Abstract
Regulators of G-protein signaling (RGS) proteins are scaffolds that control diverse signaling pathways by modulating signalosome formation and by accelerating the GTPase activity of heterotrimeric G proteins. Although expression of many RGS proteins is relatively low in quiescent cells, transcriptional and post-translational responses to environmental cues regulate both their abundance and activity. We found previously that RGS13, one of the smallest RGS proteins in the family, inhibited cyclic AMP-dependent protein kinase (PKA)-induced gene expression through interactions with the transcription factor cAMP-response element-binding (CREB) protein. Here, we show that PKA activation also leads to increased steady-state RGS13 expression through RGS13 phosphorylation, which inhibits RGS13 protein degradation. RGS13 turnover was significantly reduced in cells stimulated with cAMP, which was reversed by expression of the PKA-specific inhibitory peptide PKI. RGS13 phosphorylation was diminished by mutation of an N-terminal Thr residue (T41) identified as a phosphorylation site by mass spectrometry. Mutation of Thr41 in RGS13 to Ala (T41A) reduced steady-state RGS13 levels and its ability to inhibit M2 muscarinic receptor-mediated Erk phosphorylation compared with wild-type RGS13 by attenuating the protective effect of cAMP on RGS13 degradation. RGS13 underwent ubiquitylation, indicating that it is a likely target of the proteasome. These studies are the first to demonstrate post-translational mechanisms controlling the expression of RGS13. Stabilization of RGS13 through PKA-mediated phosphorylation could enhance RGS13 functions, providing negative feedback regulation that promotes cellular desensitization.
Keywords: RGS proteins, cAMP, protein kinase, phosphorylation
Introduction
G-protein-coupled receptors (GPCR), which comprise the largest family of cell membrane receptors, are activated by ubiquitous physiological ligands including hormones, peptides, neurotransmitters and lipids (Marinissen and Gutkind, 2001). The GPCR signal is transduced by agonist-induced exchange of guanosine diphosphate for guanosine triphosphate (GTP) on the α subunit of its cognate heterotrimeric G protein, whereas signal termination is dependent upon GTP hydrolysis by the Gα subunit (Gilman, 1987). In general, most regulators of G-protein signaling (RGS) proteins inhibit GPCR-mediated signal transduction. Through a signature motif termed the RGS box, they bind GTP-bound Gα and enhance its intrinsic GTPase activity (termed ‘GAP activity’; Hollinger and Hepler, 2002; Willars, 2006; Bansal et al., 2007). More than 30 members of the RGS family have been identified, many of which demonstrate selectivity for various Gα proteins and also contain additional scaffolding domains. RGS proteins function in cell proliferation, cancer progression, opioid receptor signaling, hypertension and immune regulation, among others (Ding et al., 2006; Georgoussi et al., 2006; Bansal et al., 2008b; Cho et al., 2008; James et al., 2009; Liang et al., 2009). GPCR-independent functions of RGS proteins have also been described. For example, our studies showed that RGS13, one of the smallest RGS proteins enriched in mast cells and B cells, suppressed allergic responses in mice by inhibiting IgE receptor-mediated degranulation of mast cells. RGS13 reduced IgE-antigen-dependent phosphoinositide-3 kinase (PI3K) activation, a critical downstream event in the degranulation pathway, by interacting with the PI3K p85α regulatory subunit (Bansal et al., 2008b).
The striking ability of RGS proteins to curb signal transmission by GPCR probably explains their low basal amounts, and numerous studies have documented wide variations in RGS gene transcription in response to environmental cues (Willars, 2006; Cheng et al., 2008; Hu et al., 2008; Stuebe et al., 2008). Post-transcriptional modifications of several RGS proteins of the R4 subfamily, which includes RGS1—5, 8, 13, 16, 18 and 21, have also been demonstrated (Hollinger and Hepler, 2002). Src kinase phosphorylates a conserved Tyr residue in RGS16, which reduces RGS16 turnover (Derrien et al., 2003). GAIP (RGS19) is phosphorylated by Erk kinase, which stimulates its GAP activity on Gαi3 in vitro (Ogier-Denis et al., 1997), while protein kinase A (PKA)-mediated phosphorylation of RGS10 induces RGS10 translocation to the nucleus (Burgon et al., 2001).
We showed recently that RGS13 regulates gene expression independent its GAP activity (Xie et al., 2008). Stimulation of Gαs-coupled GPCR such as the β-adrenergic receptor with hormones like epinephrine and norepinephrine leads to cAMP formation, PKA activation and cAMP-response element-binding (CREB) protein phosphorylation in the nucleus. RGS13 translocated to the nucleus upon PKA activation, where it formed a trimolecular complex with phosphorylated CREB (pCREB) and its co-factor CREB-binding protein (CBP/p300) to inhibit gene transcription. In the course of these investigations, we also discovered that cells expressing active PKA had much higher RGS13 protein concentrations.
In this report, we explored the mechanisms underlying the increased steady-state expression of RGS13 associated with PKA activation. We found that RGS13 undergoes ubiquitylation and proteasomal degradation while cAMP stimulation of cells protected RGS13 from degradation independently of ubiquitylation. Phosphorylation of RGS13(T41A) by PKA was reduced in vitro, which correlated with diminished steady-state expression and function of this mutant in vivo due to increased degradation. These studies are the first to address the post-translational regulation of RGS13, which could have therapeutic implications in the light of its regulatory role in immunity and gene transcription.
