ratajczak MZ, Zuba-Surma EK, Shin DM, Ratajczak J, Kucia M. Very small embryonic-like stem cells in adult organs and their potential role in rejuvenation of tissues and longevity. Exp Gerontol. 2008;43:1009–1017. doi: 10.1016/j.exger.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kucia M, Reca R, Campbell FR, Zuba-Surma E, Majka M, Ratajczak J, Ratajczak MZ. A population of very small embryonic-like CXCR4+ SSEA-1+ Oct-4+ stem cells identified in adult bone marrow. Leukemia. 2006;20:857–869. doi: 10.1038/sj.leu.2404171. [DOI] [PubMed] [Google Scholar]
- 33.Zuba-Surma E, Kucia M, Ratajczak J, Ratajczak MZ. ‘Small stem cells’ in adult tissues: very small embryonic-like stem cells stand up! Cytometry A. 2009;75:4–13. doi: 10.1002/cyto.a.20665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kucia M, Halasa M, Wysoczynski M, Baskiewicz-Masiuk M, Moldenhawer S, Zuba-Surma E, Czajka R, Wojakowski W, Machalinski B, Ratajczak MZ. Morphological and molecular characterization of novel population of CXCR4+ SSEA-4+ Oct-4+ very small embryonic-like cells purified from human cord blood: preliminary report. Leukemia. 2006;21:297–303. doi: 10.1038/sj.leu.2404470. [DOI] [PubMed] [Google Scholar]
- 35.Wojakowski W, Tendera M, Kucia M, Zuba-Surma E, Paczkowska E, Ciosek J, Halasa M, Król M, Kazmierski M, Buszman P, Ochala A, Ratajczak J, Machalinski B, Ratajczak MZ. Mobilization of bone marrow-derived Oct-4+ SSEA-4+ very small embryonic-like stem cells in patients with acute myocardial infarction. J Am Coll Cardiol. 2009;53:1–9. doi: 10.1016/j.jacc.2008.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paczkowska E, Kucia M, Koziarska D, Halasa M, Safranow K, Masiuk M, Karbicka A, Nowik M, Nowacki P, Ratajczak MZ, Machalinski B. Clinical evidence that very small embryonic-like stem cells are mobilized into peripheral blood in patients after stroke. Stroke. 2009;40:1237–1244. doi: 10.1161/STROKEAHA.108.535062. [DOI] [PubMed] [Google Scholar]
- 37.Shin DM, Zuba-Surma EK, Wu W, Ratajczak J, Wysoczynski M, Ratajczak MZ, Kucia M. Novel epigenetic mechanisms that control pluripotency and quiescence of adult bone marrow-derived Oct-4+ very small embryonic-like stem cells. Leukemia. 2009;23:2042–2051. doi: 10.1038/leu.2009.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reik W, Walter J. Genomic imprinting: parental influence on the genome. Nat Rev Genet. 2001;2:21–32. doi: 10.1038/35047554. [DOI] [PubMed] [Google Scholar]
- 39.Itier JM, Tremp GL, Leonard JF, Multon MC, Ret G, Schweighoffer F, Tocque B, Bluet-Pajot MT, Cormier V, Dautry F. Imprinted gene in postnatal growth role. Nature. 1998;393:125–126. doi: 10.1038/30120. [DOI] [PubMed] [Google Scholar]
- 40.Font de Mora J, Esteban LM, Burks DJ, Nunez A, Garces C, Garcia-Barrado MJ, Iglesias-Osma MC, Moratinos J, Ward JM, Santos E. Ras-GRF1 signaling is required for normal β-cell development and glucose homeostasis. EMBO J. 2003;22:3039–3049. doi: 10.1093/emboj/cdg280. [DOI] [PMC free article] [PubMed] [Google Scholar]