Abstract
Streptococcus pneumoniae asymptomatically colonizes the upper respiratory tract of children and is a frequent cause of otitis media. Patterns of microbial colonization likely influence S. pneumoniae colonization and otitis media susceptibility. This study compared microbial communities in children with and without otitis media. Nasal swabs and clinical and demographic data were collected in a cross-sectional study of Philadelphia, PA, children (6 to 78 months) (n = 108) during the 2008-2009 winter respiratory virus season. Swabs were cultured for S. pneumoniae. DNA was extracted from the swabs; 16S rRNA gene hypervariable regions (V1 and V2) were PCR amplified and sequenced by Roche/454 Life Sciences pyrosequencing. Microbial communities were described using the Shannon diversity and evenness indices. Principal component analysis (PCA) was used to group microbial community taxa into four factors representing correlated taxa. Of 108 children, 47 (44%) were colonized by S. pneumoniae, and 25 (23%) were diagnosed with otitis media. Microbial communities with S. pneumoniae were significantly less diverse and less even. Two PCA factors were associated with a decreased risk of pneumococcal colonization and otitis media, as follows: one factor included potentially protective flora (Corynebacterium and Dolosigranulum), and the other factor included Propionibacterium, Lactococcus, and Staphylococcus. The remaining two PCA factors were associated with an increased risk of otitis media. One factor included Haemophilus, and the final factor included Actinomyces, Rothia, Neisseria, and Veillonella. Generally, these taxa are not considered otitis media pathogens but may be important in the causal pathway. Increased understanding of upper respiratory tract microbial communities will contribute to the development of otitis media treatment and prevention strategies.
IMPORTANCE
Otitis media (middle ear infection) is the most common reason for pediatric sick visits in the United States. Streptococcus pneumoniae is a leading otitis media pathogen. S. pneumoniae must colonize the upper respiratory tract and compete with a complex community of nonpathogenic bacteria before infecting the middle ear. We compared microbial communities in the upper respiratory tract of children who had otitis media and those who did not. Members of the normal flora, i.e., Corynebacterium and Dolosigranulum, were protective for S. pneumoniae colonization and otitis media. As expected, the genera Haemophilus was associated with otitis media. Surprisingly, Actinomyces, Rothia, Neisseria, and Veillonella were associated with an increased risk of otitis media. These bacteria are not otitis media pathogens but may be associated with antibiotic use or involved in the causal pathway to disease. Increased understanding of upper respiratory tract microbial communities will lead to new ways to prevent middle ear infections, including probiotics.
INTRODUCTION
Otitis media (OM) is associated with over 10 million office visits in the United States each year and is the leading diagnosis for the prescription of antibiotics for children. OM arises in the complex microbial community of the upper respiratory tract. The bacteria most often associated with OM are Streptococcus pneumoniae, Haemophilus influenzae, and Moraxella catarrhalis (1). S. pneumoniae asymptomatically colonizes up to 50% of children (2–7). Colonization of the upper respiratory tract is the first step in infection; even transient colonization provides an opportunity for S. pneumoniae to invade the middle ear space.
Shifts in the composition of microbial communities of the upper respiratory tract are associated with age, vaccination, antibiotic use, and upper respiratory tract infection (7–12). Bacterial interactions can impact microbial community composition as well as disease incidence (13–15). Studies of children’s upper respiratory tract microbial flora often rely on culture to identify taxa of interest (16–19). In our bodies, bacterial cells outnumber human cells (20, 21), and a majority of the bacteria that colonize and infect humans are not routinely cultured or cannot be cultured (22). Therefore, it is likely that the members of the microbial flora that influence S. pneumoniae colonization and subsequent OM are not limited to taxa routinely identified by culture.
The use of 16S rRNA full- and partial-gene sequencing provides an alternative to culture-based methods (23–29). The 16S rRNA gene sequence contains both conserved and hypervariable regions, which can be used for taxonomic classification (30). Hypervariable regions V1 and V2 have been shown to provide a high level of resolution in species discrimination, including identification of the most common OM pathogens (31).
Currently, our knowledge of the nasal microbial flora and its association with OM is limited. In this cross-sectional study of children aged 6 months to 6 years, we describe the complex microbial communities present during upper respiratory tract infection, when children are most susceptible to developing OM (8). We also examine associations among members of microbial communities, S. pneumoniae colonization, and OM.
RESULTS
Study population.
Of 108 children, 20 (18.5%) were 6 to 12 months of age, 29 (26.9%) were 12 to 24 months, 19 (17.6%) were 24 to 36 months, and 40 (37.0%) were over 36 months. A total of 60 (55.6%) children were female, 75 (69.4%) were African-American, and 74 (68.5%) lived in households with other children. Forty-seven (43.5%) of the children were culture positive for S. pneumoniae. Nearly one-fourth (23.2%) of the children were diagnosed with OM. S. pneumoniae was isolated by culture in 15 (60%) of the OM cases. Most children (n = 95; 88.0%) had received the appropriate number of heptavalent pneumococcal conjugate vaccinations for their age. Logistic regression was used to assess the association between OM and demographic risk factors, including age, race, and gender. No significant associations were found, perhaps because all study subjects presented with upper respiratory tract infection, a major OM risk factor (8).
Sequence diversity.
