Abstract
A family of bacterial effectors including CHBP from Burkholderia pseudomallei and Cif from Enteropathogenic E. Coli (EPEC) adopt a functionally important papain-like hydrolytic fold. Here, CHBP is shown to be a potent inhibitor of the eukaryotic ubiquitination pathway. CHBP acts as a deamidase that specifically and efficiently deamidates Gln-40 in ubiquitin and NEDD8 both in vitro and during Burkholderia infection. Deamidated ubiquitin is impaired in supporting ubiquitin-chain synthesis. Cif selectively deamidates NEDD8, which abolishes rather than stimulates the activity of Cullin-RING ubiquitin ligases (CRLs). Ubiquitin-dependent degradation of multiple CRL substrates including key cell cycle regulators and small GTPase RhoA was impaired by Cif in EPEC-infected cells. Mutations of substrate-contacting residues in Cif abolish or attenuate Cif-induced cytopathic phenotypes of cell cycle arrest and actin stress fibers formation. Thus, NEDD8 deamidation might be the molecular mechanism underlying EPEC-induced cytopathic effect.
Gram-negative bacterial pathogens use a conserved type III secretion system (TTSS) to translocate effectors into eukaryotic host cells (1). These effectors manipulate various host functions, serving as an important virulence mechanism (2–3). Several effectors from a diverse spectrum of bacteria inhibit host cell cycle progression (4–7), thereby termed cyclomodulins (8). Cif (cycle inhibiting factor) from Enteropathogenic E. Coli (EPEC) arrests host cell cycle either at G2/M (5) or G1/S transition (9). Cif homolog in Burkholderia pseudomallei (CHBP) also blocks cell cycle progression when directly transduced into eukaryotic cells (10). Cif and CHBP belong to a growing family of TTSS effectors that adopt a papain-like hydrolytic fold with a Cys-His-Asp/Asn/Glu/Gln catalytic triad (10–13). While biochemical strategies of manipulating eukaryotic cell cycle machinery/signaling are documented for other cyclomodulins, the host target and underlying mechanism for the Cif/CHBP family are completely unknown.
Infection of HeLa cells with Cif-harboring EPEC strain stabilizes cyclin-dependent kinase inhibitors p21 and p27 (9) (also see below). Progression of eukaryotic cell cycle is driven by activities of the ubiquitin-proteasome system (UPS) that mediates timed degradation of key cell cycle regulators. We hypothesized that the Cif/CHBP family might target the host ubiquitin-proteasome system. Purified CHBP was directly transduced into HeLa cells that express UbG76V-GFP or Ub-R-GFP, two sensitive reporters of the UPS (14). Similarly to the effect of proteasome inhibitor MG132, wild-type CHBP, but not the catalytic mutant (C156S), efficiently blocked degradation of UbG76V-GFP and Ub-R-GFP (Fig. 1A). In contrast, levels of the UPS-insensitive reporter Ub-M-GFP remained constant (Fig. 1A). TNFα induces UPS-dependent degradation of IκBα, resulting in nuclear translocation of p65 to turn on NF-κB-regulated gene transcription. In wild-type CHBP, but not the C156S mutant-transduced cells, both IκBα degradation and nuclear translocation of p65 were blocked (fig. S1). CHBP did not inhibit the proteasome activity in vitro (not shown). This directed us to the ubiquitination process, a sequential three-enzyme cascade comprised of ubiquitin-activation enzyme E1, ubiquitin-conjugating enzyme E2 and ubiquitin ligase E3. In vitro ubiquitin-chain synthesis catalyzed by a Cullin-based E3 complex Cul1/ROC1 (15) was abolished by purified CHBP (Fig. 1B). Notably, CHBP was also highly effective in blocking substrate-free ubiquitin-chain synthesis catalyzed by different E3-E2 pairs including a RING-domain E3 gp78c/Ube2g2 (16) (Fig. 1C), another RING E3 GRAIL/UbcH6 (17) (fig. S2A), a distinct bacterial E3 IpaH/UbcH5c (18) and a U-box E3 LubX/UbcH5c (19) (fig. S2B). CHBP inhibited Ubc13/Uev1a-catalyzed ubiquitin-chain synthesis (fig. S2C). The activity of blocking ubiquitination required the catalytic cysteine in CHBP. Thus, CHBP is a potent and general inhibitor of the eukaryotic ubiquitination pathway.
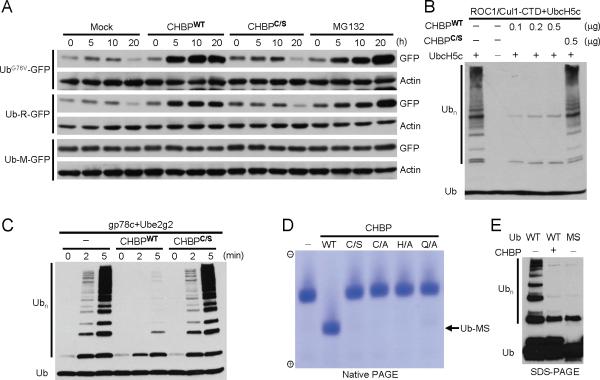
Fig. 1. CHBP blocks the ubiquitination pathway by covalently modifying ubiquitin.
