Abstract
Micro RNAs are evolutionarily conserved, single stranded molecules of about 22 nucleotides in length and function post-transcriptionally by partial binding (partial complementarity) to the mRNA of genes. Binding of a specific miRNA to its target on an mRNA can inhibit its expression by a variety of mechanisms. Although the most common mechanism is translational repression as a result of miRNA binding to the 3'UTR of an mRNA, mechanisms involving mRNA degradation and destabilization have also been described. Micro RNAs are currently considered as "master regulators" of gene expression. Since a single miRNA can bind and consequently regulate the expression of more than 100 different transcripts it has been estimated that miRNAs may be able to regulate up to 30% of the protein-coding genes in the human genome. As a result, miRNAs receive widespread attention on their potential role in complicated biological processes and multifactorial diseases. In this review we are discussing the biogenesis of miRNAs, their mode of action as well as their role in human diseases through genetic variations on their target sites.
Keywords: micro-RNA, mirSNPs, gene regulation, human diseases, review
Several different classes of small RNAs appear to have significant regulatory roles in various cellular processes. These classes include small interference RNAs (siRNAs), micro RNAs (miRNAs), repeat-associated siRNAs (rasiRNAs) and piwi (P-element induced wimpy testis) interacting RNAs (piRNAs). Small RNAs act like repressors of gene expression in plants, animals and many fungi. They function by guiding sequence-specific gene silencing at the transcriptional and/or post-transcriptional level, through a mechanism known as RNA interference (RNAi) (Figure 1). During this process naturally occurring or external small RNA duplexes are processed into specific short single-stranded RNA endproducts that can drive gene silencing by specific and distinct mechanisms. In the case of natural small RNA duplexes, they are either encoded in the genome or are generated by replication of viral intruders1, 2.
Figure 1: Overview of the RNA interference pathways.The RNAi pathway can be divided into the siRNA and miRNA pathways. The RNAi pathway involves two phases. During the initiation phase the double stranded RNA molecules (ds RNA molecules or pre-miRNAs) are cleaved by the enzyme Dicer. During the execution phase the gene specific inactivation occurs by hybridization to and degradation of the target mRNA (siRNA) or by translation inhibition (miRNA). Ribonucleoprotein (RNP) complexes (e.g. RISC) are essential for this phase.

Distinct sequence and/or structural elements in the precursor transcripts of small RNAs, recruit RNA-processing enzymes and proteins which are responsible for small RNA maturation and for the subsequent assembly of the small RNAs into effector complexes that mediate small RNA function2. The RNAi pathway involves two phases. During the initiation phase the double stranded RNA molecules, which are either in the form of a hairpin or a longer dsRNA, are cleaved by the enzyme Dicer. During the execution phase the gene specific inactivation occurs by hybridization to and degradation of the target mRNA or by translation inhibition. Ribonucleoprotein (RNP) complexes (e.g RISC) are essential for this phase1.
RNP complexes contain at their center an Argonaute/ Piwi protein family member and are loaded with distinct classes of small RNAs to form target-recognizing complexes. The Argonaute/Piwi family is well conserved and members have been identified in many species. Argonaute/Piwi proteins demonstrate bilobal architecture where the first lobe contains the N-terminal PAZ-domain responsible for binding the 3'-end of the guide small RNA. The second lobe contains the MIDdomain, responsible for binding the 5'-phosphate of the guide RNA, and the RNase H endonuclease domain, also known as the PIWI-domain2.
The different classes of small RNAs are distinguished from each other by their length, with peak lengths varying from 21 to 30 nucleotides. The lengths of the different classes of small RNAs vary due to distinct mechanisms of biogenesis. In addition, other characteristics such as the presence of a 5'-uridine, phosphorylation at the 5'-end and 2'-O-methylation at the 3' end of the RNA molecule determine the loading of small RNAs onto effector ribonucleoprotein (RNP) complexes2.
