Abstract
Parathyroid hormone-like hormone (PTHLH) is an important chondrogenic regulator; however, the gene has not been directly linked to human disease. We studied a family with autosomal-dominant Brachydactyly Type E (BDE) and identified a t(8;12)(q13;p11.2) translocation with breakpoints (BPs) upstream of PTHLH on chromosome 12p11.2 and a disrupted KCNB2 on 8q13. We sequenced the BPs and identified a highly conserved Activator protein 1 (AP-1) motif on 12p11.2, together with a C-ets-1 motif translocated from 8q13. AP-1 and C-ets-1 bound in vitro and in vivo at the derivative chromosome 8 breakpoint [der(8) BP], but were differently enriched between the wild-type and BP allele. We differentiated fibroblasts from BDE patients into chondrogenic cells and found that PTHLH and its targets, ADAMTS-7 and ADAMTS-12 were downregulated along with impaired chondrogenic differentiation. We next used human and murine chondrocytes and observed that the AP-1 motif stimulated, whereas der(8) BP or C-ets-1 decreased, PTHLH promoter activity. These results are the first to identify a cis-directed PTHLH downregulation as primary cause of human chondrodysplasia.
INTRODUCTION
Brachydactyly literally means short fingers and toes relative to the length of other long bones and other parts of the body. Five types (A–E) have been described, each containing of three subgroups, occurring as an isolated trait or as part of a syndrome (1). Brachydactyly Type E (BDE: MIM 113300) involves interfamilial variably shortened metacarpals, metatarsals and/or phalanges with frequent bilateral asymmetry (2,3). At least three autosomal-dominant BDE subtypes have been described, E1 in which shortening is limited to the fourth metacarpals and/or metatarsals, E2, in which variable combinations of metacarpals are involved with shortening also of the first and third distal and the second and fifth middle phalanges and E3, a less-well defined category, which may have a variable combination of short metacarpals without phalangeal involvement. BDE also occurs with Bilginturan syndrome (HTNB: MIM 112410), pseudohypoparathyroidism type IA (PHP1A: MIM 103580), pseudopseudohypoparathyroidism (PPHP: MIM 612463), Turner syndrome and other conditions (4). Isolated BDE has been attributed to two different missense mutations in HOXD13; however, no other isolated BDE genes have been identified to our knowledge (5). We studied a family with autosomal-dominant BDE and found a balanced translocation t(8;12)(q13;p11.2) with a der(8) breakpoint (BP) upstream of parathyroid hormone-like hormone (PTHLH) in a conserved non-coding chromosomal region. We found that the translocation caused a novel mechanism of PTHLH downregulation. Although PTHLH is known to be important in chondrogenesis, to our knowledge this report is the first to implicate PTHLH in a human genetic disease.
RESULTS
Clinical BDE characteristics and cytogenetic analyses
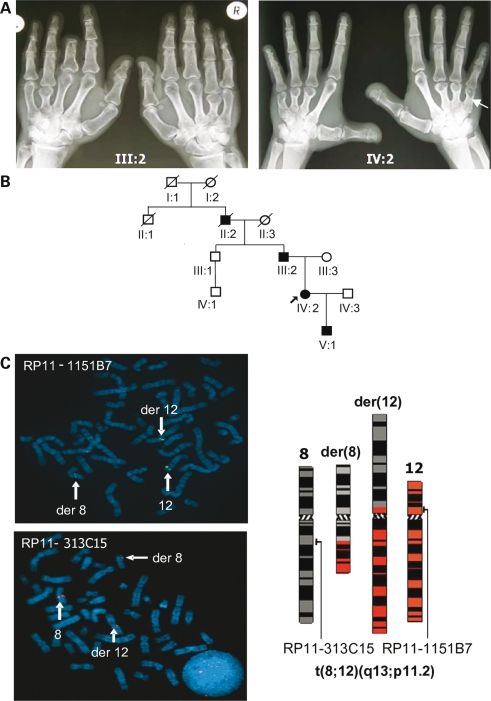
Clinical and radiographic features of the affected BDE family members are given in Table 1. The pedigree showed autosomal-dominant transmission of an isolated BDE variant characterized in hand roentgenograms from subjects III:2 and IV:2 (Fig. 1A and B) by shortened metacarpals, most pronounced on the fourth rays, with variable involvement of proximal, middle and distal phalanges and cone-shaped epiphysis (Table 1, Fig. 1A). The affected individuals' heights were in the range of lower normal. A relative shortening of the extremities in comparison to the trunk was evident. The arm span to height ratio was reduced (norm: ∼1.01–1.02, Table 1). Moreover, the affected patients showed a dysmorphic facies with macrocephaly, prominent forehead and a depressed nasal root (data not shown). They were neither hypertensive nor mentally retarded. The index patient (IV:2) had no abnormalities in breast development or nursing her son (lactation: 11 months).
Table 1.
Clinical data of BDE affected subjects
| Person | Height (centile) | Ratio arm span: Height | Head circumference (centile) |
|
|---|---|---|---|---|
| III:2 | 178.0 cm (10–25th) | 0.95 | 58 cm (98th) | |
| IV:2 | 163.8 cm (10–25th) | 0.88 | 57 cm (97th) | |
| V:1 | 162.7 cm (>97th) | 0.92 | 58.5 cm (>98th) | |
| Person | Hands: shortened bones | Feet: shortened bones | ||
| III:2 | IV:2 | III:2 | IV:2 | |
| Telephalanges | R/l: I, II, III, V | r/l: I | ||
| Mesophalanges | r/l: V | r/l: II, III, IV | r/l: II, III, IV, V | |
| Basophalanges | r: V/l: V, (I) | r/l: III, IV | r/l: II, III, IV, V | |
| Metacarpals/-tarsals | r/l: IV, (III, V) | r: III, IV, V/l: I, II, IV, V | r/l: IV (l > r) | r/l: III, IV |
Phenotypic characteristics of investigated patients with BDE. Height, arm span to height ratio and head circumference (top). Involved phalangeal, metacarpal and metatarsal bones of hands and feet (bottom). l: left, r: right.
Figure 1.
BDE characteristics and cytogenetic karyotyping. (A) The hands of patient III:2 and index patient IV:2 displayed type E brachydactyly with shortened metacarpals, particularly 3, 4 and 5. Amputation of distal phalanges in digit 3, patient III:2. (B) Family tree; solid symbols are affected. (C) Metaphase FISH with BACs spanning breakpoints (BPs) revealed t(8;12)(q13;p11.2) translocation; BAC RP11-1151B7 for chromosome 12 (green, FITC) and BAC RP11-313C15 for chromosome 8 (red, TMR), present on the normal and derivative chromosomes, der(12) and der(8).
BDD and BDE are associated with HOXD13 mutations (5). To exclude HOXD13 genetically as primary cause for the BDE, we chose four informative microsatellite markers flanking the gene locus (D2S2981, D2S2314, D2S1245, D2S138, UCSC Genome Browser Mar. 2006). Haplotype construction resulted in a cosegregating haplotype with the BDE phenotype (haplotypes Supplementary Material, Fig. S1). Due to the small pedigree size, this might be a result of chance. Therefore, we sequenced HOXD13 in BDE-affected and non-affected family members. We found no mutation (data not shown) and excluded the HOXD13 locus.
YACs and BACs in metaphase FISH analysis spanning the BPs demonstrated a balanced translocation by generating split signals on both derivative chromosomes, der(8) and der(12) (Fig. 1C). BPs were assigned to chromosome bands 8q13 and 12p11.2. Bioinformatics (NCBI Map Viewer) identified PTHLH on chromosome 12 and KCNB2 (potassium voltage-gated channel, Shab-related subfamily, member 2) on chromosome 8 as candidate genes (Fig. 2A).
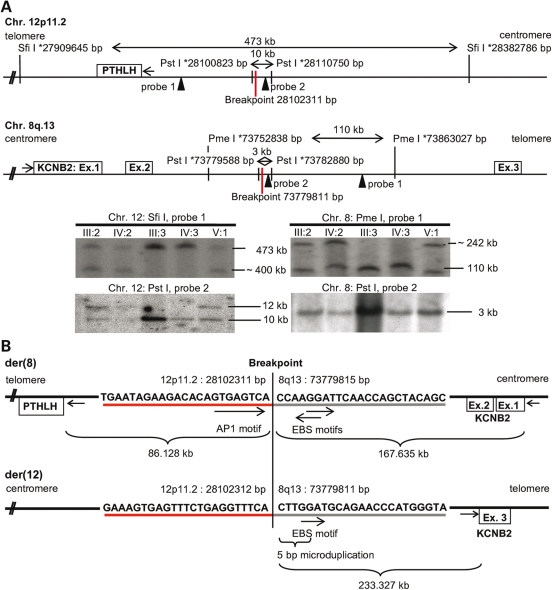
Figure 2.