(A) GFP reporter assays of effects of CHBP on the host ubiquitination pathway. HeLa cells were transfected with UbG76V-GFP, Ub-R-GFP or Ub-M-GFP reporter plasmid. Purified CHBP was delivered into transfected cells using the anthrax lethal factor system. UbG76V-GFP and Ub-R-GFP are rapidly degraded in cells, while Ub-M-GFP (a negative control) is not sensitive to the ubiquitin-proteasome pathway. CHBPWT, wild-type CHBP; CHBPC/S, the catalytic cysteine mutant (C156S). MG132 treatment serves as a positive control. Shown are anti-GFP and anti-actin immunoblots of lysates of cells harvested at indicated time points following CHBP delivery.
(B and C) Effects of CHBP on in vitro ubiquitin-chain synthesis. Ubiquitination reaction, using bacterially purified ROC1/Cul1-CTD complex as the E3 and UbcH5c as the E2, was carried out in the presence of indicated amounts of recombinant CHBP (B). In (C), gp78c and Ube2g2 were used as the E3 and E2, respectively, and reactions carried out in the presence/absence of CHBP were stopped at indicated time points. Shown are anti-ubiquitin immunoblots to examine ubiquitin-chain (Ubn) formation.
(D) Native PAGE analysis of free ubiquitin treated with purified CHBP. WT, wild-type CHBP; C/S, C/A, H/A and Q/A are mutations in the catalytic triad of CHBP. Coomassie blue stained native gel is shown.
(E) Chain formation activities of CHBP-modified ubiquitin. In vitro ubiquitination reaction was carried out as that in (C). WT, wild-type ubiquitin; MS, mobility-shifted ubiquitin generated by CHBP treatment and recovered from the native gel shown in (D) by electro-elution. Reactions were subjected to immunoblotting analysis for ubiquitin chain (Ubn) formation.
Recombinant CHBP could not disassemble E3-synthesized or synthetic polyubiquitin chains (fig. S2D and data not shown); purified Cif showed no activities towards ubiquitin-AMC, a sensitive substrate for deubiquitination enzymes (DUBs). These suggest that the CHBP/Cif family is unlikely to act as a DUB. We then analyzed each individual step in the ubiquitination process. CHBP did not affect formation of E1~Ub and E2~Ub thioester intermediates (fig. S3). However, a remarkable inhibition of chain formation was observed when CHBP was added into the ubiquitination reaction performed with E3 (gp78c) and the pre-formed E2~Ub thioester, and this inhibition required and was promoted by pre-incubation of the E2~Ub thioester with CHBP (fig. S4). These results indicate that CHBP inactivates the E2~Ub thioester, possibly through hydrolysis of or covalent modification of the E2~Ub thioester.
SDS-PAGE (reducing or non-reducing) was first employed to analyze CHBP-incubated E2-charge reaction mixtures containing the inactivated E2~Ub thioester. None of E1, E2, ubiquitin and E2~Ub thioester exhibited any changes on the SDS gel. However, when analyzed on a native PAGE gel, the E2~Ub thioester, but not E1 and E2, migrated faster towards the anode (fig. S5A). DTT treatment recovered E2 from the mobility-shifted E2~Ub thioester, which showed no mobility changes on the native gel (fig. S5B). Therefore, it is likely the ubiquitin moiety, rather than E2 or the thioester linkage, that is responsible for the mobility shift observed with CHBP-treated E2~Ub thioester. Significantly, when free ubiquitin was directly incubated with CHBP, a faster migration towards the anode also occurred, and the mobility shift was more significant than that of the E2~Ub thioester, as expected from the smaller size of free ubiquitin (Fig. 1D). None of CHBP catalytic-triad mutants generated the mobility-shifted ubiquitin (Ub-MS) (Fig. 1D). Ub-MS recovered from the native gel was impaired in supporting ubiquitin-chain synthesis, resembling the effect of CHBP on ubiquitination performed with wild-type ubiquitin (Fig. 1E). Thus, CHBP blocks the ubiquitination by covalently modifying ubiquitin.
