Abstract
Background
The mammalian suprachiasmatic nucleus (SCN), located in the ventral hypothalamus, is a major regulator of circadian rhythms in mammals and birds. However, the role of the SCN in lower vertebrates remains poorly understood. Zebrafish cyclops (cyc) mutants lack ventral brain, including the region that gives rise to the SCN. We have used cyc embryos to define the function of the zebrafish SCN in regulating circadian rhythms in the developing pineal organ. The pineal organ is the major source of the circadian hormone melatonin, which regulates rhythms such as daily rest/activity cycles. Mammalian pineal rhythms are controlled almost exclusively by the SCN. In zebrafish and many other lower vertebrates, the pineal has an endogenous clock that is responsible in part for cyclic melatonin biosynthesis and gene expression.
Results
We find that pineal rhythms are present in cyc mutants despite the absence of an SCN. The arginine vasopressin-like protein (Avpl, formerly called Vasotocin) is a peptide hormone expressed in and around the SCN. We find avpl mRNA is absent in cyc mutants, supporting previous work suggesting the SCN is missing. In contrast, expression of the putative circadian clock genes, cryptochrome 1b (cry1b) and cryptochrome 3 (cry3), in the brain of the developing fish is unaltered. Expression of two pineal rhythmic genes, exo-rhodopsin (exorh) and serotonin-N-acetyltransferase (aanat2), involved in photoreception and melatonin synthesis, respectively, is also similar between cyc embryos and their wildtype (WT) siblings. The timing of the peaks and troughs of expression are the same, although the amplitude of expression is slightly decreased in the mutants. Cyclic gene expression persists for two days in cyc embryos transferred to constant light or constant dark, suggesting a circadian clock is driving the rhythms. However, the amplitude of rhythms in cyc mutants kept in constant conditions decreased more quickly than in their WT siblings.
Conclusion
Our data suggests that circadian rhythms can be initiated and maintained in the absence of SCN and other tissues in the ventral brain. However, the SCN may have a role in regulating the amplitude of rhythms when environmental cues are absent. This provides some of the first evidence that the SCN of teleosts is not essential for establishing circadian rhythms during development. Several SCN-independent circadian rhythms have also been found in mammalian species. Thus, zebrafish may serve as a model system for understanding how vertebrate embryos coordinate rhythms that are controlled by different circadian clocks.
Background
Circadian rhythms are biological cycles in behavior, physiology, and biochemistry that occur approximately every 24 hours. These oscillations are present in almost every organism, from cyanobacteria to plants to humans [1]. All circadian rhythms are regulated by a timing system composed of intracellular clocks with periods of approximately 24 hours, environmental cues, and clock-controlled outputs. An important characteristic is that circadian clocks are able to drive output rhythms even in the absence of environmental cues. However, environmental influences such as light and temperature are required to entrain or re-set the circadian oscillators so that they stay in synchronization with the organism's surroundings.
Regulation of vertebrate circadian rhythms is best understood in mammals. The SCN, a group of neurons in the hypothalamus, is among the most intensely studied cellular site of circadian oscillators. Targeted bilateral lesion of the rodent SCN abolished circadian oscillations in activity and drinking rhythms [2]. Conversely, transplantation of a donor SCN into a host with a lesioned SCN restored circadian cycling [3]. Further, the new locomotor activity rhythms of the host matched the circadian cycle of the donor SCN, indicating that the SCN was the major influence on the phase of the restored rhythms [3]. The SCN controls rhythms in other tissues through a variety of mechanisms, including the secretion of peptide hormones and synaptic signaling [4]. For instance, rhythmic production of the circadian hormone melatonin in mammals is controlled almost entirely by a multi-synaptic pathway leading from the SCN to the cells of the pineal gland [5].
Although it is clear that the mammalian SCN is important in controlling many rhythms in numerous organs, mammals also have circadian oscillators in other tissues as well as rhythms that are SCN-independent [6,7]. For instance, the mammalian retina has an endogenous oscillator that controls local rhythms such as visual sensitivity and retinal melatonin synthesis [8]. This indicates that the mammalian circadian system may have similarities to the circadian systems of many lower vertebrates, which often have many oscillators and many sites of photoreception [9-11].
A structure anatomically equivalent to the SCN has been described in zebrafish embryos and adults [12-17]. However, the function of the zebrafish SCN in regulating circadian rhythms is unknown [18]. This question is particularly interesting as many different isolated zebrafish cells and organs, such as the pineal, eyes, heart, spleen, and kidney, have endogenous oscillators and circadian photoreceptors that are able to generate cyclic gene expression [18-21]. For example, in adult zebrafish the pineal organ contains an endogenous circadian oscillator that is sufficient to drive rhythms in melatonin synthesis as well as photoreceptive neurons that entrain this oscillator [21]. However, whether pineal rhythms are also influenced by signals from other tissues, such as the SCN, is unknown in zebrafish and many other lower vertebrates. Interestingly, in some avian species, pineal rhythms are controlled both by an endogenous pineal oscillator and by input from the SCN, raising the possibility that multiple tissues could regulate pineal circadian rhythms in zebrafish [11,22,23].
Here, we provide evidence that the SCN is not required for pineal rhythms in developing zebrafish. To do this, we took advantage of zebrafish cyc mutants, which have a mutation in one of three zebrafish nodal genes [24,25]. Lack of Cyc/Nodal signaling results in a complete absence of the hypothalamus, including the regions that give rise to the SCN, the retro-chiasmatic nucleus, and the infundibulum [17,24-26]. Consistent with this earlier work, we find that expression of the avpl gene, which is typically expressed in and around the SCN [27,28], is absent in cyc mutants. Despite this, expression of the putative clock component genes cry1b and cry3, as well as the structure and size of the pineal organ was indistinguishable between cyc embryos and their WT siblings. The phase of gene expression of two pineal rhythmic genes, exorh and aanat2 persisted in the mutants. However, semi-quantitative analysis suggested that the amplitude of the rhythms was slightly reduced. aanat2 mRNA levels maintained their rhythmic changes in cyc embryos placed in constant conditions. This suggests that transcriptional rhythms were not being controlled by the environment, but rather by an internal cellular clock, perhaps the endogenous clock within the pineal cells. Since cyc mutants never have an SCN, this work indicates daily rhythms within the developing zebrafish pineal can be initiated in the absence of the developing hypothalamus, including the SCN anlage.
Results
cyc mutants lack avpl gene expression in the ventral brain
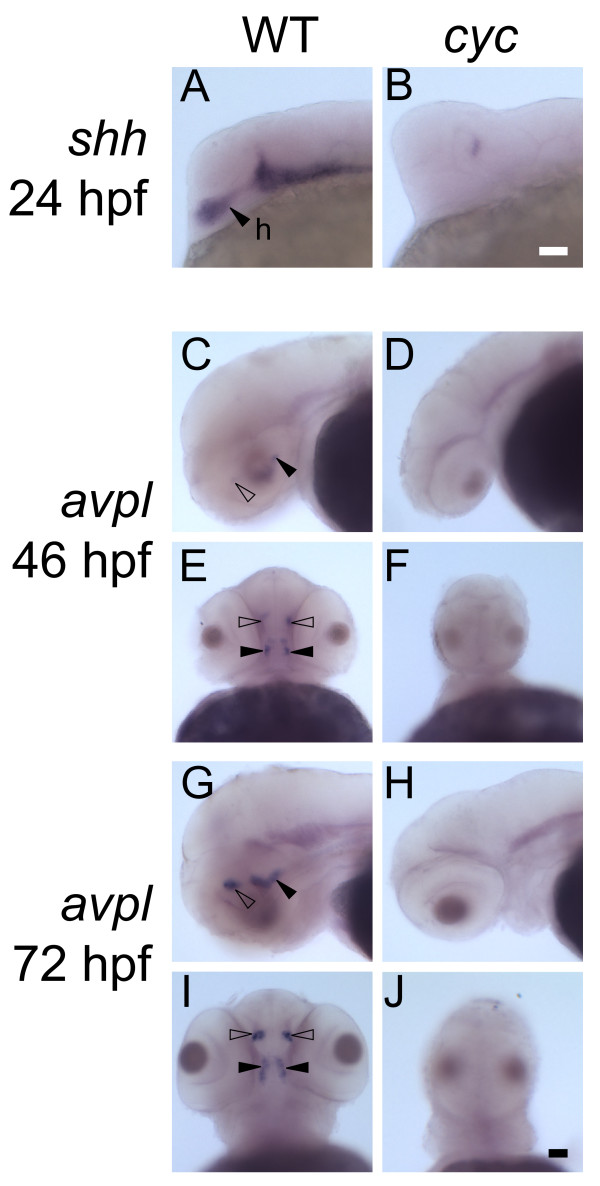
We sought to define the function of the zebrafish SCN in regulating circadian rhythms during embryogenesis. Zebrafish cyc mutants are missing the ventral brain and the spinal cord, including expression of the genes sonic hedgehog (shh), emx2, and nkx2.1 in the hypothalamus [17,25,26,29]. This strongly suggests that cyc mutants could also be lacking the hypothalamus-derived precursors to the SCN. Consistent with previous data, we found that cyc mutants are missing the shh-positive neurons in the anterior hypothalamus that are proposed to be the precursors to the SCN (Figure 1A, B) [17,30].