A mean of 1,087.5 sequences per sample (standard deviation [SD], 496.01) was included in the analyses (114,397 total sequences). The average sequence length was 234 bp. The mean Shannon diversity and evenness indices for all samples were 3.0 (SD, 0.83) and 0.62 (SD, 0.12), respectively. Both indices were significantly lower in S. pneumoniae culture-positive samples than in culture-negative samples (2.66 versus 3.25, P = 0.0002, and 0.58 versus 0.66, P = 0.002, respectively). Of 108 children, 7 (6.5%) had used antibiotics within the past 7 days, 15 (13.9%) within the past 14 days, 17 (15.7%) within the past 21 days, and 32 (29.6%) within the past 6 months. Separate analyses of variance (ANOVAs) examined the relationship between the Shannon diversity and evenness indices, pneumococcal colonization, and antibiotic use and the interaction between them. Pneumococcal colonization had a significant effect on diversity (F test, P = 0.0002 for Shannon diversity, P = 0.002 for evenness), but antibiotic use (within the past 7 days, 14 days, 21 days, or 6 months) did not. There was no significant interaction between pneumococcal colonization and antibiotic use. This suggests that antibiotic use within the past 6 months does not confound the relationship between diversity indices and pneumococcal colonization. The Shannon diversity and evenness indices were not significantly different by OM diagnosis (t test, P = 0.6 and 0.64, respectively).
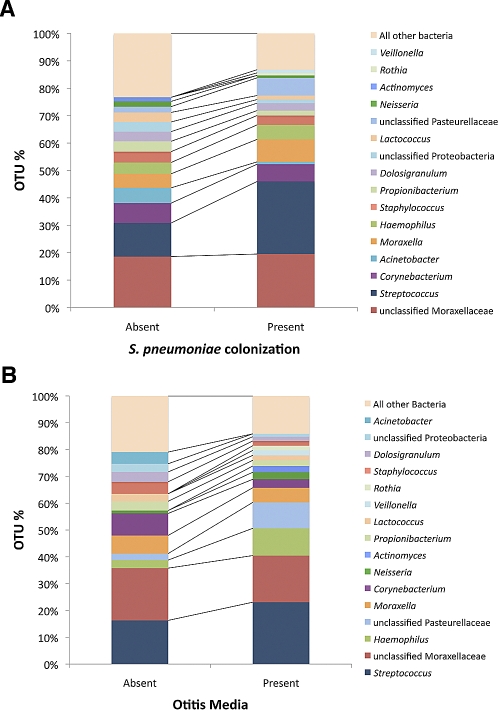
Using the Ribosomal Database Project (RDP) Classifier tool, pyrosequencing provided sufficient information to classify 68.5% of the sequences at the genus and subgenus levels with 90% confidence, 25.7% at the family and subfamily levels, 1.8% at the order and suborder levels, 0.6% at the class and subclass levels, and 3.4% at the phylum level. We identified 389 operational taxonomic units (OTUs). OTU proportions of ≥1% in the overall population (n = 16) are listed in Table 1. OTU proportions were compared by S. pneumoniae colonization (Fig. 1A) and by OM diagnosis (Fig. 1B). The most frequent OTU proportions included potential OM pathogens Streptococcus, Moraxella, and Haemophilus. In the S. pneumoniae-colonized group, we observed fewer “rare” bacteria than in the S. pneumoniae-negative group, as evidenced by the difference in the “all other bacteria” bar, suggesting lower levels of overall diversity in the S. pneumoniae-colonized group (Fig. 1A). The OTU proportion of potential OM pathogens Streptococcus (23.1%) and Haemophilus (10.2%) was greater in the OM group than in the OM-absent group (16.3% and 3.0%, respectively).
TABLE 1 .
Most frequent nasal swab OTUsa
| OTU | Frequency (%)b |
|---|---|
| Unclassified Moraxellaceae | 19.00 |
| Streptococcus | 17.86 |
| Corynebacterium | 7.04 |
| Moraxella | 6.46 |
| Haemophilus | 4.66 |
| Unclassified Pasteurellaceae | 4.09 |
| Staphylococcus | 3.84 |
| Acinetobacter | 3.44 |
| Dolosigranulum | 3.21 |
| Propionibacterium | 3.13 |
| Unclassified Proteobacteria | 2.59 |
| Lactococcus | 2.58 |
| Neisseria | 1.45 |
| Actinomyces | 1.24 |
| Rothia | 1.13 |
| Veillonella | 1.05 |
Frequency of ≥1%.
Percentage of total sequences per nasal microbial community, i.e., per child.
FIG 1 .
Comparison of the most frequent operational taxonomic unit (OTU) distributions grouped by S. pneumoniae colonization (determined by culture) (A) and by otitis media (OM) diagnosis (B). Percentages of total sequences per nasal microbial community, i.e., per child, that each OTU represented are shown.
PCA.
OTUs representing ≥1% of the microbial flora were used for principal component analysis (PCA), which grouped microbial community members into independent factors. The four resulting PCA factors (factors A to D) represent linear relationships among correlated taxa. Factor A includes Corynebacterium, Dolosigranulum, and Streptococcus (negatively correlated); factor B includes Lactococcus, Propionibacterium, unclassified Proteobacteria, and Staphylococcus; factor C includes unclassified Pasteurellaceae, Moraxella, and Haemophilus; and factor D includes Veillonella, Neisseria, Rothia, and Actinomyces.
