Abstract
Fine mapping of calcineurin (PPP3CA) gene identified single nucleotide polymorphisms (SNPs) and simple sequence repeat polymorphisms that are associated with addiction vulnerability. A trinucleotide repeat marker, located in the 5′ untranslated region (5′UTR) of the PPP3CA mRNA, exhibited significantly different genotype and allele frequencies between abusers and controls in the NIDA African–American sample. The polymorphism showed allelic-specific expression in mRNA extracted from postmortem brain specimens. Novel alternatively spliced isoforms of PPP3CA were identified and their expressions were found altered in brain regions of postmortem Alzheimer's disease patients. These data underscore the importance of calcineurin gene in the molecular mechanism of addiction and Alzheimer's diseases.
Keywords: phosphatase, SNP association, gene regulation, alternative splicing
Introduction
Genome wide association using denser and denser single nucleotide polymorphism (SNP) sets has allowed identification of a number of genes that display SNPs whose allelic frequencies distinguish individuals with substance dependence from control individuals (Johnson et al., 2006; Liu et al., 2005a,b; Uhl, Liu, Walther, Hess, & Naiman, 2001). rs1395475, an SNP whose allele frequencies distinguished dependent from control individuals, lies at the locus for the PPP3CA gene (Liu et al., 2005a,b) that encodes the alpha catalytic subunit of a Ca2+/calmodulin(CaM)-dependent serine/threonine phosphatase, also called calcineurin or PP2B.
PPP3CA/calcineruin acts as a calcium-activated heteromeric complex that contains calcium, calmodulin, a catalytic subunit and a regulatory subunit (Klee, Crouch, & Krinks, 1979). Other PPP3CA gene family members include the additional catalytic subunits, PPP3CB and PPP3CC and the regulatory subunits, PPP3R1 and PPP3R2.
Calcineurin functions include (a) regulation of presynaptic phosphorylation of dynamin and the PP1 inhibitor DARPP-32, (b) postsynaptic phosphoregulation via actions from its anchored localization to scaffolding proteins in postsynaptic densities, and (c) regulation of transcriptional modulators that include the transcription factor NFAT (Yakel, 1997). Calcineurin plays roles in multiple brain circuits important for reward, memory, and aging in ways thought to link dopaminergic and glutamatergic signals (Gerber et al., 2003). Altered calcineurin expression or function can alter rewarding effects of abused substances (Biala, Betancur, Mansuy, & Giros, 2005; Gerdjikov and Beninger, 2005), alter long-term potentiation (LTP; Winder, Mansuy, Osman, Moallem, & Kandel, 1998) and change memory functions (Malleret et al., 2001; Mansuy, Mayford, Jacob, Kandel, & Bach, 1998). Evidence that directly implicates calcineurin in the memory-associated affliction, Alzheimer's disease (AD), include observations that overall calcineurin expression levels and calcineurin activities are reduced in AD brains (Gong, Liu, Grundke-Iqbal, & Iqbal, 2006). PPP3CA activity is reduced and its aggregation is increased in the aging brain (Agbas, Zaidi, & Michaelis, 2005).
Although PPP3CA is thus a strong candidate to play central roles in addiction and memory-associated disorders, the details of PPP3CA associations with these disorders, the detailed structure of the PPP3CA gene and its variants have not all been elucidated. We have thus sought to follow up on initial evidence for association of PPP3CA markers with substance dependence. We assemble, for the first time, a complete profile of the PPP3CA gene structure, delineating novel splice variants and polymorphisms. We define isoform-selective differences in PPP3CA expression in mRNAs extracted from AD brains. These observations point to selective roles for PPP3CA and its variants in the pathogenesis of addiction, AD and, perhaps, other disorders that involve reward and/or memory-like features.
Materials and Methods
Substance Abuser and Control Research Subjects
Genomic DNAs came from NIH IRP (NIDA) research volunteers of self-reported European and African–American descent whose drug use and dependence were characterized as described (Liu et al., 2005a,b; Uhl et al., 2001). A total of 389 unrelated European-American “abusers,” averaged 35 years of age, scored 3+ on a total drug use scale of drug use of addictive substances, and demonstrated DSMIII-R or DSMIV dependence on at least one illegal abused substance. Two hundred forty “control” European-Americans with average age of 32 years reported no significant lifetime histories of use of any addictive substance and 0–1+ DUS (drug use scale) total drug use scores. DNAs from “abusers” of self-reported African–American descent numbered two hundred forty in total, while one hundred “control” African-Americans were analyzed for the current study.
Human Brain Tissue
Brain tissue from 57 pathologically confirmed AD patients and 20 non-AD “control” individuals were obtained postmortem from the Department of Pathology, Johns Hopkins Medical Institutions. For each individual sample, portions of medial temporal gyrus (Brodmann area 21) and medial frontal gyrus (Brodmann area 46) were subjected to extraction of RNA using RNAzolB (Tel-test, Friendswood, TX), as previously described (Liu et al., 2006). cDNA was prepared using SuperScript First strand cDNA synthesis (Invitrogen, Carlsbad, CA) for reverse transcription-PCR reaction. cDNA samples were used to analyze allelic and alternative splicing isoform expression (described in the following section). Additional cDNA was synthesized from total RNA from amygdala, caudate, putamen, cerebellum, cortex, hippocampus, hypothalamus, substantial nigra, heart, intestine, kidney, leukocyte, liver, lung, muscle, spleen, and testis purchased from BD Bioscience Clontech (Palo Alto, CA).
Simple Sequence Repeat Genotyping
Simple sequence length polymorphisms were identified throughout the PPP3CA gene by alignment of genomic with expressed sequence tag (EST) and mRNA sequences available from NCBI (www.ncbi.nlm.nih.gov) using Sequencher software (Version 3.0, Gene Code Co., Ann Arbor, MI).