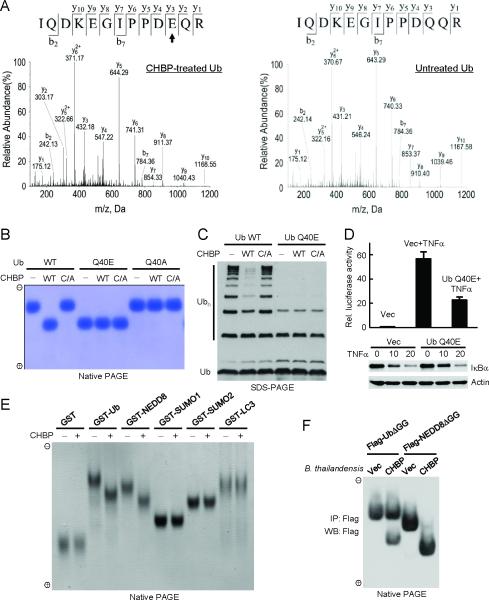
Mass spectrometric analysis was carried out to reveal the nature of CHBP-modified ubiquitin. A total of 15 tryptic peptides detected by mass spectrometry covered the entire ubiquitin sequence except for the C-terminal two glycine residues (fig. S6). Among all the recovered peptides, three overlapping ones (28AKIQDKEGIPPDQQR42, 30IQDKEGIPPDQQR42, and 30IQDKEGIPPDQQRLIFAGK48) from CHBP-treated ubiquitin showed one-dalton mass increase compared with corresponding peptides from untreated ubiquitin (fig. S6). Tandem mass spectrometry further revealed that the one-dalton increase occurred on Gln-40 in ubiquitin (Fig. 2A), indicating deamidation of the glutamine into glutamic acid. The adjacent Gln-41 was not deamidated (Fig. 2A). Treatment of ubiquitin with CHBP C156S did not generate the one-dalton mass increase (not shown). Substitution of Gln-40 with Glu, but not Ala, resulted in a mobility change indistinguishable from that of CHBP-modified ubiquitin (Fig. 2B). Further treatment of ubiquitin Q40E or Q40A with CHBP caused no additional mobility shifts on the native gel (Fig. 2B). Thus, CHBP modifies ubiquitin by deamidating its Gln-40.
Fig. 2. CHBP deamidates Gln-40 in ubiquitin/NEDD8 in vitro and during infection.
(A) Electrospray ionization (ESI) tandem mass spectrometry (MS/MS) spectrum of a Gln-40-containing tryptic peptide from CHBP-treated ubiquitin (left) or the control untreated ubiquitin (right). b and y ions are marked in the spectrum. The fragmentation patterns that generate the observed b and y ions are illustrated along the peptide sequence shown on top of the spectrum. The arrow marks the residue that shows one-dalton mass increase after CHBP treatment and is converted from glutamine into glutamic acid. (B) Native PAGE analysis of ubiquitin Gln-40 mutants and effects of further CHBP treatment. Wild-type (WT) and indicated ubiquitin mutants were loaded onto the native gel either directly or after incubation with CHBP (wild-type or the catalytic cysteine mutant (C/A)). The gel was stained with Coomassie blue.
(C) Chain formation activities of ubiquitin Q40E and comparison with effects of CHBP on wild-type ubiquitin. In vitro ubiquitination reaction was carried out as that in Fig. 1C. Reactions were subjected to immunoblotting analysis for ubiquitin chain (Ubn) formation. (D) Effects of ectopic expression of ubiquitin Q40E on TNFα-induced IκBα degradation and NF-κB pathway activation. Intact HeLa cells (for the lower immunoblot panel) or HeLa cells expressing NF-κB dual-luciferase reporters were transfected with ubiquitin Q40E expression plasmid or a vector control and then stimulated with TNFα. Shown are mean relative luciferase activities from duplicate determinations with error bars indicating standard deviation. The lower panel shows anti-IκBα and anti-actin immunoblots of total cell lysates.
(E) Native PAGE analysis of GST-tagged ubiquitin and indicated UBLs following incubation with CHBP. Coomassie blue stained native gel is shown.
(F) Native PAGE assay of ubiquitin and NEDD8 deamidation by type III-secreted CHBP. 293T cells transfected with Flag-UbΔGG or Flag-NEDD8ΔGG expression plasmid were infected with B. thailandensis or B. thailandensis expressing CHBP. UbΔGG and NEDD8ΔGG immunopurified from infected cells were subjected to native gel electrophoresis followed by anti-Flag immunoblotting. UbΔGG and NEDD8ΔGG (deletion of the last two glycine residues) were used to avoid polyconjugation and conjugation to other cellular proteins.
Similarly to the effect of CHBP, ubiquitin Q40E was compromised in supporting in vitro ubiquitin-chain ligation (Fig. 2C) without affecting E1 and E2 charging of ubiquitin (fig. S3). Mass spectrometry analysis confirmed that CHBP-inactivated E2~Ub thioester also contained a deamidated Gln-40 (fig. S7). These data suggest that ubiquitin Q40E is defective in being transferred from E2 to the acceptor ubiquitin during E3-catalyzed chain synthesis. This analysis is supported by the structural evidence that Gln-40 in ubiquitin makes a close contact with E3 (NEDD4L) in the NEDD4L/E2~Ub oxyester complex structure (20). When ubiquitin Q40E was ectopically expressed in HeLa cells, TNFα-induced IκBα degradation was delayed and the downstream NF-κB-dependent luciferase reporter activations was significantly impaired (Fig. 2D). Consistently, ectopic expression of ubiquitin Q40E also resulted in notable (but not remarkable) stabilization of substrates of the ubiquitin-proteasome pathway (fig. S8). Thus, deamidation of Gln-40 in ubiquitin by CHBP attenuates the normal function of the ubiquitination pathway both in vitro and in cells.