Genomic view on candidate genes and BP sequences. (A) PTHLH on chromosome (Chr.) 12p11.2 and KCNB2 on Chr. 8q13. Endonucleases (SfiI, PmeI, PstI) for Southern blotting (* indicate restriction sites; probes indicated as arrows) narrowed down the BP regions; fragments are shown in kilobases (kb). The Chr. 12p11.2 BP was at bp 28.102.311 and the Chr. 8q13 BP in KCNB2 intron 2 at bp 73.779.815 (UCSC Genome Browser Mar. 2006). Southern blotting: Chr.12 probes 1, 2 (left) and Chr. 8 probes 1, 2 (right). Non-affected persons (III:3, IV:3) showed the expected restriction fragments (refer to top). In BDE affected patients (III:2, IV:2, V:1) unexpected bands displayed the affected allele. (B) BP sequences on the derivative chromosomes der(8) and der(12). Der(8) BP is 86.128 kb upstream of PTHLH and 167.635 kb downstream of KCNB2, disrupted by the translocation in intron 2. The der(8) BP is surrounded by AP-1 (TGAGTCA) and bidirectional EBS binding motifs. At der(12) BP a 5-bp micro-duplication generated the core consensus sequence for EBS (GGAA/T); KCNB2 exon 3 (Ex.) is 233.327 kb upstream of der(12) BP.
BP identification
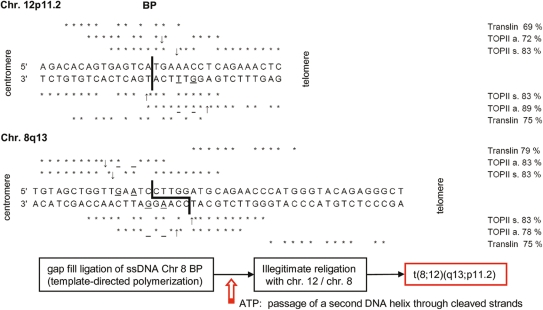
Southern blotting of DNA digested with SfiI, PmeI and PstI narrowed the critical BP regions on chromosome 12 from 473 kb (27909645–28382786 bp) to 10 kb (28100823–28110750 bp) and on chromosome 8 from 110 kb (73752838–73863027 bp) to 3 kb (73779588–73782880 bp; UCSC Genome Browser March 2006; Fig. 2A). PCR amplification of the identified PstI-fragments and their subsequent sequence analysis identified the derivative chromosome 8 breakpoint [der(8) BP] 86.128 kb upstream of PTHLH exon 1 (Chr. 12p11.2, bp 28016183), whereas KCNB2 was disrupted by the translocation in intron 2 with a distance of 167.635 kb between der(8) BP and exon 1 (Chr. 8q13, bp 73779815 UCSC Genome Browser Mar. 2006). The remaining exon 3 was localized 233.327 kb downstream of der(12) BP (Fig. 2B). Der(8) BP was located in a highly conserved region with more than 70% conservation through mammals and features a bioinformatically (rVISTA) predicted Activator protein 1 (AP-1) binding site (TGAGTCA), in contrast to the der(12) BP sequence, which is non-conserved, but displays a 5-bp microduplication containing a v-ets erythroblastosis virus E26 oncogene homolog 1 (C-ets-1/EBS binding site) core consensus sequence (GGAA). The translocation approximated a bidirectional EBS core consensus sequence from 8q13 near the AP-1 site on der(8) BP (Fig. 2B). The identification of highly homologous binding sites (69–89%) for topoisomerase II (TOPII) and translin (TSN) on chromosomes 12p11.2 (bp 28102311, ±100 bp) and 8q13 (bp 73779815, ±100 bp, Fig. 3) in the human RefSeq suggested a plausible mechanism for the t(8;12)(q13;p11.2). TSN recognizes and binds ssDNA or, in complex with Trax (6), dsDNA ends after dsDNA breaks located either 3′ or 5′ of the BP. The protein function has been described in DNA repair, translational regulation and meiotic recombination, particularly in translocations (7). TOPII catalyses ss/dsDNA breaks, ligates dsDNA and is responsible for the recognition of DNA crossover points, illegitimate recombination and DNA relaxation (8,9). Its association with TSN and micromutations was shown in different cancer studies (10–12). The constellation of the highly homologous binding sites for TOPII and TSN in the BP regions on chromosomes 12p11.2 and 8q13, and the 5 bp-microduplication at der(12) BP suggests the following translocation mechanism: a blunt dsDNA break at chromosome 12 occurred, although the sticky ssDNA ends on chromosome 8 were polymerized in a template-directed manner, generating the 5-bp-microduplication at der(12) BP. The sequence homologies (<70%) strongly hypothesize, that TSN binds the DNA ends after the dsDNA breaks, either alone or with Trax. After the religation in the presence of ATP with chromosome 12, the illegitimate religation may have created the balanced translocation t(8;12)(q13;p11.2). The potential mechanism described could be the cause for the translocation, although whether or not TOPII or crossing-over generated the BPs remains unclear (Fig. 3).
Figure 3.
Analysis of the putative cause of the translocation by asymmetrical and symmetrical topoisomerase (TOPII a, s) and translin binding. Wild-type sequences are from RefSeq (NCBI) with homology matching bp (*); BPs as bold lines. TOPII asymmetric (a) consensus sequence: 5′-A/GNc/TNNCNNGC/T^NGG/TTNC/TN-3′; symmetric (s): 5′-A/GNNNNC/TA/GNA/GC/T^A/TVBT/AA/GC/TNC/TA/GNNNNC/T-3′. Two different translin recognition motifs: 5′-ATGCAG-0–4 bp-GCCCA/TG/CG/CA/T-3′ and 5′-GCNCA/TG/CCT-0–2 bp-GCCCA/TG/CG/CA/T-3′. ↑↓ Potential TOPII restriction sites, * verified +2 (guanine)/+4 (thymine) TOPII at a position behind the restriction site (72).
AP-1 and C-ets-1 bind at the der(8) BP
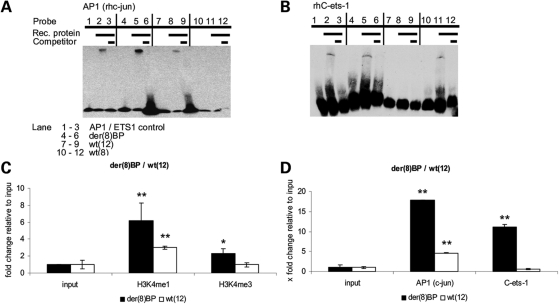
We used electromobility shift assays (EMSA) to test whether or not the transcription factors AP-1 (c-Jun) and C-ets-1 bind in vitro at the TFBS identified in silico. We found that AP-1 (c-Jun) and C-ets-1 bound to der(8) BP (Fig. 4A and B, Lane 5). Since the AP-1 site is present on wild type allele chromosome 12 [wt(12)], c-Jun binding was detected (Fig. 4A, Lane 8), but not C-ets-1 (Fig. 4B, Lane 8). On der(12) BP, C-ets-1 binding was detected due to a 5-bp microduplication (Fig. 4B, Lane 11), which generated the EBS core consensus sequence, whereas c-Jun did not bind (Fig. 4A, Lane 11). These results confirm that der(8) BP contains AP-1 and C-ets-1 binding sites and suggest that introduction of the novel C-ets-1 site may influence differential regulation of PTHLH downstream.
Figure 4.
Electrophoretic mobility shift analysis (EMSA) and Chromatin immunoprecipitations (ChIP) at der(8) BP and wt(12). (A) EMSA of the wild-type (wt) and translocation BPs far upstream of PTHLH. Recombinant human (rh) AP-1 (c-Jun) and rhC-ets-1 were incubated with 3′-biotin-labeled oligonucleotides representing der(8) BP and wt(8) and wt(12). rhAP-1 bound der(8) BP (lane 5) and wt(12) (lane 8), but not wt(8) (lane 11). (B) rhC-ets-1 bound der(8) BP (lane 5) and wt(8) (lane 11), but not wt(12) (lane 8). Competition with 400-fold excess unlabeled oligo (lanes 3, 6, 9, 12) prevented binding in all cases; AP-1 or C-ets-1 consensus oligos as controls (lane 2); probes without protein (lanes 1, 4, 7, 10). (C) ChIP to determine chromatin stage at der(8) BP. The regulatory histone modifications H3K4me1 and H3K4me3 enrichments in fibroblasts of a BDE patient between der(8) BP affected and wt(12) unaffected allele were 2-fold higher at der(8) BP. Epigenetic modifications occurred only at the translocation BP. (D) The transcription factors AP-1 (c-Jun) and C-ets-1 in vivo binding at der(8) BP and wt(12) allele in chondrogenically differentiated fibroblasts of a BDE patient. AP-1 and C-ets-1 were significantly enriched at der(8) BP, AP-1 was also found at wt(12) (n = 3, **P < 0.01, *P < 0.05).