Logistic regression analysis was used to examine associations between PCA factors and S. pneumoniae colonization or OM diagnosis. Two of the four factors were significantly associated with a decreased risk of S. pneumoniae colonization (odds ratio [OR] [95% confidence interval {CI}]), as follows: factor A (0.55 [0.35, 0.86]) and factor B (0.34 [0.20, 0.59]) (Table 2). These same two factors were also associated with a decreased risk of OM (OR [95% CI]), as follows: factor A (0.54 [0.30, 0.96]) and factor B (0.44 [0.21, 0.91]) (Table 3). Factors A and B include bacteria that potentially are interfering members of the normal flora (Tables 2 and 3). Furthermore, the presence of Corynebacterium, Dolosigranulum, and Lactococcus was negatively correlated with the genus Streptococcus, with r values of −0.25 (P = 0.01), −0.26 (P = 0.006), and −0.19 (P = 0.05), respectively. Two other factors were significantly associated with an augmented risk for OM (OR [95% CI]), as follows: factor C (1.73 [1.10, 2.73]) and factor D (2.24 [1.26, 3.97]) (Table 3). Factor C includes Haemophilus and unclassified Pasteurellaceae, and factor D includes Veillonella, Rothia, Actinomyces, and Neisseria (Tables 2 and 3).
TABLE 2 .
Associations between S. pneumoniae colonization determined by culture and factors obtained by PCA as well as individual taxa within factors
| Factor | OR (95% CI)b |
|
|---|---|---|
| PCA factor | Component | |
| Factor A | 0.55 (0.35, 0.86) | |
| Corynebacterium | 0.99 (0.95, 1.03) | |
| Dolosigranulum | 0.99 (0.91, 1.06) | |
| Streptococcusa | 1.06 (1.03, 1.09) | |
| Factor B | 0.34 (0.20, 0.56) | |
| Lactococcus | 0.73 (0.61, 0.89) | |
| Propionibacterium | 0.89 (0.79, 1.00) | |
| Unclassified Proteobacteria | 0.94 (0.88, 1.02) | |
| Staphylococcus | 0.98 (0.93, 1.02) | |
| Factor C | 0.94 (0.64, 1.39) | |
| Unclassified Pasteurellaceae | 1.08 (1.00, 1.16) | |
| Moraxellaa | 1.03 (0.99, 1.07) | |
| Haemophilus | 1.02 (0.98, 1.05) | |
| Factor D | 0.61 (0.37, 1.02) | |
| Veillonella | 0.98 (0.83, 1.15) | |
| Neisseria | 0.85 (0.70, 1.04) | |
| Rothia | 0.82 (0.62, 1.08) | |
| Actinomyces | 0.77 (0.58, 1.02) | |
The OTU was negatively correlated with the factor; factor loadings were otherwise positive.
OR and 95% CI values were obtained by separate logistic regression analyses of each factor and each taxa within each factor.
TABLE 3 .
Associations between OM diagnosis and factors obtained by PCA as well as individual taxa within factors
| Factor | OR (95% CI)b |
|
|---|---|---|
| PCA factor | Component | |
| Factor A | 0.54 (0.30, 0.96) | |
| Corynebacterium | 0.90 (0.81, 0.99) | |
| Dolosigranulum | 0.91 (0.80, 1.04) | |
| Streptococcusa | 1.02 (1.00, 1.05) | |
| Factor B | 0.44 (0.21, 0.91) | |
| Staphylococcus | 0.86 (0.71, 1.06) | |
| Lactococcus | 0.92 (0.78, 1.09) | |
| Unclassified Proteobacteria | 0.93 (0.83, 1.05) | |
| Propionibacterium | 0.97 (0.87, 1.07) | |
| Factor C | 1.73 (1.10, 2.73) | |
| Unclassified Pasteurellaceae | 1.08 (1.01, 1.15) | |
| Haemophilus | 1.04 (1.00, 1.08) | |
| Moraxellaa | 0.99 (0.96, 1.06) | |
| Factor D | 2.24 (1.26, 3.97) | |
| Veillonella | 1.37 (1.07, 1.76) | |
| Rothia | 1.35 (1.03, 1.77) | |
| Actinomyces | 1.34 (1.07, 1.68) | |
| Neisseria | 1.13 (0.99, 1.29) | |
The OTU was negatively correlated with the factor; factor loadings were otherwise positive.
OR and 95% CI values were obtained by separate logistic regression analyses of each factor and each taxa within each factor.
As factor D taxa are not generally recognized as OM pathogens, we explored additional associations and found that the OTU proportion of each factor D taxon (Veillonella, Rothia, Actinomyces, and Neisseria) in a sample was significantly higher in swabs obtained from children with OM (t test, P = 0.001, 0.02, 0.002, and 0.03, respectively). In addition, samples obtained from children had significantly higher factor D scores if antibiotics had been prescribed within the past 21 days or the past 6 months (t test, P = 0.02 or 0.05, respectively).
DISCUSSION
We describe nasal microbial communities in children with upper respiratory tract infection with and without concurrent OM. We observed lower microbial diversity when S. pneumoniae is present. Using principal component analysis to group microbial community members into factors revealed taxa associated with a decreased risk for S. pneumoniae colonization and OM. These taxa may be useful for the development of multispecies probiotics for OM. We also identified taxa associated with an increased risk for OM. Further study is warranted to define their role in OM pathogenesis.