Figure 1.
avpl expression is absent in cyc mutants. Embryos were fixed at (A, B) 24 hours post-fertilization (hpf), (C-F) 46 hpf, and (G-I) 72 hpf and then processed for WISH with antisense probes for the indicated mRNAs. shh-expression in the ventral brain, including the hypothalamus (h, closed arrowhead) is (A) apparent in WT embryos but (B) absent in cyc mutants. (C, E, G, and I) avpl is expressed in the dorsal preoptic area (open arrowheads) and the ventral hypothalamus (closed arrowheads). (D, F, H, and J) Both avpl expression domains are eliminated in cyc mutants. (A-D, G, and H) are lateral views and (E, F, I, and J) are ventral views. The images in panels (C-J) are representative embryos of three independent experiments (n = 3 fish per experiment). avpl expression was also completely absent in cyc mutants at 47 hpf, Zeitgeber Time (ZT) 23.5; 52 hpf, ZT 5.5; 62 hpf, ZT 15.5, and 71.5 hpf, ZT 23.5. No circadian rhythm in avpl expression was detected in the WT larva processed in parallel (n>4 larva per time point). Scale bars = 50 μm.
To further characterize the ventral brain defect in cyc mutants, we examined the expression of the avpl gene. In mammals, vasopressin is expressed by the SCN neurons with a strong circadian rhythm, and has been linked to changes in hormone secretion from the pituitary, regulation of reproduction in females, and behaviors such as daily rhythms in wheel running activity in nocturnal rodents [28]. In non-mammalian vertebrates, vasopressin is replaced by Avpl, which is expressed in many ventral brain neurons including cells in and around the SCN [27]. In zebrafish, avpl mRNA is found in two domains of the ventral brain: the dorsal preoptic area and the anterior aspect of the ventral hypothalamus [31]. Based on the location of the avpl positive cells in the ventral hypothalamus, these cells likely correspond to the zebrafish SCN. In cyc embryos, both avpl expression domains were undetectable (Figure 1C-1J). This suggests that neurons expressing avpl, including those in the SCN, are missing.
exorh mRNA is expressed rhythmically in the pineal organs of cyc mutants
We next wanted to examine the expression of rhythmic genes in a tissue outside of the SCN (Figure 2). The developing pineal organ was ideal for this analysis. The pineal contains an endogenous oscillator and photoreceptors that are able to entrain, or reset, this oscillator in response to photic input [21]. Further, pineal rhythms can be easily followed by assaying the expression of circadian-regulated pineal genes [32-39].
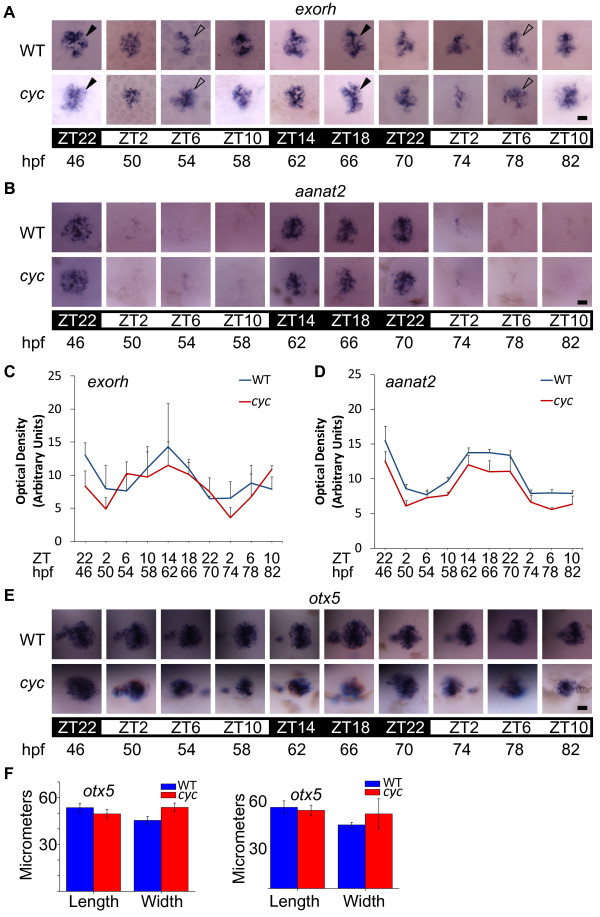
Figure 2.
Rhythmic expression of pineal genes persists in cyc mutants. Embryos were raised in a 14:10 h L/D cycle and then sets of embryos were fixed at the indicated stages and ZT and processed for WISH for expression of (A) exorh or (B) aanat2. Note the time of peak (marked with closed arrowheads for exorh) and trough (marked with open arrowheads for exorh) expression is the same between both sets of embryos. (C, D) Quantification of the WISH signal indicates that the amplitude of aanat2 and exorh gene expression is slightly lower in cyc mutants than in their WT siblings. Two-way ANOVA reveals that the difference in the strength of the aanat2 signals are significantly different between cyc and WT (p < 0.01), while the difference in the strength of the exorh signals does not reach significance (p = 0.14). (E, F) In contrast, the expression of otx5, a gene expressed in all pineal cells, is comparable between WT and cyc mutants. (F) At 54 hpf, ZT6 (left graph) and 78 hpf, ZT6 (right graph), length of otx5 gene expression domain is similar between WT and cyc mutants, while the width of the pineal at each stage is slightly bigger in the mutants, perhaps due to changes in the morphology of the cyc head (54 hpf, pineal length, p = 0.11; 54 hpf, pineal width, p = 0.0045; 78 hpf, pineal length, p = 0.52; 78 hpf, pineal width, p = 0.14, n≥3 embryos per time point). In A, B, and E, light conditions are indicated by the white (light period) and black (dark period) bars. All images are dorsal views, anterior to the top. Each experiment was repeated three times, and representative images are shown. Scale bars = 20 μm.
To determine what happens to pineal rhythms in cyc mutants, we first assayed the embryos for expression of exorh mRNA and protein. Exorh is a putative light-sensing, G-protein coupled receptor expressed in the pineal [40-43]. exorh mRNA is synthesized with a daily rhythm, with highest levels during the night and lower levels during the day [43,44].
Embryos were raised in a standard 14:10 h light/dark (L/D) cycle, with all other environmental parameters held constant. Rhythms in exorh mRNA were present in the pineal organs of both WT and cyc fish (Figure 2A, C). Further, the timing of the troughs and peaks of expression were similar between both sets of embryos (Figure 2A, C).
However, the amplitude of exorh expression was slightly lower at almost every time point tested in the cyc embryos (Figure 2C). To rule out the possibility that this was due to smaller pineal organs in the mutants, we measured the length and width of the pineal organs labeled for expression of orthodenticle homeobox 5 (otx5), a gene that is constitutively expressed in all pineal cells [35]. We found that the pineal size was not significantly different between WT and cyc fish (Figure 2E, F). This suggests that the decrease in the amplitude of exorh expression in cyc embryos was not due to smaller pineal organs.
A gene involved in melatonin biosynthesis is expressed rhythmically in cyc mutants
aanat2 encodes the penultimate enzyme in the melatonin biosynthetic pathway [39]. In zebrafish and many other vertebrates, aanat genes are expressed in the pineal with dramatic differences between day and night levels [45]. Thus, aanat2 expression serves as a very sensitive readout of pineal circadian cycling. In WT and cyc embryos maintained in a L/D cycle, aanat2 transcripts cycled with indistinguishable periods and phases (Figure 2B, D). However, as was the case for exorh, the amplitude of aanat2 expression in cyc mutants was slightly diminished compared to WT siblings raised in parallel (Figure 2D). Thus, the nocturnal expression of aanat2 in the pineal was largely unaffected by the loss of the putative SCN in cyc mutant embryos.
Rhythmic expression of aanat2 persists in cyc embryos placed in constant environmental conditions
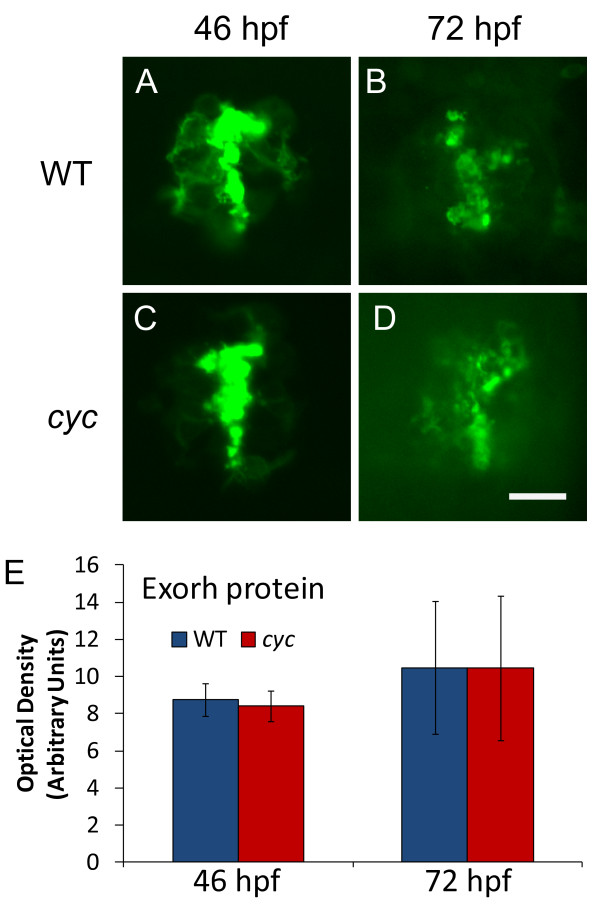
The pineal organs of zebrafish, as well as other tissues, contain functional photoreceptors [18-21,46]. Further, we found that the Exorh protein was expressed normally in cyc mutants, suggesting that the pineal photoreceptors could be functional in these mutants (Figure 3)[Additional File 1]. Thus, one possibility was that the mRNA rhythms in the pinealocytes of cyc embryos were due to direct responses to changing light conditions, not to the presence of a circadian clock. To rule out this possibility, cyc embryos and their WT siblings were raised for two or three days in a standard L/D cycle, and then transferred to either constant light or constant dark for an additional two days (Figure 4)[Additional Files 2 and 3].