Chromatin immunoprecipitation to characterize the der(8) BP
We then asked whether AP-1 and C-ets-1 could be involved in regulating the PTHLH gene in the context of chondrogenesis and under the influence of der(8) BP. We investigated the transcriptionally active euchromatin stage at the der(8) BP in BDE skin fibroblasts. Enriched H3K4me3 appears in promoter regions and H3K4me1 is known to occur at cis-regulatory elements (CRE) with enhancing gene regulatory action (13). We performed chromatin immunoprecipitation (ChIP) assays to analyze der(8) BP and wt(12) alleles for differences in enriched H3K4me1 and H3K4me3. At der(8) BP, H3K4me1 and H3K4me3 were 2-fold enriched compared with wt(12) allele (Fig. 4C). These results indicate that modified euchromatin acts as a cis-acting enhancing regulator and that epigenetic modification occurs at der(8) BP in a highly conserved region.
Since AP-1 and C-ets-1 are known to cooperate and interact in a combinatorial fashion (14,15), we employed ChIP to determine if they bind at the der(8) BP and wt(12) allele in fibroblasts from a BDE patient in patterns similar to those found by EMSA. For this purpose, we chondrogenically differentiated BDE fibroblasts for 14 days. The ChIP resulted in strong enrichment of AP-1 (c-Jun) and C-ets-1 at der(8) BP, the affected allele of the BDE patient. At the BP position of the wt(12) unaffected allele of the same BDE sample, AP-1 binding was reduced and due to the absence of the EBS motif, C-ets-1 was not enriched (Fig. 4D). These in vivo results reproduce and strengthen the EMSA results. Both transcription factors bound at the der(8) BP in BDE fibroblasts. The characteristic euchromatin histone modifications, H3K4me1 and H3K4me3, at der(8) BP enable the suggested AP-1 and C-ets-1 binding and identify der(8) BP as a cis-regulatory element.
KCNB2 disruption and PTHLH dysregulation
Because of its role in chondrogenesis, PTHLH was considered as candidate despite the fact that the gene itself was not disrupted by the translocation. KCNB2 was directly affected by the translocation, suggesting haploinsufficiency based on truncated protein of 193 amino acids (out of 911) or unstable mRNA. However, in situ hybridization studies in rat detected unique Kcnb2 gene expression in brain, namely in granule cells of the olfactory bulb, cerebellum, hippocampus and amygdale (16). In Xenopus laevis immunohistochemical studies, Kv2.2, the coded protein, was localized to ventrolateral regions of the brain and spinal cord, especially in areas of growing axonal tracts, in the retina, inner ear and olfactory epithelium (17). Functional analysis of Kv2.2 channels found mediation of a slow delayed rectifier and maintenance of high frequency action potential firing in medial nucleus of trapezoid body neurons (18). Due to these data, we performed clinical tests of vestibular and auditory function in the BDE patients resulting in normal function (data not shown). We suspected that the function of the disrupted KCNB2 gene must be complemented by the intact allele. Additionally, we excluded KCNB2 by in situ hybridization of fetal mouse sections and whole mounts at embryonic days 12.5, 13.5 and 14.5, respectively, where KCNB2 mRNA was not detectable in developing limbs (Supplementary Material, Fig. S2). To test KCNB2 expression in patient tissue, we used undifferentiated control and BDE patient fibroblasts, as well as chondrogenically induced fibroblasts, and the human chondrocytes C28/I2 and found no KCNB2 expression in qRT-PCR, in contrast to the positive controls from human brain and hippocampus. KCNB2 was highly expressed in brain tissue, verifying the in situ hybridizations. No significant difference between unaffected controls and the BDE affected patient was detected (Supplementary Material, Fig. S2).
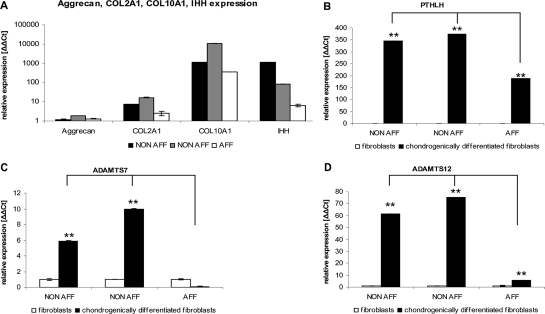
We therefore hypothesized that a deregulation of PTHLH was associated with impaired chondrogenic differentiation and BDE development, either by disrupting or by generating a regulatory element directly at the der(8) BP or by uncoupling cis-acting regulatory elements upstream of the BP. The identification of AP-1 and EBS binding sites at der(8) BP supported the former possibility. The coding sequence of PTHLH was sequenced to exclude any mutations. Since patient chondrocytes were not obtainable, we used the multilineage differentiation potential of skin fibroblasts to induce chondrogenic differentiation at passage four in BDE and control fibroblasts (19–22). The pelleted micromass cultures were differentiated for 4 weeks and stained with Safranin O or Alcian blue to detect proteoglycans in the extracellular matrix (Supplementary Material, Fig. S3). Total RNA was prepared and qRT-PCR determined the successful chondrogenic differentiation of the upregulated chondrocyte markers, Aggrecan, COL2A1, COL10A1 and IHH, relative to undifferentiated skin fibroblasts (Fig. 5A). As expected for chondrogenic differentiation, PTHLH was upregulated in non-affected controls compared with the undifferentiated fibroblast samples confirming the tissue specific regulation of PTHLH. However, PTHLH expression was similar in non-affected differentiated cultures, but PTHLH was downregulated in the differentiated fibroblasts derived from the BDE patient. This suggested that the normal chondrocyte-specific PTHLH control mechanism was impaired (Fig. 5B). Furthermore, we analyzed the PTHrP protein level in skin fibroblasts and also observed a downregulation of PTHrP in cells from the BDE patient (data not shown). We then analyzed the expression of important chondrocyte-related downstream targets of PTHrP. We selected ADAMTS-7 and ADAMTS-12 (disintegrin and metalloproteinases with thrombosponding type 1 motifs), recently described as chondrogenic regulators and direct targets of PTHrP (23,24). Expression analysis performed by qRT-PCR in the same samples showed that ADAMTS-7 and ADAMTS-12 were highly expressed in chondrogenically differentiated non-affected fibroblasts, whereas they were downregulated in the BDE cultures (Fig. 5C and D). This result coincides with the observed PTHLH downregulation, and suggests its involvement in ADAMTS-7 and ADAMTS-12 downregulation.
Figure 5.
Gene expression in BDE and unaffected fibroblasts undergoing chondrogenic induction in pelleted micromass cultures. (A) The cartilage markers Aggrecan (AGC), COL2A1, COL10A1 and IHH were upregulated in the non-affected control individuals (NON-AFF) and in the BDE affected patient (AFF) and verified the chondrogenic differentiation. (B–D) PTHLH, ADAMTS-7 and ADAMTS-12 expressions were similar in differentiated NON-AFF, but significantly downregulated in differentiated BDE cultures (independent differentiations, n = 4, **P < 0.01, *P < 0.05).
AP-1 and C-ets-1 at der(8) BP negatively regulate PTHLH in vitro
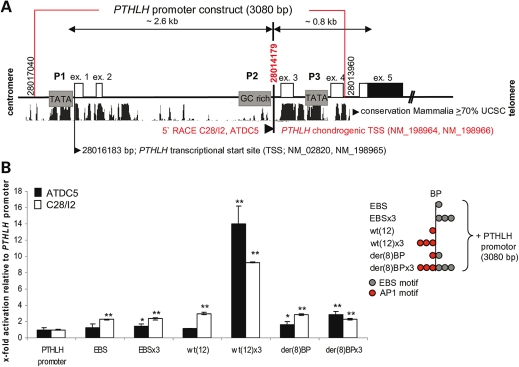
We next addressed the functional analysis of the der(8) BP by in vitro reporter assays. To mimic the strong tissue specificity for PTHLH regulation, we chose two different chondrogenic cell lines; murine ATDC5 cells (25), which can undergo hypertrophic differentiation, and the immortalized human chondrocytes, C28/I2, which reflect proliferating chondrocytes in postnatal cartilage (26). PTHLH transcription starts from three different promoters. P1 and P3 contain TATA boxes, whereas P2 is a GC-rich promoter region (27,28). Several studies have shown a prevalent P2/P3 promoter usage in many cancers such as breast and bone cancers in addition to Adult T-cell leukaemia/lymphoma (ATLL) (29–31). The promoter utilized in chondrocytes has not yet been defined. We determined by 5′ RACE PCR the PTHLH transcription start site (TSS; bp 28014179 UCSC Genome browser Mar. 2006) in both ATDC5 and C28/I2 cells downstream of the GC-rich promoter (P2) within the most highly conserved mammalian PTHLH promoter region (Fig. 6A). To determine the functionality of der(8) BP, we performed reporter gene assays with luciferase reporter constructs in ATDC5 and C28/I2 chondrocytes. We chose the complete promoter region of 3080-bp from distal (P1) to proximal (P3) PTHLH TATA box (bp 28013960-28017040 UCSC Genome browser Mar. 2006) to maintain the fully functional promoter. This region includes the three known PTHLH promoters and the chondrogenic PTHLH TSS identified in 5′ RACE PCR (Fig. 6A). We cloned the following upstream of the PTHLH promoter, consistent with their genomic orientation, as single motifs or as three-time repeats (x3): an EBS core consensus sequence (EBS), wt(12), containing the AP-1 motif and the der(8) BP containing both AP-1 and EBS sites due to the translocation. EBS, EBSx3, wt(12) and der(8) BP slightly increased the basal PTHLH promoter activity by 2-fold or less. In contrast, the three AP-1 binding sites in wt(12)x3 strongly stimulated the PTHLH-reporter activity by more than 10-fold in both ATDC5 and C28/I2 cells, whereas the der(8) BPx3 showed a reduction of the AP-1-mediated PTHLH transactivation by 5-fold. This finding indicates that the additive effect of the three AP-1 binding sites in the der(8) BPx3-PTHLH-Luc construct was inhibited by the presence of the EBS (Fig. 6B) and suggests that, while AP-1 had a positive potential to transactivate PTHLH, C-ets-1 was negatively modulating AP-1-driven PTHLH transcription.
