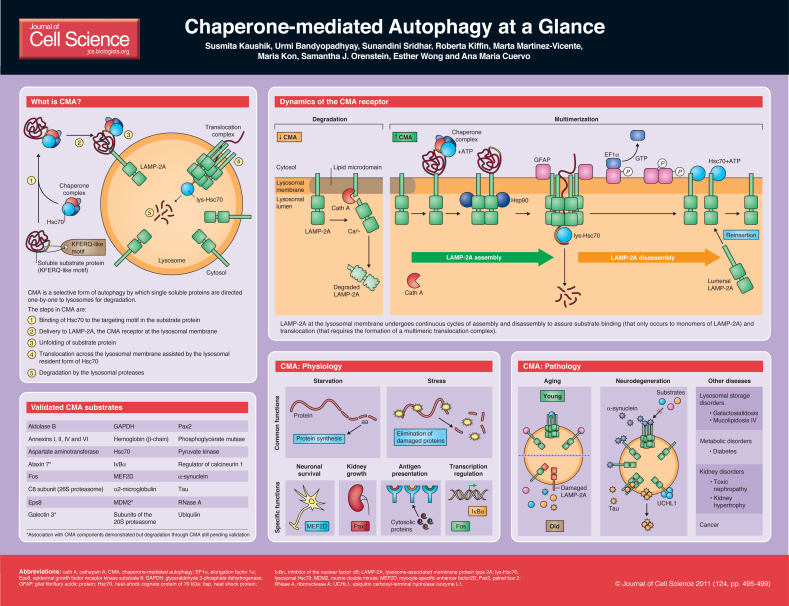
Chaperone-mediated autophagy (CMA) is an intracellular catabolic pathway that mediates the degradation of a selective subset of cytosolic proteins in lysosomes (Dice, 2007; Cuervo, 2010; Kon and Cuervo, 2010; Orenstein and Cuervo, 2010). The term autophagy (or self-eating) is broadly used to designate the lysosomal delivery and degradation of intracellular components (Mizushima et al., 2008; Mizushima and Levine, 2010; Yang and Klionsky, 2010). Various types of autophagy co-exist in almost all cells, and they can be differentiated by the mechanisms that mediate the delivery of cargo (the substrates to be degraded) to lysosomes. Macroautophagy and microautophagy are variants of the autophagic process, in which entire regions of cytosol (in ‘bulk’ autophagy) or selective cytosolic components (organelles, protein complexes, protein aggregates, pathogens, etc.) are sequestered in vesicular compartments. Lysosomal enzymes can gain access to the enclosed cargo through direct fusion of the vesicles with lysosomes (in macroautophagy), or by internalization of cargo-containing vesicles that form at the lysosomal membrane (in microautophagy). A third form of autophagy, solely dedicated to degradation of soluble proteins can also be detected in most cell types in mammals. This autophagic process, known as chaperone-mediated autophagy, differs from the other forms of autophagy in both the way in which cargo proteins are recognized for lysosomal delivery and the way in which these proteins reach the lysosomal lumen (Dice, 2007; Cuervo, 2010). In this article and the accompanying poster, we summarize the main steps involved in degradation of cytosolic proteins by CMA, the essential components of this pathway both in the cytosol and at the lysosomal membrane and the basis for the regulation of this autophagic process. We also include a synopsis of the described physiological functions of CMA and some
of the connections established between malfunctioning of CMA and disease.
CMA step by step
For a protein to be amenable for lysosomal degradation via CMA, the presence of a pentapeptide motif biochemically related to KFERQ in its amino acid sequence is absolutely necessary (Dice, 1990). This motif is recognized by a cytosolic chaperone, the heat shock cognate protein of 70 kDa (Hsc70) that along with its modulatory co-chaperones (Bag1, Hip, Hop and Hsp40) brings the substrate protein to the surface of the lysosomes (Chiang et al., 1989). After this targeting step, the substrate protein–chaperone complex docks at the lysosomal membrane through interaction with the cytosolic tail of a single-span membrane protein, the lysosome-associated membrane protein type 2A (LAMP-2A), which acts as a receptor for this autophagic pathway (Cuervo and Dice, 1996). Internalization of the substrate protein is preceded by its unfolding (Salvador et al., 2000), a step not required in the other types of autophagy. Translocation of the substrate across the lysosomal membrane also requires the presence of a luminal form of Hsc70 (lys-Hsc70), which assists in substrate translocation into the lyosomal lumen (Agarraberes et al., 1997; Cuervo et al., 1997). After tanslocation, substrate proteins are rapidly degraded by the abundant array of lysosomal hydrolases.