Figure 3.
Pineal Exorh protein expression is indistinguishable between cyc mutant and WT embryos. Embryos were fixed and processed for whole mount antibody staining with the anti-Rhodopsin antibody 4D2. Quantification of fluorescent antibody signal shows that optical density of the pineal immunostaining is not significantly different at either time point (46 hpf, ZT 22, n = 10 embryos, p = 0.41; 72 hpf, ZT 0, n = 10 larvae, p = 0.67). Images are dorsal views with anterior to the top. Experiment was repeated two times with similar results, and representative embryos are shown. Scale bar = 20 μm.
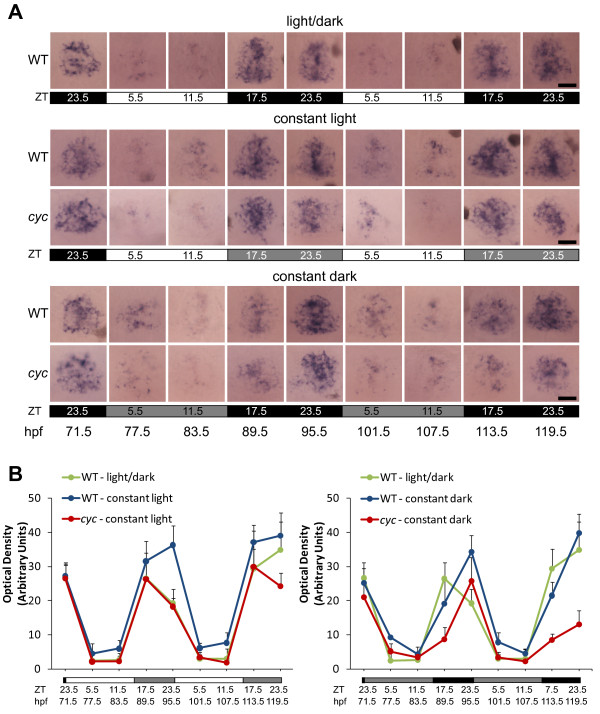
Figure 4.
Rhythmic expression of aanat2 persists in constant conditions. Embryos were raised in a 14:10 h L/D cycle. At 71.5 hpf, ZT 23.5, a set of embryos was transferred to constant dark, constant temperature conditions. At 77.5 hpf, ZT 5.5 a set of embryos were transferred to constant light, constant temperature conditions. (A) Embryos were fixed at the indicated stages and ZT and processed for WISH for expression of aanat2. Note the time of peak expression is similar between the cyc embryos and their WT siblings. All images are dorsal views, anterior to the top. (B) Quantification of the WISH signal indicates that the amplitude of aanat2 gene expression is slightly lower in cyc mutants than in their WT siblings in constant conditions. Two-way ANOVA reveals that the differences between aanat2 signal is significantly different between cyc and WT (p < 0.01 for both constant light and constant dark, n>10 larva per time point, images of 4 larva per each experimental condition and time point used for statistical analysis). For the samples in a L/D cycle, position within the photoperiod is indicated by ZT and light conditions by the white (light period) and black (dark period) bars. For the constant light samples, the original L/D cycle is indicated by the ZT, and the white (original light period) and light grey (original dark period) bars. For the constant dark experiment, the original L/D cycle is indicated by the ZT, and the black (original dark period) and grey (original light period) bars. Experiment was repeated two times with similar results, and representative images from one of the experiments are shown in A. Scale bars = 20 μm.
When the embryos were exposed to a L/D cycle for only two days before transfer to constant conditions, the amplitude of the rhythmic gene expression was very low in both WT and cyc mutants, suggesting that circadian cycling was not fully established by such a short period of entrainment [Additional File 2]. In contrast, when embryos were maintained instead for three days in a L/D cycle, rhythmic expression of aanat2 persisted in WT and in cyc larva after transfer to either constant light or constant dark conditions (Figure 4). Under constant dark conditions, a slight shift in the peak of expression to the right was apparent, consistent with previous studies that showed the endogenous period of the clock that controls zebrafish pineal rhythms is slightly longer than 24 hours (Figure 4B) [10,35,39,47]. However, this shift was not readily apparent in fish in constant light (Figure 4B). The persistence in cyclic changes in expression after transfer to constant lighting conditions suggests that the rhythms are not due to responses to rhythmic environmental changes, but instead due to a functioning circadian clock in the cyc embryos.
Although the overall rhythm in gene expression persisted in constant conditions, there were some significant differences between the cyc mutants and their WT siblings. As in the L/D experiments (Figure 2), the levels of mRNA were slightly lower in the cyc embryos at most time points (Figure 4). Further, after approximately 48 hours in constant conditions, for both constant light and constant dark, the aanat2 expression was notably lower in the cyc embryos than in their WT siblings (Figure 4).
cry gene expression is present in cyc mutants
To gain insight into other potential changes in cyc embryos, we examined the expression of putative clock components during embryogenesis. The vertebrate circadian clock is composed of complex positive and negative feedback loops that take approximately 24 hours to go through one cycle. Cry proteins are essential components of this clock, in which they act as transcriptional repressors in the negative feedback loop. Zebrafish have six cry genes, all of which are expressed with a daily rhythm [48,49].
We found that the expression of cry1b and cry3 was indistinguishable between cyc embryos and their WT siblings (Figures 5 and 6). In both types of embryos, cry1b was expressed widely in the developing brain (Figure 5). cry3 was also expressed in the brain, and more strongly in the ear, liver, and in retinal cells near the lens (Figures 5 and 6). These results indicate that transcription of cry1b and cry3 is present even when the ventral brain is absent.
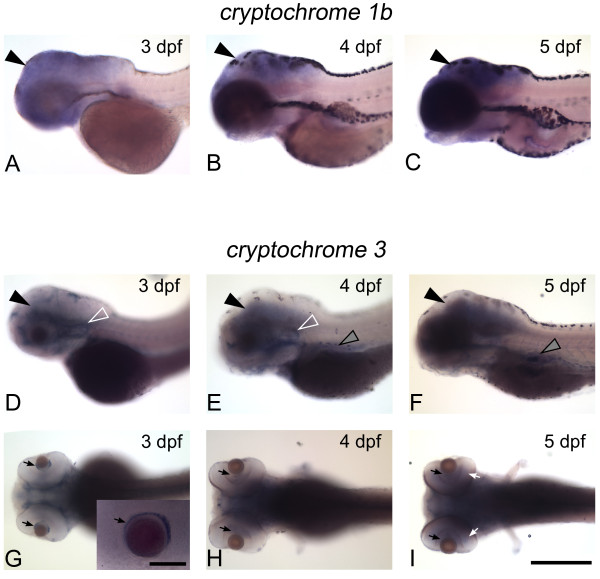
Figure 5.
Expression of cry genes in zebrafish larvae. WT larvae were fixed at the indicated stages and then processed for WISH with an antisense probe for cry1b or cry3. (A-C) cry1b is expressed in the brain (closed arrowhead) from 3 to 5 dpf. (D, G) At 3 dpf, cry3 is expressed in the brain (closed arrowhead), ear (open arrowhead) and in a region of the retina around the lens (black arrows). Inset in panel G is a higher magnification of the area around the lens. (E, H) At 4 dpf, cry3 is expressed in the brain (closed arrowhead), ear (open arrowhead), and liver (gray arrowhead). (F, I) By 5 dpf, cry3 transcripts are present in the ganglion cell (black arrows) and inner nuclear layers (white arrows) of the retina, and persist in the brain (closed arrowhead) and liver (gray arrowhead). WISH at each stage was repeated two times and representative images are shown (n≥5 embryos for the experiment shown). Panels (A-F) are lateral views, anterior to the left and panels (G-I) are dorsal views, anterior to the left. Scale bar in panel I (for all images except G inset) = 500 μm, Scale bar for G inset = 100 μm.