A group of ubiquitin-like proteins (UBLs) share a three-dimensional fold with ubiquitin (21). Among UBLs, NEDD8 harbors about 80% sequence similarity with ubiquitin while other UBLs are generally not similar to ubiquitin in primary sequence (fig. S9A). Gln-40 is conserved in ubiquitin, NEDD8, SUMO2/3 and LC3 and occupies nearly the same position in three-dimensional structures of ubiquitin and NEDD8 (fig. S9). When equal amounts of a panel of UBLs were incubated with CHBP, only NEDD8 underwent a mobility shift on the native gel, similarly to that observed with ubiquitin (Fig. 2E). The shifted NEDD8 also had a one-dalton mass increase on its Gln-40, but not the two flanking glutamine (fig. S10). Several non-UBL proteins including p27 were not deamidated by the CHBP family (fig. S11). These results establish NEDD8 as another specific deamidation substrate of CHBP.
CHBP is only present in B. pseudomallei and its function in B. pseudomallei pathogenesis is not defined. The closely related B. thailandensis is used as a model to study the virulence-associated TTSS system of B. pseudomallei (22). B. thailandensis transformed with a CHBP expression plasmid was used to infect 293T cells that express Flag-tagged UbΔGG or NEDD8ΔGG. Nearly 100% of NEDD8ΔGG and about 50% of UbΔGG in infected cells were deamidated in a CHBP-dependent manner (Fig. 2F). This is consistent with the potent in vitro activity of CHBP and its slight preference for NEDD8 over ubiquitin (see below). This result also demonstrates that type III-secreted CHBP from Burkholderia deamidates both NEDD8 and ubiquitin in infected cells.
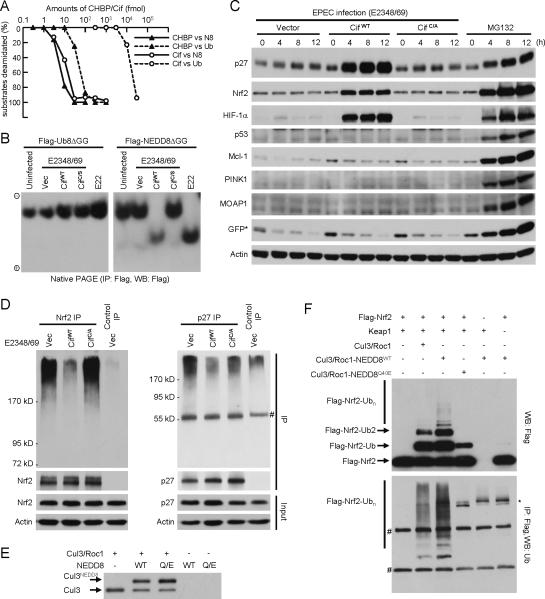
To gain insights into the enzymatic property of CHBP and compare it with its EPEC homologue Cif, 350 pmol of NEDD8 or ubiquitin in a 20-μl reaction (18 μM), a concentration slightly higher than the estimated cellular ubiquitin concentration (13 μM) (23), was titrated with recombinant CHBP or Cif (fig. S12). Following a 20-min reaction, as little as 0.3 pmol and 0.03 pmol of CHBP were sufficient to deamidate nearly all 350 pmol of ubiquitin and NEDD8, respectively (Fig. 3A and fig. S12A). Notably, while the activity of Cif on NEDD8 was comparably robust as that of CHBP on NEDD8, the activity of Cif on ubiquitin was about 1000 times lower than that on NEDD8 (Fig. 3A and fig. S12B). The high selectivity of Cif for NEDD8 over ubiquitin suggests that NEDD8 is likely its preferred host target.
Fig. 3. Cif selectively inactivates CRLs by deamidating NEDD8 in vitro and in vivo.
(A) Enzyme-titration measurements of the deamidase activity of CHBP/Cif towards ubiquitin and NEDD8. 350 pmol (3 μg) of ubiquitin or NEDD8 were subjected to 20-min incubation with increasing amounts of purified CHBP or Cif in a 20-μl reaction at 37°C. The whole reaction mixtures were analyzed by native gel electrophoresis followed by Coomassie blue staining as shown in fig. S12. Intensity of native ubiquitin/NEDD8 bands on native gels was quantified and plotted versus the amount of CHBP/Cif used in each reaction.