Vaccine-driven immunological pressures and increasing antibiotic resistance may result in shifts in the upper respiratory tract microbial community that affect the distribution and pathogenic potential of OM-associated bacteria. We used complete-linkage clustering in our diversity calculations through the RDP pyrosequencing pipeline. Average-linkage clustering may yield more accurate estimates of diversity with regard to identifying rare microbiota in short amplicon (60-bp) data sets (32, 33). However, Quince et al. found that diversity inflation was mitigated in data sets of longer sequences (250 bp), and our sequences are of similar lengths (average, 234 bp) (33).
In culture-positive S. pneumoniae samples, diversity and evenness indices were lower than those in culture-negative samples. Children with OM have decreased levels of healthy normal flora bacteria like viridans group streptococci and diphtheroids, while the OTU proportion of OM pathogens increases by 2- to 3-fold (34). The association between diversity indices and OM was not significant here. However, all children in the study were seen for a sick visit, which in this population may have attenuated the differences in microbial diversity by OM status.
The two factors associated with a reduced risk of OM were dominated by the genera Corynebacterium, Dolosigranulum, Propionibacterium, Lactococcus, and Staphylococcus. Together, these two factors (factors A and B) (Tables 2 and 3) identify potential protective members of the normal flora that may interfere with OM pathogens. Streptococcus loaded negatively on factor A and is negatively correlated with both Corynebacterium and Dolosigranulum, suggesting a competitive interaction between this potential OM pathogen and these two genera. Lemon et al. also observed negative correlations between members of the normal flora such as the Corynebacteriaceae and Propionibacteriaceae families and the Staphylococcaceae family, which includes potential OM pathogens, in nasal samples obtained from healthy adults by PhyloChip analysis (35).
Samples obtained from children with high factor C loading are characterized by OM diagnosis as well as by the presence of Haemophilus and the absence of Moraxella. This is consistent with previous literature linking Haemophilus with OM (1). In addition, H. influenzae has been negatively correlated with M. catarrhalis during upper respiratory tract infection (36). It is important to note that as sequence data could not classify at the species level, factor C likely contains other species of Moraxella in addition to the OM pathogen M. catarrhalis.
Several mechanisms may account for the association between factor D and an increased risk of OM. Although the bacteria associated with factor D are not normally recognized as OM pathogens, they may be important to the causal pathway. Higher levels of colonization by anaerobes such as Actinomyces and Veillonella have been identified during acute OM in children up to 2 years of age (37). Neisseria has also been isolated from children with chronic OM (38). In this study, the OTU proportion of Neisseria was significantly higher in swabs obtained from children with OM. Previous studies show that bacterial species can impact the colonization and pathogenicity of other species (39, 40). Therefore, individual species or even specific strains, which cannot cause disease themselves, can interact in a synergistic fashion when in the presence of other coinfecting species. Sibley et al. also found a synergistic effect on pathogenicity between chronic pulmonary infection pathogen Pseudomonas aeruginosa and both Actinomyces and Rothia spp. in the Drosophila model of polymicrobial infections (41). Factor D taxa (Veillonella, Neisseria, Rothia, and Actinomyces) may or may not be pathogenic themselves but may be able to increase the pathogenicity of OM pathogens. Further study is needed to better elucidate the associations among these taxa, known OM pathogens, and the development of OM.
Antibiotic resistance may also play a role in the observed association between factor D taxa and OM. Samples in this study obtained from children who were prescribed antibiotics had significantly higher factor D scores. Both Neisseria and Veillonella have been shown to have widespread antibiotic resistance (42–44). Fluoroquinolone resistance has emerged recently in Neisseria (44), and Veillonella spp. are among the most prevalent tetracycline-resistant bacteria in the oral cavity (42). The survival of Streptococcus mutans increases during antibiotic treatment when cocultured with Veillonella parvula in a biofilm (45). As a result of repeated OM-related courses of antibiotics, children with a history of recurrent OM certainly have the potential to develop a more resistant microbial flora.
One limitation of our study is that data on viral pathogens, although important in OM, were not collected. In addition, as this was a cross-sectional study, we could not characterize within-subject variability. All children presented with upper respiratory tract infection symptoms. Additional studies comparing healthy and sick children are necessary. The Roche/454 Life Sciences pyrosequencing platform is one of several methods available for high-throughput sequencing and circumvents the need for creating clone libraries. However, a limitation of the short-read sequencing technology is that it does not provide sufficient sequence data for the classification of species. Thus, the majority of sequences could be classified only at the genus level. In addition, due to potential PCR and sequencing biases, the relationship between the number of sequences per OTU and the bacteria in the population may not be linear.
The present study is based on nasal sampling, which has the advantage of being easier and is more comfortable for children than nasopharyngeal sampling (46). Other studies have used data obtained from nasopharyngeal sampling to characterize pneumococcal colonization patterns in children (6, 47). However, Rapola et al. have shown that the rates of OM pathogens S. pneumoniae and H. influenzae isolation do not differ by site (48). Although regions of the upper respiratory tract harbor distinct microbial communities, they are physically connected, and thus, a high level of overlap is observed between sites (35).
Our culture-independent classification of bacteria present in children’s nasal mucosa during upper respiratory tract infection contributes to the understanding of nasal microbial communities, development of OM, and polymicrobial interactions. Increased understanding of complex microbial communities will advance the development of efficacious prevention and treatment protocols for OM, including probiotic therapies that target high-risk individuals, which in turn can reduce the risk of permanent damage caused by OM.