Definition of microRNAs
MicroRNAs (miRNAs) belong to the most abundant class of small RNAs in animals2. It is a recently discovered class of eukaryotic, endogenous, non-coding RNAs that play a key role in the regulation of gene expression. When mature, they are short, single-stranded RNA molecules approximately 21-23 nucleotides in length, which usually have an uridine at their 5'-end and they are partially complementary to one or more messenger RNA (mRNA) molecules (target mRNAs)2. Their main function is to down-regulate gene expression by inhibiting translation or by targeting the mRNA for degradation or deadenylation3. However, it was recently found that some miRNAs, like miR-369-3, can upregulate the translation of the tumor necrosis factor alpha4.
Each miRNA is thought to regulate multiple genes. Since hundreds of miRNA genes are predicted to be present in higher eukaryotes ('1000 unique miRNAs), the potential regulatory circuitry afforded by miRNA is huge5, 6. Acting at the post-transcriptional level, these molecules may alter the expression of as much as 20-30% of all mammalian protein-encoding genes7. Several research groups have verified that miRNAs may act as key regulators of processes as diverse as embryonic development, cell proliferation, cell growth, tissue differentiation and apoptosis. Recent studies of miRNA expression involve miRNAs in cellular signaling network, cross-species gene expression variation and co-regulation with transcription factors. Consequently, mutation on miRNAs, dysfunction of miRNA biogenesis and disregulation may result in a broad spectrum of diseases. Furthermore, components required for miRNA processing and/or function have also been implicated in various disorders such as fragile-X mental retardation, DiGeorge syndrome and cancer. Currently, there have been reported '70 diseases which are associated with miRNAs8, 9.
Biogenesis of microRNAs
MiRNAs are first transcribed as primary transcripts (pri-miRNA) with a cap and poly-A tail by RNA polymerase II or RNA polymerase III15,16. A typical pri-miRNA is composed of a double stranded stem of '33 base pairs, a terminal loop and two flanking unstructured single-stranded segments as shown in (Figure 3)17. PrimiRNA is processed to a short 70-nucleotide stem-loop structure known as pre-miRNA by a protein complex known as the Microprocessor complex which consists of the ribonuclease Drosha, an RNase III enzyme18 and a double stranded-RNA binding protein called DGCR8 (DiGeorge syndrome critical region 8 gene)15, 17.
Figure 3: Overview of microRNA processing. MiRNAs are first transcribed as primary transcripts (pri-miRNA) with a cap and poly-A tail by RNA polymerase II or RNA polymerase III . A pri-miRNA is composed of a double stranded stem of '33 base pairs, a terminal loop and two flanking unstructured single-stranded segments. Pri-miRNA is processed to a short 70-nucleotide stem-loop structure known as pre-miRNA by a protein complex known as the Microprocessor complex which consists of the ribonuclease Drosha, an RNase III enzyme and a double stranded-RNA binding protein called DGCR8. After the pre-miRNA is generated in the nucleus, it is exported by Exportin-5 (Exp- 5) to the cytoplasm by the action of RanGTPase. In the cytoplasm, an RNAse III endonuclease called Dicer cleaves pre-miRNAs into short RNA duplexes termed miRNA duplexes. The RNA strand of the miRNA duplex that is complementary to the mature miRNA is shown with a star symbol (*). After cleavage, the miRNA duplex is unwound by an unidentified RNA helicase and the mature miRNA strand binds to an Argonaute (Ago) protein into an RNP complex. The miRNA* strand is degraded.
The cleavage of a pri-miRNA by microprocessor begins with DGCR8 recognizing the ssRNA-dsRNA junction typical of a pri-miRNA. Drosha is then brought close to its substrate through interaction with DGCR8 and cleaves the stem of a pri-miRNA '11 nt away from the two single stranded segments ('22 nts away from the loop). Drosha removes the double-stranded stem from the remainder of the pri-miRNA by cleaving proximal and distal of the stem, generating a pre-miRNA that has a 5'-monophosphate and a 3'-2-nt overhang (Figure 3). Although microprocessing is already sufficient for conversion of a pri-miRNA into a pre-miRNA in vitro, cleavage of pri-miRNA in vivo does not depend on Drosha and DGCR8 only, but also on other accessory proteins, such as the RNA binding protein hnRNP A1 and the p68 and p72 RNA helicases17.