(B) Native PAGE assay of Cif deamidation of NEDD8 during EPEC infection of HeLa cells. EPEC E22 strain bears a functional Cif while EPEC E2348/49 strain harbors a naturally truncated and nonfunctional Cif. Experiments were performed and data are presented similarly as those in Fig. 2F.
(C) Effects of Cif on steady levels of CRL and non-CRL substrates in EPEC-infected cells. HeLa cells were infected with EPEC strains expressing wild-type Cif (CifWT) or the catalytic mutant (CifC/A) for indicated time durations. Endogenous levels of indicated proteins were shown by immunoblotting of total cell lysates using specific antibodies. p27, HIF-1α, and Nrf2 are ubiquitination substrates of Cul1, Cul2 and Cul3 CRL complex, respectively. p53, MOAP1, PINK1 and Mcl-1 are unstable proteins whose degradation is independent of CRLs in HeLa cells. GFP* indicates transfected Ub-R-GFP reporter. MG132 treatment was included as a control.
(D) Effects of Cif on ubiquitination of Nrf2 and p27 in infected cells. Endogenous Nrf2 or p27 was immunoprecipitated under denaturing conditions from EPEC-infected and MG132-treated HeLa cells (expressing Flag-ubiquitin) using the specific or a control antibody. The immunoprecipitates (IP) were analyzed by anti-Flag (upper panels) and anti-Nrf2 (or p27) immunoblotting. Total cells lysates (Input) were also blotted with anti-Nrf2 (or anti-p27) and anti-actin antibodies as shown. # marks IgG.
(E and F) Effects of NEDD8 deamidation on neddylation-stimulated CRL activity of catalyzing substrate ubiquitination. Purified Cul3/GST-Roc1 complex was first subjected to neddylation reaction with NEDD8 (WT) or NEDD8 Q40E and neddylated Cul3 is shown by anti-Cul3 immunoblot in (E). The Cul3/GST-Roc1 complex present in the left three reactions in (E) was immobilized onto glutathione beads and then used to ubiquitinate Flag-Nrf2 (1–97) in the presence of Keap1 as indicated in (F). Reaction mixtures were analyzed by anti-Flag immunoblotting (upper) or subjected to anti-Flag immunoprecipitation under denaturing conditions followed by anti-ubiquitin immunoblotting (lower). Flag-Nrf2-Ub, Flag-Nrf2-Ub2 and Flag-Nrf2-Ubn denote mono-, di-, and poly-ubiquitinated Nrf2, respectively. #, IgG heavy and light chains; *, a nonspecific band.
The deamidase activity of Cif was further examined using EPEC infection of HeLa cells. NEDD8ΔGG was completely deamidated in cells infected with Cif-bearing EPEC strain (E22), but not the Cif-deficient strain (E2348/49) (Fig. 3B). Complete NEDD8 deamidation was restored when the Cif-deficient strain was complemented with wild-type Cif, but not the catalytic mutant. Consistent with the extremely poor in vitro activity of recombinant Cif on ubiquitin (Fig. 3A), deamidation of UbΔGG was not detected in cells infected with any of the EPEC strains (Fig. 3B). Thus, Cif efficiently and specifically deamidates NEDD8, but not ubiquitin, during EPEC infection.
NEDD8 is mainly conjugated to Cullin proteins that mediate the assembly of a large repertoire of Cullin-RING ubiquitin ligase (CRL) complexes (24–25). Conjugation by NEDD8 (neddylation) stimulates the activity of CRLs (26). Among a panel of ubiquitin-proteasome substrates, substrates of CRLs including p27, Nrf2 and HIF-1α were significantly accumulated in Cif catalytic cysteine-dependent manner (Fig. 3C and fig. S13) while their mRNA levels were not affected by Cif over the infection course (fig. S14). Less ubiquitination of CRL substrates (p27 and Nrf2) was observed in cells infected with Cif-harboring EPEC when cellular proteasome activity was blocked (Fig. 3D). In contrast, non-CRL substrates including p53, Mcl-1(27), PINK1(28), MOAP1(29) as well as the Ub-R-GFP reporter were not stabilized by Cif despite that they were sensitive to proteasome inhibition (Fig. 3C and fig. S13). These results agree with the observation that Cif only deamidated NEDD8, but not ubiquitin, during EPEC infection (Fig. 3B), and also demonstrate that NEDD8 deamidation by Cif downregulates the activity of CRLs in infected host cells.