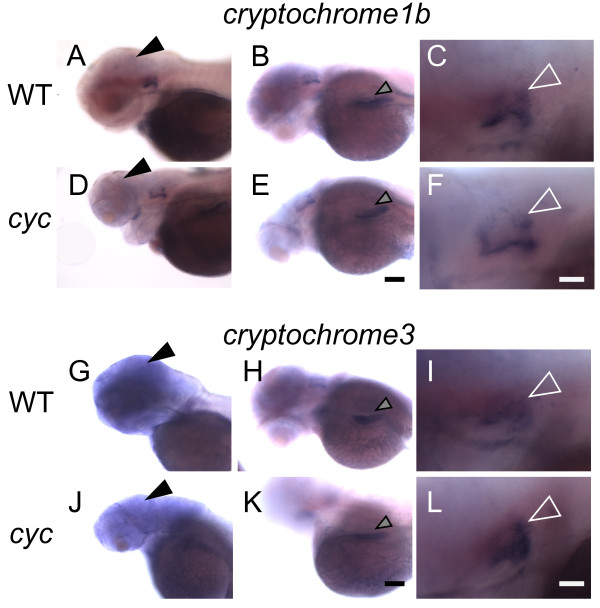
Figure 6.
cry gene expression is present in cyc mutants. cyc embryos and their WT siblings were raised in a 14:10 h L/D cycle, fixed at 72 hpf, ZT 0, and then processed for WISH. In both (A-C, G-I) WT embryos and (D-F, J-L) cyc mutants, cry1b and cry3 are expressed in the (A, D, G, J) brain (closed arrowheads), (B, E, H, K) liver (gray arrowheads), and (C, F, I, L) ear (open white arrowheads). Experiment was repeated three times, and representative images are shown. All images are lateral views, anterior to the left. Scale bars = 40 μm for (A-B, D-E, G-H, J-K) and 100 μm for (C, F, I, L).
Discussion
In mammals and birds, the SCN has an important role in regulating circadian rhythms throughout the organism. Retinal photoreceptors entrain circadian oscillators within the SCN, and the SCN then regulates pineal rhythms through a multisynaptic pathway [50]. Although the retina-SCN-pineal pathway is present in fish, amphibians, and reptiles, its function is not well understood [15,51-53]. Here we provide evidence that pineal circadian rhythms are established and have normal phase in zebrafish cyc mutants, which lack ventral brain. This suggests that neither the SCN nor the retinal-hypothalamic pathway is essential for the onset of pineal circadian rhythms during embryogenesis. However, the amplitude of the rhythms is slightly, but significantly decreased, in cyc mutants. This suggests that the SCN or other tissues that are missing in these mutants, which include other regions of the ventral brain, may have a role in regulating rhythms during development.
Pineal rhythms persist in cyc mutants
The SCN has been defined anatomically in zebrafish embryos and adults [15]. However, the function of the zebrafish SCN is unknown. We used cyc mutants to test the function of the zebrafish SCN. These mutants are missing the whole ventral brain including the entire hypothalamus, and thus, the precursors to the SCN [17,26]. We now demonstrate that expression of avpl, which is usually present in and around the SCN, is lacking in cyc mutants (Figure 1). This provides strong evidence that the SCN is completely absent in cyc mutants.
We found that rhythmic expression of aanat2 and exorh are present in cyc mutants during the first few days of development (Figure 2). The phases of expression of these genes are indistinguishable between cyc mutants and WT siblings raised in parallel in a L/D cycle. Further, the aanat2 expression rhythms persist after the embryos are transferred to constant dark or constant light, indicating they are being driven by a circadian clock (Figure 4).
Diminished amplitude of gene expression in cyc mutants
The expression amplitude of aanat2 and exorh mRNA levels are slightly reduced in the cyc mutants compared to WT siblings raised in parallel. This was true for fish in a L/D cycle, and after transfer to constant dark or constant light conditions (Figures 2 and 4). In addition, the decrease in amplitude of expression became more pronounced when the fish approached the third day in constant conditions (Figure 4).
cyc mutants have many developmental defects, making it difficult to precisely identify the cause of the decrease in gene expression. It is unlikely that the change is due to defects in pineal development, as the size of the pineal was not significantly decreased in cyc mutants (Figure 2F). However, the lower amplitude of gene expression could be due in part to poor health or subtle changes in the pineal that we have not detected [24-26].
A more interesting possibility is that the SCN or other missing tissues in the ventral brain play a role in regulating the amplitude of pineal rhythms. Because the phenotype of cyc mutants becomes more severe as the fish get older, we kept the fish in a L/D cycle for only a short time before being assayed. Thus, one possibility is that the SCN/ventral brain functions to help establish self-sustaining rhythms during development. This role for the SCN could explain why the amplitude of rhythms were lower in the cyc pineal overall, and why the rhythms in the cyc pineal started to dampen sooner after transfer to constant conditions. Testing this hypothesis will require a mutant that more specifically affects the SCN or a way to specifically inhibit the function of SCN cells.
Tissues that could be promoting circadian rhythms in cyc mutants
We do not completely understand what is driving pineal rhythms in cyc mutants. One possibility is that these rhythms are driven by clocks within the zebrafish pineal. Persistent circadian rhythms in melatonin synthesis were observed previously in isolated adult zebrafish pineals cultured for several days [21]. This suggests that the pineal oscillators in adult zebrafish are self-sustained. Clocks in the embryonic pineal may work in the same way. In support of this, a previous study showed that light input very early in development, long before the differentiation of an SCN, is necessary for onset of pineal rhythms [54].
Oscillator proteins present in the pineal are also in many other zebrafish cells and tissues. Likewise, circadian rhythms have been detected in isolated cultured zebrafish embryonic and adult tissues [18-20]. Thus, it is possible that other tissues present in cyc mutants are interacting to promote circadian cycling in the pineal.
We also cannot rule out a role for the lateral eyes. The first zebrafish retinal photoreceptors differentiate at approximately 2.5 dpf [55]. Thus, detection of environmental light conditions by the retina could explain why circadian rhythms in pineal gene expression were better established when embryos were maintained for three rather than two days in a L/D cycle (compare Figure 4 with Additional file 2). Previous work suggested that the lateral eyes are not required for establishing rhythms in the pineal. Rhythms in pineal aanat2 expression are present in zebrafish chokh (chk) mutants, which lack lateral eyes [56]. Further, chokh mutants can re-entrain to a shifted L/D cycle [56]. However, the persistence of pineal rhythms in chokh fish after transfer to constant lighting conditions has not yet been tested, raising the possibility that they could have defects similar to those we found in cyc mutants.
A final possibility is that pineal rhythms are present in cyc mutants because there are some residual SCN neurons. However, this is unlikely, as the whole ventral brain is absent in cyc mutants (Figure 1A-1B) [17,24-26,29]. chk mutants are missing both eyes and hypothalamic avpl expression [57]. Despite this, they have rhythms in gene expression of two clock components (clock and period4) and pineal aanat2 [56,58]. These results are consistent with our findings that loss of neurons expressing avpl, including those in the SCN, does not abolish pineal rhythms.
Normal cry gene expression in cyc mutants
Cry proteins are required for regulation of circadian rhythms in both plants and animals [59]. These proteins were shown to play two major roles in regulation of circadian rhythms: as a photoreceptor to entrain the clock and as a repressor of Clock/BMAL-induced circadian transcription [59]. Six Cry proteins are present in zebrafish [60]. By heterologous expression, it was shown that Cry1b, but not Cry3, is capable of blocking Clock/BMAL-dependent transcription [60]. This suggests that Cry1b and Cry3 have different functions. We find that cry1b and cry3 mRNAs are found in specific tissues of zebrafish embryos and larvae (Figures 5 and 6). Further, we demonstrate that cry1b and cry3 expression in developing brain, ear, and liver are present in cyc mutants (Figures 5 and 6). Although the function of the cry genes in zebrafish is not yet fully understood, the presence of cry1b and cry3 transcripts in the cyc brain suggests that the circadian clock may not be severely disrupted.
Exorh protein expression is normal in cyc mutants
exorh was originally identified as a rhodopsin class gene expressed in the zebrafish and salmon pineal organs [40,61]. Exorh has been predicted to be a G-protein coupled receptor with ~70% amino acid sequence identity with vertebrate Rhodopsin [40]. Our group and others have previously demonstrated the rhythmic expression of exorh mRNA [43,44]. In contrast to exorh mRNA, we find that Exorh protein expression does not follow a significant daily rhythm in WT embryos, nor is its expression affected in cyc mutants (Figure 3)[Additional File 1].
The functional significance of this difference in temporal expression pattern is unknown. There are other cases where mRNA displays cyclic expression, while the corresponding protein does not. In the chicken pineal gland, the mRNA encoding the photopigment Pinopsin shows a rhythm that is dependent upon activation by light [62]. In zebrafish, interphotoreceptor retinoid-binding protein (irbp) mRNA expression is circadian-regulated, while and IRBP protein levels are constant [37,63]. In the case of IRBP, it is proposed that the higher expression of mRNA during the day is necessary to maintain the constant levels of protein, as protein turnover is much higher during the day than during the night [37,63]. This could be the explanation for the rhythmic pattern of exorh and pinopsin mRNA expression as well. Consistent with this, Pinopsin protein and mRNA levels decline rapidly when the embryos are placed in constant darkness [62].