Figure 6.
PTHLH promoter characterization and PTHLH reporter gene assays in chondrocyte cell lines. (A) 5′ RACE PCR of PTHLH-transcripts in ATDC5 and C28/I2 cells. Two TATA boxes (P1, P3), one GC-rich promoter (P2), the UCSC mammalian conservation with approximate genomic kilobases distances and the PTHLH exons 1–5 (ex.) are shown. Identical chondrogenic transcription start site (TSS, bp 28014179) were revealed in ATDC5 and C28/I2 cells downstream of GC-rich promoter (bottom). Two further PTHLH transcripts start at bp 28016183 (see NCBI RefSeq). The red box represents 3080 bp of the PTHLH promoter used to generate the reporter construct. (B) PTHLH-luciferase reporter gene assays with EBS (C-ets-1 core consensus sequence), wt(12) (AP-1 motif), der(8) BP (C-ets-1 and AP-1 motif) or a three time repeats (×3) inserted upstream of the 3080 bp-PTHLH promoter (scheme right). Only the wt(12) × 3 permitted strong stimulation of PTHLH promoter activity and the presence of the C-ets-1 motifs in der(8) BP × 3 prevented activation by endogenous factors (n = 3, **P < 0.01, *P < 0.05).
DISCUSSION
There are several important findings from this research. First, our report is the first to implicate PTHLH in a genetic human disease, namely isolated BDE. Second, we present evidence that altered PTHLH modulation is associated with ADAMTS-7 and ADAMTS-12 downregulation in chondrogenically induced BDE fibroblasts. Finally, we show insight into epigenetic dysregulation by AP-1 and C-ets-1 as a result of the translocation. We believe that our findings shed novel insight into the function of PTHLH and resultant disease through unexpected, unconventional mechanisms.
Position effects are known to cause disease. For instance, chromosomal rearrangements cause disturbances of chromosome structure and long-range gene regulation. Decoupling, deletions and duplications of regulatory elements, chromatin alterations or subsequent processes can cause disease (32,33). The SOX9 and the BMP2 loci provide excellent examples for long-range regulation of the gene in development and disease and the importance of CRE for a strong tissue-specific expression (34,35). Chromosomal BPs up- and downstream of SOX9 cause campomelic dysplasia, whereas different aberrations in complex regulatory regions in non-coding elements >1 Mb on both sides of SOX9 are associated with cleft palates (32,36). Duplications of a ∼2 Mb interval 5-prime of SOX9 were found in patients with brachydactyly anonchia (37). Several kilobases duplicated downstream of BMP2 cause brachydactyly type A2 (35). Another example is the uncoupling of CRE due to an inversion upstream of Shh resulting in dysregulation with further influences on Ihh and Pthlh expression and brachydactyly in Dsh/Dsh mice (38). Pthrp over-expressing, as well as Pthlh knockout, mice are small and have short extremities (39–42). These phenotypes have brachydactyly similarities; however, the direct link of PTHrP to a human phenotype has not been elucidated. Aside from Gli transcription factors, the transcription factors controlling PTHLH in chondrogenesis are unknown (43). AP-1 or C-ets-1 are known to regulate PTHLH-expression in T-lymphocytes, breast cancer cells and prostate cancer cells (31,44). Members of the AP-1 transcription factor family also regulate cartilage and bone development (45–48). Furthermore, prechondrogenic nuclear extracts of differentiated dermal fibroblasts show increased binding to AP-1 motifs in CRE (49).
Our studies implicated malfunction in PTHLH. A strong and finely tuned cell- and tissue-specific, developmental regulation is essential so that PTHrP can carry out its diverse biological roles. During chondrogenesis, IHH and PTHrP interact through a negative feedback loop. Disturbances in PTHLH expression or the IHH feedback loop lead to disruption of the sensitive balance between proliferating and hypertrophic chondrocytes in the growth plate (42,50,51). We determined the translocation-mediated direct downregulation of PTHLH and its correlation with the reduced expression of its target genes ADAMTS-7 and ADAMTS-12 in chondrogenically induced BDE fibroblasts. ADAMTS-7 negatively regulates chondrogenic differentiation (23). Inhibited metalloproteinases repress COL2A1 and COL10A1 expression in chondrogenic differentiated mesenchymal stromal cells (52,53). Our observations indicated that downregulation of PTHLH, ADAMTS-7 and ADAMTS-12 could alter further downstream chondrogenic regulators, such as COL10A1 and IHH, and thereby lead to delayed cartilage formation. The differential expression of chondrogenic markers Aggrecan, COL2A1, COL10A1 and IHH in chondrogenically differentiated non-affected fibroblast cultures, compared with BDE cultures, suggests that this disturbance may delay the normal differentiation mechanisms in BDE chondrocytes. The altered chondrogenic fibroblast differentiation could be the clue in the elucidation of dysregulated chondrogenesis leading to the abnormalities in BDE.
We provide strong evidence that AP-1 and C-ets-1 are positive and negative PTHLH regulators in BDE chondrogenically differentiated cells. The der(8) BP upstream of PTHLH generates a new binding site that binds C-ets-1 in addition to AP-1 and inhibits PTHLH promoter activity in these cells. The data are consistent with increased pre-chondrogenic AP-1 binding to the wild-type non-translocated sequence, which activates PTHLH promoter activity, and the known capacity of AP-1 to regulate the maturation of chondrocytes. Moreover c-fos and Pthrp knockout mice show distinct similarities in endochondral growth plates (39,47). AP-1 activity is necessary but unchanged during chondrogenic differentiation of embryonic mesenchymal cells (54). The AP-1 repeated sequence motif as showed in reporter gene assays illustrates the potential to activate the PTHLH transcription. Thus, AP-1 and C-ets-1 bind at der(8) BP, but do not synergistically trans-activate the PTHLH, as it was described elsewhere for the human urokinase-type plasminogen activator, the tissue inhibitor of metalloproteinase-1 promoters and the granulocyte-macrophage colony stimulating factor (55–57). C-ets-1 seems to regulate the strong AP-1-directed PTHLH activation in a negative manner and C-ets-1 was found to activate PTHLH expression in tumorigenic cells exclusively (31). However, ETS1 in its diverse roles in biological control and cancer can either induce or prevent apoptosis (58–60), which highlights the importance of cellular- and sequence-specific context in regulating the outcomes and interactions between different transcription factors. Our data suggest that AP-1-directed PTHLH expression are silenced by C-ets-1 in BDE patients during chondrogenesis, altering chondrocyte proliferation and differentiation and leading to abnormal ossification and length reduction in affected bones. Altered PTHrP-directed expression of its targets could potentially affects chondrocyte differentiation resulting in malformations or delayed cartilage formations and alterations in the ECM produced by hypertrophic chondrocytes (53). Since AP-1 and C-ets-1 protein interactions are also known (61–63), we propose that the distance between the AP-1 and C-ets-1 binding sites at der(8) BP (2 nt) is either too short for the described cooperative activation, or the AP-1-directed PTHLH promoter activity is inhibited directly due to binding of C-ets-1 at the bidirectional EBS core consensus sequence in a competitive manner. The genomic orientation, the bidirectional EBS motif, the chondrogenic tissue or the insufficient core consensus sequence (GGAA/T) may support a configuration that does not favor co-transcriptional activation.
We also analyzed the chromatin status of BDE fibroblasts. We found that both alleles have distinct histone modifications at wt(12) and der(8) BP that predict regulatory activity and show epigenetic modifications when chromosomal translocations occur. High levels of H3K4me1 and low levels of H3K4me3 were found as characteristic in regulating and possibly enhancing histone modifications in HeLa cells (13). Furthermore, H3K4me1 and H3K4me3 were enriched at chromosomal BPs associated with lymphomas and leukemias (64). The CRE-characteristic histone modifications, highly enriched at der(8) BP, force the AP-1 and C-ets-1 binding compared with the wt(12) allele and confirm that the der(8) BP is a cis-acting regulator that plays a major role in PTHLH regulation due to the translocation.