To understand the mechanism of CRL inactivation by Cif during EPEC infection, neddylation-stimulated CRL activation was in vitro reconstituted using the Cul3/Roc1/Keap1 complex-catalyzed Nrf2 ubiquitination system (Fig. 3, E and F). As expected, significantly increased Nrf2 ubiquitination appeared when Nrf2 was reacted with the neddylated Cul3/Roc1 complex. However, replacement of NEDD8 with NEDD8 Q40E in the two-step reconstitution abolished the effect of neddylation-stimulated Nrf2 ubiquitination (Fig. 3F) without compromising the efficiency of Cul3 neddylation (Fig. 3E). More importantly, the activity of NEDD8 Q40E-conjugated Cul3 complex was even lower than the unneddylated counterpart (Fig. 3F). Thus, deamidation of NEDD8 directly impairs the ubiquitination ligase activity of the neddylated CRL complex, which likely accounts for stabilization of CRL substrates by Cif observed during infection. We also noticed a Cif deamidase activity-dependent increase of neddylated levels of Cul1 and Cul3 (in relation to the unmodified form) in infected cells (fig. S15A). This is likely an indirect consequence of NEDD8 deamidation as Cif did not affect Cullin neddylation in vitro (not shown) and the deamidated NEDD8 was as competent as wild-type NEDD8 for conjugation onto Cullins (Fig. 3E).
Cif-induced cytopathic effect also features formation of striking actin stress fibers (5). Mutation of the deamidase catalytic residues completely abolished the development of actin stress fibers in EPEC-infected HeLa cells (fig. S16A). Actin stress fibers formation is controlled by RhoA, a member of Rho-family small GTPases (30) and also a specific substrate of the Cul3/BACURD CRL complex (31). Consistently, among a panel of Rho GTPases examined, only RhoA was significantly stabilized by Cif in its deamidase activity-dependent manner (fig. S16B). Thus, Cif-induced formation of prominent actin stress fibers in EPEC infection is likely attributed to dysfunction of CRLs as a result of NEDD8 deamidation.
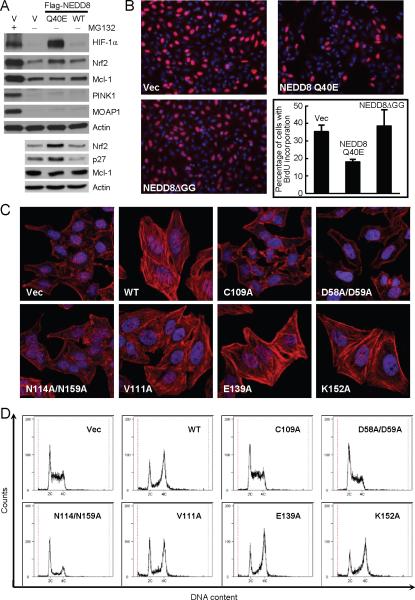
Ectopic expression of NEDD8 Q40E resulted in accumulation of CRL substrates such as p27, Nrf2 and HIF-1α, but not non-CRL substrates (Fig. 4A and fig. S17). HeLa cells expressing NEDD8 Q40E showed considerably decreased bromodeoxyuridine (BrdU) incorporation (Fig. 4B) (the extent is less than that induced by Cif during infection (9)), indicating a defect in cell cycle progression. A significant portion of HeLa cells became enlarged with strong actin stress fibers (not shown), morphologically resembling EPEC-infected cells. Also similarly to that observed with EPEC infection, percentages of neddylated Cul1 and Cul3 were increased in cells expressing NEDD8 Q40E, compared with those in intact HeLa cells or cells expressing wild-type NEDD8 (fig. S15B). Thus, ectopic expression of NEDD8 Q40E partially recapitulates the effects of Cif on impairing the CRL function.
Fig. 4. NEDD8 deamidation is linked to Cif-induced cytopathic effect during infection.
(A) Effects of ectopic expression of NEDD8 Q40E on steady levels of CRL and non-CRL substrates. HeLa cells were transfected with indicated Flag-NEDD8 expression plasmid. Endogenous levels of indicated proteins were shown by immunoblotting of total cell lysates using specific antibodies.
(B) Effects of ectopic expression of NEDD8 Q40E on cell cycle progression. 10 μM of BrdU were added into medium of HeLa cells transfected with indicated Flag-NEDD8 expression plasmid for 1 h. Cells were stained with anti-BrdU antibody and DAPI. Statistics of BrdU-positive cells are presented in the graph as means ± SD of four independent counting of about 200 cells each. The experiment was repeated for at least three times.
(C and D) Effects of mutations in the substrate-contacting surface in Cif on stress fibers formation and cell cycle progression. HeLa cells were infected with EPEC strains expressing wild-type Cif (WT) or indicated Cif mutants. Fluorescence images of actin stress fibers stained by Rhodamine-phalloidin and DAPI-stained nuclei are shown (C) and cell cycle profiles of infected cells were determined by flow cytometry analysis of DNA contents (D).