CMA substrates
Putative CMA substrates were identified by the presence of a KFERQ-like motif in their sequence (Dice, 1990) and, using this criterion, it was estimated that ~30% of cytosolic proteins are candidates for CMA (Wing et al., 1991). However, to classify a protein as a bona fide CMA substrate, additional experimental validation is required (Kaushik and Cuervo, 2009). Basic criteria that a CMA substrate needs to fulfill include: (1) presence of a KFERQ-like motif; (2) association with lysosomes, preferentially with those that have higher CMA activity (positive for lys-Hsc70); (3) reduced degradation rates when lysosomal proteolytic activity is blocked; (4) interaction with cytosolic Hsc70; (5) interaction with LAMP-2A through its cytosolic tail; (6) increase in the intracellular levels of the candidate protein in cells that lack LAMP-2A (although activation of other autophagic pathways to compensate for reduced CMA prevents substrate accumulation in many instances); (7) capability to directly translocate into isolated lysosomes. The latter probably being the most definitive evidence of a protein being a CMA substrate, as in this type of in vitro assay there is no contribution of any other proteolytic system to the observed lysosomal translocation and degradation (Kaushik and Cuervo, 2009).
About 25 proteins have been validated as CMA substrates thus far and five more have been shown to fulfill one or two of the above criteria, and are pending further validation. The spectrum of CMA substrates includes – among others – several glycolytic enzymes (Aniento et al., 1993; Cuervo et al., 1994), transcription factors and inhibitors of transcription factors (Aniento et al., 1996; Cuervo et al., 1998; Sooparb et al., 2004; Liu et al., 2009), Ca2+-binding and lipid-binding proteins (Cuervo et al., 1999; Cuervo et al., 2000), and components of other proteolytic systems (Cuervo et al., 1995b; Rothenberg et al., 2010).
Molecular components of CMA
The cytosolic chaperones and co-chaperones that participate in CMA are also involved in other intracellular pathways (Chiang et al., 1989). The chaperones located in the lysosomal lumen or associated to the lysosomal membrane – namely lys-Hsc70 (Cuervo et al., 1997), membrane associated Hsc70 (Agarraberes et al., 1997) and lys-Hsp90 (Bandyopadhyay et al., 2008) – appear to be exclusively dedicated to CMA. However, as these are post-translational variations of cytosolic chaperones rather than independent gene products, regulating their expression does not provide a mean for selectively affecting CMA activity.
LAMP-2A, the membrane protein that acts as a receptor for the CMA substrates (Cuervo and Dice, 1996) has often been manipulated to regulate CMA activity. In fact, levels of LAMP-2A at the lysosomal membrane directly determine rates of CMA activity, because substrate binding to the cytosolic tail of LAMP-2A is a limiting step in CMA (Cuervo and Dice, 2000c). LAMP-2A is a spliced variant of a single Lamp2 gene, which also encodes two other variants, LAMP-2B and LAMP-2C (Eskelinen et al., 2005), with identical luminal regions but different transmembrane and cytosolic tails. Substrate binding to the LAMP-2A cytosolic tail does not occur at the KFERQ-targeting region and a designated LAMP-2A-binding motif in the substrate has not been identified yet. However, the fact that substrate binding requires the four positive charges in the LAMP-2A cytosolic tail suggests that electrostatic interactions, rather than specific amino acid residues, mediate substrate binding (Cuervo and Dice, 2000c).
The function of LAMP-2A extends beyond that of a receptor as this protein is also an essential component of the CMA translocation complex (Bandyopadhyay et al., 2008). Binding of substrate proteins to LAMP-2A monomers drives its organization into a 700 kDa multimeric complex at the lysosomal membrane. The GxxG motif present in the transmembrane region of LAMP-2A is important for multimerization. We have shown that mutations that prevent multimerization abolish substrate translocation but not substrate binding to LAMP-2A (Bandyopadhyay et al., 2008). In fact, substrate proteins only bind to LAMP-2A monomers that then ‘drag’ the substrate towards the translocation complex during multimerization, whereas their binding to preformed translocation complexes is not possible (Bandyopadhyay et al., 2008). Different lines of evidence support the idea that LAMP-2A undergoes conformational changes upon substrate binding and during the process that results in its multimerization. The presence of a lysosome-specific form of Hsp90 on the luminal side of the lysosomal membrane is essential to preserve the stability of LAMP-2A while it undergoes these conformational changes at the lysosomal membrane (Bandyopadhyay et al., 2008).