On the basis of the sequence binding motifs for TSN and TOPII and the unique chromatin signature at wt(12) and der(8), we propose that the BP region is a conserved euchromatin and putative recombination hotspot. Chromatin organization and the involved TOPII play a major role in generating translocations (65,66). The TSN relevance in chromatin signatures and organization should be a topic of future studies. We also demonstrated that AP-1 and C-ets-1 are novel PTHLH transcriptional regulators during chondrogenesis and propose that either the sequence at der(8) BP or, further upstream, the highly conserved sequence with AP-1 and C-ets-1 binding motifs regulates PTHLH promoter activity under single or combinatorial conditions. We suggest that the downregulated PTHLH, ADAMTS-7 and ADAMTS-12 chondrocyte expression that we demonstrated in chondrogenically induced BDE fibroblasts is the responsible mechanism for BDE in our patients in vivo. Due to the translocation, PTHLH is negatively regulated and its altered regulation correlates with both impaired chondrogenesis and also abnormal expression of PTHrP downstream targets. This state-of-affairs results in shortened extremities and clinically apparent BDE. The mechanism coincides with suggested interpretations and confirmed data explaining the regulatory processes during chondrogenesis (50,67–70).
We have highlighted the importance of characterizing intergenic non-coding sequences, particularly in the background of chromosomal aberrations. We have shown that alterations in conserved elements with transcription factor binding sites (TFBS) cause positional effects and result in alterations in phenotype. We believe that our report provides an example of how these issues can be approached. However, the long-range control of gene regulation must be investigated and verified further with high-throughput methods to determine transcription factor interactions, as well as epigenetic and chromatin modifications.
MATERIALS AND METHODS
Patients
Three affected (III:2; IV:2; V:1) and two non-affected (III:3; IV:3) individuals participated in the study. Skin fibroblasts from one affected (IV:2) and two non-affected (III:3; IV:3) were obtained for research. Familial history, physical examination, growth measurements, chemistries and roentgenograms were performed for all five individuals. The institutional ethics committees of Charité Universitätsmedizin Berlin and Faculty of Medicine Carl Gustav Carus at the Technical University Dresden approved the study and written, informed consent was obtained.
Tissue culture, histochemistry, FISH, in situ and whole-mount hybridization
Lymphoblastoid cell lines (LCL) were generated by EBV immortalization and cultured in RPMI medium 1640 with 20% FCS, 100 U/ml penicillin and 100 µg/ml streptomycin. Fibroblasts were cultured in Eagle's MEM 199 with 10% FCS, 100 U/ml penicillin and 100 µg/ml streptomycin, C28/I2 and ATDC5 cells were grown in DMEM/Hanks F12 (1:1), supplemented with 10 and 5% FCS, respectively. The chondrogenic differentiation of fibroblasts was done in pellet culture with DMEM, supplemented with 10% FCS, 100 U/ml penicillin, 100 µg/ml streptomycin, 1× ITS+ (Invitrogen), 10 ng/ml hTGF-β1 (R&D Systems), 500 ng/ml rhIGF-1 (R&D Systems), 50 µm scorbate-2-phosphate (Sigma) and l-glutamine (PAA), for 14–28 days. For ChIP experiments fibroblasts (14 days) were stimulated further for 48 h with PMA (1:1000, Sigma). Fibroblasts were fixed for 10 min with paraformaldehyde (Sigma), were stained in 1% Alcian blue (Chroma, 3% acetic acid) for 30 min or 6% Safranin O (Sigma, dH2O) for 2 min. Prior to microscopic documentation the cells were washed twice with 90% ethanol. Metaphase FISH analysis was performed with LCL using standard protocols. BACs for FITC and TMR labeling were ordered at RZPD (www.imagenes-bio.de). In situ hybridizations were done using digoxygenin-UTP-labeled murine Kcnb2 riboprobe (chromosome 1: bp 15704673–15705436 UCSC Genome browser March 2006) according to standard protocols. KCNB2 qRT-PCR, fw-primer GCTAATTGGCGGTTGTCATT, rv-primer CGAGAGCGAGAAGGAGAAGA. For conditions refer to qRT-PCR section.
Pulse field gelelectrophoresis and Southern blotting
A total of 4 × 107 LCL cells were harvested by routine methods and mixed with 2% Agarose and poured in plug molds (BioRad). After polymerization, the cells were lysed by incubation in 100 mm EDTA pH 8.0, 1% N-lauroylsarcosine, 1 mg/ml proteinase K two times for 24 h at 50°C, 1 mm PMSF was added and agarose blocks were stored in 50 mm EDTA pH 8.0, 20 mm Tris pH 8.0 at 4°C. One third of each agarose block was used for overnight endonuclease digestion (60 U enzyme) with SfiI, PmeI, PstI, SacI, EcoRI or BclI, respectively. Fragments between 4 and 500 kb were separated by pulsed-field electrophoresis (22 h with 6 V/cm, angle 120°, pulse 6–60 s at 14°C; 1% agarose) in 0.5% TBE buffer by CHEF MAPPER™ (BioRad). Southern blotting was performed with hybridization buffer, Perfect Hyb™ Plus (Sigma, primers for probes in Supplementary Material, Table S1).
Amplification and sequence analysis
Four primer pairs were designed to amplify der(8) BP, der(12) BP, wt(8) and wt(12) at BP positions (Supplementary Material, Table S2). The two wild-type forward-orientated primers amplified der(8) BP and the two wild-type reverse primers amplified der(12) BP. Microsatellite markers for the HOXD13 locus were PCR-amplified and analyzed on 3130xl Genetic Analyzer (Applied Biosystems) using Gene Mapper® Software Version 4.0 (Supplementary Material, Table S7). HOXD13 PCR-amplified products (Supplementary Material, Table S8) were sequenced using the Big Dye® Terminator Cycle Sequencing Kit v1.1 (Applied Biosystems) and analyzed on 3130xl Genetic Analyzer. SeqMan software (Lasergene Version 7.0; DNAStar) was used to evaluate the traces.
Bioinformatics
We identified conserved TFBS on derivative chromosomes by using rVISTA. Murine homologous sequences were compared with human sequences. Regulatory sequences from Transfac Professional V10.2 and rVISTA results were further analyzed using the UCSC Genome Browser (March, 2006) with 70% conservation through mammals set as the threshold. Microduplication on der(12) BP were identified by NCBI BLAST and UCSC BLAT.
Electromobility shift assays
Double-stranded oligonucleotides (Supplementary Material, Table S3) were labeled with 3′-biotin-11-dUTP according to manufacturer's instructions (Pierce). For EMSA, we preincubated 150 ng of recombinant c-Jun (Promega) for 15 min at room temperature in 10× binding buffer (LightShift Chemiluminescent EMSA Kit, Pierce), to which were added 200 ng poly (dI-dC), 0.05% NP40, 10 mm MgCl2 and 10 mm DTT. About 175 ng C-ets-1 (RayBiotech) was preincubated in 20 mm Hepes pH 7.9, 60 mm KCl, 200 ng poly (dI-dC), 8% Ficoll 400, 1.5 µg BSA, 1 µg phenolyzed, sonicated salmon sperm DNA and 10 mm DTT. The total reaction volume was 20 µl containing 12.5 fmol/µl of the labeled probe and the loading buffer was 8% Ficoll 400. For competition, 400-fold molar excess of unlabelled oligos were added and incubation was continued for an additional 30 min. We separated free probes from protein-bound DNA complexes on a native 5% acrylamide gel in 1× TBE buffer. After transferring the samples to a positively charged nylon membrane (Amersham), signals were detected by autoradiography following the manufacturer's instructions (Pierce).
Chromatin immunoprecipitation
ChIP assays were performed according a previously described protocol (71). Anti- H3K4me1, -H3K4me3, -c-Jun and -C-ets-1 ChIP-tested antibodies were obtained from Abcam. QPCR was done using SYBR Green (Eurogentech) on an ABI Prism® 7700 (Applied Biosystems). Each reaction contained 1 µl of precipitated chromatin, 0.75 µl of each primer (10 pmol/µl) and 12.5 µl of SYBR Green master mix. Products were amplified for 1 cycle at 95°C, 45 cycles at 95°C for 30 s, 60°C for 1 min and 72°C for 30 s. For each sample Ct was normalized to the Ct of the input sample (10%). The relative enrichment was determined by 2−(Ct1–Ct2). Primers for negative and positive controls and specific amplifications in QPCR are found in Supplementary Material, Table S4.