SSLPs were genotyped using PCR amplifications in which one primer of the pair was labeled with carboxylfluorescein (6-FAM) at the 5′-terminal end of the primer (Table S1). For the PPP3CA_Prmt and PPP3CA_Int2 primer pairs, genomic DNA samples (10 ng/μl) were added to the reaction using 1× Gold buffer, 1.5 mM MgCl2, 0.8 mM dNTPs, 20 μM of each oligonucleotide primer, and 0.13 U of AmpliTaq Gold polymerase. Each 5 μl reaction used a procedure of 94°C for 5 min, 35 cycles of 94°C for 0.5 min, 55°C for 0.5 min, 72°C for 0.75 min, followed by 72°C for 7 min. The PPP3CA_EX1 primer pair required use of Herculase Hotstart DNA polymerase (Stratagene, La Jolla, CA) and 5% dimethylsulfoxide (DMSO) to amplify this GC-rich region. For the 5 μl reaction, genomic DNA samples were amplified using a procedure of 95°C for 10 min, 45 cycles of 94°C for 0.5 min, 62°C for 0.5 min, 72°C for 0.75 min, followed by 72°C for 10 min. The PCR products were denatured with Hi-Di Formamide (Applied Biosystems, Foster City, CA) at 95° for 5 min. The fragments were electrophoresed using a DNA Sequencer (ABI 3100, Applied Biosystems) with a fluorescent ladder ROX350 (Applied Biosystems) as an internal size marker and analyzed with GENOTYPER software (Version 3.7).
SNP Genotyping With Taqman Assays. SNPs rs2851060, rs713455, and rs3730251 were genotyped using Allelic Discrimination software (SDS v2.1) and Taqman assays for ABI Prism 7900HT Sequence Detection System (Applied Biosystems). To distinguish between each SNP allele, one Taqman minor groove binding (MGB) probe was labeled with VIC reporter dye and another labeled with FAM reporter dye included with SNP sequence-specific forward and reverse primers. Reactions were performed in a 384-well format in a total reaction volume of 5 μl containing 6.0 ng of genomic DNA and 1× TaqMan Universal PCR Master Mix without AmpErase UNG (Applied Biosystems) using 50°C for 2 min, 95°C for 10 min, and 40 cycles of 95°C for 0.25 min, 60°C for 1 min. Fluorescent signals between 500—660 nm were analyzed for each plate and genotypes were scored using automated software.
SNP Genotyping Using Multiplex PCR and SNaPshot Assays
PCR primers (Table S2) were generated using Primer3 software (http://www-genome.wi.mit.edu/cgi-bin/primer/primer3_www.cgi). Amplicons were designed to display sizes of 80 bp–400 bp. Five microliters PCR reactions were performed using 3 ng–10 ng of genomic DNA, 2.5 mM MgCl2, 0.1 mM dNTP, 0.025 U AmpliTaq Gold DNA polymerase, and 1 pmol of eight mixed primers. Initial denaturation at 95° for 10 min was followed by 40 cycles of 95° for 30 s, 55° for 15 s, and 72° for 30 s using GeneAmp PCR Biosystem 9700 in 384-well reaction plates. PCR conditions were verified by direct DNA sequencing using mixed PCR products after treatment with exonuclease (USB), 0.5 units per sample, and shrimp alkaline phosphatase (SAP, USB), 0.50 U/sample for 30 min at 37°, and 5 min at 95°. Following multiplex DNA amplification, unincorporated PCR primers and dNTPs were degraded by adding 0.5 units per sample exonuclease and 0.50 U/sample SAP, incubating for 30 min at 37° and incubating for a further 5 min at 95°. After purification, 1.5 μl of cleaned PCR product was mixed with 5.5 μl of water, 2.0 μl of SNaPshot ready Reaction Mix and 1.0 μl of mixed SNaPshot extension primers (2 pmol/μl). Twenty-five thermocycles of 96° for 10 s, 50° for 5 s, 60° for 30 s were followed by treatment of postextension products 0.50 U SAP/sample, incubation for 30 min at 37° and for 5 min at 95°. One microliter of final product was denatured with 8.5 μl Hi-Di Formamide (ABI) and 0.5 μl Liz-120 Size Standards (ABI) at 95° for 5 min. Fragments were then electrophoresed using a 3100 DNA Analyzer (ABI) and analyzed using ABI GeneScan version 3.7 software. Genotypes were scored manually, using size standards for peak verification.
Allele-Specific Expression
cDNA samples from human brain region tissues and genomic samples, both from individuals who were heterozygous for the PPP3CA 5′UTR tri-nucleotide repeat (GCC), were used for allele-specific expression assays. Genomic DNA and cDNA samples from medial temporal gyrus (n = 13) and/or medial frontal gyrus (n = 14) were added to 384-well plates containing 1× reaction buffer (Stratagene), 0.8 mM dNTPs (Stratagene), CYBR-1 nucleic acid green stain dye (Biowhittaker Molecular Applications), 1× Rox reference dye (Invit-rogen), 5% DMSO, 20 uM each of PPP3CA_EX1F4 and PPP3CA_EX1R5, and Herculase enzyme (Stratagene). Reactions used an ABI Prism 7900HT Sequence Detection System with 95°C denaturation for 10 min followed by 45 cycles of 94°C for 0.5 min, 62°C for 0.5 min, 72°C for 0.75 min, and then terminal 10 min 72°C incubation. Dissociation, involved a hold at 50°C for 5 min, followed by one cycle of 95°C for 0.25 min, 60°C for 0.25 min, and 95°C for 0.25 min, and was used to degrade short fragment primer dimers. The linear range for each sample was identified using SDS v2.1 software. Using the determined linear range, each sample was analyzed in duplicate by PCR using the FAM-labeled primer for PPP3CA_EX1. Products were resolved using an ABI 3100 sequence analyzer, as described. Peak area measurements were averaged. Relative peak areas for each allele were compared to produce an assessment of relative peak areas for each heterozygous sample for brain cDNAs and heterozygote genomic DNA standards.
Isoform-Specific Expression
PPP3CA isoforms were identified by aligning genomic sequence with human PPP3CA ESTs using Sequencher software (Version 3.0, Gene Code Co.). As described previously (Liu et al., 2006), exon-specific primers and fluorescent FAM-labeled MGB probes were designed (Table S3) across two alternatively spliced exons using Primer Express (Applied Biosystems) to avoid nonspecific amplification of genomic DNA. The cDNA templates were amplified using ABI Prism 7900HT Sequence Detection System (Applied Biosystems) with a cycling program of 50°C for 2 min, 95°C for 10 min, followed by 40 cycles of 95°C for 0.25 min and 60°C for 1 min. Predeveloped human glyceraldehyde-3-phosphate dehydrogenase (GAPD) (Applied Biosystems) provided a control. Quantification used methods in user bulletin #2 (ABI Prism 7700 Sequence Detection System) and graphs used Prism 3.0 (GraphPad Software, Inc. San Diego, CA).