Results
PKA activation is associated with increased RGS13 expression
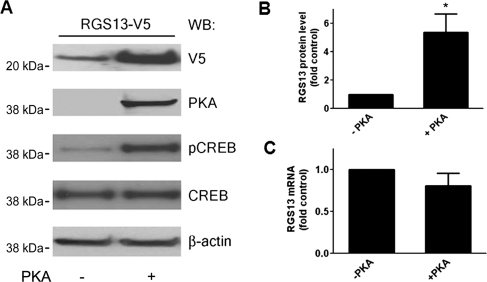
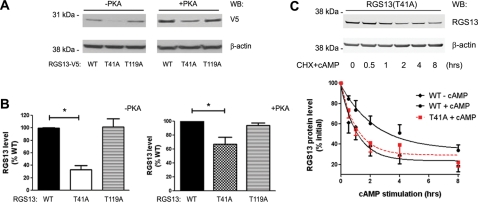
We observed increased steady-state expression of transfected RGS13 in human embryonic kidney (HEK) 293T cells co-expressing a plasmid encoding the catalytic subunit of PKA compared with cells expressing empty vector (Xie et al., 2008; Figure 1A). Cells expressing active PKA had on average five times more RGS13 than control cells under steady-state conditions (Figure 1B). In contrast, the levels of exogenous RGS13 mRNA (driven by a CMV promoter) were not affected by the expression of PKA (Figure 1C). PKA overexpression and activation were demonstrated by immunoblotting with antibodies specific for PKA and pCREB(Ser133), respectively, as CREB is a well-known substrate of active PKA (Mayr and Montminy, 2001) (Figure 1A).
Figure 1.
PKA activation is associated with increased RGS13 protein expression. (A) Increased steady-state levels of RGS13 in cells expressing PKA. HEK293T cells were transfected with pcDNA3.1/RGS13-V5 plasmid together with empty vector or a plasmid encoding the catalytic subunit of PKA. After 24 h, cell lysates were prepared and analyzed by immunoblotting using anti-V5 to detect RGS13 and anti-β-actin to assess protein loading. The membrane was also blotted with anti-PKA or anti-phospho-CREB(Ser133) to assess PKA expression and activity, respectively. (B) Densitometric scanning of RGS13 protein levels (normalized to β-actin) from three independent experiments presented as mean ± SEM of the control, set as ‘1’. *P<0.05, Student's t-test. (C) Co-transfection of PKA does not affect RGS13 mRNA levels. Transfection was performed as in (A). Twenty-four hours later, total RNA was isolated followed by cDNA synthesis and analysis of RGS13 mRNA by real-time qPCR. Data are from three independent experiments and expressed as mean ± SEM.
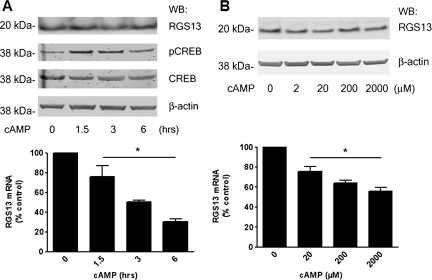
To determine how PKA activation affected expression of endogenous RGS13, we examined LAD2 mast cells exposed to a cell-permeable cAMP analogue (8-pCPT-cAMP). RGS13 mRNA decreased by ∼50% over a 3-h period in cAMP-stimulated cells compared with control and remained low for up to 6 h (Figure 2A, lower panel). Surprisingly, however, RGS13 protein amounts did not vary substantially in cells treated with cAMP during this period (Figure 2A, upper panel). Cells stimulated with cAMP also had increased CREB phosphorylation at Ser133 compared with unstimulated cells, indicating PKA activation (Figure 2A, middle panels). To confirm these results, we stimulated cells with a range of concentrations of 8-pCPT-cAMP for 3 h and measured RGS13 amounts by qPCR. cAMP significantly reduced RGS13 mRNA amounts in a dose-dependent fashion. In contrast, RGS13 protein levels remained unchanged after treatment with all concentrations of cAMP (Figure 2B). These results suggest that activation of the PKA pathway by cAMP has a dual effect on RGS13 expression. In addition reducing RGS13 transcription, the invariant RGS13 protein levels in cAMP-treated cells indicated that cAMP could also regulate RGS13 expression through post-transcriptional and/or translational mechanisms.
Figure 2.
Regulation of endogenous RGS13 expression by the PKA activator cAMP in LAD2 mast cells. (A) Cells were stimulated with 2 mM 8-pCPT-cAMP for the time periods indicated. Cell lysates were prepared and analyzed by immunoblotting with anti-RGS13 antibody (upper panel) and anti-phosphoCREB(Ser133) to assess PKA activation (middle panel). Total RNA was isolated from a portion of the cells, followed by cDNA synthesis and analysis of RGS13 mRNA by real-time qPCR (lower panel). (B) Dose-response of RGS13 protein and mRNA levels after 3-h stimulation by cAMP. Cells were treated with a range of 8-pCPT-cAMP concentrations for 3 h followed by preparation of cell lysates and immunoblotting (upper panel) or extraction of RNA, cDNA synthesis and evaluation of RGS13 mRNA by qPCR (lower panel). RGS13 mRNA levels (bar graphs in both A and B) are presented as the fold control (mean ± SEM from three independent experiments;*P<0.001, one-way ANOVA).
PKA activation slows RGS13 degradation
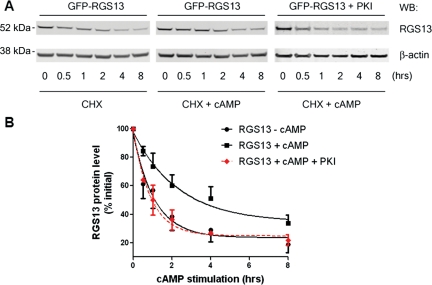
To determine how PKA activation regulated RGS13 protein turnover, we stimulated RGS13-transfected HEK293T cells with 8-pCPT-cAMP or vehicle control in the presence of the protein synthesis inhibitor cycloheximide (CHX). In unstimulated cells, RGS13 had a short half-life of about 1 h (Figure 3B). In cells treated with cAMP, we observed a substantially reduced rate of RGS13 degradation. The half-life of RGS13 in cAMP-treated cells was nearly three times that of the protein in untreated cells (Figure 3B). Forskolin, which activates adenylyl cyclase directly and leads to cAMP generation and PKA activation, also slowed RGS13 decay (data not shown). The enhanced RGS13 stability in the presence of cAMP was completely abolished by co-expression of an inhibitory peptide (PKI), which specifically binds to and inhibits the kinase activity of PKA (Day et al., 1989; Gudi et al., 1996; Castellone et al., 2005; Figure 3B). These results clearly demonstrate that PKA activation by cAMP inhibits RGS13 degradation.