Several point mutations in Cif were generated according to the co-crystal structure of CHBP/ubiquitin complex (unpublished). D58A/D59A and N114A/N159A are mutations at the two enzyme/substrate contact interfaces (but away from the catalytic pocket); the former completely abolished the NEDD8 deamidation activity of Cif and activity of the latter mutant is also significantly attenuated (fig. S18). When translocated into HeLa cells by EPEC infection, both mutants failed to induce actin stress fibers formation (Fig. 4C). The D58A/D59A mutant completely lost the activity of blocking cell cycle progression and the N114A/N159A mutant only had some marginal weak activity of delaying cell cycle progression (Fig. 4D). In contrast, mutations of residues not involved in substrate binding, such as V111A, E139A and K152A, had little effects on the in vitro NEDD8 deamidation activity (fig. S18), and these mutants behaved identically as wild-type Cif in producing stress fibers and cell cycle arrest phenotypes (Fig. 4, C and D). These results suggest that deamidation of NEDD8 is closely linked to Cif-induced cytopathic effects of actin stress fibers formation and cell cycle arrest.
In summary, we discover that the CHBP/Cif family of papain-like TTSS effectors harbor a specific deamidase activity towards Gln-40 in ubiquitin/NEDD8. Different from the two known bacterial deamidase toxins (E. coli cytotoxic necrotizing factor 1 and Pasteurella multocida toxin) that activate their G protein substrates by targeting a glutamine residue critical for GTP hydrolysis (32–34), deamidation by the Cif family inactivates ubiquitin and NEDD8. Deamidated ubiquitin is impaired in supporting ubiquitin ligase-catalyzed ubiquitin-chain synthesis while deamidated NEDD8, when conjugated to Cullins, abolishes rather than stimulates the ubiquitin ligase activity of CRLs. For cyclomodulin Cif, selective deamidation of NEDD8 is closely linked to EPEC infection-induced cytopathic effects of cell cycle arrest and formation of actin stress fibers. Given the universal role of ubiquitination and Cullin-mediated ubiquitination in many important cellular processes (35), our discovery further predicts a possible pleiotropic function of the Cif/CHBP family of effectors in bacterial pathogenesis.
It is increasingly appreciated that bacterial pathogens have evolved to hijack the host ubiquitin system as an effective virulence mechanism (36–38). Known effectors in this regard generally mimic functions of the host ubiquitin system, and several bacterial effectors are shown to harbor the ubiquitin ligase or DUB activity. The CHBP/Cif family represents an unprecedented mode of hijacking the host ubiquitination pathway by deamidating a specific glutamine residue in ubiquitin/NEDD8. Such kind of regulation on ubiquitin and UBLs signaling has not been documented even for the eukaryote itself. Thus, the CHBP/Cif family of deamidases are powerful toxins that could escape from potential host counteracting mechanisms. Moreover, the role of Gln-40 in ubiquitin/NEDD8 in ubiquitination and CRL-mediated ubiquitination has not been probed. Bacterial pathogens have left us interesting questions how such a single charge substitution significantly comprises the activity of ubiquitin in supporting chain formation and how the same conversion on NEDD8 blocks the ubiquitin ligase activity of neddylated CRL complexes that otherwise would be stimulated. Further efforts in this regard will promote new understating of ubiquitin and UBL biochemistry.
One-sentence summaries.
Bacterial effectors deamidate ubiquitin/NEDD8 and interfere with the host ubiquitination pathway.
Supplementary Material
Acknowledgments
We thank Drs. Yihong Ye, C. Garrison Fathman, Zhen-Qiang Pan, Dieter Haas, Victor C. Yu, Edgar C. Boedeker, and Zhijian Chen for providing reagents. We are grateful to Drs. Fenghe Du and Xiaodong Wang for their assistance in obtaining reagents. We also thank members of the Shao lab for helpful discussions and technical assistance. This work was supported by the Chinese Ministry of Science and Technology Grant 2008AA022309 and National Basic Research Plan of China 973 grants to F.S.