Genetic Data and Statistical Analyses
GOLD software (http://www.sph.umich.edu/csg/abecasis/GOLD) allowed graphical display of LD between SNP and microsatellite markers (Abecasis and Cookson, 2000). PHASE version 2.1 (http://www.stat.washington.edu/stephens/software.html) provided haplotype estimates (Stephens and Donnelly, 2003; Stephens, Smith, & Donnelly, 2001). Chi-square tests (Microsoft Excel, MS Office 2003) identified genotype or allele frequency association for PPP3CA microsatellite and SNP markers. Statistical analyses for PPP3CA brain region expression differences used Prism 3.0 software (GraphPad, Inc., San Diego, CA).
Results
rs1395475 Genotypes in Individually Genotyped Substance Dependence versus Control Samples
We sought verification of the microarray-based genome scan data that used pooled DNA samples (Liu et al., 2005a,b) to identify association of rs1395475 [Figure 1(A)], by genotyping this SNP in 379 individual African–American abuser and control samples (a subset of the individuals genotyped in pools in Liu et al., 2005a,b). These results produced significant allele frequency differences in this marker, which lies in PPP3CA intron 3(χ2 = 6.07, p = .014).
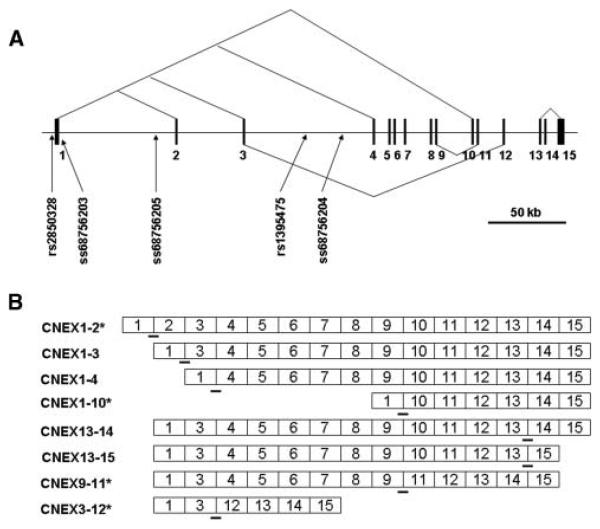
Figure 1.
Calcineurin (PPP3CA) marker map. Physical representation of polymorphisms and splicing isoforms throughout the 330 kb calcineurin A subunit gene on chromosome 4. (A) Physical map of the PPP3CA genomic locus with lines depicting various alternative splicing sites. Also shown are novel polymorphisms identified in genetic analysis: rs2850328, 5′UTR trinucleotide repeat (ss#68756203), variable nucleotide repeat (ss#68756205), penta-nucleotide repeat (ss#68756204), as well as the SNP originally identified in “10K SNP chip” association genome scan, rs1395475 (Liu et al., 2005a,b). (B) Alternative splicing schematic depicting possible isoforms for the 15 exon transcript. Dashes illustrate the location primer and probe to detect each transcript by RT-PCR. Asterisks indicate novel isoforms identified.
Genomic Assembly and Identification of Novel PPP3CA Variants and Splicing Isoforms
To identify potential functional variants that might contribute to associations with the phenotype, we reassembled >300 kb sequences at the PPP3CA locus. We searched these sequences for simple sequence repeats and SNPs, relying on alignments of genomic BAC and EST sequences from NCBI and Celera. We identified several simple sequence length poly-morphism/microsatellite markers. A trinucleotide repeat lies in PPP3CA's 5′untranslated region (UTR) (ss68756203). We observed six and seven GCC allellic variants, containing between 2 and 10 repeat, in European and African–American samples, respectively. A 16 nucleotide tandem repeat (ss68756205) with eight nucleotide insertion lies in intron 1. A pentanucleotide repeat (ss68756204) lies in intron 3. The SNP now termed rs2850328 lies in 5′ flanking putative promoter sequences [Figure 1(A)]. Several of these markers were used in genotyping assays (see above and below) to identify allele frequency differences between disease and control samples.
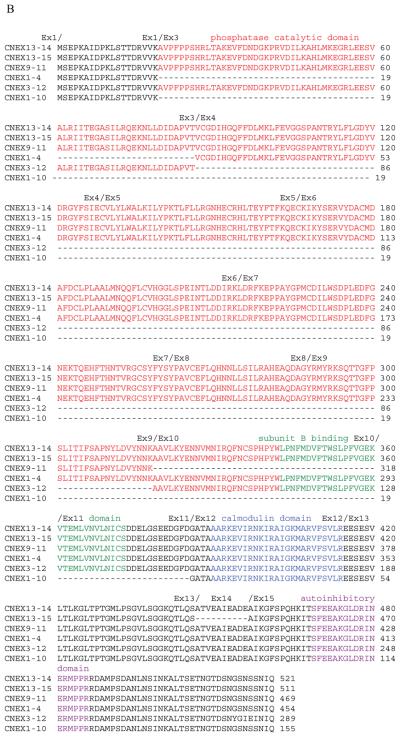
By aligning 15 exons of PPP3CA gene with ESTs (dbEST: http://www.ncbi.nlm.nih.gov/dbEST/) and sequences of the RT-PCR products generated here, we were able to identify novel, alternatively spliced PPP3CA isoforms that can be all translated in frame. Exon 1 can be individually spliced to four different downstream exons [Figure 1(A)], generating isoforms that we term CNEX1-2, 1-3, 1-4, and 1-10 [Figure 1(B)]. Different translation initiation sites for CNEX1-2 and 1-3 create different N-terminal sequences of 6 and 19 amino acids; these are followed by the translation products of exon 3 and other exons [Figure 2(A)]. CNEX1-4 and 1-10 delete 67 and 366 amino acids, respectively. These “deletions” include part or whole the phosphatase catalytic domain [Figure 2(B)], respectively. CNEX9-11 and 13-15 skip the single exons 10 and 14, respectively. Such skipped exons delete the domains responsible for binding subunit B as well as amino acids that lie N-terminal to calcineurin's autoinhibitory domain [Figure 2(B)]. CNEX3-12 skips exon 4-11. The resultant 274 amino acid deletion removes both phosphatase catalytic and subunit B binding domains [Figure 2(B)]. More than two dozens of calcineurin isoforms can thus be formed by these alternative-splicing events. CNEX1-3, 1-4, 13-14, and 13-15 isoforms have been previously described (Guerini and Klee, 1989; Kincaid et al., 1990; Reuter, Mi, Sehrsam, Ludolph, & Volkel, 2001). CNEX1-2, 1-10, 3-12, and 9-11 isoforms are reported here for the first time.