Figure 3.
Activation of the PKA pathway by cAMP protects RGS13 from degradation. (A) HEK293T cells were transfected with pEGFP-RGS13 together with empty vector or a plasmid encoding PKI. After 24 h, cells were stimulated with 8-pCPT-cAMP (2 mM) or vehicle in the presence of CHX for the indicated time periods. Cell lysates were prepared and analyzed by immunoblotting using anti-RGS13 and anti-β-actin antibodies. (B) Densitometric analysis of RGS13 levels normalized by β-actin levels (mean ± SEM from three to five independent experiments; P < 0.001 for RGS13 in the presence and absence of cAMP, Student's t-test).
Phosphorylation of RGS13 by PKA in vitro
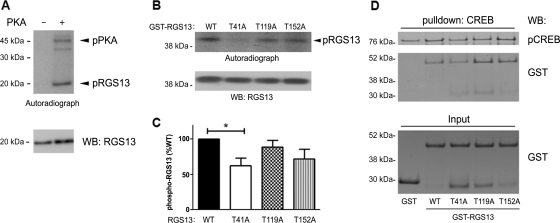
Because cAMP regulated RGS13 protein expression through PKA kinase activity, we hypothesized that RGS13 was a direct substrate of PKA. To test this possibility, we incubated recombinant, purified RGS13 with active PKA in the presence of radiolabeled [γ-32P]-ATP and measured incorporation of radioactivity by autoradiography. We found that PKA directly phosphorylated RGS13 in vitro (Figure 4A). To identify putative phosphorylation sites in RGS13, we performed in-gel proteinase digestion of recombinant RGS13 phosphorylated by PKA followed by mass spectrometric analysis. This study uncovered three residues in RGS13 that were putative PKA phosphorylation sites: Thr41, Thr119 and Thr152. To determine whether any or all of these amino acid residues were required for PKA-mediated phosphorylation, we generated recombinant GST-RGS13 fusion protein as well as mutant proteins in which each individual Thr residue was mutated to Ala (T41A, T119A and T152A). We expressed the proteins in bacteria and purified them by glutathione Sepharose chromatography. Incubation of these proteins with PKA revealed that the phosphorylation of RGS13(T41A) was reduced ∼40% compared with WT RGS13, confirming that Thr41 is a site of PKA-mediated phosphorylation (Figure 4B). Phosphorylation of the T119A and T152A mutants in vitro was not different from phosphorylation of the WT protein, suggesting that these sites do not contribute significantly to PKA-mediated phosphorylation of RGS13 (Figure 4C). To exclude the possibility that these mutations altered RGS13 conformation and/or function, we evaluated their ability to bind pCREB in pull-down assays, as we showed previously that RGS13 binds the pCREB–CBP complex (Xie et al., 2008). Recombinant His6-CREB phosphorylated by PKA and immobilized on Ni2+ beads was incubated with CBP together with GST or GST-RGS13 (WT and mutants). We evaluated the binding of RGS13 to pCREB by immunoblotting after bead washing to eliminate non-specifically bound proteins. Each T → A mutant bound pCREB similar to WT RGS13 (Figure 4D). This result suggests that the overall structure of these proteins remained intact.
Figure 4.
Phosphorylation of RGS13 by PKA in vitro. (A) Recombinant RGS13 was incubated for 1 h at 30°C in the presence or absence of the catalytic subunit of PKA in kinase buffer containing [γ-32P]-ATP, and protein phosphorylation was assessed by autoradiography. Recombinant RGS13 was detected with anti-RGS13 antibody. (B) Recombinant GST-RGS13 (WT or mutants) was incubated with PKA as in (A), and the reaction mixtures were purified by glutathione Sepharose chromatography prior to SDS–PAGE, autoradiography and immunoblotting (upper panel). (C) The phosphorylation of each mutant relative to WT RGS13 is presented in the bar graph (mean ± SEM of four independent experiments; *P<0.05, one way ANOVA). (D) Recombinant His6MBP-CREB phosphorylated by PKA was incubated with p300 and GST or GST-RGS13 (WT or mutants) followed by collection of the mixture with Ni2+ beads and immunoblotting as indicated. Input amounts of each GST fusion protein are shown in the lower panel. Image represents three independent experiments.
Role of Thr41 in RGS13 protein expression
Since mutation of Thr41 significantly decreased RGS13 phosphorylation by PKA in vitro, we examined how this mutation affected steady-state RGS13 levels. We evaluated expression of RGS13(T119A) in parallel since this mutation did not reduce PKA-mediated RGS13 phosphorylation. We expressed RGS13 WT or mutants in HeLa cells together with an empty vector or a plasmid encoding active PKA and assessed protein levels by immunoblotting. The steady-state amounts of RGS13 WT and T119A were similar, and abundance of each protein was increased by the presence of PKA (Figure 5A). In contrast, the expression of RGS13(T41A) was significantly reduced in the presence and absence of PKA compared with WT (Figure 5A and B) although their mRNA levels were similar (Supplementary Figure S1). These results suggest a critical role for Thr41 in the steady-state expression of RGS13. Consistent with this interpretation, the decay of RGS13(T41A) protein in the presence of CHX and cAMP was similar to that of WT RGS13 in the absence of cAMP (Figure 5C). This loss of protection from degradation suggests that cAMP regulates RGS13 turnover by phosphorylating RGS13 at Thr41.
Figure 5.
Crucial role of Thr41 in the maintenance of RGS13 protein expression in vivo. (A and B) pcDNA3.1/V5-His RGS13 (WT or mutants) plasmids were transfected into HEK293T or HeLa cells in the presence or absence of a plasmid encoding the catalytic subunit of PKA. After 24 h, cell lysates were prepared and analyzed by immunoblotting as indicated. Bar graphs in (B) show the mean ± SEM (fold of RGS13 WT) of three independent experiments (*P < 0.005, one-way ANOVA, RGS13 WT vs. T41A). (C) Critical role of Thr41 in cAMP-induced inhibition of RGS13 decay. HEK293T cells were transfected with pEGFP-RGS13-T41A. Twenty-four hours later, cells were stimulated with 8-pCPT-cAMP for the indicated time periods in the presence of CHX followed by collection of cell lysates and immunoblotting by anti-RGS13 and anti-β-actin antibodies (upper panel). Densitometric analysis of RGS13 levels (normalized to β-actin) are shown in from the lower panel (mean ± SEM of four independent experiments; P < 0.01, RGS13 WT + cAMP vs. RGS13(T41A) + cAMP, Student's t-test. The decay curve of RGS13(T41A) is overlaid with curves from Figure 3B for comparison.