Conclusion
More than two decades ago, a structure anatomically equivalent to the SCN was first described in a teleost fish (the goldfish) [64,65]. Several years later, a morphologically similar group of neurons was described in zebrafish [15]. However, no experimental evidence exists for a pacemaker role of the SCN in zebrafish or in other teleosts [18]. Here, we establish that the zebrafish SCN is not required to establish circadian rhythms in the embryonic pineal. However, the amplitude of circadian gene expression was slightly reduced, and so we cannot rule out a role for the SCN or other tissues missing in cyc mutants in helping to establish or maintain rhythms in developing zebrafish. This will form a strong foundation for future studies that explore the communication between different circadian tissues during development, and for comparative studies of pineal biology and circadian regulation between vertebrate species.
Methods
Zebrafish
Zebrafish embryos and larvae were obtained by natural matings and were raised at 28.5°C at 14:10 hour (h) L/D cycle according to standard procedures [66]. Developing fish were placed in a Sanyo MIR-153 incubator (Amsterdam, The Netherlands) with heating and cooling capabilities for maximum temperature stability. Light consisted of a single 60 watt Globe EnerSaver light bulb placed within the incubator. Stocks used were Oregon AB (WT), ZDR (WT) (Aquatica Tropicals, Plant City, FL), and cycm294 [25,66-68].
Whole mount RNA in situ hybridization (WISH)
WISH was carried out as described by Liang et al. [69]. Antisense RNA probes included aanat2 [39], exorh [40], shh [69,70], cry1b [49], cry3 (Open Biosystems, Huntsville, AL), otx5 [35], and avpl [31].
To generate cry1b probe, cry1b cDNA in plasmid pME-18S-FL3 was subcloned into plasmid pBSK(+) using a 5' EcoRI and a 3' NotI restriction sites. Plasmid pBSK+ cry1b was linearized with EcoRI and was used for probe synthesis with T3 RNA polymerase. cry3 probe was PCR amplified from the cry3 cDNA in plasmid pME-18S-FL3 using primers and cycling conditions in ZFIN [http://zfin.org/cgi-bin/webdriver?MIval=aa-markerview.apg&OID=ZDB-CDNA-040425-188]. cry3 mRNA was made with T3 RNA polymerase.
Whole mount antibody staining
Whole mount antibody staining was done as described [71]. A mouse monoclonal antibody (4D2) against the N-terminus of bovine Rhodopsin [72,73] was used to detect Exorh protein at a dilution of 1:60. A goat anti-mouse secondary antibody coupled to Oregon Green 488 (Invitrogen Molecular Probes, Carlsbad, CA) was used at a dilution of 1:2000.
Morpholino and mRNA injections
Control and translation blocking morpholinos (MO) against exorh (exorh atg MO) were obtained from Gene Tools (Philomath, OR). Sequences of the MO used were: control MO, 5'-CCT CTT ACC TCA GTT ACA ATT TATA-3' and exorh atg MO, 5'-AGT TGG GTC CCT CCG TCC CGT TCAT-3'. One-cell stage embryos were injected with either 1.5 nanograms (ng) of control or exorh atg MO using a Harvard Instruments PLI-90 Pico-Injector (Holliston, MA).
To generate exorh mRNA for overexpression, the entire coding sequence of the zebrafish exorh gene (from plasmid pCR2.1-full-length exorh) [GenBank Accession Number: AB025312] was subcloned into pGEMHE plasmid [40,74]. Donor and host plasmids were digested with EcoRI. The resulting EcoRI-digested full-length exorh coding sequence was ligated non-directionally into pGEMHE (plasmid pGEMHE-full-length exorh hereafter pGEMHE-flex) using the TAKARA DNA Ligation Kit version 1 (Madison, WI). The ligation reaction was transformed into Z-Competent E. coli cells (Zymo Research, Orange, CA). Clone orientation was verified via restriction digestion (double digestion with SacI and BanII) and DNA sequencing. DNA sequencing was done using forward and reverse sequencing primers (forward primer: 5'-TTT TTG CAG AAG CTC AGA ATA-3'; reverse primer: 5'-CAT TTT GTA AAG TGT AAG TTG GTAT-3'). DNA sequences were verified by doing a BLAST search [75].
To synthesize full-length exorh mRNA, pGEMHE-flex plasmid was linearized with SphI. Linearized pGEMHE-flex was used for mRNA synthesis with an Ambion mMESSAGE mMACHINE kit (Austin, TX). Full-length exorh mRNA was verified through RNA formaldehyde agarose gel-electrophoresis. One blastomere of 8-16 cell stage embryos were injected with 400 picograms (pg) of beta-galactosidase (control) or full-length exorh mRNA and were fixed 7 h post-injection. Injected and fixed embryos were processed for whole mount antibody staining as described above.
Photography and Image Analysis
Embryonic and larval zebrafish samples were imaged using a Zeiss Axioplan 2 Imaging Microscope (Thornwood, NY) or Nikon Eclipse 801 Epifluorescent Microcope (Melville, NY) connected to a Spot RTke7.4 Slider digital camera together with Spot 4.5.9.1 software (Diagnostic Instruments, Sterling Heights, MI). Resulting images were processed and quantified for optical density using ImageJ 1.36b and 1.42q software [Rasband, W.S., ImageJ, U. S. National Institutes of Health, Bethesda, Maryland, USA, http://rsb.info.nih.gov/ij/, 1997-2009]. Calibration was performed following developer's instructions [http://rsb.info.nih.gov/ij/docs/examples/calibration/]. For in situ hybridization data, images were first converted to 8-bit grayscale and an oval encompassing the expression domain was drawn using the Specify tool. Optical Density (OD) was calculated by multiplying the oval area with the average intensity, both of which were obtained through the Analyze tool. Two-way Analysis of Variance (ANOVA) statistical analysis was carried out using Microsoft Excel.
For quantification of aanat2 expression in the constant light and constant dark experiments, the background value was subtracted as follows. A small circle with diameter of 20 pixels was selected close to, but not touching the anterior edge of the WISH expression signal. The circular area and optical density was determined as above. Background-corrected OD values were calculated by subtracting the (OD value of small circle × pineal oval area/small circle area) from the OD of oval around the pineal expression domain. All OD data are means ± standard deviation. Representative images with ODs closest to the mean were chosen for figures.
Fluorescent images were analyzed as above with the exception that 8-bit grayscale images were first processed using the Inverted tool prior to choosing the oval area to be quantified for OD. For Exorh protein temporal expression, fluorescent image data was tested for significance using One-way ANOVA and Tukey's test using Origin Software Version 7.5 SR4 (Northampton, MA).
To measure the dimensions of the otx5 expression domain, lines covering the length or the width of the pineal were drawn using ImageJ and the corresponding pixel length or width were converted to micrometers by calibrating the number of pixels in a 20 micrometer line.
Authors' contributions
RRN conceived of and designed the study, performed experiments on Exorh protein expression and antibody staining, and studies on aanat2 rhythms, shh, cry, and otx5 expression in cyc mutants, and wrote the first draft of the manuscript. PL helped optimize the antibody staining protocol and carried out studies on cry1b and cry3 expression during development, exorh mRNA rhythms in cyc mutants, and parts of the constant light and constant dark experiments. LGK assisted in designing and in performing the experiments on aanat2 rhythms and otx5 expression in cyc mutants raised in L/D. EG cloned and generated the probe for avpl and performed WISH on cyc mutants. JOL helped design and coordinated the study, carried out parts of the constant light and constant dark studies and the avpl in situs. All authors helped write the manuscript and have approved the final version.
Supplementary Material
Exorh protein is expressed without a significant rhythm (A, B) Embryos were injected with (A) control or (B) exorh atg MO, fixed at 64 hpf, and processed for fluorescent whole mount immunostaining with the anti-bovine Rhodopsin antibody 4D2. (A) Control embryos have robust fluorescent signal in the pineal organ that is (B) severely reduced in Exorh depleted embryos. (C-C') Embryos injected with beta-galactosidase mRNA have undetectable levels of immunoreactivity with the 4D2 antibody at 8 hpf. (D-D') In contrast, embryos injected with exorh mRNA show strong antibody staining at 8 hpf. (E, F) Embryos were fixed in a circadian time course and then processed for 4D2 antibody staining. One-way Analysis of Variance (ANOVA) and Tukey's analysis revealed no significant changes in pineal Exorh protein levels that followed a daily rhythm (n≥9 embryos per time point). However, a few time points were significantly different (p ≤ 0.05) from each other including 72 and 108 hpf, 72 and 116 hpf, 76 and 116 hpf, and 80 and 116 hpf. (A-B, and E) are dorsal views, anterior to the top and (C-D') are lateral views. (C') and (D') are higher magnification images of the regions boxed in (C) and (D), respectively. Scale bars = 20 μm for (A-B, C', D', E) and 100 μm for (C, D).