Figure 2.
Amino acid sequences of calcineurin (PPP3CA) isoforms. (A) CNEX1–2 and 1–3 isoform amino acid sequences. (B) CNEX13–14, 13–15, 1–4, 3–12, and 1–10 isoform amino acid alignment. The isoform names are listed on the left-hand side of the amino acid sequences and exon junctions and functional domains are marked above amino acid sequences. Protein domains: red refers to phosphatase catalytic domain; green refers to calcineurin B subunit binding domain; blue refers to calmodulin binding domain; and purple refers to calcineurin autoinhibitory domain.
Genotyping Additional PPP3CA Variants in Substance Dependent and Control Samples
To seek additional information from these PPP3CA variants in substance abuse vulnerability, we genotyped additional simple sequence repeat and SNP markers in abuser/control populations of European and/or African-American descent (Tables 1 and 2, respectively). In African–American samples, there was a nominally significant genotype frequency difference for the 5′ 16 nucleotide repeat intron 1 VNTR, ss68756205 (p = .024) [Figure 1(A) and Table 2].
Table 1.
NIDA Caucasian genotyping results
| SNP/Marker | n | Allele frequencies (%) | p value | Genotype frequencies (%) | p Value | HWE | |||
|---|---|---|---|---|---|---|---|---|---|
| rs2850328 | A | G | AA | AG | GG | ||||
| Control | 157 | 66.9 | 33.1 | 0.962 | 43.3 | 47.1 | 9.55 | 0.968 | 0.758 |
| Abuser | 270 | 67.0 | 33.0 | 44.1 | 45.9 | 10.0 | |||
| SS68756205 (intronl_VNTR) | Allele 1 (1 rep.) | Allele2 (2 rep.) | 1 1 | 1 2 | 2 2 | ||||
| Control Abuser | 195 | 75.9 | 24.1 | 0.660 | 59.0 | 33.8 | 7.18 | 0.893 | 0.614 |
| Abuser | 323 | 77.1 | 22.9 | 60.4 | 33.4 | 6.19 | |||
| rs713455 | C | G | CC | CG | GG | ||||
| Control | 210 | 72.9 | 27.1 | 0.958 | 52.9 | 40.0 | 7.14 | 0.956 | 0.901 |
| Abuser | 350 | 73.0 | 27.0 | 52.6 | 40.9 | 6.57 | |||
| SS68756204 (intron3_CTTTT) | Allele 1 (1 rep.) | Allele2 (2 rep.) | 1 1 | 1 2 | 2 2 | ||||
| Control | 232 | 60.3 | 39.7 | 0.782 | 38.4 | 44.0 | 17.7 | 0.880 | 0.561 |
| Abuser | 372 | 59.5 | 40.5 | 36.6 | 46.0 | 17.5 | |||
| rs2851060 | C | T | CC | CT | TT | ||||
| Control | 219 | 23.1 | 76.9 | 0.068 | 6.39 | 33.3 | 60.3 | 0.082 | 0.999 |
| Abuser | 360 | 18.6 | 81.4 | 2.78 | 31.7 | 65.6 | |||
| rs7840 | A | T | AA | AT | TT | ||||
| Control | 240 | 79.6 | 20.4 | 0.944 | 63.5 | 32.0 | 4.41 | ||
| Abuser | 389 | 79.7 | 20.3 | 64.2 | 31.2 | 4.66 | 0.977 | 0.923 | |
Table 2.
NIDA African–American genotyping results
| SNP/Marker | n | Allele frequencies (%) | p Value | Genotype frequencies (%) |
p Value | HWE | |||
|---|---|---|---|---|---|---|---|---|---|
| rs2850328 | A | G | AA | AG | GG | ||||
| Control | 62 | 20.2 | 79.8 | 0.313 | 11.3 | 17.7 | 71.0 | 0.540 | 1.2 × 10−4 |
| Abuser | 237 | 24.5 | 75.5 | 12.7 | 23.6 | 63.7 | |||
| SS68756205 (intronl_VNTR) | Allelel (1 rep.) | Allele2 (2 rep.) | 1 1 | 12 | 22 | ||||
| Control | 95 | 52.6 | 47.4 | 0.100 | 25.3 | 55.8 | 18.9 | 0.024 | 0.183 |
| Abuser | 234 | 59.6 | 40.4 | 39.7 | 40.6 | 19.7 | |||
| rs713455 | C | G | CC | CG | GG | ||||
| Control | 95 | 64.7 | 35.3 | 0.466 | 42.1 | 45.3 | 12.6 | 0.757 | 0.999 |
| Abuser | 234 | 61.7 | 38.3 | 37.9 | 47.7 | 14.5 | |||
| SS68756204 (intron3_CTTTT) | Allelel (1 rep.) | Allele2 (2 rep.) | 1 1 | 1 2 | 2 2 | ||||
| Control | 97 | 41.8 | 58.2 | 0.104 | 17.5 | 48.5 | 34.0 | 0.237 | 0.731 |
| Abuser | 243 | 35.0 | 65.0 | 14.1 | 41.9 | 44.0 | |||
| rs2851060 | C | T | CC | CT | TT | ||||
| Control | 97 | 11.3 | 88.7 | 0.135 | 2.06 | 18.6 | 79.4 | 0.180 | 0.950 |
| Abuser | 237 | 15.8 | 84.2 | 1.69 | 28.3 | 70.0 | |||
| rs7840 | A | T | AA | AT | TT | ||||