RGS13 is a target of the ubiquitin–proteasome pathway
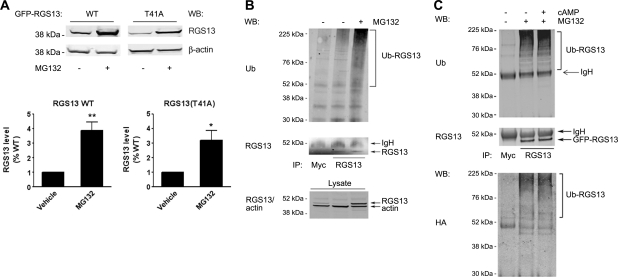
To explore how cAMP–PKA regulated RGS13 expression, we investigated the molecular mechanism of RGS13 degradation. Several proteins of the R4 RGS family that are highly related to RGS13 undergo ubiquitylation and proteasome-mediated proteolysis (Davydov and Varshavsky, 2000; Krumins et al., 2004; Lee et al., 2005; Bodenstein et al., 2007). To determine whether RGS13 was also processed by the proteasome, we evaluated its steady-state amounts in the presence or absence of the proteasome inhibitor MG132. RGS13 quantities were ∼4-fold higher in cells treated with MG132 than in untreated cells (Figure 6A), indicating that RGS13 was degraded by the proteasome. The levels of RGS13(T41A) increased by a similar amount after MG132 treatment (Figure 6A), confirming that degradation of RGS13 occurs through the proteasome.
Figure 6.
RGS13 is ubiquitylated and targeted for proteasome-mediated degradation. (A) Increased RGS13 protein levels in the presence of a proteasome inhibitor MG132. pEGFP-RGS13 WT or T41A was expressed in HEK293T cells. After 24 h, cells were treated with vehicle (DMSO) or MG132 (10 μM) overnight. Cell lysates were extracted and subjected to immunoblotting with antibodies indicated (upper panel). Densitometric analysis of three independent experiments are shown in the bar graph [mean ± SEM of RGS13 relative to control (untreated) cells; *P<0.05; **P<0.01, Student's t-test]. (B) RGS13 is ubiquitylated. pEGFP-RGS13 and HA-Ub plasmids were expressed in HEK293T cells. After 24 h, cells were treated with DMSO vehicle or MG132 overnight. Cell lysates were extracted and immunoprecipitated with the indicated antibodies prior to SDS–PAGE. Total cell lysates were analyzed in parallel and immunoblotted with the indicated antibodies to assess input protein amounts. (C) cAMP does not affect RGS13 ubiquitylation. HEK293T cells were transfected as in (B) and treated overnight with MG132. Twenty-four hours later, cells were treated with vehicle alone or 8-pCPT-cAMP for an additional 4 h. Cell lysates were prepared, and immunoprecipitation performed as in (B) followed by immunoblotting as indicated. Images are representative of three independent experiments.
Since proteasomal processing requires covalent modification by poly-ubiquitin (Ub) chains, we determined whether RGS13 underwent ubiquitylation. We co-expressed plasmids encoding RGS13 and HA-Ub in HEK293T cells and treated cells with MG132 to allow accumulation of Ub-containing proteins. Immunoprecipitation of RGS13 with the RGS13 antibody (but not a control antibody) followed by immunoblotting with anti-Ub demonstrated that RGS13 was ubiquitylated to a greater degree in MG132-treated cells than in untreated cells (Figure 6B). Regulation of protein turnover and proteasome function by cAMP has been described (Hoang et al., 2004; Zhang et al., 2007). For example, PKA activation inhibits the ubiquitylation of β-catenin (Hino et al., 2005). However, we found that RGS13 was ubiquitylated to a similar extent in untreated and cAMP-treated cells (Figure 6C). This result suggests that PKA activation prevents RGS13 degradation through mechanism(s) other than inhibition of its ubiquitylation.
Functional consequences of Thr41 mutation in RGS13
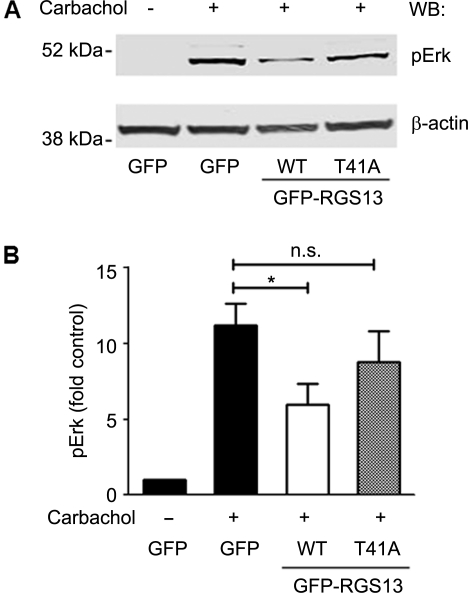
The reduced steady-state expression and increased turnover of RGS13(T41A) compared with the WT protein suggested that Thr41 has crucial role in determining its capacity to regulate GPCR signaling. We showed previously that RGS13 inhibits M2 muscarinic receptor (M2R)-induced Erk phosphorylation (Johnson and Druey, 2002). To examine the importance of Thr41 for RGS13 function, we transfected fibroblast A9L cells stably expressing M2 muscarinic receptors (A9L-M2R) with RGS13 WT or RGS13(T41A). After 24 h, we stimulated cells with the M2 agonist carbachol and examined Erk phosphorylation by immunoblotting. We observed a >10-fold increase in Erk phosphorylation in carbachol-treated cells compared with untreated cells (Figure 7). Expression of WT RGS13, but not RGS13(T41A), significantly inhibited carbachol-stimulated Erk phosphorylation. Thus, mutation of Thr41 in RGS13 reduced its ability to regulate M2R-evoked Erk activation.