Recently, a subclass of pre-miRNAs, pre-miRNA/introns (known as mirtrons), has been shown to depend on the RNA splicing machinery for their biogenesis in Drosophila melanogaster, Caenorhabditis elegans and mammals. Mirtrons are derived from certain debranched introns that fold into hairpin structures, mimic the structural hallmarks of pre-miRNAs and enter the miRNA-processing pathway. The discovery of mirtrons suggests that any RNA, with a size analogous to a premiRNA and all the structural features of a pre-miRNA, can be used as a substrate by the miRNA processing machinery, and give rise to a functional miRNA17.
After the pre-miRNA is generated in the nucleus, it is exported by Exportin-5 (Exp-5) to the cytoplasm by the action of RanGTPase. RanGTP is hydrolyzed by RanGAP to RanGDP, and the pre-miRNA is released from Exp-5 which is also important for stabilizing premiRNAs in the nucleus. When Exp-5 is knocked down by siRNAs, the levels of pre-miRNAs are reduced not only in the cytoplasm, but also in the nucleus, suggesting that binding of pre-miRNAs to Exp-5 protects them from degradation17.
In the cytoplasm, an RNAse III endonuclease called Dicer cleaves pre-miRNAs into short RNA duplexes termed miRNA duplexes (Figure 3). The RNA strand of the miRNA duplex that is complementary to the mature miRNA is shown with a star symbol (miRNA*). Mammalian Dicer has an N-terminal ATPase/helicase domain, a PAZ domain, two RNase III catalytic domains and a C-terminal dsRNA binding domain (dsRBD)18. Biochemical experiments have revealed that both PAZ domain and dsRBD are essential for the interaction of Dicer with pre-miRNAs. PAZ domain recognizes and binds the 2-nt 3' -overhang at the base of the pre-miRNA hairpin generated by Drosha17,18. After that, Dicer dimerizes its two RNase III domains intramolecularly, to form a single processing center and cuts the stem of pre-miRNAs '22 nt away from their termini at positions separated by 2 nts, thereby generating 3' -2-nt overhang. After cleavage, the miRNA duplex is unwound by an unidentified RNA helicase and the mature miRNA strand binds to an Argonaute (Ago) protein into an RNPcomplex in a process called "miRNA loading" or assembly. The miRNA* strand is degraded (Figure 3)17,18. A primary determinant as to which of the two strands of a miRNA duplex will be loaded on Ago proteins is the inherent thermodynamic asymmetry of the miRNA duplex. The RNA strand whose 5'- end is less stably bound to the opposite strand will be loaded to Ago proteins and form the mature miRNA17.
MicroRNA targets and mechanism of action
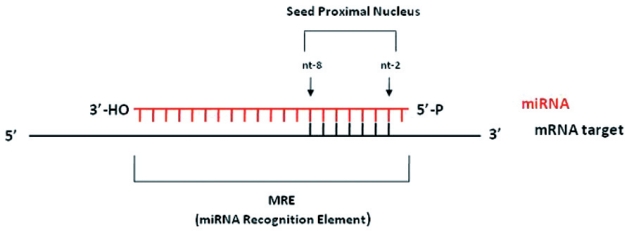
The mature miRNA binds to the target mRNA and typically in the 3'-untranslated region (3'- UTR). Thus, it interferes with translation of the mRNA. MiRNAs base-pair with miRNA recognition elements (MREs) located on their mRNA targets, usually on the 3'-UTR, through a critical region called the 'seed region' which includes nucleotides 2-8 from the 5-end of the miRNA as shown in figure 419. Asymmetry is the general rule for matches between a microRNA and its target. The 5'-end of the microRNA tends to have more bases complementary to the target than the 3'-end does. Cohen et al., concluded that the complementarity of seven or more bases to the 5'-end miRNA is sufficient for regulation. Basepairing between the 3'-segment of the miRNA and the mRNA target is not always essential for repression, but strong base-pairing within this region can partially compensate for weaker seed matches or enhance repression. As a matter of fact, miR-24 regulates expression of E2F2, MYC, AURKB, CCNA2, CDC2, CDK4, and FEN1 by recognizing seedless but highly complementary sequences20. Additionally, multiple MREs for the same or different miRNAs within the same 3'UTR can function cooperatively to enhance repression. Spacing of the seed sites within the 3'UTR may play a significant role in the action of miRNAs. Finally, sequences adjacent to MREs and the secondary structure of the 3'UTR of the target mRNA may affect binding of miRNAs; miRNPs cannot efficiently "unwind" structured RNA areas, and thus miRNAs cannot bind to sites that are embedded in such structured areas.