The CMA translocation complex is not present in a stable form at the lysosomal membrane but rather forms actively upon substrate binding to LAMP-2A, and is rapidly disassembled once the substrate is translocated. LAMP-2A undergoes continuous cycles of assembly and disassembly that facilitate binding of substrate proteins (when it is monomeric) and their translocation across the lysosomal membrane (when organized as a multimer) (Bandyopadhyay et al., 2008). Hsc70 actively contributes to dissociation of LAMP-2A from the multimeric complex in a process that depends on the ATPase activity of Hsc70 and the absence of any substrate proteins bound to it (Bandyopadhyay et al., 2008).
Regulation of CMA
The signaling mechanisms that contribute to the regulation of CMA activity are currently poorly understood. The main target for CMA regulation appears to be LAMP-2A, whose levels at the lysosomal membrane directly correlate with CMA activity and, consequently, are subjected to tight regulation (Cuervo and Dice, 2000b). Under specific circumstances (i.e. oxidative stress), lysosomal LAMP-2A levels increase through transcriptional induction and de novo synthesis (Kiffin et al., 2004). However, in most cases, changes in the lysosomal levels of LAMP-2A are regulated directly at the lysosomal membrane and do not require de novo synthesis of LAMP-2A. LAMP-2A is also subjected to a tightly regulated degradation at the lysosomal membrane through sequential cleavage by cathepsin A and a membrane-associated metalloprotease, the nature of which remains unknown (Cuervo and Dice, 2000b; Cuervo et al., 2003). The cleaved form of LAMP-2A is then released in the lysosomal lumen where it is rapidly degraded. Under conditions that require maximal CMA activation, the regulated degradation of LAMP-2A decreases (doubling its half-life) with the consequent marked increase of its lysosomal levels without requiring synthesis of new LAMP-2A protein. In addition, a luminal pool of intact LAMP-2A (Jadot et al., 1996) can also be retrieved to the lysosomal membrane during CMA activation (Cuervo and Dice, 2000b). This LAMP-2A retrieval is independent of the luminal pH and depends on the lysosomal membrane potential, the presence of CMA substrates and the levels of membrane-associated Hsc70 (Cuervo and Dice, 2000b). Retrieval of LAMP-2A to the lysosomal membrane is increased during persistent activation of CMA, for example in response to prolonged starvation. Although CMA is already activated after 10 hours of starvation – presumably through a blockage of LAMP-2A degradation – maximal activation is attained after 24 hours through the recruitment of the luminal resident pool of LAMP-2A to the membrane (Cuervo and Dice, 2000b).
The regulation of CMA through changes in lysosomal LAMP-2A highlights the importance of lateral mobility within the membrane, which has been shown to be determined by its dynamic association with lysosomal lipid microdomains (Kaushik et al., 2006). Under conditions of low CMA activity, part of LAMP-2A is recruited into regions of defined lipid composition, whereas the number of LAMP-2A molecules in these lipid microdomains is markedly reduced when CMA is activated. Accordingly, an increase in microdomain size by augmenting lysosomal cholesterol results in reduced CMA, whereas cholesterol-extracting drugs increase membrane levels of LAMP-2A and, thus, activate CMA (Kaushik et al., 2006). In fact, the regulated degradation of LAMP-2A described above occurs in these lipid microdomains, as luminal cathepsin A preferentially associates to the lysosomal membrane in these regions. By contrast, binding of substrates to LAMP-2A and its assembly into and disassembly from the multimeric CMA translocation complex only pertains to LAMP-2A molecules outside these microdomains (Kaushik et al., 2006).