5′ RACE PCR and qRT-PCR
RNA was prepared from ATDC5 and C28-I2 cells using Trizol® Reagent (Invitrogen) and reverse transcribed using Superscript™ III RT (Invitrogen). The GeneRacer® Kit (Invitrogen) was used for 5′ RACE PCR. PCR fragments were subcloned (TOPO TA Cloning® Kit; Invitrogen) and subsequently sequenced. UCSC BLAT was used to map the sequenced transcripts. qRT-PCR was performed on RNA isolated with Trizol® (Invitrogen) from fibroblasts, using primer sequences shown in Supplementary Material, Table S5. B-Actin amplification served for normalization. Expression was quantified applying the ΔΔCt method.
Transfection
Transfections were performed using 500 ng each of EBS, EBSx3-, wt(12)-, wt(12)x3-, der(8) BP- and der(8) BPx3-PTHLH-promoter-luciferase constructs (oligo sequences in Supplementary Material, Table S6). The vector backbone was pGL2-basic (Promega). Twenty nanogram of pRL-TK (Promega) were added to control transfection efficiency. A total of 8 x 104 ATDC5 and 1 × 105 C28/I2 cells were seeded 12–24 h prior transfection with Lipofectamine and Plus reagent (Invitrogen) in 24 well plates. Cells were transfected at 70% confluency for 48 h, lysed in Passive Lysis buffer (Promega) and analyzed using the Dual-Luciferase® Reporter Assay System (Promega) in a Berthold luminometer.
Statistics
Functional analysis by EMSA, reporter assays, ChIP and qRT-PCR were reproduced at least three times. Numbers (n) of experiments are mentioned in figure legends. Significance was determined by Student's t-test (**P < 0.01, *P < 0.05).
WEB RESOURCES
http://www.ncbi.nlm.nig.gov/Omim; http://rvista.dcode.org; http://www.gene-regulation.com/pub/databases.html; http://genome.ucsc.edu; www.imagenes-bio.de.
ACCESSION CODES
PTHLH NM_198965, Pthlh NM_008970, Kcnb2 XM_001473804, IHH NM_002181, COL2A1 NM_001844, COL10A1 NM_000493, GAPDH NM_002046, ALDOA NM_000034, ACTG1 NM_001614, SAT2 NM_133491, c-JUN NM_002228, ETS1 NM_005238, ADAMTS–7 NM_014272.3, ADAMTS–12 NM_030955.2, HOXD13 NM_000523.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
FUNDING
This work was supported by Deutsche Forschungsgemeinschaft (DFG) (grant number BA1773/5-1 to S.B.). P.G.M. was supported by a Max-Delbrück-Center scholarship.
ACKNOWLEDGEMENT
Sincere thanks are given to all family members for their participation in this study. We thank I. Nitz, A. Kobelt and K. Hoffmann for the detailed dysmorphological description of the phenotype. We are grateful to M.B. Goldring's staff for their advice and for providing the cell lines. We thank E. Klein, A. Mühl and Y. Neuenfeld for expert technical assistance.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Bell J. Brachydactyly and symphalangism. In: Penrose L.S., editor. The Treasury of Human Inheritance. Vol. 5. University of Cambridge Press, Cambridge: Hereditary digital anomalies; 1951. pp. 1–31. [Google Scholar]
- 2.Schuster H., Wienker T.E., Bähring S., Bilginturan N., Toka H.R., Neitzel H., Jeschke E., Toka O., Gilbert D., Lowe A., et al. Severe autosomal dominant hypertension and brachydactyly in a unique Turkish kindred maps to human chromosome 12. Nat. Genet. 1996;13:98–100. doi: 10.1038/ng0596-98. [DOI] [PubMed] [Google Scholar]
- 3.Schuster H., Wienker T.F., Toka H.R., Bähring S., Jeschke E., Toka O., Busjahn A., Hempel A., Tahlhammer C., Oelkers W., et al. Autosomal dominant hypertension and brachydactyly in a Turkish kindred resembles essential hypertension. Hypertension. 1996;28:1085–1092. doi: 10.1161/01.hyp.28.6.1085. [DOI] [PubMed] [Google Scholar]
- 4.Schwabe G.C., Mundlos S. Genetics of congenital hand anomalies. Handchir. Mikrochir. Plast. Chir. 2004;36:85–97. doi: 10.1055/s-2004-817884. [DOI] [PubMed] [Google Scholar]
- 5.Johnson D., Kan S.H., Oldridge M., Trembath R.C., Roche P., Esnouf R.M., Giele H., Wilkie A.O. Missense mutations in the homeodomain of HOXD13 are associated with brachydactyly types D and E. Am. J. Hum. Genet. 2003;72:984–997. doi: 10.1086/374721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gupta G.D., Makde R.D., Kamdar R.P., D'Souza J.S., Kulkarni M.G., Kumar V., Rao B.J. Co-expressed recombinant human Translin-Trax complex binds DNA. FEBS Lett. 2005;579:3141–3146. doi: 10.1016/j.febslet.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 7.Aoki K., Suzuki K., Sugano T., Tasaka T., Nakahara K., Kuge O., Omori A., Kasai M. A novel gene, Translin, encodes a recombination hotspot binding protein associated with chromosomal translocations. Nat. Genet. 1995;10:167–174. doi: 10.1038/ng0695-167. [DOI] [PubMed] [Google Scholar]
- 8.Gajecka M., Pavlicek A., Glotzbach C.D., Ballif B.C., Jarmuz M., Jurka J., Shaffer L.G. Identification of sequence motifs at the breakpoint junctions in three t(1;9)(p36.3;q34) and delineation of mechanisms involved in generating balanced translocations. Hum. Genet. 2006;120:519–526. doi: 10.1007/s00439-006-0222-1. [DOI] [PubMed] [Google Scholar]
- 9.Rose D., Thomas W., Holm C. Segregation of recombined chromosomes in meiosis I requires DNA topoisomerase II. Cell. 1990;60:1009–1017. doi: 10.1016/0092-8674(90)90349-j. [DOI] [PubMed] [Google Scholar]
- 10.Obata K., Hiraga H., Nojima T., Yoshida M.C., Abe S. Molecular characterization of the genomic breakpoint junction in a t(11;22) translocation in Ewing sarcoma. Genes Chromosomes Cancer. 1999;25:6–15. [PubMed] [Google Scholar]
- 11.Lovett B.D., Lo Nigro L., Rappaport E.F., Blair I.A., Osheroff N., Zheng N., Megonigal M.D., Williams W.R., Nowell P.C., Felix C.A. Near-precise interchromosomal recombination and functional DNA topoisomerase II cleavage sites at MLL and AF-4 genomic breakpoints in treatment-related acute lymphoblastic leukemia with t(4;11) translocation. Proc. Natl. Acad. Sci. USA. 2001;98:9802–9807. doi: 10.1073/pnas.171309898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wei Y., Sun M., Nilsson G., Dwight T., Xie Y., Wang J., Hou Y., Larsson O., Larsson C., Zhu X. Characteristic sequence motifs located at the genomic breakpoints of the translocation t(X;18) in synovial sarcomas. Oncogene. 2003;22:2215–2222. doi: 10.1038/sj.onc.1206343. [DOI] [PubMed] [Google Scholar]
- 13.Heintzman N.D., Stuart R.K., Hon G., Fu Y., Ching C.W., Hawkins R.D., Barrera L.O., Van Calcar S., Qu C., Ching K.A., et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 14.Basuyaux J.P., Ferreira E., Stehelin D., Buttice G. The Ets transcription factors interact with each other and with the c-Fos/c-Jun complex via distinct protein domains in a DNA-dependent and -independent manner. J. Biol. Chem. 1997;272:26188–26195. doi: 10.1074/jbc.272.42.26188. [DOI] [PubMed] [Google Scholar]
- 15.Wasylyk B., Wasylyk C., Flores P., Begue A., Leprince D., Stehelin D. The c-ets proto-oncogenes encode transcription factors that cooperate with c-Fos and c-Jun for transcriptional activation. Nature. 1990;346:191–193. doi: 10.1038/346191a0. [DOI] [PubMed] [Google Scholar]
- 16.Hwang P.M., Glatt C.E., Bredt D.S., Yellen G., Snyder S.H. A novel K+ channel with unique localizations in mammalian brain: molecular cloning and characterization. Neuron. 1992;8:473–481. doi: 10.1016/0896-6273(92)90275-i. [DOI] [PubMed] [Google Scholar]