Figure 4: MicroRNA binding to the target mRNA. The mature miRNA binds to the target mRNA and typically in the 3'-untranslated region (3'- UTR). MiRNAs base-pair with miRNA recognition elements (MREs) located on their mRNA targets, usually on the 3'-UTR, through a critical region called the 'seed region' which includes nucleotides 2-8 from the 5-end of the miRNA.
In plants, the main mechanism of regulation by miRNAs is mRNA cleavage. However, in animals miRNAs repress the translation of their mRNA targets and/or destabilize them without endonucleolytic cleavage. Early studies in Caenorhabditis elegans showed that the small non-coding RNAs lin-4 and let-7, represented imperfect complementary matches to sequences within the 3'UTRs of their target mRNAs. Furthermore, this interaction between the miRNA and target mRNA resulted in decreased target protein levels without affecting the stability of the mRNA. This profile of a significant reduction in protein level without a proportionate reduction in target mRNA levels became a hallmark of miRNA function. We now know that the vast majority of animal miRNAs base-pair with imperfect complementarity with their mRNA targets.
The expression of a large number of predicted human miRNA genes has been confirmed (721 miRNAs, release 14 miRBase), but the predicted miRNA targets remain to be identified and verified. In order for a miRNA to regulate the expression of a specific gene it must first exhibit some partial complementarity with the target mRNA, specifically in the 3'UTR of the mRNA. However, theoretical sequence complementarity of the miRNA to the mRNA is not a reliable predictor of a real miRNA target. The energy required to overcome the secondary structure of the mRNA should also be taken into consideration. Several groups have developed algorithms for identification of mRNA sequences that could serve as target sites for known miRNAs. Some of them are MiRanda, PicTar and TargetScan10–12. In general those algorithms take into account matches between the 5'-end of the miRNA and the 3'UTRs, the free energy of the RNA-RNA duplex and the degree of conservation of the miRNA target in related genomes.
Multiple algorithms should be utilized for target prediction because these programs usually predict different miRNA-binding sites. When a certain gene was analyzed for putative miRNA-binding sites, miRanda algorithm predicted 27 miRNA-binding sites. But when TargetScan was used it predicted 37 miRNA-binding sites with an overlap of only 8 of those predicted by miRanda. Moreover, when the gene was analyzed by the PicTar algorithm no miRNA-binding site was predicted because of the strict requirement for sequence conservation of binding sites in different species19. For an accurate and efficient target prediction the miRNA-target interaction must be confirmed, the predicted miRNA and mRNA target gene must be coexpressed, a given miRNA must have a predictable effect on the expression of the target protein and the miRNA-mediated regulation of target gene expression should equate with altered biological function19. The initial hypothesized interaction between a specific miRNA and its targer mRNA, is of paramount importance and it can be tested with in vitro systems with the use of sensor constructs, such as Luciferase constructs with 3'UTR of the target gene20.
Polymorphisms within miRNA binding sites
Because of the stringent recognition requirement between the miRNA seed region and its target, sequence variations such as SNPs in the miRNA-binding seed region (mirSNPs) can disrupt the miRNA-mRNA target site interaction affecting the expression of the miRNA targets. The occurrence of SNPs in pre-miRNA sequences is relatively rare. Only '10% of human premiRNAs contain documented SNPs and less than 1% of miRNAs have SNPs in the functional seed region. In PolymiRTS (database for Polymorphism in microRNA Target Site) there are already 15791 records for SNPs in microRNA target sites that have been spotted in humans [2]. A SNP can eliminate or weaken the binding to a miRNA target or increase the binding by creating a perfect sequence match to the seed of a miRNA that normally is not associated with the given mRNA. These can lead to a significant alteration of protein expression. Moreover SNPs may reduce the stability of the pri-miR, the efficiency of of pri-miR processing into pre-miR, and the efficiency of pre-miR processing into the mature miRNA1,22.