Intrinsic properties of LAMP-2A are required to modulate its membrane dynamics. In addition to the GxxG motif required for multimerization (Bandyopadhyay et al., 2008), a proline residue that is present at the interface between its transmembrane and luminal regions is absolutely required for the mobilization of LAMP-2A into the lipid microdomains (Kaushik et al., 2006). Other components at the lysosomal membrane that modulate LAMP-2A dynamics are the intermediate filament protein glial fibrillary acidic protein (GFAP) and elongation factor 1α (EF1α) – a pair of interacting proteins that modify the stability of the multimeric LAMP-2A complex and the association of LAMP-2A with the lipid microdomains in a GTP-dependent manner (Bandyopadhyay et al., 2010). A lysosome-specific variant of GFAP associates with LAMP-2A multimers, therefore enhancing the stability of the complex and counteracting the disassembly-promoting effect of Hsc70. Lysosomal GFAP partitions into two subpopulations; unphosphorylated GFAP that binds to multimers of LAMP-2A and phosphorylated GFAP (GFAP-P), the latter of which is usually bound to the GTP-binding protein EF1α. Unphosphorylated GFAP has higher affinity for GFAP-P than for LAMP-2A, but formation of GFAP–GFAP-P dimers is usually prevented by the presence of EF1α bound to GFAP-P. In the presence of GTP, EF1α is released from the lysosomal membrane allowing the dissociation of GFAP from the translocation complex and its binding to GFAP-P (Bandyopadhyay et al., 2010). This dissociation favors the rapid disassembly of the LAMP-2A multimeric complex and its active mobilization to lipid microdomains for degradation. Changes in the levels of GFAP–GFAP-P, EF1-α present at the lysosomal membrane, as well as of intracellular GTP or intra-lysosomal Ca2+ (facilitating association of cathepsin A to lipid microdomains) can all contribute to modulation of CMA activity.
Physiological roles of CMA
Analyses of cellular conditions that activate CMA, substrates degraded under these conditions and the consequences of blocking this pathway in cultured cells have helped to understand the cellular functions of CMA (Dice, 2007; Cuervo, 2010). CMA is activated in response to nutrient deprivation in almost all cell types. In contrast to other autophagic processes that are upregulated as early as 30 minutes after access to nutrients has been limited, activation of CMA starts later (after more than 10 hours into the starvation process) (Backer and Dice, 1986), reaches a plateau of maximal activation ~36 hours following the onset of starvation and remains active for up to 3 days (Cuervo et al., 1995a). The broad range of CMA substrates that are degraded during starvation suggests that most of them are degraded to provide free amino acids for the synthesis of essential cellular proteins. The selectivity of CMA for individual soluble proteins might allow cells to degrade proteins that are no longer needed without affecting levels of proteins that are required under these stress conditions. In support of the idea that CMA contributes free amino acids for protein synthesis and/or energy, reduced ATP levels have been detected during starvation of cells with compromised CMA (Massey et al., 2006).
The other function of CMA that is common to all cell types is the selective removal of altered or damaged proteins. This function becomes particularly important during exposure to stressors that generate protein damage, such as mild oxidative stress. In fact, CMA is upregulated during oxidative stress (Kiffin et al., 2004; Finn and Dice, 2005) and the inability to upregulate CMA renders cells susceptible to oxidative agents (Massey et al., 2006). Cells also upregulate CMA during exposure to toxic compounds that target and denature cytosolic proteins (Cuervo et al., 1999), which are then selectively removed by lysosomes via CMA.
Also described have been a growing number of specialized functions for CMA that are linked to the cell type in which CMA activation occurs or the specific protein degraded by this pathway. CMA contributes, among others, to the regulation of neuronal survival through the degradation of the neuronal survival factor MEF2D (Yang et al., 2009), regulation of growth of tubular kidney cells through degradation of the transcription factor Pax2 (Sooparb et al., 2004), antigen presentation in dendritic cells (Zhou et al., 2005), and control of NF-κB-mediated transcription in response to nutritional stress through the degradation of IκB (Cuervo et al., 1998).
Pathology of CMA
Decreased CMA activity has been described in numerous cells types and tissues of old rodents and in cells of older adult human subjects (Dice, 1982; Cuervo and Dice, 2000a). The functional decline of CMA occurs gradually with age and has been attributed primarily to the age-dependent reduction of LAMP-2A levels at the lysosomal membrane (Cuervo and Dice, 2000a). These lower levels of LAMP-2A do not result from transcriptional downregulation of the Lamp2 gene with age, altered splicing, reduced synthesis or problems with targeting of the LAMP-2A protein to the lysosomal membrane during lysosomal biogenesis. Instead, a reduced stability of LAMP-2A at the lysosomal membrane with increasing age appears to contribute to the lower LAMP-2A content in aged organisms (Kiffin et al., 2007). The observed switch from a regulated degradation at the lysosomal membrane to a randomly enhanced degradation of LAMP-2A in the lysosomal lumen probably results from undesired post-translational modifications of lysosomal membrane components or changes in the lipid composition of the lysosomal membrane. Recent studies in a transgenic mouse model, in which normal levels of LAMP-2A are preserved until late in life, have confirmed that the functional decline of CMA contributes to different aspects of the aging phenotype, such as alterations in cellular homeostasis and in the response of the cell and organ to stress, which contribute to the functional compromise of aged organisms (Zhang and Cuervo, 2008).