Two days in a L/D cycle is not sufficient to initiate robust circadian cycling of aanat2 expression. Embryos were raised in a 14:10 h L/D cycle. At 47.5 hpf, ZT 23.5, a set of embryos was transferred to constant dark, constant temperature conditions. Embryos were fixed at the indicated stages and ZT and (A) processed for WISH for expression of aanat2 and (B) the WISH signal was quantified. Note the time of peak (closed arrowheads) expression is similar between the cyc embryos and their WT siblings. All images are dorsal views, anterior to the top. For the samples in a L/D cycle, position within the photoperiod is indicated by ZT and light conditions by the white (light period) and black (dark period) bars. For the constant dark samples, the original L/D cycle is indicated by the ZT and the black (original dark period) and dark grey (original light period) bars. Experiment was repeated two times with similar results, and representative images from one of the experiments are shown. Scale bars = 20 μm.
Comparison between larva processed for WISH with antisense and sense probes reveals low background staining. Embryos were raised in a 14:10 h L/D cycle and fixed and processed for WISH using aanat2 antisense or sense probe. Note that the sense probe produces no detectable signal, as it would recognize antisense mRNA, which should not be present. The brown regions are melanocytes in the skin, which have a natural pigment. All images are dorsal views, anterior to the top, with the pineal indicated (closed arrowheads). Position within the photoperiod is indicated by ZT and light conditions by the white (light period) and black (dark period) bars. Representative images are shown. Scale bar = 30 μm.
Contributor Information
Ramil R Noche, Email: ramil_noche@hms.harvard.edu.
Po-Nien Lu, Email: pxl89@case.edu.
Lauren Goldstein-Kral, Email: ltg1@williams.edu.
Eric Glasgow, Email: eg239@georgetown.edu.
Jennifer O Liang, Email: joliang@d.umn.edu.
Acknowledgements
We would like to thank Drs. Josh Gamse (Vanderbilt University, Nashville, TN), Eric Mintz (Kent State University, Kent, OH), and Neal Peachey (Cleveland Clinic, OH) for their helpful comments on this manuscript, Dr. Yoshitaka Fukada (University of Tokyo, Japan) for providing the full-length exorh cDNA, Dr. Michael Romero (Mayo Clinic, Rochester, MN) for sharing the pGEMHE plasmid and sequencing primers and advice on subcloning, Dr. Robert Molday (University of British Columbia, Canada) for providing the 4D2 antibody, Sandra Albro (Case Western Reserve University) for assistance with statistical analysis, and Drs. David Klein (National Institutes of Health), Yoav Gothilf (Tel-Aviv University, Israel), and Bernard and Christine Thisse (University of Virginia) for plasmids. Allisan Aquilina-Beck, Adelle Schuman, Jessica Clay, Stephan Brannan, Kristine DiMonte, Lain Pierce, Olga Ponomareva, Perry Hwang, Gerry Babcock, Andy Schreiner, and Brian Chen provided expert technical assistance. Supported in part by research grant No. 5-FY02-259 from the March of Dimes Birth Defect Foundation (to JOL), an Edward Mallinckrodt, Jr. Foundation grant (to JOL), 1 RO1 HD054523 (to JOL and Joshua Gamse), the Hathaway Brown Student Research Program (to LGK), and a Phi Beta Kappa Student Research Grant Award (to RRN).
References
- Pando MP, Sassone-Corsi P. Unraveling the mechanisms of the vertebrate circadian clock: zebrafish may light the way. Bioessays. 2002;24(5):419–426. doi: 10.1002/bies.10091. [DOI] [PubMed] [Google Scholar]
- Stephan FK, Zucker I. Circadian rhythms in drinking behavior and locomotor activity of rats are eliminated by hypothalamic lesions. Proc Natl Acad Sci USA. 1972;69(6):1583–1586. doi: 10.1073/pnas.69.6.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralph MR, Foster RG, Davis FC, Menaker M. Transplanted suprachiasmatic nucleus determines circadian period. Science. 1990;247(4945):975–978. doi: 10.1126/science.2305266. [DOI] [PubMed] [Google Scholar]
- Piggins HD, Loudon A. Circadian biology: clocks within clocks. Curr Biol. 2005;15(12):R455–457. doi: 10.1016/j.cub.2005.06.019. [DOI] [PubMed] [Google Scholar]
- Korf HW, Schomerus C, Stehle JH. The pineal organ, its hormone melatonin, and the photoneuroendocrine system. Adv Anat Embryol Cell Biol. 1998;146:1–100. doi: 10.1007/978-3-642-58932-4. [DOI] [PubMed] [Google Scholar]
- Honma K, Honma S. The SCN-independent clocks, methamphetamine and food restriction. Eur J Neurosci. 2009;30(9):1707–1717. doi: 10.1111/j.1460-9568.2009.06976.x. [DOI] [PubMed] [Google Scholar]
- Bell-Pedersen D, Cassone VM, Earnest DJ, Golden SS, Hardin PE, Thomas TL, Zoran MJ. Circadian rhythms from multiple oscillators: lessons from diverse organisms. Nat Rev Genet. 2005;6(7):544–556. doi: 10.1038/nrg1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan GX, Zhang DQ, Zhou T, Yamazaki S, McMahon DG. Circadian organization of the mammalian retina. Proc Natl Acad Sci USA. 2006;103(25):9703–9708. doi: 10.1073/pnas.0601940103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tosini G, Bertolucci C, Foa A. The circadian system of reptiles: a multioscillatory and multiphotoreceptive system. Physiol Behav. 2001;72(4):461–471. doi: 10.1016/S0031-9384(00)00423-6. [DOI] [PubMed] [Google Scholar]
- Cahill GM. Clock mechanisms in zebrafish. Cell Tissue Res. 2002;309(1):27–34. doi: 10.1007/s00441-002-0570-7. [DOI] [PubMed] [Google Scholar]
- Underwood H, Steele CT, Zivkovic B. Circadian organization and the role of the pineal in birds. Microsc Res Tech. 2001;53(1):48–62. doi: 10.1002/jemt.1068. [DOI] [PubMed] [Google Scholar]
- Rink E, Wullimann MF. Connections of the ventral telencephalon (subpallium) in the zebrafish (Danio rerio) Brain Res. 2004;1011(2):206–220. doi: 10.1016/j.brainres.2004.03.027. [DOI] [PubMed] [Google Scholar]
- Yanez J, Busch J, Anadon R, Meissl H. Pineal projections in the zebrafish (Danio rerio): overlap with retinal and cerebellar projections. Neuroscience. 2009;164(4):1712–1720. doi: 10.1016/j.neuroscience.2009.09.043. [DOI] [PubMed] [Google Scholar]
- Wulliman MF, Rupp B, Reichert H. Neuroanatomy of the Zebrafish Brain: A Topological Atlas. Birkhauser; 1996. [Google Scholar]
- Burrill JD, Easter SS Jr. Development of the retinofugal projections in the embryonic and larval zebrafish (Brachydanio rerio) J Comp Neurol. 1994;346(4):583–600. doi: 10.1002/cne.903460410. [DOI] [PubMed] [Google Scholar]
- Rink E, Guo S. The too few mutant selectively affects subgroups of monoaminergic neurons in the zebrafish forebrain. Neuroscience. 2004;127(1):147–154. doi: 10.1016/j.neuroscience.2004.05.004. [DOI] [PubMed] [Google Scholar]
- Mathieu J, Barth A, Rosa FM, Wilson SW, Peyrieras N. Distinct and cooperative roles for Nodal and Hedgehog signals during hypothalamic development. Development. 2002;129(13):3055–3065. doi: 10.1242/dev.129.13.3055. [DOI] [PubMed] [Google Scholar]
- Kaneko M, Hernandez-Borsetti N, Cahill GM. Diversity of zebrafish peripheral oscillators revealed by luciferase reporting. Proc Natl Acad Sci USA. 2006;103(39):14614–14619. doi: 10.1073/pnas.0606563103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitmore D, Foulkes NS, Sassone-Corsi P. Light acts directly on organs and cells in culture to set the vertebrate circadian clock. Nature. 2000;404(6773):87–91. doi: 10.1038/35003589. [DOI] [PubMed] [Google Scholar]
- Whitmore D, Foulkes NS, Strahle U, Sassone-Corsi P. Zebrafish Clock rhythmic expression reveals independent peripheral circadian oscillators. Nat Neurosci. 1998;1(8):701–707. doi: 10.1038/3703. [DOI] [PubMed] [Google Scholar]
- Cahill GM. Circadian regulation of melatonin production in cultured zebrafish pineal and retina. Brain Res. 1996;708(1-2):177–181. doi: 10.1016/0006-8993(95)01365-2. [DOI] [PubMed] [Google Scholar]
- Brandstatter R, Abraham U. Hypothalamic circadian organization in birds. I. Anatomy, functional morphology, and terminology of the suprachiasmatic region. Chronobiol Int. 2003;20(4):637–655. doi: 10.1081/CBI-120023343. [DOI] [PubMed] [Google Scholar]