Newly developed methods that identify miRNAs and their targets and genomic databases of SNPs, provide an opportunity to explore human evolution at miRNA target level. To identify variations in the target sites that are related to a biological/pathological event in epidemiologic studies, a number of factors should be considered. First, the target genes should be directly or indirectly related to the disease under study. In addition, the polymorphisms should be within the seed region. Furthermore, there should be specific data relating to the population frequency of the SNPs in the ethnic group being studied and should have a preferred frequency of not <5% for the variant allele19. The frequency of polymorphisms in the miRNAs sequences is low, apparently because of their small length and their highly conserved sequences. SNPs in miRNAs or miRNAs targets have been associated with the development of several diseases, such as the early age of breast cancer onset21, papillary thyroid carcinoma22, breast/ovarian cancer23, and hepatocellular carcinoma24. MirSNPs have also been correlated with Schizophrenia and autism25. Up to date, there is only one report that indicates the presence of a causative mutation on the miRNA seed itself. Specifically, the authors demonstrated that mutations found in the seed region of human miR-96 are responsible for nonsyndromic progressive hearing loss26.
As mentioned before, miRNAs have multiple targets and regulate the expression of hundreds of genes that have important cellular functions like for example cell differentiation, development, proliferation and progression of human diseases. As a result, a variation in miRNAs may have an extensive and often deleterious impact with implications for both evolutionary studies and biomedical research19, 27.
Figure 2: Transcription of miRNAs. MicroRNAs are transcribed as individual miRNA genes, from introns of coding genes classified as intronic or from the genomic region between the clusters of genes classified as intergenic. Some microRNAs are clustered in polycistronic transcripts.
References
- 1.Felekkis K, Deltas C. RNA interference: A powerful laboratory tool and its therapeutic implications. Hippokratia. 2006;10:112–115. [PMC free article] [PubMed] [Google Scholar]
- 2.Farazi AT, Juranek AS, Tuschl T. The growing catlog of small RNAs and their association with distinct Argonaute/Piwi family members. Development. 2008;135:1201–1214. doi: 10.1242/dev.005629. [DOI] [PubMed] [Google Scholar]
- 3.Wu L, Fa J, Belasco GJ. MicroRNAs direct rapid deadenylation of mRNA. PNAS. 2006;103:4034–4039. doi: 10.1073/pnas.0510928103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vasudevan S, Tong Y, Steitz AJ. Switching from Repression to Activation:MicroRNAs can Up-Regulate Translation. Science. 2007;318:1931–1933. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- 5.Berezicov E, Guryev V, Belt Jvd, Wienholds E, Plasterk HAR, Cuppen E. Phylogenetic Shadowing and Computational Identification of Human microRNA Genes. Cell. 2005;120:21–24. doi: 10.1016/j.cell.2004.12.031. [DOI] [PubMed] [Google Scholar]
- 6.Xie X, Lu J, Kulbokas EJ, Golub RT, Mootha V, Lindblad-Toh K, et al. Systematic discovery of regulatory motifs in human promoters and 3UTRs by comparison of several mammals. Nature. 2005;434:338–345. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lewis B, Burge C, DP B. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 8.Lu M, Zhang Q, Deng M, Miao J, Guo Y, Gao W, et al. An analysis of human microRNA and disease associations. Plos ONE. 2008;3:e3420. doi: 10.1371/journal.pone.0003420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wienholds E, Plasterk HAR. MicroRNA function in animal development. FEBS Letters. 2005;579:5911–5922. doi: 10.1016/j.febslet.2005.07.070. [DOI] [PubMed] [Google Scholar]
- 10.Ying S-Y, Lin S-L. Current perspectives in intronic micro RNAs. Journal of Biochemical Science. 2006;13:5–15. doi: 10.1007/s11373-005-9036-8. [DOI] [PubMed] [Google Scholar]