Figure 7.
Thr41 mutation reduces RGS13 inhibition of a GPCR response. (A) A9L-M2R cells were transfected with pEGFP, GFP-RGS13(WT) or GFP-RGS13(T41A) plasmids. After 24 h, serum-starved cells were stimulated with carbachol (100 nM) for 4 min at 37°C followed by preparation of cell lysates and immunoblotting to assess Erk phosphorylation. (B) Bar graph of densitometric analysis of pErk levels (normalized to β-actin) from three independent experiments (mean ± SEM; *P<0.05, Student's t-test; n.s. = not significant).
Discussion
We discovered that RGS13 is phosphorylated by PKA at Thr41 and that activation of the cAMP–PKA pathway promoted RGS13 protein expression in cells dramatically. PKA activation may reduce RGS13 protein turnover through a mechanism dependent on the phosphorylation of RGS13 at Thr41. Mutation of this residue not only decreased PKA-mediated phosphorylation in vitro, but also significantly reduced the steady-state expression of RGS13 protein. Phosphorylation appeared to protect RGS13 from degradation downstream of ubiquitylation as RGS13 ubiquitylation was not affected by cAMP. These studies provide the first insight into mechanisms of post-translational modulation of RGS13 abundance.
PKA has been shown to phosphorylate other RGS proteins such as RGS10, RGS9-1 and RGS14 (Balasubramanian et al., 2001; Hollinger et al., 2003). RGS10 phosphorylation by PKA induced its nuclear translocation and crippled its ability to inhibit inwardly rectifying K+ (GIRK) channels in Xenopus laevis oocytes by sequestration from membrane channels (Burgon et al., 2001; Bender et al., 2008). In contrast to RGS10, RGS13 phosphorylation by PKA seems to promote RGS13 function through stabilization of its expression. Whereas mutation of the PKA target residue in RGS13 (Thr41) did not affect its ability to interact with other signaling proteins [pCREB (Figure 4) and Gα (data not shown)], the RGS13(T41A) had reduced steady-state expression (Figure 5), increased turnover in the presence of cAMP (Figure 5) and decreased capacity to inhibit GCPR signaling (Figure 7).
Although RGS13 lacks a clear consensus PKA phosphorylation site, we identified Thr41 as a PKA phosphorylation site in RGS13 by mass spectrometry (MS). Mutation of this residue significantly decreased, but did not abolish, RGS13 phosphorylation by PKA, suggesting that additional PKA phosphorylation sites may exist in RGS13 and could contribute to the inhibition of RGS13 degradation by PKA activation. On the other hand, protection of RGS13 degradation by cAMP was completely reversed both by mutation of Thr41 and by expression of PKI, a peptide that specifically inhibits PKA kinase activity in vivo but not the activity of closely related kinases (Gudi et al., 1996). These results indicated that cAMP inhibited RGS13 degradation solely by activation of PKA but not other cAMP-activated mediators such as Epac (Gloerich and Bos, 2010) and that such inhibition most likely occurs through phosphorylation of Thr41 in RGS13 by PKA.
Although Ser/Thr phosphorylation may affect protein ubiquitylation and proteasomal degradation of some proteins (Hino et al., 2005), RGS13 phosphorylation by PKA did not inhibit RGS13 ubiquitylation. Ubiquitylated proteins may be relatively stable unless they also contain an unstructured region, which is recognized by the 19S regulatory component of the proteasome complex (Schrader et al., 2009). Thus, phosphorylation of RGS13 at Thr41 by PKA could render the N-terminus more structured overall, impairing recognition and/or handling of ubiquitylated RGS13 by the proteasomal machinery. We hypothesized that this mechanism could underlie the stabilization of the related protein RGS16 by phosphorylation of a conserved Tyr residue at its C-terminus, which is in close proximity to the N-terminus (Derrien et al., 2003). PKA-induced phosphorylation could also protect RGS13 from proteasomal targeting indirectly by affecting protein–protein interactions. PKA activation induces RGS13 translocation to the nucleus and binding to pCREB. We found previously that extinction of CREB expression by RNAi also reduced RGS13 nuclear localization in cells activated by PKA (Xie et al., 2008). Therefore, the amount of free cytosolic RGS13 available for proteasome-mediated degradation could be reduced in the presence of PKA.
Owing to their powerful ability to modulate numerous cellular signaling pathways, RGS protein concentrations are often regulated by induction or repression of their transcription by many environmental stimuli. As our studies with RGS13 demonstrate, RGS protein degradation, as is true for many signaling molecules, is also rapidly and precisely coordinated by post-transcriptional and post-translational events. This study lays the foundation for further discovery of unique molecular components that modulate RGS13 longevity and possibly other mechanisms that affect its expression and/or function.
Materials and methods
Reagents and cell culture
HEK293T and HeLa cells were obtained from ATCC, and mouse fibroblast A9L cells stably expressing M2 muscarinic receptors (A9L-M2R) were the generous gift of Dr Jurgen Wess (NIDDK/NIH). Cells were maintained in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum at 37°C in 5% CO2. LAD2 mast cells (Bansal et al., 2008a) were grown in Stem-Pro medium containing Stem-Pro supplement (Invitrogen) and 100 ng/ml human stem cell factor (R&D Systems). The polyclonal RGS13 antibody, which does not detect the appropriate-sized band in RGS13-deficient cells, was described previously (Bansal et al., 2008a). Other antibodies were purchased from the following sources: mouse anti-β-actin (Sigma-Aldrich); goat anti-GST and rabbit anti-myc (Santa Cruz Biotechnology); rabbit anti-CREB, anti-PKA, mouse anti-phospho-CREB, anti-Ub, anti-phospho-Erk1/2 and anti-Erk1/2 (Cell Signaling Technology); mouse anti-V5 (Invitrogen). 8-(4-Chlorophenylthio)-adenosine 3′,5′-cyclic monophosphate (8-pCPT cAMP), MG132 and CHX were from Sigma. Nickel-nitriloacetic acid agarose beads were from Qiagen and Lipofectamine 2000 was purchased from Invitrogen. [γ-32P]-ATP was from MP Biomedicals. Recombinant PKA (catalytic subunit), His6-mannose-binding protein (MBP), CREB and FLAGp300 were purchased (Xie et al., 2008). GST-RGS13 expression and purification were done as previously described (Bansal et al., 2008b).