- Abraham U, Albrecht U, Brandstatter R. Hypothalamic circadian organization in birds. II. Clock gene expression. Chronobiol Int. 2003;20(4):657–669. doi: 10.1081/CBI-120022414. [DOI] [PubMed] [Google Scholar]
- Rebagliati MR, Toyama R, Haffter P, Dawid IB. cyclops encodes a nodal-related factor involved in midline signaling. Proc Natl Acad Sci USA. 1998;95(17):9932–9937. doi: 10.1073/pnas.95.17.9932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampath K, Rubinstein AL, Cheng AM, Liang JO, Fekany K, Solnica-Krezel L, Korzh V, Halpern ME, Wright CV. Induction of the zebrafish ventral brain and floorplate requires cyclops/nodal signalling. Nature. 1998;395(6698):185–189. doi: 10.1038/26020. [DOI] [PubMed] [Google Scholar]
- Hatta K, Kimmel CB, Ho RK, Walker C. The cyclops mutation blocks specification of the floor plate of the zebrafish central nervous system. Nature. 1991;350(6316):339–341. doi: 10.1038/350339a0. [DOI] [PubMed] [Google Scholar]
- Balment RJ, Lu W, Weybourne E, Warne JM. Arginine vasotocin a key hormone in fish physiology and behaviour: a review with insights from mammalian models. Gen Comp Endocrinol. 2006;147(1):9–16. doi: 10.1016/j.ygcen.2005.12.022. [DOI] [PubMed] [Google Scholar]
- Caldwell HK, Lee HJ, Macbeth AH, Young WS. Vasopressin: behavioral roles of an "original" neuropeptide. Prog Neurobiol. 2008;84(1):1–24. doi: 10.1016/j.pneurobio.2007.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohr KB, Barth KA, Varga ZM, Wilson SW. The nodal pathway acts upstream of hedgehog signaling to specify ventral telencephalic identity. Neuron. 2001;29(2):341–351. doi: 10.1016/S0896-6273(01)00210-0. [DOI] [PubMed] [Google Scholar]
- Puelles L. A segmental morphological paradigm for understanding vertebrate forebrains. Brain Behav Evol. 1995;46(4-5):319–337. doi: 10.1159/000113282. [DOI] [PubMed] [Google Scholar]
- Eaton JL, Holmqvist B, Glasgow E. Ontogeny of vasotocin-expressing cells in zebrafish: selective requirement for the transcriptional regulators orthopedia and single-minded 1 in the preoptic area. Dev Dyn. 2008;237(4):995–1005. doi: 10.1002/dvdy.21503. [DOI] [PubMed] [Google Scholar]
- Gothilf Y, Toyama R, Coon SL, Du SJ, Dawid IB, Klein DC. Pineal-specific expression of green fluorescent protein under the control of the serotonin-N-acetyltransferase gene regulatory regions in transgenic zebrafish. Dev Dyn. 2002;225(3):241–249. doi: 10.1002/dvdy.10152. [DOI] [PubMed] [Google Scholar]
- Triqueneaux G, Thenot S, Kakizawa T, Antoch MP, Safi R, Takahashi JS, Delaunay F, Laudet V. The orphan receptor Rev-erbalpha gene is a target of the circadian clock pacemaker. J Mol Endocrinol. 2004;33(3):585–608. doi: 10.1677/jme.1.01554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appelbaum L, Toyama R, Dawid IB, Klein DC, Baler R, Gothilf Y. Zebrafish serotonin-N-acetyltransferase-2 gene regulation: pineal-restrictive downstream module contains a functional E-box and three photoreceptor conserved elements. Mol Endocrinol. 2004;18(5):1210–1221. doi: 10.1210/me.2003-0439. [DOI] [PubMed] [Google Scholar]
- Gamse JT, Shen YC, Thisse C, Thisse B, Raymond PA, Halpern ME, Liang JO. Otx5 regulates genes that show circadian expression in the zebrafish pineal complex. Nat Genet. 2002;30(1):117–121. doi: 10.1038/ng793. [DOI] [PubMed] [Google Scholar]
- Appelbaum L, Anzulovich A, Baler R, Gothilf Y. Homeobox-clock protein interaction in zebrafish. A shared mechanism for pineal-specific and circadian gene expression. J Biol Chem. 2005;280(12):11544–11551. doi: 10.1074/jbc.M412935200. [DOI] [PubMed] [Google Scholar]
- Stenkamp DL, Cunningham LL, Raymond PA, Gonzalez-Fernandez F. Novel expression pattern of interphotoreceptor retinoid-binding protein (IRBP) in the adult and developing zebrafish retina and RPE. Mol Vis. 1998;4:26. [PubMed] [Google Scholar]
- Ziv L, Levkovitz S, Toyama R, Falcon J, Gothilf Y. Functional development of the zebrafish pineal gland: light-induced expression of period2 is required for onset of the circadian clock. J Neuroendocrinol. 2005;17(5):314–320. doi: 10.1111/j.1365-2826.2005.01315.x. [DOI] [PubMed] [Google Scholar]
- Gothilf Y, Coon SL, Toyama R, Chitnis A, Namboodiri MA, Klein DC. Zebrafish serotonin N-acetyltransferase-2: marker for development of pineal photoreceptors and circadian clock function. Endocrinology. 1999;140(10):4895–4903. doi: 10.1210/en.140.10.4895. [DOI] [PubMed] [Google Scholar]
- Mano H, Kojima D, Fukada Y. Exo-rhodopsin: a novel rhodopsin expressed in the zebrafish pineal gland. Brain Res Mol Brain Res. 1999;73(1-2):110–118. doi: 10.1016/S0169-328X(99)00242-9. [DOI] [PubMed] [Google Scholar]
- Asaoka Y, Mano H, Kojima D, Fukada Y. Pineal expression-promoting element (PIPE), a cis-acting element, directs pineal-specific gene expression in zebrafish. Proc Natl Acad Sci USA. 2002;99(24):15456–15461. doi: 10.1073/pnas.232444199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falcon J, Gothilf Y, Coon SL, Boeuf G, Klein DC. Genetic, temporal and developmental differences between melatonin rhythm generating systems in the teleost fish pineal organ and retina. J Neuroendocrinol. 2003;15(4):378–382. doi: 10.1046/j.1365-2826.2003.00993.x. [DOI] [PubMed] [Google Scholar]
- Vuilleumier R, Besseau L, Boeuf G, Piparelli A, Gothilf Y, Gehring WG, Klein DC, Falcon J. Starting the zebrafish pineal circadian clock with a single photic transition. Endocrinology. 2006;147(5):2273–2279. doi: 10.1210/en.2005-1565. [DOI] [PubMed] [Google Scholar]
- Pierce LX, Noche RR, Ponomareva O, Chang C, Liang JO. Novel functions for Period 3 and Exo-rhodopsin in rhythmic transcription and melatonin biosynthesis within the zebrafish pineal organ. Brain Res. 2008;1223:11–24. doi: 10.1016/j.brainres.2008.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein DC. Evolution of the vertebrate pineal gland: the AANAT hypothesis. Chronobiol Int. 2006;23(1-2):5–20. doi: 10.1080/07420520500545839. [DOI] [PubMed] [Google Scholar]
- Carr AJ, Whitmore D. Imaging of single light-responsive clock cells reveals fluctuating free-running periods. Nat Cell Biol. 2005;7(3):319–321. doi: 10.1038/ncb1232. [DOI] [PubMed] [Google Scholar]
- Kazimi N, Cahill GM. Development of a circadian melatonin rhythm in embryonic zebrafish. Brain Res Dev Brain Res. 1999;117(1):47–52. doi: 10.1016/S0165-3806(99)00096-6. [DOI] [PubMed] [Google Scholar]
- Lahiri K, Vallone D, Gondi SB, Santoriello C, Dickmeis T, Foulkes NS. Temperature regulates transcription in the zebrafish circadian clock. PLoS Biol. 2005;3(11):e351. doi: 10.1371/journal.pbio.0030351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi Y, Ishikawa T, Hirayama J, Daiyasu H, Kanai S, Toh H, Fukuda I, Tsujimura T, Terada N, Kamei Y. et al. Molecular analysis of zebrafish photolyase/cryptochrome family: two types of cryptochromes present in zebrafish. Genes Cells. 2000;5(9):725–738. doi: 10.1046/j.1365-2443.2000.00364.x. [DOI] [PubMed] [Google Scholar]
- Liu Q, Frey RA, Babb-Clendenon SG, Liu B, Francl J, Wilson AL, Marrs JA, Stenkamp DL. Differential expression of photoreceptor-specific genes in the retina of a zebrafish cadherin2 mutant glass onion and zebrafish cadherin4 morphants. Exp Eye Res. 2007;84(1):163–175. doi: 10.1016/j.exer.2006.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertolucci C, Sovrano VA, Magnone MC, Foa A. Role of suprachiasmatic nuclei in circadian and light-entrained behavioral rhythms of lizards. Am J Physiol Regul Integr Comp Physiol. 2000;279(6):R2121–2131. doi: 10.1152/ajpregu.2000.279.6.R2121. [DOI] [PubMed] [Google Scholar]
- Foa A, Brandstatter R, Bertolucci C. The circadian system of ruin lizards: a seasonally changing neuroendocrine loop? Chronobiol Int. 2006;23(1-2):317–327. doi: 10.1080/07420520500521954. [DOI] [PubMed] [Google Scholar]
- Kramer BM, Song JY, Westphal NJ, Jenks BG, Roubos EW. Regulation of neurons in the suprachiasmatic nucleus of Xenopus laevis. Comp Biochem Physiol B Biochem Mol Biol. 2002;132(1):269–274. doi: 10.1016/S1096-4959(01)00539-5. [DOI] [PubMed] [Google Scholar]