MATERIALS AND METHODS
Study design and participants.
Nasal swabs were collected as part of a cross-sectional study of Philadelphia, PA, children presenting with upper respiratory tract infection symptoms at one of six primary care clinics within the Pediatric Research Consortium of the Children’s Hospital of Philadelphia during the 2008-2009 winter respiratory virus season. Samples obtained for this study (n = 108) were randomly selected from swabs collected between 9 December 2008 and 2 January 2009 from participants aged 6 months to 6 years (n = 237). Demographic data collected from each participant through a case report form at the clinic visit included age, gender, race, ethnicity, and number of children in household. Clinical data, including diagnosis at visit, comorbidities, vaccination history, and recent antibiotic use, were obtained from electronic medical record extraction, including International Classification of Diseases, 9th Edition (ICD-9) codes. Trained research assistants obtained informed consent for participation in the study during the clinic visit. The Institutional Review Board (IRB) of the University of Pennsylvania approved the study protocol.
Nasal swab collection and processing.
Anterior nasal swabs (49) were collected from each child by inserting a rayon-tipped swab moistened with saline approximately 1 cm into the anterior nares. Swabs were placed in STGG (skim milk, Oxoid tryptone soya broth, glucose, and glycerol) transport media until processed (19). Pneumococcal colonization was determined by standard microbiological methods. DNA was extracted using the QIAamp DNA minikit (Qiagen, Valencia, CA) protocol for extraction of Gram-positive bacteria from nasal swabs, with one modification: DNA was eluted into 50 µl.
Roche/454 Life Sciences pyrosequencing of a 16S rRNA gene fragment targeting hypervariable regions V1 and V2 was used to describe the taxa of bacteria colonizing the nasal mucosa. 16S rRNA gene amplification was performed as described (23). The forward primer sequence included the Roche/454 Life Sciences adaptor primer B, TC-linker, and the 27F conserved bacterial 16S rRNA forward primer: 5′ GCCTTGCCAGCCCGCTCAGTCAGAGTTTGATCCTGGCTCAG 3′ (23). The reverse primers carried the Roche/454 Life Sciences adaptor primer A, a unique eight-base barcode (denoted here as 8 N), CA-linker, and the 338R conserved bacterial primer: 5′ GCCTCCCTCGCGCCATCAGNNNNNNNNCACTGCTGCCTCCCGTAGGAGT 3′ (23). PCR reactions were done in triplicate, and products were cleaned by QIAquick PCR purification (Qiagen). Samples were pooled in equimolar amounts and submitted for pyrosequencing.
Pyrosequencing results analysis.
Amplicons were sequenced at the Yale University Center for Genomics and Proteomics using the SLR70 Roche/454 Life Sciences pyrosequencing platform. The sequences were first searched for the linker, primers, and their reverse complements using in-house programs (http://graphics.med.yale.edu/trim/). Two errors were allowed, and the identified primer sequences were trimmed from each sequence read. Sequence reads that did not contain the 5′-end primer were removed from the data set. The same program was also used for barcode identification. Barcodes were identified within the first 15 bases of the reads, with one error allowed. Sequence reads were binned into separate FASTA files based on the individual barcodes, and the barcode sequences were trimmed. Current methods for chimera detection are designed for longer sequences (~500 bp or longer), such as ChimeraSlayer and Bellerophon (50, 51; http://microbiomeutil.sourceforge.net/). In an effort to minimize the number of chimeras, we set strict alignment criteria; all sequences had to align to at least 100 bp of the 16S rRNA gene, and any sequence aligning outside the 27 and/or 338 positions of the 16S rRNA gene was discarded.
Sequences were analyzed using tools available from the Ribosomal Database Project (RDP). Individual sequences were aligned using the Aligner tool. Aligned sequence files for each child were processed by complete-linkage clustering using distance criteria. We used a maximum cluster distance cutoff of 3% (97% identity). These data were used to calculate the Shannon diversity and evenness indices (25, 26, 52, 53). Shannon’s index measures the degree of nonredundancy within a microbial community, and the evenness index measures the distribution of species or groups within a sample.
Sequences were classified at 90% by the RDP Classifier tool (52). The RDP website recommends using a 50% bootstrap value for short pyrosequences to increase the number of sequences classified (54). Claesson et al. (54) and Wang et al. (52) note that the proportion of sequences classified correctly increases as the bootstrap cutoff is increased. Therefore, we selected a 90% cutoff in order to classify the sequences with greater accuracy. The sequence data could not be classified at the species level; OTUs (operational taxonomic units) were defined by grouping together all sequences belonging to the same genus. Sequences that were unclassified at the genus level were classified and grouped at the next lowest taxonomic level. In total, there were 389 OTUs. We normalized the data by calculating the OTU proportion—the proportion of total sequences per nasal microbial community, i.e., per child, that each OTU represented. These data were used for principal component analysis (PCA) and all subsequent analyses.
Data analysis.
Associations among community diversity indices, the presence of S. pneumoniae by culture, OM diagnosis, and antibiotic use within the past 7 days, 14 days, 21 days, or 6 months were explored using Student’s t test, Fisher’s exact chi-square test, and analysis of variance (ANOVA). PCA was used to group microbial community members into four factors representing linear relationships among selected component taxa (SAS 9.1.3; SAS Institute, Cary, NC). OTUs that were more frequent than 1% of the microbial community were included as component taxa (n = 16) in the PCA. An eigenvalue of 1 and an orthogonal rotation were specified for PCA. PCA factor components had significant loadings of at least ±0.4. Associations among PCA factors, PCA factor scores, and the presence of S. pneumoniae by culture and OM diagnosis were explored using Student’s t test, correlation, and logistic regression.