A primary defect in CMA activity has also been described in some neurodegenerative disorders, such as Parkinson's disease and certain tauopathies (Cuervo et al., 2004; Martinez-Vicente et al., 2008; Wang et al., 2009). In both cases, the basis for the CMA dysfunction is the aberrant binding of pathogenic proteins that are known to accumulate in cells affected by these disorders to CMA components at the lysosomal membrane. For example, α-synuclein (Cuervo et al., 2004; Martinez-Vicente et al., 2008; Xilouri et al., 2009; Mak et al., 2010) and UCHL1 (Kabuta et al., 2008), proteins associated with Parkinson's disease pathogenesis, bind with abnormally high affinity to LAMP-2A at the lysosomal membrane. In the case of α-synuclein, this tight binding has been shown to inhibit CMA of other cytosolic proteins, thus rendering cells more susceptible to stressors and unable to accommodate stress or the energetic demands of nutritional deprivation (Massey et al., 2006). CMA is also perturbed by mutant forms of Tau, a cytoskeleton-associated protein responsible for cellular toxicity in tauopathies and in Alzheimer's disease (Wang et al., 2009). A particular mutant of Tau is targeted to lysosomes for CMA degradation but, despite its high-affinity binding to the lysosomal membrane, fails to completely translocate. The part of the protein that has already gained access to the lysosomal lumen undergoes sequential cleavage, which generates highly amyloidogenic peptides that oligomerize (Wang et al., 2009). These irreversible oligomeric complexes of fragmented Tau that form at the lysosomal membrane interfere with normal CMA activity and, eventually, destabilize lysosomes. Dysfunctional CMA has also been described in some lysosomal storage disorders (Cuervo et al., 2003; Venugopal et al., 2009), in the diabetic kidney (Sooparb et al., 2004), different types of toxic nephropathy (Cuervo et al., 1999) and in oncogenic processes (Welsch et al., 2010).
Perspectives
Although CMA was identified as an autophagic process more than 20 years ago, its molecular dissection, physiological relevance and the links between CMA malfunctioning and disease have only been established in recent years. The identification of proteins dedicated to this pathway, such as LAMP-2A, now permits genetic manipulation of this autophagic process and direct analysis of the consequences at the cellular and organism level. As in any developing field, there are still many questions that require further clarification regarding CMA. For example, the specific roles of each of the co-chaperones that associate to the substrate–Hsc70 complex in the cytosol and at the lysosomal membrane remain poorly characterized, as are the energetic requirements for CMA. Although ATP is required, it is not clear whether it is necessary for substrate unfolding or translocation across the membrane. Other outstanding issues include the signaling mechanisms that connect the different stressors to CMA activation and the possible contribution of CMA to the degradation of proteins located in other subcellular compartments during their transit through the cytosol. The molecular players that mediate coordinated activity of CMA with different autophagic pathways (for example, a blockage of CMA results in the compensatory activation of macroautophagy) and with other proteolytic pathways, such as the proteasome, are also not fully elucidated. Finally, alterations in CMA activity in other pathologies also require further investigations.
Acknowledgments
Work in our laboratory is supported by NIH grants from NIA (AG021904, AG031782), NIDKK (DK041918), NINDS (NS038370) and a Hirsch/Weill-Caulier Career Scientist Award. S.K. is supported by a NIA Training Grant and R.K. by a Ruth L. Kirschstein fellowship. Deposited in PMC for release after 12 months.