| Control | 100 | 89.2 | 10.8 | 0.752 | 80.0 | 18.5 | 1.54 | 0.532 | 0.188 |
| Abuser | 240 | 90.2 | 9.8 | 83.3 | 13.7 | 2.99 | |||
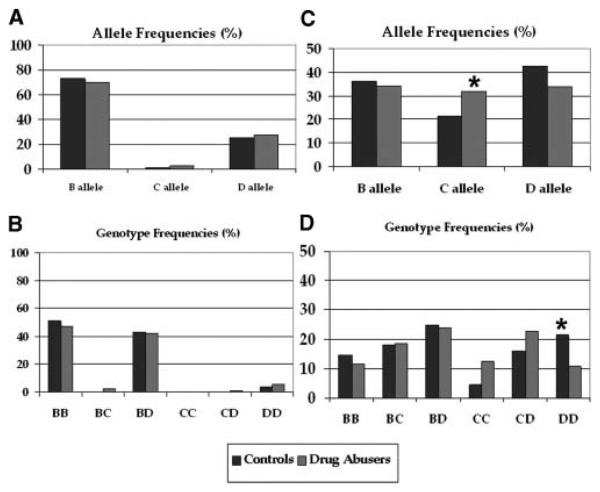
These data received support from genotypes for the trinucleotide repeat in the PPP3CA 5′UTR. The most frequent alleles for the 5′UTR trinucleotide repeat are distinguished by 8, 9, or 10 GGC repeats, forming the alleles that we denote as “B,” “C,” and “D,” respectively (Figure 3). In African–American samples, in which these three alleles display similar frequencies (B 0.35, C 0.20, D 0.42), there were significant, ca 10% “C” allele frequency differences between substance abuser and control samples (p = .03 after Bonferroni correction; Figure 3). D alleles were correspondingly more frequent in controls than in abusers, though the p-value was only 0.09 after Bonferroni correction. Corresponding odds ratios and confidence intervals were: OR = 1.75; CI = 1.16–2.63, for the C, and OR = 0.69; CI = 0.49–0.99, for the D alleles.
Figure 3.
5′UTR repeat allele and genotype frequencies in NIDA populations. (A, B): NIDA European–American (“Abusers” n = 389; “Controls” n = 240). (C, D): NIDA African–American (“Abusers” n = 240; “Control” n = 100). C * refers to significant allele frequency difference between drug abuser population and controls, p value = .031 (Bonferroni correction). D * refers to significant genotype frequency difference between control and drug abuser populations, p value = .093 (Bonferroni correction).
For the other markers (Tables 1 and 2), allele or genotypic frequency differences did not reach statistical significance. Distributions of allele and genotype frequencies for the promoter SNP rs2850328 deviate from Hardy-Weinberg equilibriums, especially in the African–American substance dependent samples (Table 2; χ2 p-value = 9.95 × 10−4).
Allele-Specific Expression Based on PPP3CA 5′ Utr Trinucleotide Repeat Alleles
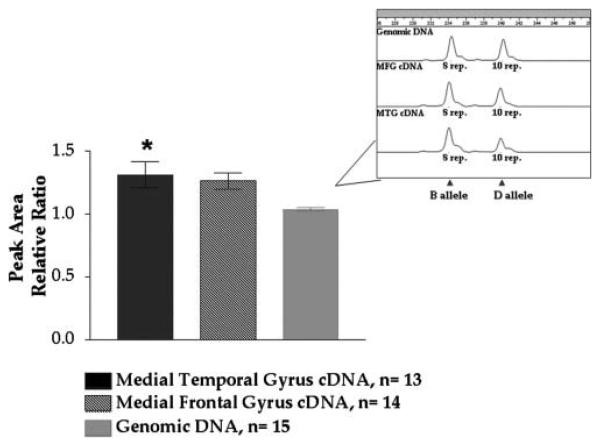
Based on the data noted, haplotypes marked by variants that tag mRNA and lie at the 5′ end of the PPP3CA gene are thus candidates to play roles in some of the association findings noted here. To seek allele-specific expression based on 5′ PPP3CA haplotypes, we compared effects of haplotypes marked by “B” versus “D” alleles in mRNAs extracted from postmortem brain samples, using genomic DNA from heterozygotes as a control. Using the linear range for amplification of cDNA and genomic DNA for each sample, we were able to determine the relative trinucleotide repeat expression by quantifying peak area ratios (i.e., B allele peak area to D allele peak area) produced using GENESCAN. In mRNA extracted from 13 medial temporal gyrus samples, we observed significant differences in the B/D allele ratio in comparison to control values observed in studies of genomic DNA (Figure 4) (p = .0154). Such data support the idea that the tri-nucleotide repeats identify 5′ PPP3CA haplotypes that can alter PPP3CA expression in ways that are preserved in postmortem cerebral cortical samples. Conceivably, the repeat sequences itself might even contribute to such differential expression.
Figure 4.
Allele-specific expression of calcineurin 5′UTR tri-nucleotide repeat alleles. Black, medial temporal gyrus cDNA (n = 13); Hatched, medial frontal gyrus cDNA (n = 14); Gray, genomic DNA. Values represent the average ratio of the area for B allele (8 repeats) relative to the area for the D allele (10 repeats). Medial temporal gyrus peak area ratio is significantly higher than genomic DNA peak area ratio, p value = .0154. Inset: Genotyping profile depicting peaks for B and D alleles.