- Ziv L, Gothilf Y. Circadian time-keeping during early stages of development. Proc Natl Acad Sci USA. 2006;103(11):4146–4151. doi: 10.1073/pnas.0600571103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kljavin IJ. Early development of photoreceptors in the ventral retina of the zebrafish embryo. J Comp Neurol. 1987;260(3):461–471. doi: 10.1002/cne.902600311. [DOI] [PubMed] [Google Scholar]
- Kennedy BN, Stearns GW, Smyth VA, Ramamurthy V, van Eeden F, Ankoudinova I, Raible D, Hurley JB, Brockerhoff SE. Zebrafish rx3 and mab21l2 are required during eye morphogenesis. Dev Biol. 2004;270(2):336–349. doi: 10.1016/j.ydbio.2004.02.026. [DOI] [PubMed] [Google Scholar]
- Tessmar-Raible K, Raible F, Christodoulou F, Guy K, Rembold M, Hausen H, Arendt D. Conserved sensory-neurosecretory cell types in annelid and fish forebrain: insights into hypothalamus evolution. Cell. 2007;129(7):1389–1400. doi: 10.1016/j.cell.2007.04.041. [DOI] [PubMed] [Google Scholar]
- Dickmeis T, Lahiri K, Nica G, Vallone D, Santoriello C, Neumann CJ, Hammerschmidt M, Foulkes NS. Glucocorticoids play a key role in circadian cell cycle rhythms. PLoS Biol. 2007;5(4):e78. doi: 10.1371/journal.pbio.0050078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cashmore AR. Cryptochromes: enabling plants and animals to determine circadian time. Cell. 2003;114(5):537–543. doi: 10.1016/j.cell.2003.08.004. [DOI] [PubMed] [Google Scholar]
- Ishikawa T, Hirayama J, Kobayashi Y, Todo T. Zebrafish CRY represses transcription mediated by CLOCK-BMAL heterodimer without inhibiting its binding to DNA. Genes Cells. 2002;7(10):1073–1086. doi: 10.1046/j.1365-2443.2002.00579.x. [DOI] [PubMed] [Google Scholar]
- Philp AR, Bellingham J, Garcia-Fernandez J, Foster RG. A novel rod-like opsin isolated from the extra-retinal photoreceptors of teleost fish. FEBS Lett. 2000;468(2-3):181–188. doi: 10.1016/S0014-5793(00)01217-5. [DOI] [PubMed] [Google Scholar]
- Takanaka Y, Okano T, Iigo M, Fukada Y. Light-Dependent Expression of Pinopsin Gene in Chicken Pineal Gland. Journal of Neurochemistry. 1998;70(3):908–913. doi: 10.1046/j.1471-4159.1998.70030908.x. [DOI] [PubMed] [Google Scholar]
- Rajendran RR, Van Niel EE, Stenkamp DL, Cunningham LL, Raymond PA, Gonzalez-Fernandez F. Zebrafish interphotoreceptor retinoid-binding protein: differential circadian expression among cone subtypes. J Exp Biol. 1996;199(12):2775–2787. doi: 10.1242/jeb.199.12.2775. [DOI] [PubMed] [Google Scholar]
- Springer AD, Gaffney JS. Retinal projections in the goldfish: a study using cobaltous-lysine. J Comp Neurol. 1981;203(3):401–424. doi: 10.1002/cne.902030306. [DOI] [PubMed] [Google Scholar]
- Springer AD, Mednick AS. Selective innervation of the goldfish suprachiasmatic nucleus by ventral retinal ganglion cell axons. Brain Res. 1984;323(2):293–296. doi: 10.1016/0006-8993(84)90300-7. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of Embryonic Development of the Zebrafish. Developmental Dynamics. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Malicki J, Neuhauss SC, Schier AF, Solnica-Krezel L, Stemple DL, Stainier DY, Abdelilah S, Zwartkruis F, Rangini Z, Driever W. Mutations affecting development of the zebrafish retina. Development. 1996;123:263–273. doi: 10.1242/dev.123.1.263. [DOI] [PubMed] [Google Scholar]
- Schier AF, Neuhauss SC, Harvey M, Malicki J, Solnica-Krezel L, Stainier DY, Zwartkruis F, Abdelilah S, Stemple DL, Rangini Z. et al. Mutations affecting the development of the embryonic zebrafish brain. Development. 1996;123:165–178. doi: 10.1242/dev.123.1.165. [DOI] [PubMed] [Google Scholar]
- Liang JO, Etheridge A, Hantsoo L, Rubinstein AL, Nowak SJ, Izpisua Belmonte JC, Halpern ME. Asymmetric nodal signaling in the zebrafish diencephalon positions the pineal organ. Development. 2000;127(23):5101–5112. doi: 10.1242/dev.127.23.5101. [DOI] [PubMed] [Google Scholar]
- Krauss S, Concordet JP, Ingham PW. A functionally conserved homolog of the Drosophila segment polarity gene hh is expressed in tissues with polarizing activity in zebrafish embryos. Cell. 1993;75(7):1431–1444. doi: 10.1016/0092-8674(93)90628-4. [DOI] [PubMed] [Google Scholar]
- Pierce LX, Harrison D, Liang JO. The Time Reaper 5-Channel Automatic Liquid Dispenser: a new tool for studying zebrafish development. Zebrafish. 2007;4(3):169–177. doi: 10.1089/zeb.2007.0511. [DOI] [PubMed] [Google Scholar]
- Laird DW, Molday RS. Evidence agains the role of rhodopsin in rod outer segment binding to RPE cells. Invest Ophthalmol Vis Sci. 1988;29:419–428. [PubMed] [Google Scholar]
- Hicks D, Molday RS. Differential immunogold-dextran labeling of bovine and frog rod and cone cells using monoclonal antibodies against bovine rhodopsin. Exp Eye Res. 1986;42(1):55–71. doi: 10.1016/0014-4835(86)90017-5. [DOI] [PubMed] [Google Scholar]
- Liman ER, Tytgat J, Hess P. Subunit stoichiometry of a mammalian K+ channel determined by construction of multimeric cDNAs. Neuron. 1992;9(5):861–871. doi: 10.1016/0896-6273(92)90239-A. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Exorh protein is expressed without a significant rhythm (A, B) Embryos were injected with (A) control or (B) exorh atg MO, fixed at 64 hpf, and processed for fluorescent whole mount immunostaining with the anti-bovine Rhodopsin antibody 4D2. (A) Control embryos have robust fluorescent signal in the pineal organ that is (B) severely reduced in Exorh depleted embryos. (C-C') Embryos injected with beta-galactosidase mRNA have undetectable levels of immunoreactivity with the 4D2 antibody at 8 hpf. (D-D') In contrast, embryos injected with exorh mRNA show strong antibody staining at 8 hpf. (E, F) Embryos were fixed in a circadian time course and then processed for 4D2 antibody staining. One-way Analysis of Variance (ANOVA) and Tukey's analysis revealed no significant changes in pineal Exorh protein levels that followed a daily rhythm (n≥9 embryos per time point). However, a few time points were significantly different (p ≤ 0.05) from each other including 72 and 108 hpf, 72 and 116 hpf, 76 and 116 hpf, and 80 and 116 hpf. (A-B, and E) are dorsal views, anterior to the top and (C-D') are lateral views. (C') and (D') are higher magnification images of the regions boxed in (C) and (D), respectively. Scale bars = 20 μm for (A-B, C', D', E) and 100 μm for (C, D).
Two days in a L/D cycle is not sufficient to initiate robust circadian cycling of aanat2 expression. Embryos were raised in a 14:10 h L/D cycle. At 47.5 hpf, ZT 23.5, a set of embryos was transferred to constant dark, constant temperature conditions. Embryos were fixed at the indicated stages and ZT and (A) processed for WISH for expression of aanat2 and (B) the WISH signal was quantified. Note the time of peak (closed arrowheads) expression is similar between the cyc embryos and their WT siblings. All images are dorsal views, anterior to the top. For the samples in a L/D cycle, position within the photoperiod is indicated by ZT and light conditions by the white (light period) and black (dark period) bars. For the constant dark samples, the original L/D cycle is indicated by the ZT and the black (original dark period) and dark grey (original light period) bars. Experiment was repeated two times with similar results, and representative images from one of the experiments are shown. Scale bars = 20 μm.
Comparison between larva processed for WISH with antisense and sense probes reveals low background staining. Embryos were raised in a 14:10 h L/D cycle and fixed and processed for WISH using aanat2 antisense or sense probe. Note that the sense probe produces no detectable signal, as it would recognize antisense mRNA, which should not be present. The brown regions are melanocytes in the skin, which have a natural pigment. All images are dorsal views, anterior to the top, with the pineal indicated (closed arrowheads). Position within the photoperiod is indicated by ZT and light conditions by the white (light period) and black (dark period) bars. Representative images are shown. Scale bar = 30 μm.