DNA constructs
pEGFP-RGS13 and pGEX4T-1-RGS13 have been described elsewhere (Johnson and Druey, 2002; Xie et al., 2008). pRSV-PKI plasmid was the kind gift of Dr J. Silvio Gutkind (NIDCR/NIH) and pCR3.1-HA-Ub was obtained from Dr John Kehrl (NIAID/NIH). pcDNA3.1/V5-His-RGS13 was generated by cloning the PCR product of human RGS13 (using primer set 5′-GCC ACC ATG GCA AGC AGG CGG AAT TG-3′ and 5′-GAA ACT GTT GTT GGA CTG CAT AG-3′) into pcDNA3.1/V5-His TOPO expression vector (Invitrogen). T41 → A41 (T41A), T119 → A119 (T119A) and T152 → A152 (T152A) point mutations were introduced into pEGFP-RGS13, pcDNA3.1/V5-His and pGEX4T1 backgrounds using the QuikChange II site-directed mutagenesis kit (Stratagene/Agilent).
Transfection and immunoblotting
Cells were transfected using Lipofectamine 2000 according to the manufacturer's instructions. Cell lysates were prepared using radioimmunoprecipitation (RIPA) lysis buffer containing 50 mM Tris, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.5% sodium deoxycholate and 0.1% SDS supplemented with protease inhibitor cocktail (Complete, Roche) and a phosphatase inhibitor cocktail (PhosSTOP, Roche) for 15 min at 4°C. Proteins were separated on SDS–PAGE, blotted with primary antibodies as indicated in the figure legends followed by incubation with Infrared IRDye-labeled secondary antibodies (Li-Cor Biosciences). Signals were detected and quantitated using the Li-Cor Odyssey Imaging System.
In vitro phosphorylation assay
One hundred and fifty nanograms recombinant, untagged RGS13 or 500 ng GST-RGS13 (WT and mutants) was incubated with 20 units PKA catalytic subunit at 30°C for 1 h in kinase buffer (25 mM Tris, pH 7.5, 10 mM MgCl2, 0.5 mM EDTA) supplemented with 10 µCi [γ-32P]-ATP, 2 mM DTT, 0.1 mg/ml bovine serum albumin and 50 µM unlabeled ATP. Reaction mixtures were then subject to SDS–PAGE for autoradiography and immunoblotting.
Mass spectrometry
Recombinant RGS13 was phosphorylated by PKA in vitro followed by SDS–PAGE gel separation. The phosphoRGS13 band was excised from the gel and analyzed for post-translational modification using in-gel reduction, alkylation and enzymatic digestion (Tufts University Core Facility). Liquid chromatography and MS analysis was performed on the in-gel-digested extracts, and the MS/MS spectra were searched against the NCBI non-redundant protein sequence database using the SEQUEST computer algorithm.
Real-time TaqMan PCR analysis
Total RNA was isolated using the RNeasy kit (Qiagen), and 1 µg RNA was reverse transcribed into cDNA using SuperScriptIII reverse transcriptase (Invitrogen) following the manufacturer's instructions. RNA incubated without reverse transcriptase was used as a negative control for the PCR reaction. One microliter synthesized cDNA was then used for amplification of human RGS13 using the real-time TaqMan gene expression assay kit (Applied Biosystems). GAPDH was used as an endogenous housekeeping gene for normalization, and relative quantifications were based on the amount of RGS13 divided by the GAPDH result.
Analysis of RGS13 decay and ubiquitylation
For CHX chase assays, HEK293T or HeLa cells were transfected with pEGFP-RGS13 together with empty vector or pRSV-PKI for 24 h. Cells were then stimulated for the indicated times with 2 mM 8-pCPT-cAMP or vehicle alone in the presence of CHX (15 μg/ml) before harvest in lysis buffer and immunoblot analysis. For proteasomal inhibition, cells were transfected with pEGFP-RGS13 constructs for 24 h followed by incubation with MG132 (10 µM) or vehicle control (DMSO) in cell growth medium overnight at 37°C prior to cell lysis and immunoblotting. For analysis of ubiquitylation, cells transfected with pEGFP-RGS13 and HA-Ub were treated overnight with MG132 prior to additional incubation with vehicle or cAMP for an additional 4 h at 37°C. Cells were then harvested in RIPA buffer followed by either immunoblot analysis or immunoprecipitation with polyclonal anti-myc (control) or anti-RGS13 antibodies and immunoblotting.
Analysis of Erk activation
A9L-M2R cells were transfected with pEGFP or pEGFP-RGS13 (WT or T41A mutant). Twenty-four hours later, cells were stimulated with 100 nM carbachol or vehicle control for 4 min at 37°C. The reaction was stopped by adding ice-cold PBS containing protease and phosphatase inhibitors. Cell lysates were prepared and analyzed by immunoblotting with indicated antibodies.
Statistical analysis
Data were analyzed using the GraphPad Prism 5 software package. Student's t-test or one way ANOVA was used for statistical analysis. P-values of <0.05 were considered significant.
Supplementary data
Supplementary data are available online at http://jmcb.oxfordjournals.org.
Conflicts of interest: none declared.
Funding
This study was carried out as part of the authors' work for the National Institute of Allergy and Infectious Diseases (NIAID). It supported by the Intramural Research Program of the NIH, NIAID (grant no. AI000939-06 LAD).