References and Notes
- 1.Galan JE, Wolf-Watz H. Nature. 2006;444:567. doi: 10.1038/nature05272. [DOI] [PubMed] [Google Scholar]
- 2.Roy CR, Mocarski ES. Nat Immunol. 2007;8:1179. doi: 10.1038/ni1528. [DOI] [PubMed] [Google Scholar]
- 3.Bhavsar AP, Guttman JA, Finlay BB. Nature. 2007;449:827. doi: 10.1038/nature06247. [DOI] [PubMed] [Google Scholar]
- 4.Lara-Tejero M, Galan JE. Science. 2000;290:354. doi: 10.1126/science.290.5490.354. [DOI] [PubMed] [Google Scholar]
- 5.Marches O, et al. Mol Microbiol. 2003;50:1553. doi: 10.1046/j.1365-2958.2003.03821.x. [DOI] [PubMed] [Google Scholar]
- 6.Iwai H, et al. Cell. 2007;130:611. doi: 10.1016/j.cell.2007.06.043. [DOI] [PubMed] [Google Scholar]
- 7.Huang J, Lesser CF, Lory S. Nature. 2008 doi: 10.1038/nature07355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nougayrede JP, Taieb F, De Rycke J, Oswald E. Trends Microbiol. 2005;13:103. doi: 10.1016/j.tim.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 9.Samba-Louaka A, et al. Cell Microbiol. 2008;10:2496. doi: 10.1111/j.1462-5822.2008.01224.x. [DOI] [PubMed] [Google Scholar]
- 10.Yao Q, et al. Proc Natl Acad Sci U S A. 2009;106:3716. doi: 10.1073/pnas.0900212106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shao F, Merritt PM, Bao Z, Innes RW, Dixon JE. Cell. 2002;109:575. doi: 10.1016/s0092-8674(02)00766-3. [DOI] [PubMed] [Google Scholar]
- 12.Hsu Y, et al. J Mol Biol. 2008;384:465. doi: 10.1016/j.jmb.2008.09.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jubelin G, et al. PloS one. 2009;4:e4855. doi: 10.1371/journal.pone.0004855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dantuma NP, Lindsten K, Glas R, Jellne M, Masucci MG. Nat Biotechnol. 2000;18:538. doi: 10.1038/75406. [DOI] [PubMed] [Google Scholar]
- 15.Wu K, Chen A, Pan ZQ. J Biol Chem. 2000;275:32317. doi: 10.1074/jbc.M004847200. [DOI] [PubMed] [Google Scholar]
- 16.Li W, Tu D, Brunger AT, Ye Y. Nature. 2007;446:333. doi: 10.1038/nature05542. [DOI] [PubMed] [Google Scholar]
- 17.Anandasabapathy N, et al. Immunity. 2003;18:535. doi: 10.1016/s1074-7613(03)00084-0. [DOI] [PubMed] [Google Scholar]
- 18.Zhu Y, et al. Nat Struct Mol Biol. 2008;15:1302. doi: 10.1038/nsmb.1517. [DOI] [PubMed] [Google Scholar]
- 19.Kubori T, Hyakutake A, Nagai H. Mol Microbiol. 2008;67:1307. doi: 10.1111/j.1365-2958.2008.06124.x. [DOI] [PubMed] [Google Scholar]
- 20.Kamadurai HB, et al. Mol Cell. 2009;36:1095. doi: 10.1016/j.molcel.2009.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kerscher O, Felberbaum R, Hochstrasser M. Annu Rev Cell Dev Biol. 2006;22:159. doi: 10.1146/annurev.cellbio.22.010605.093503. [DOI] [PubMed] [Google Scholar]
- 22.Haraga A, West TE, Brittnacher MJ, Skerrett SJ, Miller SI. Infection and immunity. 2008;76:5402. doi: 10.1128/IAI.00626-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Inomata K, et al. Nature. 2009;458:106. doi: 10.1038/nature07839. [DOI] [PubMed] [Google Scholar]
- 24.Merlet J, Burger J, Gomes JE, Pintard L. Cell Mol Life Sci. 2009;66:1924. doi: 10.1007/s00018-009-8712-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rabut G, Peter M. EMBO Rep. 2008;9:969. doi: 10.1038/embor.2008.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saha A, Deshaies RJ. Mol Cell. 2008;32:21. doi: 10.1016/j.molcel.2008.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhong Q, Gao W, Du F, Wang X. Cell. 2005;121:1085. doi: 10.1016/j.cell.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 28.Zhou C, et al. Proc Natl Acad Sci U S A. 2008;105:12022. doi: 10.1073/pnas.0802814105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fu NY, Sukumaran SK, Yu VC. Proc Natl Acad Sci U S A. 2007;104:10051. doi: 10.1073/pnas.0700007104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hall A. Science. 1998;279:509. doi: 10.1126/science.279.5350.509. [DOI] [PubMed] [Google Scholar]
- 31.Chen Y, et al. Mol Cell. 2009;35:841. doi: 10.1016/j.molcel.2009.09.004. [DOI] [PubMed] [Google Scholar]
- 32.Orth JH, et al. Proc Natl Acad Sci U S A. 2009;106:7179. doi: 10.1073/pnas.0900160106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schmidt G, et al. Nature. 1997;387:725. doi: 10.1038/42735. [DOI] [PubMed] [Google Scholar]
- 34.Flatau G, et al. Nature. 1997;387:729. doi: 10.1038/42743. [DOI] [PubMed] [Google Scholar]
- 35.Bhoj VG, Chen ZJ. Nature. 2009;458:430. doi: 10.1038/nature07959. [DOI] [PubMed] [Google Scholar]
- 36.Angot A, Vergunst A, Genin S, Peeters N. PLoS Pathog. 2007;3:e3. doi: 10.1371/journal.ppat.0030003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Munro P, Flatau G, Lemichez E. Curr Opin Microbiol. 2007;10:39. doi: 10.1016/j.mib.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 38.Rytkonen A, Holden DW. Cell Host Microbe. 2007;1:13. doi: 10.1016/j.chom.2007.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.