Tissue-Specific Expression of PPP3CA Alternatively Spliced Isoforms
To supplement fragmentary prior descriptions of the human tissue-specific expression of PPP3CA isoforms, we studied expression patterns in eight brain regions and nine peripheral tissues using relatively quantitative Taqman assays. MGB probes were designed according to junction sequences of two alternatively spliced exons [Figure 1(B) and S3] to avoid detecting genomic DNA. We selected CNEX1-3 expression in amygdala as “100%” reference, and compare isoform expression data from other brain regions and peripheral tissues to this value (Table 3). The most prominent isoforms, CNEX1-3 and 13-14, are expressed at the highest levels in caudate and putamen (200%–400% of amygdala levels), high levels in cortex, amygdala, cerebellum, and hippocampus (100%–200%), and lower levels in hypothalamus and substantia nigra (5%–20%). CNEX1-3 and 13-15 were expressed at intermediate levels in leukocytes, muscle, spleen, and testis (7%–36%). CNEX13-14 contains a 10 amino acid insertion [Figure 2(B), 427-ATVEAIEADE-436] as compared to CNEX13-15. CNEX13-14 is more concentrated in brain, whereas CNEX13-15 is more concentrated in peripheral tissues (Table 3). CNEX1-2, CNEX1-4, and CNEX9-11 with truncated N-terminus, partial deletion of phosphatase domain, and complete deletion of subunit b binding domain, respectively, are expressed at low levels in brain regions and peripheral tissues (<2%). CNEX3-12 and 1-10, which have completely deleted phosphatase domains, were expressed at very low levels (<0.2%) and only detected in brain (Table 3).
Table 3.
Relative expression of PPP3CA isoforms (expressed in percentage relative to Amygdala CNEX1-3 isoform)
| CNEX1-3 | CNEX1-2 | CNEX14 | CNEX13-14 | CNEX13-15 | CNEX9-11 | CNEX1-10 | CNEX3-12 | |
|---|---|---|---|---|---|---|---|---|
| Tissues | (%) | (%) | (%) | (%) | (%) | (%) | (%) | (%) |
| Amygdala | 100 | 4.1910−3 | 0.624 | 101 | 7.76 | 0.574 | 1.74 × 10−2 | 2.01 × 10−2 |
| Caudate | 262 | 5.01 × 10−3 | 1.47 | 404 | 28.3 | 0.969 | 0.118 | 0.200 |
| Putaman | 169 | 4.67 × 10−3 | 0.873 | 207 | 6.67 | 0.502 | 6.89 × 10−2 | 5.46 × 10−2 |
| Cerebellum | 111 | 2.77 × 10−2 | 0.381 | 103 | 22.8 | 0.906 | 1.33 × 10−2 | 1.00 × 10−2 |
| Cortex | 137 | 7.38 × 10−3 | 0.722 | 166 | 9.09 | 1.01 | 4.81 × 10−2 | 3.96 × 10−2 |
| Hippocampus | 114 | 1.08 × 10−3 | 0.593 | 108 | 14.5 | 0.536 | ND | 3.65 × 10−2 |
| Hypothalumus | 5.05 | 7.09 × 10−4 | 3.19 × 10−2 | 11.0 | 3.36 | 9.97 × 10−2 | 3.89 × 10−3 | ND |
| Substantia | ||||||||
| Nigra | 19.9 | 4.71 × 10−3 | 6.44 × 10−2 | 11.9 | 5.07 | 0.237 | ND | ND |
| Heart | 2.49 | 6.18 × 10−4 | 1.89 × 10−3 | 1.40 | 0.994 | 1.78 × 10−2 | ND | ND |
| Intestine | 4.29 | 4.50 × 10−3 | 3.80 × 10−3 | 0.174 | 2.40 | 0.216 | ND | ND |
| Kidney | 3.54 | 1.96 × 10−3 | 5.66 × 10−3 | 1.37 × 10−2 | 1.53 | 9.99 × 10−2 | ND | ND |
| Leukocytes | 22.0 | 3.16 × 10−2 | 1.89 × 10−2 | 0.493 | 8.01 | 1.80 | ND | ND |
| Liver | 2.30 | ND | 9.73 × 10−3 | 1.16 × 10−2 | 1.21 | ND | ND | ND |
| Lung | 1.04 | 1.25 × 10−3 | 5.35 × 10−4 | 0.897 | 6.56 × 10−2 | 2.21 × 10−2 | ND | ND |
| Muscle | 32.8 | 2.87 × 10−2 | 8.30 × 10−2 | 1.43 | 16.0 | 0.390 | ND | ND |
| Spleen | 36.1 | 2.35 × 10−2 | 0.141 | 1.59 | 15.4 | 0.504 | ND | ND |
| Testis | 19.0 | 1.73 × 10−2 | 7.14 × 10−2 | 3.25 | 7.31 | 0.156 | ND | ND |
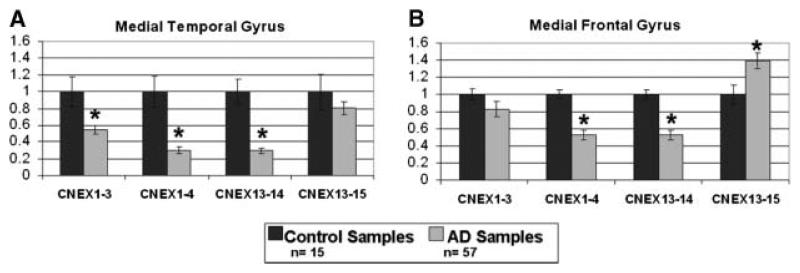
We compared PPP3CA expression in brain samples from 57 pathologically verified AD patients and 15 non-AD control using real time PCR/Taqman assays. We focused on PPP3CA isoforms with intact functional domains, namely CNEX1-3, 1-4, 13-14, and 13-15. There was significant reduction of levels of CNEX1-3, 1-4, and 13-14 isoforms in medial temporal gyrus. Reduction in CNEX1-4 and CNEX13-14 expression also reached significance in mRNA from medial frontal gyrus samples (Figure 5). Expression of the CNEX13-15 isoform, by contrast, was not significantly reduced in medial temporal gyrus and was actually significantly increased in AD medial frontal gyri (Figure 5, Mann-Whitney p = .032).
Figure 5.
PPP3CA isoform expression analysis in Alzheimer's disease (AD). Expression profile for four major isoforms compared between control samples (n = 15) and AD samples (n = 57) in medial temporal gyrus (A) and medial frontal gyrus (B). After normalizing to GAPDH, the expression level for each isoform was determined relative to control tissue samples. * refers to significant values by nonparametric Mann–Whitney test (p value <.05).
Discussion
In this report, we describe a more complete image of the structure of the PPP3CA gene. This picture can be linked to convergent data from association, genomic, and expression studies that support roles for PPP3CA, also known as the calcineurin catalytic subunit A, in addiction and in AD.