References
- Balasubramanian N., Levay K., Keren-Raifman T., Faurobert E., Slepak V.Z. Phosphorylation of the regulator of G protein signaling RGS9-1 by protein kinase A is a potential mechanism of light- and Ca2+-mediated regulation of G protein function in photoreceptors. Biochemistry. 2001;40:12619–12627. doi: 10.1021/bi015624b. doi:10.1021/bi015624b. [DOI] [PubMed] [Google Scholar]
- Bansal G., Druey K.M., Xie Z. R4 RGS proteins: regulation of G-protein signaling and beyond. Pharmacol. Ther. 2007;116:473–495. doi: 10.1016/j.pharmthera.2007.09.005. doi:10.1016/j.pharmthera.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bansal G., DiVietro J.A., Kuehn H.S., Rao S., Nocka K.H., Gilfillan A.M., Druey K.M. RGS13 controls g protein-coupled receptor-evoked responses of human mast cells. J. Immunol. 2008a;181:7882–7890. doi: 10.4049/jimmunol.181.11.7882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bansal G., Xie Z., Rao S., Nocka K.H., Druey K.M. Suppression of immunoglobulin E-mediated allergic responses by regulator of G protein signaling 13. Nat. Immunol. 2008b;9:73–80. doi: 10.1038/ni1533. doi:10.1038/ni1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender K., Nasrollahzadeh P., Timpert M., Liu B., Pott L., Kienitz M.C. A role for RGS10 in beta-adrenergic modulation of G-protein-activated K+ (GIRK) channel current in rat atrial myocytes. J. Physiol. 2008;586:2049–2060. doi: 10.1113/jphysiol.2007.148346. doi:10.1113/jphysiol.2007.148346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodenstein J., Sunahara R.K., Neubig R.R. N-terminal residues control proteasomal degradation of RGS2, RGS4, and RGS5 in human embryonic kidney 293 cells. Mol. Pharmacol. 2007;71:1040–1050. doi: 10.1124/mol.106.029397. doi:10.1124/mol.106.029397. [DOI] [PubMed] [Google Scholar]
- Burgon P.G., Lee W.L., Nixon A.B., Peralta E.G., Casey P.J. Phosphorylation and nuclear translocation of a regulator of G protein signaling (RGS10) J. Biol. Chem. 2001;276:32828–32834. doi: 10.1074/jbc.M100960200. doi:10.1074/jbc.M100960200. [DOI] [PubMed] [Google Scholar]
- Castellone M.D., Teramoto H., Williams B.O., Druey K.M., Gutkind J.S. Prostaglandin E2 promotes colon cancer cell growth through a Gs-axin-beta-catenin signaling axis. Science. 2005;310:1504–1510. doi: 10.1126/science.1116221. doi:10.1126/science.1116221. [DOI] [PubMed] [Google Scholar]
- Cheng Y.S., Lee T.S., Hsu H.C., Kou Y.R., Wu Y.L. Characterization of the transcriptional regulation of the regulator of G protein signaling 2 (RGS2) gene during 3T3-L1 preadipocyte differentiation. J. Cell Biochem. 2008;105:922–930. doi: 10.1002/jcb.21893. doi:10.1002/jcb.21893. [DOI] [PubMed] [Google Scholar]
- Cho H., Park C., Hwang I.Y., Han S.B., Schimel D., Despres D., Kehrl J.H. Rgs5 targeting leads to chronic low blood pressure and a lean body habitus. Mol. Cell. Biol. 2008;28:2590–2597. doi: 10.1128/MCB.01889-07. doi:10.1128/MCB.01889-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davydov I.V., Varshavsky A. RGS4 is arginylated and degraded by the N-end rule pathway in vitro. J. Biol. Chem. 2000;275:22931–22941. doi: 10.1074/jbc.M001605200. doi:10.1074/jbc.M001605200. [DOI] [PubMed] [Google Scholar]
- Day R.N., Walder J.A., Maurer R.A. A protein kinase inhibitor gene reduces both basal and multihormone-stimulated prolactin gene transcription. J. Biol. Chem. 1989;264:431–436. [PubMed] [Google Scholar]
- Derrien A., Zheng B., Osterhout J.L., Ma Y.C., Milligan G., Farquhar M.G., Druey K.M. Src-mediated RGS16 tyrosine phosphorylation promotes RGS16 stability. J. Biol. Chem. 2003;278:16107–16116. doi: 10.1074/jbc.M210371200. doi:10.1074/jbc.M210371200. [DOI] [PubMed] [Google Scholar]
- Ding J., Guzman J.N., Tkatch T., Chen S., Goldberg J.A., Ebert P.J., Levitt P., Wilson C.J., Hamm H.E., Surmeier D.J. RGS4-dependent attenuation of M4 autoreceptor function in striatal cholinergic interneurons following dopamine depletion. Nat. Neurosci. 2006;9:832–842. doi: 10.1038/nn1700. doi:10.1038/nn1700. [DOI] [PubMed] [Google Scholar]
- Georgoussi Z., Leontiadis L., Mazarakou G., Merkouris M., Hyde K., Hamm H. Selective interactions between G protein subunits and RGS4 with the C-terminal domains of the mu- and delta-opioid receptors regulate opioid receptor signaling. Cell. Signal. 2006;18:771–782. doi: 10.1016/j.cellsig.2005.07.003. doi:10.1016/j.cellsig.2005.07.003. [DOI] [PubMed] [Google Scholar]
- Gilman A.G. G proteins: transducers of receptor-generated signals. Annu. Rev. Biochem. 1987;56:615–649. doi: 10.1146/annurev.bi.56.070187.003151. doi:10.1146/annurev.bi.56.070187.003151. [DOI] [PubMed] [Google Scholar]
- Gloerich M., Bos J.L. Epac: defining a new mechanism for cAMP action. Annu. Rev. Pharmacol. Toxicol. 2010;50:355–375. doi: 10.1146/annurev.pharmtox.010909.105714. doi:10.1146/annurev.pharmtox.010909.105714. [DOI] [PubMed] [Google Scholar]
- Gudi T., Huvar I., Meinecke M., Lohmann S.M., Boss G.R., Pilz R.B. Regulation of gene expression by cGMP-dependent protein kinase. Transactivation of the c-fos promoter. J. Biol. Chem. 1996;271:4597–4600. doi: 10.1074/jbc.271.9.4597. doi:10.1074/jbc.271.9.4597. [DOI] [PubMed] [Google Scholar]