ACKNOWLEDGMENTS
We thank Linda Crossette for managing the field component of this study.
The National Institute of Allergy and Infectious Diseases provided funding for this research (grant R01 A1068043 to M.M.P. and grants R21 AI078189 and K24 073957 to J.P.M.).
Footnotes
Citation Laufer, A. S., J. P. Metlay, J. F. Gent, K. P. Fennie, Y. Kong, et al. 2011. Microbial communities of the upper respiratory tract and otitis media in children. mBio 2(1):e00245-10. doi:10.1128/mBio.00245-10.
REFERENCES
- 1. Revai K., Mamidi D., Chonmaitree T. 2008. Association of nasopharyngeal bacterial colonization during upper respiratory tract infection and the development of acute otitis media. Clin. Infect. Dis. 46:e34–e37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Watson K., Carville K., Bowman J., Jacoby P., Riley T. V., Leach A. J., Lehmann D., and the Kalgoorlie Otitis Media Research Project Team. 2006. Upper respiratory tract bacterial carriage in Aboriginal and non-Aboriginal children in a semi-arid area of Western Australia. Pediatr. Infect. Dis. J. 25:782–790 [DOI] [PubMed] [Google Scholar]
- 3. Dagan R., Givon-Lavi N., Zamir O., Fraser D. 2003. Effect of a nonavalent conjugate vaccine on carriage of antibiotic-resistant Streptococcus pneumoniae in day-care centers. Pediatr. Infect. Dis. J. 22:532–540 [DOI] [PubMed] [Google Scholar]
- 4. Syrjanen R. K., Kilpi T. M., Kaijalainen T. H., Herva E. E., Takala A. K. 2001. Nasopharyngeal carriage of Streptococcus pneumoniae in Finnish children younger than 2 years old. J. Infect. Dis. 184:451–459 [DOI] [PubMed] [Google Scholar]
- 5. Bogaert D., De Groot R., Hermans P. W. 2004. Streptococcus pneumoniae colonisation: the key to pneumococcal disease. Lancet Infect. Dis. 4:144–154 [DOI] [PubMed] [Google Scholar]
- 6. Gray B. M., Converse G. M., III, Dillon H. C., Jr. 1980. Epidemiologic studies of Streptococcus pneumoniae in infants: acquisition, carriage, and infection during the first 24 months of life. J. Infect. Dis. 142:923–933 [DOI] [PubMed] [Google Scholar]
- 7. Faden H., Duffy L., Wasielewski R., Wolf J., Krystofik D., Tung Y. 1997. Relationship between nasopharyngeal colonization and the development of otitis media in children. Tonawanda/Williamsville Pediatrics. J. Infect. Dis. 175:1440–1445 [DOI] [PubMed] [Google Scholar]
- 8. Chonmaitree T., Revai K., Grady J. J., Clos A., Patel J. A., Nair S., Fan J., Henrickson K. J. 2008. Viral upper respiratory tract infection and otitis media complication in young children. Clin. Infect. Dis. 46:815–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chung M. H., Griffith S. R., Park K. H., Lim D. J., DeMaria T. F. 1993. Cytological and histological changes in the middle ear after inoculation of influenza A virus. Acta Otolaryngol. 113:81–87 [DOI] [PubMed] [Google Scholar]
- 10. Regev-Yochay G., Dagan R., Raz M., Carmeli Y., Shainberg B., Derazne E., Rahav G., Rubinstein E. 2004. Association between carriage of Streptococcus pneumoniae and Staphylococcus aureus in children. JAMA 292:716–720 [DOI] [PubMed] [Google Scholar]
- 11. Madhi S. A., Adrian P., Kuwanda L., Cutland C., Albrich W. C., Klugman K. P. 2007. Long-term effect of pneumococcal conjugate vaccine on nasopharyngeal colonization by Streptococcus pneumoniae—and associated interactions with Staphylococcus aureus and Haemophilus influenzae colonization—in HIV-infected and HIV-uninfected children. J. Infect. Dis. 196:1662–1666 [DOI] [PubMed] [Google Scholar]
- 12. Klepac-Ceraj V., Lemon K. P., Martin T. R., Allgaier M., Kembel S. W., Knapp A. A., Lory S., Brodie E. L., Lynch S. V., Bohannan B. J., Green J. L., Maurer B. A., Kolter R. 2010. Relationship between cystic fibrosis respiratory tract bacterial communities and age, genotype, antibiotics and Pseudomonas aeruginosa. Environ. Microbiol. 12:1293–1303 [DOI] [PubMed] [Google Scholar]
- 13. Armbruster C. E., Hong W., Pang B., Weimer K. E., Juneau R. A., Turner J., Swords W. E. 2010. Indirect pathogenicity of Haemophilus influenzae and Moraxella catarrhalis in polymicrobial otitis media occurs via interspecies quorum signaling. mBio 1:e00102-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lysenko E. S., Ratner A. J., Nelson A. L., Weiser J. N. 2005. The role of innate immune responses in the outcome of interspecies competition for colonization of mucosal surfaces. PLoS Pathog. 1:e1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kuramitsu H. K., He X., Lux R., Anderson M. H., Shi W. 2007. Interspecies interactions within oral microbial communities. Microbiol. Mol. Biol. Rev. 71:653–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Brogden K. A., Guthmiller J. M. 2002. Polymicrobial diseases. ASM Press, Washington, DC: [PubMed] [Google Scholar]
- 17. Jacoby P., Watson K., Bowman J., Taylor A., Riley T. V., Smith D. W., Lehmann D., and the Kalgoorlie Otitis Media Research Project Team. 2007. Modelling the co-occurrence of Streptococcus pneumoniae with other bacterial and viral pathogens in the upper respiratory tract. Vaccine 25:2458–2464.