The PPP3CA gene encodes a number of alternative calcineurin isoforms, some previously described (Guerini and Klee, 1989; Kincaid et al., 1990; Reuter et al., 2001) and some described here for the first time, which are likely to acquire alternative functions as a result of deleting functional amino acid domains due to differential splicing of the calcineurin primary transcript. Many of these splice variants delete portions or all of the catalytic domain [Figures 2(A) and (B)] and/or regulatory subunit b binding domain. Removal of exon 14 (e.g., in CNEX13-15) removes a 10 amino acid domain whose function is currently unknown [Figure 2(B)]. Interestingly, each PPP3CA isoform appears to retain its calmodulin-binding domain and thus retain the potential for calcium/calmodulin regulation. Expression studies demonstrate that CNEX1-3, 13-14, and 13-15 represent the major PPP3CA isoforms in brain and peripheral tissues. Other isoforms appear to be expressed at less than 1% of the levels of expression of these major isoforms. CNEX1-10 and 3-12 isoforms are expressed in brain only, where they are likely to participate in regulation of brain-specific calcineurin isoform activity. However, isoforms that are expressed at low levels in whole tissue samples might be selectively expressed in small cell subpopulations where they might play more substantial roles.
The 5′ portions of the PPP3CA gene harbor haplotypes that display nominally significant association with polysubstance abuse vulnerability in an African–American poly-substance abuse versus control comparison. The evidence is consistent with modest, polygenic contributions from variants at the PPP3CA locus in addiction. Nevertheless, roles for variants in levels of calcineurin expression in reward- and memory-related phenotypes are well documented in animal model studies (Biala et al., 2005; Gerdjikov and Beninger, 2005; Mansuy et al., 1998). Data in the present report supports prior observations of altered PPP3CA expression in postmortem samples from AD brains. The present haplotype-specific expression data, based on 5′ UTR markers, provides a direct link between association data and prior data that describes behavioral effects of altered PPP3CA expression and/or calcineurin activities.
The alternative spliced PPP3CA variants include or delete exons in this region of that might exert significant differences in PPP3CA function with unique patterns of brain and/or peripheral tissue expression, in manners that suggest specific regulation. CNEX13-15 appears to be selectively preserved in mRNAs from postmortem AD brains in ways that support the specificity of effects on other PPP3CA transcripts and might even suggest that this CNEX13-15 isoform might be expressed by different cell types.
Calcineurin upregulation has been postulated in AD brain due to its involvement in calcium homeostasis (Liu et al., 2005a,b) and inflammatory responses AD (Norris et al., 2005), whereas calcineurin downregulation has been postulated based on oxidative stress (Celsi et al., 2007), pathological cell losses and/or other pathological cellular damage (Brion, Couck, & Conreur, 1995). Alterations in phosphoregulation do provide major pathways by which cytoskeleton proteins, especially tau, (Lian, Ladner, Magnuson, & Lee, 2001) contribute to the neurofibrillary pathology characteristic of AD. Our observations that majority of PPP3CA isoform expression is reduced in AD postmortem brains thus supports much, though not all, prior work. Evidence for the specificity of this observation also comes from the study of expression of specific isoforms. Identification of the isoform that excludes exon 14 and is expressed at levels that are higher in AD than in control samples is of special interest, despite the fact that the function of the amino acids deleted in this isoform has not been well documented. Conceivably, the alternative splicing factors that produce CNEX13-14 and 13-15 might be altered in AD brain. Alternatively, it may be that this relative preservation could indicate that the CNEX13-14 and 13-15 isoforms might be differentially expressed by cell types (e.g., microglia) whose abundance can be increased in AD brains. Understanding the functions of the individual PPP3CA isoforms and understanding the mechanisms whereby the differential splicing is regulated might each provide unique insights into specific AD pathogenetic features.
Messenger RNAs from brain samples can provide evidence for effects of agonal state, effects of acute gene regulation, consequences of illness, death, developmental underrepresentation, developmental overrepresentation, or trait differences, among other features, in the relative levels of expression. Studies that document features of the clinical course and brain handling that could influence mRNA levels and work that uses the “haplotype” (or allele) specific expression approaches employed in the present study provide some of the best current means to control for these issues. Studies of relative expression levels of specific splice variants of the same primary transcript also provide significant control for several of these issues. Nevertheless, replication of these findings in independent samples of brains from different sources will also help solidify the observations made in postmortem RNA.
Despite these and other limitations, the convergence between the molecular biologic and association data presented here, when added to prior information about the behavioral consequences of altering calcineurin and/or PPP3CA activities, provides a collective strength. Taken together, these findings support careful examination of effects of polygenic PPP3CA variations that might include effects of 5′ haplotypes on levels of expression and 3′ haplotypes on relative expression of different splicing variants on the effects of this interesting gene in brain systems and disorders of memory and, perhaps, memory-related phenomena in addiction.
Supplementary Material
Acknowledgments
We thank Judith Hess, Feely Carillo, and Brenda Campbell for assistance with collection of the NIDA clinical samples studied here and cite financial support from NIH IRP (NIDA). We also thank Dr. Florence Theberge and Dr. Maria Flavia Barbano for the abstract translations to French and Spanish, respectively.
Footnotes
Declaration of Interest
The authors report no conflict of interest. The authors alone are responsible for the content and writing of this paper.