- Hino S., Tanji C., Nakayama K.I., Kikuchi A. Phosphorylation of beta-catenin by cyclic AMP-dependent protein kinase stabilizes beta-catenin through inhibition of its ubiquitination. Mol. Cell. Biol. 2005;25:9063–9072. doi: 10.1128/MCB.25.20.9063-9072.2005. doi:10.1128/MCB.25.20.9063-9072.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang T., Fenne I.S., Cook C., Borud B., Bakke M., Lien E.A., Mellgren G. cAMP-dependent protein kinase regulates ubiquitin-proteasome-mediated degradation and subcellular localization of the nuclear receptor coactivator GRIP1. J. Biol. Chem. 2004;279:49120–49130. doi: 10.1074/jbc.M409746200. doi:10.1074/jbc.M409746200. [DOI] [PubMed] [Google Scholar]
- Hollinger S., Hepler J.R. Cellular regulation of RGS proteins: modulators and integrators of G protein signaling. Pharmacol. Rev. 2002;54:527–559. doi: 10.1124/pr.54.3.527. doi:10.1124/pr.54.3.527. [DOI] [PubMed] [Google Scholar]
- Hollinger S., Ramineni S., Hepler J.R. Phosphorylation of RGS14 by protein kinase A potentiates its activity toward G alpha i. Biochemistry. 2003;42:811–819. doi: 10.1021/bi026664y. doi:10.1021/bi026664y. [DOI] [PubMed] [Google Scholar]
- Hu W., Li F., Mahavadi S., Murthy K.S. Interleukin-1beta up-regulates RGS4 through the canonical IKK2/IkappaBalpha/NF-kappaB pathway in rabbit colonic smooth muscle. Biochem. J. 2008;412:35–43. doi: 10.1042/BJ20080042. doi:10.1042/BJ20080042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James M.A., Lu Y., Liu Y., Vikis H.G., You M. RGS17, an overexpressed gene in human lung and prostate cancer, induces tumor cell proliferation through the cyclic AMP-PKA-CREB pathway. Cancer Res. 2009;69:2108–2116. doi: 10.1158/0008-5472.CAN-08-3495. doi:10.1158/0008-5472.CAN-08-3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson E.N., Druey K.M. Functional characterization of the G protein regulator RGS13. J. Biol. Chem. 2002;277:16768–16774. doi: 10.1074/jbc.M200751200. doi:10.1074/jbc.M200751200. [DOI] [PubMed] [Google Scholar]
- Krumins A.M., Barker S.A., Huang C., Sunahara R.K., Yu K., Wilkie T.M., Gold S.J., Mumby S.M. Differentially regulated expression of endogenous RGS4 and RGS7. J. Biol. Chem. 2004;279:2593–2599. doi: 10.1074/jbc.M311600200. doi:10.1074/jbc.M311600200. [DOI] [PubMed] [Google Scholar]
- Lee M.J., Tasaki T., Moroi K., An J.Y., Kimura S., Davydov I.V., Kwon Y.T. RGS4 and RGS5 are in vivo substrates of the N-end rule pathway. Proc. Natl. Acad. Sci. USA. 2005;102:15030–15035. doi: 10.1073/pnas.0507533102. doi:10.1073/pnas.0507533102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang G., Bansal G., Xie Z., Druey K.M. RGS16 inhibits breast cancer cell growth by mitigating phosphatidylinositol 3-kinase signaling. J. Biol. Chem. 2009;284:21719–21727. doi: 10.1074/jbc.M109.028407. doi:10.1074/jbc.M109.028407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinissen M.J., Gutkind J.S. G-protein-coupled receptors and signaling networks: emerging paradigms. Trends Pharmacol. Sci. 2001;22:368–376. doi: 10.1016/s0165-6147(00)01678-3. doi:10.1016/S0165-6147(00)01678-3. [DOI] [PubMed] [Google Scholar]
- Mayr B., Montminy M. Transcriptional regulation by the phosphorylation-dependent factor CREB. Nat. Rev. Mol. Cell. Biol. 2001;2:599–609. doi: 10.1038/35085068. doi:10.1038/35085068. [DOI] [PubMed] [Google Scholar]
- Ogier-Denis E., Petiot A., Bauvy C., Codogno P. Control of the expression and activity of the Galpha-interacting protein (GAIP) in human intestinal cells. J. Biol. Chem. 1997;272:24599–24603. doi: 10.1074/jbc.272.39.24599. doi:10.1074/jbc.272.39.24599. [DOI] [PubMed] [Google Scholar]
- Schrader E.K., Harstad K.G., Matouschek A. Targeting proteins for degradation. Nat. Chem. Biol. 2009;5:815–822. doi: 10.1038/nchembio.250. doi:10.1038/nchembio.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuebe S., Wieland T., Kraemer E., Stritzky A., Schroeder D., Seekamp S., Vogt A., Chen C.K., Patten M. Sphingosine-1-phosphate and endothelin-1 induce the expression of rgs16 protein in cardiac myocytes by transcriptional activation of the rgs16 gene. Naunyn Schmiedebergs Arch. Pharmacol. 2008;376:363–373. doi: 10.1007/s00210-007-0214-2. doi:10.1007/s00210-007-0214-2. [DOI] [PubMed] [Google Scholar]
- Willars G.B. Mammalian RGS proteins: multifunctional regulators of cellular signalling. Semin. Cell Dev. Biol. 2006;17:363–376. doi: 10.1016/j.semcdb.2006.03.005. doi:10.1016/j.semcdb.2006.03.005. [DOI] [PubMed] [Google Scholar]
- Xie Z., Geiger T.R., Johnson E.N., Nyborg J.K., Druey K.M. RGS13 acts as a nuclear repressor of CREB. Mol. Cell. 2008;31:660–670. doi: 10.1016/j.molcel.2008.06.024. doi:10.1016/j.molcel.2008.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F., Paterson A.J., Huang P., Wang K., Kudlow J.E. Metabolic control of proteasome function. Physiology (Bethesda) 2007;22:373–379. doi: 10.1152/physiol.00026.2007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.