- 11.Rodriguez A, Griffiths-Jones S, Ashurst LJ, Bradley A. Identification of Mammalian microRNA Host Genes and Transcription Units. Genome Research. 2004;14:1902–1910. doi: 10.1101/gr.2722704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim Y-K, Kim VN. Processing of intronic microRNAs. The EMBO Journal. 2007;26:775–783. doi: 10.1038/sj.emboj.7601512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Seitz H, Royo H, Bortolin, ML, Lin SP, Ferguson-Smith AC, Cavaille J. Alarge imprinted microRNA gene cluster at the mouse Dlk 1-Gtl2 domain. Genome Res. 2004;14:1741–1748. doi: 10.1101/gr.2743304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Altuvia Y, Landgraf P, Lithwick G, Elefant N, Pfeffer S, Aravin A, et al. Clustering and conservation patterns of human microRNAs. Nucleic Acids Research. 2005;33:2697–2706. doi: 10.1093/nar/gki567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Denli A, Tops B, Plasterk R, Ketting R, Hannon G. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;432:231–235. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- 16.Borchert G, Lanier W, Davidson B. RNA Polymerase III transcribes human microRNAs. Nat Struct Mol Biol. 2006;12:1097–1101. doi: 10.1038/nsmb1167. [DOI] [PubMed] [Google Scholar]
- 17.Liu X, Fortin K, Mourelatos Z. MicroRNAs: Biogenesis and Molecular Functions. Brain Pathology. 2008;18:113–121. doi: 10.1111/j.1750-3639.2007.00121.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cullen RB. Transcription and Processing of Human microRNA Precursors. Molecular Cell. 2004;16:861–865. doi: 10.1016/j.molcel.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 19.Chen K, Song F, Calin AG, Qingyi W, Hao X, Zhang W. Polymorphisms in microRNA targets: a gold mine for molecular epidemiology. Carcinogenesis. 2008;29:1306–1311. doi: 10.1093/carcin/bgn116. [DOI] [PubMed] [Google Scholar]
- 20.Lal A, Navarro F, Maher CA, Maliszewski LE, Yan N, O'Day E, et al. miR-24 Inhibits cell proliferation by targeting E2F2, MYC, and other cell-cycle genes via binding to "seedless" 3'UTR microRNA recognition elements. Mol Cell. 2009;35:610–625. doi: 10.1016/j.molcel.2009.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Song F, Zheng H, Liu B, Wei S, Dai H, Zhang L, et al. An miR-502-binding site single-nucleotide polymorphism in the 3'-untranslated region of the SET8 gene is associated with early age of breast cancer onset. Clin Cancer Res. 2009;15:6292–6300. doi: 10.1158/1078-0432.CCR-09-0826. [DOI] [PubMed] [Google Scholar]
- 22.Jazdzewski K, Murray EL, Franssila K, Jarzab B, Schoenberg DR, de la Chapelle A. Common SNP in pre-miR-146a decreases mature miR expression and predisposes to papillary thyroid carcinoma. Proc Natl Acad Sci U S A. 2008;105:7269–7274. doi: 10.1073/pnas.0802682105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shen J, Ambrosone CB, DiCioccio RA, Odunsi K, Lele SB, Zhao H. A functional polymorphism in the miR-146a gene and age of familial breast/ovarian cancer diagnosis. Carcinogenesis. 2008;29:1963–1966. doi: 10.1093/carcin/bgn172. [DOI] [PubMed] [Google Scholar]
- 24.Xu T, Zhu Y, Wei QK, Yuan Y, Zhou F, Ge YY, et al. A functional polymorphism in the miR-146a gene is associated with the risk for hepatocellular carcinoma. Carcinogenesis. 2008;29:2126–2131. doi: 10.1093/carcin/bgn195. [DOI] [PubMed] [Google Scholar]
- 25.Sun G, Yan J, Noltner K, Feng J, Li H, Sarkis D. SNPs in human miRNA genes affect biogenesis and function. RNA. 2009;15:1640–1651. doi: 10.1261/rna.1560209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mencia A, Modamio-Hoybjor S, Redshaw N, Morin M, Mayo-Merino F, Olavarrieta L, et al. Mutations in the seed region of human miR-96 are responsible for nonsyndromic progressive hearing loss. Nat Genet. 2009;41:609–613. doi: 10.1038/ng.355. [DOI] [PubMed] [Google Scholar]
- 27.Saunders AM, Liang H, Li W-H. Human polymorphism at microRNAs and microRNA target sites. PNAS. 2007;104:3300–3305. doi: 10.1073/pnas.0611347104. [DOI] [PMC free article] [PubMed] [Google Scholar]