- 18. Harrison L. M., Morris J. A., Telford D. R., Brown S. M., Jones K. 1999. The nasopharyngeal bacterial flora in infancy: effects of age, gender, season, viral upper respiratory tract infection and sleeping position. FEMS Immunol. Med. Microbiol. 25:19–28 [DOI] [PubMed] [Google Scholar]
- 19. Pitkaranta A., Roivainen M., Blomgren K., Peltola J., Kaijalainen T., Raty R., Ziegler T., Ronkko E., Hatakka K., Korpela R., Poussa T., Leinonen M., Hovi T. 2006. Presence of viral and bacterial pathogens in the nasopharynx of otitis-prone children. A prospective study. Int. J. Pediatr. Otorhinolaryngol. 70:647–654 [DOI] [PubMed] [Google Scholar]
- 20. Savage D. C. 1977. Microbial ecology of the gastrointestinal tract. Annu. Rev. Microbiol. 31:107–133 [DOI] [PubMed] [Google Scholar]
- 21. Costello E. K., Lauber C. L., Hamady M., Fierer N., Gordon J. I., Knight R. 2009. Bacterial community variation in human body habitats across space and time. Science 326:1694–1697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Langendijk P. S., Schut F., Jansen G. J., Raangs G. C., Kamphuis G. R., Wilkinson M. H., Welling G. W. 1995. Quantitative fluorescence in situ hybridization of Bifidobacterium spp. with genus-specific 16S rRNA-targeted probes and its application in fecal samples. Appl. Environ. Microbiol. 61:3069–3075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hamady M., Walker J. J., Harris J. K., Gold N. J., Knight R. 2008. Error-correcting barcoded primers for pyrosequencing hundreds of samples in multiplex. Nat. Methods 5:235–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Andersson A. F., Lindberg M., Jakobsson H., Backhed F., Nyren P., Engstrand L. 2008. Comparative analysis of human gut microbiota by barcoded pyrosequencing. PLoS One 3:e2836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sogin M. L., Morrison H. G., Huber J. A., Welch D. M., Huse S. M., Neal P. R., Arrieta J. M., Herndl G. J. 2006. Microbial diversity in the deep sea and the underexplored “rare biosphere.” Proc. Natl. Acad. Sci. U. S. A. 103:12115–12120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dethlefsen L., Huse S., Sogin M. L., Relman D. A. 2008. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. 6:e280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Parameswaran P., Jalili R., Tao L., Shokralla S., Gharizadeh B., Ronaghi M., Fire A. Z. 2007. A pyrosequencing-tailored nucleotide barcode design unveils opportunities for large-scale sample multiplexing. Nucleic Acids Res. 35:e130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Worthen P. L., Gode C. J., Graf J. 2006. Culture-independent characterization of the digestive-tract microbiota of the medicinal leech reveals a tripartite symbiosis. Appl. Environ. Microbiol. 72:4775–4781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ley R. E., Turnbaugh P. J., Klein S., Gordon J. I. 2006. Microbial ecology: human gut microbes associated with obesity. Nature 444:1022–1023 [DOI] [PubMed] [Google Scholar]
- 30. Van de Peer Y., Chapelle S., De Wachter R. 1996. A quantitative map of nucleotide substitution rates in bacterial rRNA. Nucleic Acids Res. 24:3381–3391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chakravorty S., Helb D., Burday M., Connell N., Alland D. 2007. A detailed analysis of 16S ribosomal RNA gene segments for the diagnosis of pathogenic bacteria. J. Microbiol. Methods 69:330–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Huse S. M., Welch D. M., Morrison H. G., Sogin M. L. 2010. Ironing out the wrinkles in the rare biosphere through improved OTU clustering. Environ. Microbiol. 12:1889–1898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Quince C., Lanzen A., Curtis T. P., Davenport R. J., Hall N., Head I. M., Read L. F., Sloan W. T. 2009. Accurate determination of microbial diversity from 454 pyrosequencing data. Nat. Methods 6:639–641 [DOI] [PubMed] [Google Scholar]
- 34. Faden H., Stanievich J., Brodsky L., Bernstein J., Ogra P. L. 1990. Changes in nasopharyngeal flora during otitis media of childhood. Pediatr. Infect. Dis. J. 9:623–626 [PubMed] [Google Scholar]
- 35. Lemon K. P., Klepac-Ceraj V., Schiffer H. K., Brodie E. L., Lynch S. V., Kolter R. 2010. Comparative analyses of the bacterial microbiota of the human nostril and oropharynx. mBio 1(3):e00129-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pettigrew M. M., Gent J. F., Revai K., Patel J. A., Chonmaitree T. 2008. Microbial interactions during upper respiratory tract infections. Emerg. Infect. Dis. 14:1584–1591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kononen E., Syrjanen R., Takala A., Jousimies-Somer H. 2003. Nasopharyngeal carriage of anaerobes during health and acute otitis media by two years of age. Diagn. Microbiol. Infect. Dis. 46:167–172 [DOI] [PubMed] [Google Scholar]