- 17.Gravagna N.G., Knoeckel C.S., Taylor A.D., Hultgren B.A., Ribera A.B. Localization of Kv2.2 protein in Xenopus laevis embryos and tadpoles. J. Comp. Neurol. 2008;510:508–524. doi: 10.1002/cne.21804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnston J., Griffin S.J., Baker C., Skrzypiec A., Chernova T., Forsythe I.D. Initial segment Kv2.2 channels mediate a slow delayed rectifier and maintain high frequency action potential firing in medial nucleus of the trapezoid body neurons. J. Physiol. 2008;586:3493–3509. doi: 10.1113/jphysiol.2008.153734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mizuno S., Glowacki J. Low oxygen tension enhances chondroinduction by demineralized bone matrix in human dermal fibroblasts in vitro. Cells Tissues Organs. 2005;180:151–158. doi: 10.1159/000088243. [DOI] [PubMed] [Google Scholar]
- 20.Hashimoto J., Kariya Y., Miyazaki K. Regulation of proliferation and chondrogenic differentiation of human mesenchymal stem cells by laminin-5 (laminin-332) Stem Cells. 2006;24:2346–2354. doi: 10.1634/stemcells.2005-0605. [DOI] [PubMed] [Google Scholar]
- 21.Chen F.G., Zhang W.J., Bi D., Liu W., Wei X., Chen F.F., Zhu L., Cui L., Cao Y. Clonal analysis of nestin(−) vimentin(+) multipotent fibroblasts isolated from human dermis. J. Cell. Sci. 2007;120:2875–2883. doi: 10.1242/jcs.03478. [DOI] [PubMed] [Google Scholar]
- 22.Scharstuhl A., Schewe B., Benz K., Gaissmaier C., Buhring H.J., Stoop R. Chondrogenic potential of human adult mesenchymal stem cells is independent of age or osteoarthritis etiology. Stem Cells. 2007;25:3244–3251. doi: 10.1634/stemcells.2007-0300. [DOI] [PubMed] [Google Scholar]
- 23.Bai X.H., Wang D.W., Kong L., Zhang Y., Luan Y., Kobayashi T., Kronenberg H.M., Yu X.P., Liu C.J. ADAMTS-7, a direct target of PTHrP, adversely regulates endochondral bone growth by associating with and inactivating GEP growth factor. Mol. Cell. Biol. 2009;29:4201–4219. doi: 10.1128/MCB.00056-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bai X.H., Wang D.W., Luan Y., Yu X.P., Liu C.J. Regulation of chondrocyte differentiation by ADAMTS-12 metalloproteinase depends on its enzymatic activity. Cell. Mol. Life Sci. 2009;66:667–680. doi: 10.1007/s00018-008-8633-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shukunami C., Ishizeki K., Atsumi T., Ohta Y., Suzuki F., Hiraki Y. Cellular hypertrophy and calcification of embryonal carcinoma-derived chondrogenic cell line ATDC5 in vitro. J. Bone Miner. Res. 1997;12:1174–1188. doi: 10.1359/jbmr.1997.12.8.1174. [DOI] [PubMed] [Google Scholar]
- 26.Finger F., Schorle C., Zien A., Gebhard P., Goldring M.B., Aigner T. Molecular phenotyping of human chondrocyte cell lines T/C-28a2, T/C-28a4, and C-28/I2. Arthritis Rheum. 2003;48:3395–3403. doi: 10.1002/art.11341. [DOI] [PubMed] [Google Scholar]
- 27.Mangin M., Ikeda K., Dreyer B.E., Broadus A.E. Identification of an up-stream promoter of the human parathyroid hormone-related peptide gene. Mol. Endocrinol. 1990;4:851–858. doi: 10.1210/mend-4-6-851. [DOI] [PubMed] [Google Scholar]
- 28.Vasavada R.C., Wysolmerski J.J., Broadus A.E., Philbrick W.M. Identification and characterization of a GC-rich promoter of the human parathyroid hormone-related peptide gene. Mol. Endocrinol. 1993;7:273–282. doi: 10.1210/mend.7.2.8469240. [DOI] [PubMed] [Google Scholar]
- 29.Richard V., Luchin A., Brena R.M., Plass C., Rosol T.J. Quantitative evaluation of alternative promoter usage and 3' splice variants for parathyroid hormone-related protein by real-time reverse transcription-PCR. Clin. Chem. 2003;49:1398–1402. doi: 10.1373/49.8.1398. [DOI] [PubMed] [Google Scholar]
- 30.Richard V., Nadella M.V., Green P.L., Lairmore M.D., Feuer G., Foley J.G., Rosol T.J. Transcriptional regulation of parathyroid hormone-related protein promoter P3 by ETS-1 in adult T-cell leukemia/lymphoma. Leukemia. 2005;19:1175–1183. doi: 10.1038/sj.leu.2403787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hamzaoui H., Rizk-Rabin M., Gordon J., Offutt C., Bertherat J., Bouizar Z. PTHrP P3 promoter activity in breast cancer cell lines: role of Ets1 and CBP (CREB binding protein) Mol. Cell. Endocrinol. 2007;268:75–84. doi: 10.1016/j.mce.2007.01.014. [DOI] [PubMed] [Google Scholar]
- 32.Velagaleti G.V., Bien-Willner G.A., Northup J.K., Lockhart L.H., Hawkins J.C., Jalal S.M., Withers M., Lupski J.R., Stankiewicz P. Position effects due to chromosome breakpoints that map approximately 900 Kb upstream and approximately 1.3 Mb downstream of SOX9 in two patients with campomelic dysplasia. Am. J. Hum. Genet. 2005;76:652–662. doi: 10.1086/429252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kleinjan D.A., van Heyningen V. Long-range control of gene expression: emerging mechanisms and disruption in disease. Am. J. Hum. Genet. 2005;76:8–32. doi: 10.1086/426833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gordon C.T., Tan T.Y., Benko S., Fitzpatrick D., Lyonnet S., Farlie P.G. Long-range regulation at the SOX9 locus in development and disease. J. Med. Genet. 2009;46:649–656. doi: 10.1136/jmg.2009.068361. [DOI] [PubMed] [Google Scholar]
- 35.Dathe K., Kjaer K.W., Brehm A., Meinecke P., Nürnberg P., Neto J.C., Brunoni D., Tommerup N., Ott C.E., Klopocki E., et al. Duplications involving a conserved regulatory element downstream of BMP2 are associated with brachydactyly type A2. Am. J. Hum. Genet. 2009;84:483–492. doi: 10.1016/j.ajhg.2009.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Benko S., Fantes J.A., Amiel J., Kleinjan D.J., Thomas S., Ramsay J., Jamshidi N., Essafi A., Heaney S., Gordon C.T., et al. Highly conserved non-coding elements on either side of SOX9 associated with Pierre Robin sequence. Nat. Genet. 2009;41:359–364. doi: 10.1038/ng.329. [DOI] [PubMed] [Google Scholar]
- 37.Kurth I., Klopocki E., Stricker S., van Oosterwijk J., Vanek S., Altmann J., Santos H.G., van Harssel J.J., de Ravel T., Wilkie A.O., et al. Duplications of noncoding elements 5' of SOX9 are associated with brachydactyly-anonychia. Nat. Genet. 2009;41:862–863. doi: 10.1038/ng0809-862. [DOI] [PubMed] [Google Scholar]
- 38.Niedermaier M., Schwabe G.C., Fees S., Helmrich A., Brieske N., Seemann P., Hecht J., Seitz V., Stricker S., Leschik G., et al. An inversion involving the mouse Shh locus results in brachydactyly through dysregulation of Shh expression. J. Clin. Invest. 2005;115:900–909. doi: 10.1172/JCI200523675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Karaplis A.C., Luz A., Glowacki J., Bronson R.T., Tybulewicz V.L., Kronenberg H.M., Mulligan R.C. Lethal skeletal dysplasia from targeted disruption of the parathyroid hormone-related peptide gene. Genes Dev. 1994;8:277–289. doi: 10.1101/gad.8.3.277. [DOI] [PubMed] [Google Scholar]
- 40.Lanske B., Karaplis A.C., Lee K., Luz A., Vortkamp A., Pirro A., Karperien M., Defize L.H., Ho C., Mulligan R.C., et al. PTH/PTHrP receptor in early development and Indian hedgehog-regulated bone growth. Science. 1996;273:663–666. doi: 10.1126/science.273.5275.663. [DOI] [PubMed] [Google Scholar]
- 41.Kovacs C.S., Chafe L.L., Fudge N.J., Friel J.K., Manley N.R. PTH regulates fetal blood calcium and skeletal mineralization independently of PTHrP. Endocrinology. 2001;142:4983–4993. doi: 10.1210/endo.142.11.8509. [DOI] [PubMed] [Google Scholar]
- 42.Weir E.C., Philbrick W.M., Amling M., Neff L.A., Baron R., Broadus A.E. Targeted overexpression of parathyroid hormone-related peptide in chondrocytes causes chondrodysplasia and delayed endochondral bone formation. Proc. Natl. Acad. Sci. USA. 1996;93:10240–10245. doi: 10.1073/pnas.93.19.10240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mau E., Whetstone H., Yu C., Hopyan S., Wunder J.S., Alman B.A. PTHrP regulates growth plate chondrocyte differentiation and proliferation in a Gli3 dependent manner utilizing hedgehog ligand dependent and independent mechanisms. Dev. Biol. 2007;305:28–39. doi: 10.1016/j.ydbio.2007.01.031. [DOI] [PubMed] [Google Scholar]