References
- Abecasis GR, Cookson WO. GOLD–graphical overview of linkage disequilibrium. Bioinformatics. 2000;16(2):182–183. doi: 10.1093/bioinformatics/16.2.182. [DOI] [PubMed] [Google Scholar]
- Agbas A, Zaidi A, Michaelis EK. Decreased activity and increased aggregation of brain calcineurin during aging. Brain Research. 2005;1059(1):59–71. doi: 10.1016/j.brainres.2005.08.008. [DOI] [PubMed] [Google Scholar]
- Biala G, Betancur C, Mansuy IM, Giros B. The reinforcing effects of chronic D-amphetamine and morphine are impaired in a line of memory-deficient mice overexpressing calcineurin. European Journal of Neuroscience. 2005;21(11):3,089–3,096. doi: 10.1111/j.1460-9568.2005.04132.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brion JP, Couck AM, Conreur JL. Calcineurin (phosphatase 2B) is present in neurons containing neurofibrillary tangles and in a subset of senile plaques in Alzheimer's disease. Neurodegeneration. 1995;4(1):13–21. doi: 10.1006/neur.1995.0002. [DOI] [PubMed] [Google Scholar]
- Celsi F, et al. Beta-amyloid causes downregulation of calcineurin in neurons through induction of oxidative stress. Neurobiology of Disease. 2007;26(2):342–352. doi: 10.1016/j.nbd.2006.12.022. [DOI] [PubMed] [Google Scholar]
- Gerber DJ, et al. Evidence for association of schizophrenia with genetic variation in the 8p21.3 gene, PPP3CC, encoding the calcineurin gamma subunit. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(15):8,993–8,998. doi: 10.1073/pnas.1432927100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdjikov TV, Beninger RJ. Differential effects of calcineurin inhibition and protein kinase A activation on nucleus accumbens amphetamine-produced conditioned place preference in rats. European Journal of Neuroscience. 2005;22(3):697–705. doi: 10.1111/j.1460-9568.2005.04256.x. [DOI] [PubMed] [Google Scholar]
- Gong CX, Liu F, Grundke-Iqbal I, Iqbal K. Dysregulation of protein phosphorylation/dephosphorylation in Alzheimer's disease: a therapeutic target. Journal of Biomedical and Biotechnology. 2006;2006(3):31,825. doi: 10.1155/JBB/2006/31825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerini D, Klee CB. Cloning of human calcineurin A: evidence for two isozymes and identification of a polyproline structural domain. Proceedings of the National Academy of Sciences of the United States of America. 1989;86(23):9,183–9,187. doi: 10.1073/pnas.86.23.9183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson C, et al. Pooled association genome scanning for alcohol dependence using 104,268 SNPs: validation and use to identify alcoholism vulnerability loci in unrelated individuals from the collaborative study on the genetics of alcoholism. American Journal of Medical Genetics Part B: Neuropsychiatry Genetics. 2006;141B(8):844–853. doi: 10.1002/ajmg.b.30346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kincaid RL, et al. Cloning and characterization of molecular isoforms of the catalytic subunit of calcineurin using nonisotopic methods. Journal of Biological Chemistry. 1990;265(19):11,312–11,319. [PubMed] [Google Scholar]
- Klee CB, Crouch TH, Krinks MH. Calcineurin: a calcium- and calmodulin-binding protein of the nervous system. Proceedings of the National Academy of Sciences of the United States of America. 1979;76(12):6,270–6,273. doi: 10.1073/pnas.76.12.6270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian Q, Ladner CJ, Magnuson D, Lee JM. Selective changes of calcineurin (protein phosphatase 2B) activity in Alzheimer's disease cerebral cortex. Experimental Neurology. 2001;167(1):158–165. doi: 10.1006/exnr.2000.7534. [DOI] [PubMed] [Google Scholar]
- Liu F, Grundke-Iqbal I, Iqbal K, Oda Y, Tomizawa K, Gong CX. Truncation and activation of calcineurin A by calpain I in Alzheimer disease brain. Journal of Biological Chemistry. 2005a;280(45):37,755–37,762. doi: 10.1074/jbc.M507475200. [DOI] [PubMed] [Google Scholar]
- Liu QR, et al. Pooled association genome scanning: validation and use to identify addiction vulnerability loci in two samples. Proceedings of the National Academy of Sciences of the United States of America. 2005b;102(33):11,864–11,869. doi: 10.1073/pnas.0500329102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu QR, Lu L, Zhu XG, Gong JP, Shaham Y, Uhl GR. Rodent BDNF genes, novel promoters, novel splice variants, and regulation by cocaine. Brain Research. 2006;1067(1):1–12. doi: 10.1016/j.brainres.2005.10.004. [DOI] [PubMed] [Google Scholar]
- Malleret G, et al. Inducible and reversible enhancement of learning, memory, and long-term potentiation by genetic inhibition of calcineurin. Cell. 2001;104(5):675–686. doi: 10.1016/s0092-8674(01)00264-1. [DOI] [PubMed] [Google Scholar]
- Mansuy IM, Mayford M, Jacob B, Kandel ER, Bach ME. Restricted and regulated overexpression reveals calcineurin as a key component in the transition from short-term to long-term memory. Cell. 1998;92(1):39–49. doi: 10.1016/s0092-8674(00)80897-1. [DOI] [PubMed] [Google Scholar]
- Norris CM, et al. Calcineurin triggers reactive/inflammatory processes in astrocytes and is upregulated in aging and Alzheimer's models. Journal of Neuroscience. 2005;25(18):4,649–4,658. doi: 10.1523/JNEUROSCI.0365-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter A, Mi J, Sehrsam I, Ludolph AC, Volkel H. A novel calcineurin splice variant that modifies calcineurin activity. European Journal of Biochemistry. 2001;268(22):5,955–5,960. doi: 10.1046/j.0014-2956.2001.02551.x. [DOI] [PubMed] [Google Scholar]
- Stephens M, Donnelly P. A comparison of Bayesian methods for haplotype reconstruction from population genotype data. American Journal of Human Genetics. 2003;73(5):1,162–1,169. doi: 10.1086/379378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. American Journal of Human Genetics. 2001;68(4):978–989. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhl GR, Liu QR, Walther D, Hess J, Naiman D. Polysubstance abuse-vulnerability genes: genome scans for association, using 1,004 subjects and 1,494 single-nucleotide polymorphisms. American Journal of Human Genetics. 2001;69(6):1,290–1,300. doi: 10.1086/324467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winder DG, Mansuy IM, Osman M, Moallem TM, Kandel ER. Genetic and pharmacological evidence for a novel, intermediate phase of long-term potentiation suppressed by calcineurin. Cell. 1998;92(1):25–37. doi: 10.1016/s0092-8674(00)80896-x. [DOI] [PubMed] [Google Scholar]
- Yakel JL. Calcineurin regulation of synaptic function: from ion channels to transmitter release and gene transcription. Trends in Pharmacological Sciences. 1997;18(4):124–34. doi: 10.1016/s0165-6147(97)01046-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.