- 38. Jokipii A. M., Karma P., Ojala K., Jokipii L. 1977. Anaerobic bacteria in chronic otitis media. Arch. Otolaryngol. 103:278–280 [DOI] [PubMed] [Google Scholar]
- 39. Dowd S. E., Wolcott R. D., Sun Y., McKeehan T., Smith E., Rhoads D. 2008. Polymicrobial nature of chronic diabetic foot ulcer biofilm infections determined using bacterial tag encoded FLX amplicon pyrosequencing (bTEFAP). PLoS One 3:e3326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Peleg A. Y., Tampakakis E., Fuchs B. B., Eliopoulos G. M., Moellering R. C., Jr., Mylonakis E. 2008. Prokaryote-eukaryote interactions identified by using Caenorhabditis elegans. Proc. Natl. Acad. Sci. U. S. A. 105:14585–14590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sibley C. D., Duan K., Fischer C., Parkins M. D., Storey D. G., Rabin H. R., Surette M. G. 2008. Discerning the complexity of community interactions using a Drosophila model of polymicrobial infections. PLoS Pathog. 4:e1000184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lancaster H., Ready D., Mullany P., Spratt D., Bedi R., Wilson M. 2003. Prevalence and identification of tetracycline-resistant oral bacteria in children not receiving antibiotic therapy. FEMS Microbiol. Lett. 228:99–104 [DOI] [PubMed] [Google Scholar]
- 43. Seville L. A., Patterson A. J., Scott K. P., Mullany P., Quail M. A., Parkhill J., Ready D., Wilson M., Spratt D., Roberts A. P. 2009. Distribution of tetracycline and erythromycin resistance genes among human oral and fecal metagenomic DNA. Microb. Drug Resist. 15:159–166 [DOI] [PubMed] [Google Scholar]
- 44. Wu H. M., Harcourt B. H., Hatcher C. P., Wei S. C., Novak R. T., Wang X., Juni B. A., Glennen A., Boxrud D. J., Rainbow J., Schmink S., Mair R. D., Theodore M. J., Sander M. A., Miller T. K., Kruger K., Cohn A. C., Clark T. A., Messonnier N. E., Mayer L. W., Lynfield R. 2009. Emergence of ciprofloxacin-resistant Neisseria meningitidis in North America. N. Engl. J. Med. 360:886–892 [DOI] [PubMed] [Google Scholar]
- 45. Luppens S. B., Kara D., Bandounas L., Jonker M. J., Wittink F. R., Bruning O., Breit T. M., Ten Cate J. M., Crielaard W. 2008. Effect of Veillonella parvula on the antimicrobial resistance and gene expression of Streptococcus mutans grown in a dual-species biofilm. Oral Microbiol. Immunol. 23:183–189 [DOI] [PubMed] [Google Scholar]
- 46. Heikkinen T., Marttila J., Salmi A. A., Ruuskanen O. 2002. Nasal swab versus nasopharyngeal aspirate for isolation of respiratory viruses. J. Clin. Microbiol. 40:4337–4339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Greiner O., Day P., Bosshard P., Imeri F., Altwegg M., Nadal D. 2001. Quantitative detection of Streptococcus pneumoniae in nasopharyngeal secretions by real-time PCR. J. Clin. Microbiol. 39:3129–3134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Rapola S., Salo E., Kiiski P., Leinonen M., Takala A. K. 1997. Comparison of four different sampling methods for detecting pharyngeal carriage of Streptococcus pneumoniae and Haemophilus influenzae in children. J. Clin. Microbiol. 35:1077–1079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Capeding M. R., Nohynek H., Sombrero L. T., Pascual L. G., Sunico E. S., Esparar G. A., Esko E., Leinonen M., Ruutu P. 1995. Evaluation of sampling sites for detection of upper respiratory tract carriage of Streptococcus pneumoniae and Haemophilus influenzae among healthy Filipino infants. J. Clin. Microbiol. 33:3077–3079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Huber J. A., Morrison H. G., Huse S. M., Neal P. R., Sogin M. L., Mark Welch D. B. 2009. Effect of PCR amplicon size on assessments of clone library microbial diversity and community structure. Environ. Microbiol. 11:1292–1302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. DeSantis T. Z., Hugenholtz P., Larsen N., Rojas M., Brodie E. L., Keller K., Huber T., Dalevi D., Hu P., Andersen G. L. 2006. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72:5069–5072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wang Q., Garrity G. M., Tiedje J. M., Cole J. R. 2007. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73:5261–5267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Cole J. R., Wang Q., Cardenas E., Fish J., Chai B., Farris R. J., Kulam-Syed-Mohideen A. S., McGarrell D. M., Marsh T., Garrity G. M., Tiedje J. M. 2009. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 37:D141–D145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Claesson M. J., O’Sullivan O., Wang Q., Nikkila J., Marchesi J. R., Smidt H., de Vos W. M., Ross R. P., O’Toole P. W. 2009. Comparative analysis of pyrosequencing and a phylogenetic microarray for exploring microbial community structures in the human distal intestine. PLoS One 4:e6669 [DOI] [PMC free article] [PubMed] [Google Scholar]