- 44.Asadi F., Kukreja S. Parathyroid hormone-related protein in prostate cancer. Crit. Rev. Eukaryot. Gene Expr. 2005;15:15–28. doi: 10.1615/critreveukaryotgeneexpr.v15.i1.20. [DOI] [PubMed] [Google Scholar]
- 45.Kameda T., Watanabe H., Iba H. C-Jun and JunD suppress maturation of chondrocytes. Cell Growth Differ. 1997;8:495–503. [PubMed] [Google Scholar]
- 46.Thomas D.P., Sunters A., Gentry A., Grigoriadis A.E. Inhibition of chondrocyte differentiation in vitro by constitutive and inducible overexpression of the c-fos proto-oncogene. J. Cell. Sci. 2000;113:439–450. doi: 10.1242/jcs.113.3.439. [DOI] [PubMed] [Google Scholar]
- 47.Watanabe H., Saitoh K., Kameda T., Murakami M., Niikura Y., Okazaki S., Morishita Y., Mori S., Yokouchi Y., Kuroiwa A., et al. Chondrocytes as a specific target of ectopic Fos expression in early development. Proc. Natl. Acad. Sci. USA. 1997;94:3994–3999. doi: 10.1073/pnas.94.8.3994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Karreth F., Hoebertz A., Scheuch H., Eferl R., Wagner E.F. The AP1 transcription factor Fra2 is required for efficient cartilage development. Development. 2004;131:5717–5725. doi: 10.1242/dev.01414. [DOI] [PubMed] [Google Scholar]
- 49.Yates K.E. Identification of cis and trans-acting transcriptional regulators in chondroinduced fibroblasts from the pre-phenotypic gene expression profile. Gene. 2006;377:77–87. doi: 10.1016/j.gene.2006.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vortkamp A., Lee K., Lanske B., Segre G.V., Kronenberg H.M., Tabin C.J. Regulation of rate of cartilage differentiation by Indian hedgehog and PTH-related protein. Science. 1996;273:613–622. doi: 10.1126/science.273.5275.613. [DOI] [PubMed] [Google Scholar]
- 51.Gao B., Hu J., Stricker S., Cheung M., Ma G., Law K.F., Witte F., Briscoe J., Mundlos S., He L., et al. A mutation in Ihh that causes digit abnormalities alters its signalling capacity and range. Nature. 2009;458:1196–2000. doi: 10.1038/nature07862. [DOI] [PubMed] [Google Scholar]
- 52.Bertram H., Boeuf S., Wachters J., Boehmer S., Heisel C., Hofmann M.W., Piecha D., Richter W. Matrix metalloprotease inhibitors suppress initiation and progression of chondrogenic differentiation of mesenchymal stromal cells in vitro. Stem Cells Dev. 2009;18:881–892. doi: 10.1089/scd.2008.0306. [DOI] [PubMed] [Google Scholar]
- 53.Riemer S., Gebhard S., Beier F., Poschl E., von der Mark K. Role of c-fos in the regulation of type X collagen gene expression by PTH and PTHrP: localization of a PTH/PTHrP-responsive region in the human COL10A1 enhancer. J. Cell. Biochem. 2002;86:688–699. doi: 10.1002/jcb.10260. [DOI] [PubMed] [Google Scholar]
- 54.Seghatoleslami M.R., Tuan R.S. Cell density dependent regulation of AP-1 activity is important for chondrogenic differentiation of C3H10T1/2 mesenchymal cells. J. Cell. Biochem. 2002;84:237–248. doi: 10.1002/jcb.10019. [DOI] [PubMed] [Google Scholar]
- 55.Logan S.K., Garabedian M.J., Campbell C.E., Werb Z. Synergistic transcriptional activation of the tissue inhibitor of metalloproteinases-1 promoter via functional interaction of AP-1 and Ets-1 transcription factors. J. Biol. Chem. 1996;271:774–782. doi: 10.1074/jbc.271.2.774. [DOI] [PubMed] [Google Scholar]
- 56.Cirillo G., Casalino L., Vallone D., Caracciolo A., De Cesare D., Verde P. Role of distinct mitogen-activated protein kinase pathways and cooperation between Ets-2, ATF-2, and Jun family members in human urokinase-type plasminogen activator gene induction by interleukin-1 and tetradecanoyl phorbol acetate. Mol. Cell. Biol. 1999;19:6240–6252. doi: 10.1128/mcb.19.9.6240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thomas R.S., Tymms M.J., McKinlay L.H., Shannon M.F., Seth A., Kola I. ETS1, NFkappaB and AP1 synergistically transactivate the human GM-CSF promoter. Oncogene. 1997;14:2845–2855. doi: 10.1038/sj.onc.1201125. [DOI] [PubMed] [Google Scholar]
- 58.Zhang C., Kavurma M.M., Lai A., Khachigian L.M. Ets-1 protects vascular smooth muscle cells from undergoing apoptosis by activating p21WAF1/Cip1: ETS-1 regulates basal and and inducible p21WAF1/Cip: ETS-1 regulates basal and inducible p21WAF1/Cip1 transcription via distinct cis-acting elements in the p21WAF/Cip1 promoter. J. Biol. Chem. 2003;278:27903–27909. doi: 10.1074/jbc.M304328200. [DOI] [PubMed] [Google Scholar]
- 59.Pei H., Li C., Adereth Y., Hsu T., Watson D.K., Li R. Caspase-1 is a direct target gene of ETS1 and plays a role in ETS1-induced apoptosis. Cancer Res. 2005;65:7205–7213. doi: 10.1158/0008-5472.CAN-04-3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hsu T., Trojanowska M., Watson D.K. Ets proteins in biological control and cancer. J. Cell. Biochem. 2004;91:896–903. doi: 10.1002/jcb.20012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Verger A., Duterque-Coquillaud M. When Ets transcription factors meet their partners. Bioessays. 2002;24:362–370. doi: 10.1002/bies.10068. [DOI] [PubMed] [Google Scholar]
- 62.Dittmer J. The biology of the Ets1 proto-oncogene. Mol. Cancer. 2003;2:29–50. doi: 10.1186/1476-4598-2-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Oikawa T., Yamada T. Molecular biology of the Ets family of transcription factors. Gene. 2003;303:11–34. doi: 10.1016/s0378-1119(02)01156-3. [DOI] [PubMed] [Google Scholar]
- 64.Barski A., Cuddapah S., Cui K., Roh T.Y., Schones D.E., Wang Z., Wei G., Chepelev I., Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 65.Stuardo M., Martinez M., Hidalgo K., Montecino M., Javed A., Lian J.B., Stein G.S., Stein J.L., Gutierrez S.E. Altered chromatin modifications in AML1/RUNX1 breakpoint regions involved in (8;21) translocation. J. Cell. Physiol. 2009;218:343–349. doi: 10.1002/jcp.21599. [DOI] [PubMed] [Google Scholar]
- 66.Zhang Y., Strissel P., Strick R., Chen J., Nucifora G., Le Beau M.M., Larson R.A., Rowley J.D. Genomic DNA breakpoints in AML1/RUNX1 and ETO cluster with topoisomerase II DNA cleavage and DNase I hypersensitive sites in t(8;21) leukemia. Proc. Natl. Acad. Sci. USA. 2002;99:3070–3075. doi: 10.1073/pnas.042702899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kobayashi T., Soegiarto D.W., Yang Y., Lanske B., Schipani E., McMahon A.P., Kronenberg H.M. Indian hedgehog stimulates periarticular chondrocyte differentiation to regulate growth plate length independently of PTHrP. J. Clin. Invest. 2005;115:1734–1742. doi: 10.1172/JCI24397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kronenberg H.M. Developmental regulation of the growth plate. Nature. 2003;423:332–336. doi: 10.1038/nature01657. [DOI] [PubMed] [Google Scholar]
- 69.Kronenberg H.M. PTHrP and skeletal development. Ann. N Y. Acad. Sci. 2006;1068:1–13. doi: 10.1196/annals.1346.002. [DOI] [PubMed] [Google Scholar]
- 70.Goldring M.B., Tsuchimochi K., Ijiri K. The control of chondrogenesis. J. Cell. Biochem. 2006;97:33–44. doi: 10.1002/jcb.20652. [DOI] [PubMed] [Google Scholar]
- 71.Li Z., Van Calcar S., Qu C., Cavenee W.K., Zhang M.Q., Ren B. A global transcriptional regulatory role for c-Myc in Burkitt's lymphoma cells. Proc. Natl. Acad. Sci. USA. 2003;100:8164–8169. doi: 10.1073/pnas.1332764100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Muller M.T., Spitzner J.R., DiDonato J.A., Mehta V.B., Tsutsui K., Tsutsui K. Single-strand DNA cleavages by eukaryotic topoisomerase II. Biochemistry. 1988;27:8369–8379. doi: 10.1021/bi00422a012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.