References
- Agarraberes F., Terlecky S., Dice J. (1997). An intralysosomal hsp70 is required for a selective pathway of lysosomal protein degradation. J. Cell Biol. 137, 825-834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aniento F., Roche E., Cuervo A. M., Knecht E. (1993). Uptake and degradation of glyceraldehyde-3-phosphate dehydrogenase by rat liver lysosomes. J. Biol. Chem. 268, 10463-10470 [PubMed] [Google Scholar]
- Aniento F., Papavassiliou A. G., Knecht E., Roche E. (1996). Selective uptake and degradation of c-Fos and v-Fos by rat liver lysosomes. FEBS Lett. 390, 47-52 [DOI] [PubMed] [Google Scholar]
- Backer J., Dice J. (1986). Covalent linkage of ribonuclease S-peptide to microinjected proteins causes their intracellular degradation to be enhanced by serum withdrawal. Proc. Nat. Acad. Sci. USA 83, 5830-5834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandyopadhyay U., Kaushik S., Varticovski L., Cuervo A. M. (2008). The chaperone-mediated autophagy receptor organizes in dynamic protein complexes at the lysosomal membrane. Mol. Cell. Biol. 28, 5747-5763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandyopadhyay U., Sridhar S., Kaushik S., Kiffin R., Cuervo A. M. (2010). Identification of regulators of chaperone-mediated autophagy. Mol. Cell 39, 535-547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang H., Terlecky S., Plant C., Dice J. F. (1989). A role for a 70-kilodalton heat shock protein in lysosomal degradation of intracellular proteins. Science 246, 382-385 [DOI] [PubMed] [Google Scholar]
- Cuervo A. M. (2010). Chaperone-mediated autophagy: selectivity pays off. Trends Endocrinol. Metab. 21, 142-150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuervo A. M., Dice J. F. (1996). A receptor for the selective uptake and degradation of proteins by lysosomes. Science 273, 501-503 [DOI] [PubMed] [Google Scholar]
- Cuervo A. M., Dice J. F. (2000a). Age-related decline in chaperone-mediated autophagy. J. Biol. Chem. 275, 31505-31513 [DOI] [PubMed] [Google Scholar]
- Cuervo A. M., Dice J. F. (2000b). Regulation of lamp2a levels in the lysosomal membrane. Traffic 1, 570-583 [DOI] [PubMed] [Google Scholar]
- Cuervo A. M., Dice J. F. (2000c). Unique properties of lamp2a compared to other lamp2 isoforms. J. Cell Sci. 113, 4441-4450 [DOI] [PubMed] [Google Scholar]
- Cuervo A. M., Terlecky S. R., Dice J. F., Knecht E. (1994). Selective binding and uptake of ribonuclease A and glyceraldehyde-3-phosphate dehydrogenase by rat liver lysosomes. J. Biol. Chem. 269, 26374-26380 [PubMed] [Google Scholar]
- Cuervo A. M., Knecht E., Terlecky S. R., Dice J. F. (1995a). Activation of a selective pathway of lysosomal proteolysis in rat liver by prolonged starvation. Am. J. Physiol. 269, C1200-C1208 [DOI] [PubMed] [Google Scholar]
- Cuervo A. M., Palmer A., Rivett A. J., Knecht E. (1995b). Degradation of proteasomes by lysosomes in rat liver. Eur. J. Biochem. 227, 792-800 [DOI] [PubMed] [Google Scholar]
- Cuervo A. M., Dice J. F., Knecht E. (1997). A population of rat liver lysosomes responsible for the selective uptake and degradation of cytosolic proteins. J. Biol. Chem. 272, 5606-5615 [DOI] [PubMed] [Google Scholar]
- Cuervo A. M., Hu W., Lim B., Dice J. F. (1998). IkappaB is a substrate for a selective pathway of lysosomal proteolysis. Mol. Biol. Cell 9, 1995-2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuervo A. M., Hildebrand H., Bomhard E. M., Dice J. F. (1999). Direct lysosomal uptake of alpha2-microglobulin contributes to chemically induced nephropathy. Kidney Int. 55, 529-545 [DOI] [PubMed] [Google Scholar]
- Cuervo A. M., Gomes A. V., Barnes J. A., Dice J. F. (2000). Selective degradation of annexins by chaperone-mediated autophagy. J. Biol. Chem. 275, 33329-33335 [DOI] [PubMed] [Google Scholar]
- Cuervo A. M., Mann L., Bonten E., d'Azzo A., Dice J. (2003). Cathepsin A regulates chaperone-mediated autophagy through cleavage of the lysosomal receptor. EMBO J. 22, 12-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuervo A. M., Stefanis L., Fredenburg R., Lansbury P. T., Sulzer D. (2004). Impaired degradation of mutant alpha-synuclein by chaperone-mediated autophagy. Science 305, 1292-1295 [DOI] [PubMed] [Google Scholar]
- Dice J. F. (1982). Altered degradation of proteins microinjected into senescent human fibroblasts. J. Biol. Chem. 257, 14624-14627 [PubMed] [Google Scholar]
- Dice J. F. (1990). Peptide sequences that target cytosolic proteins for lysosomal proteolysis. Trends Biochem. Sci. 15, 305-309 [DOI] [PubMed] [Google Scholar]
- Dice J. F. (2007). Chaperone-mediated autophagy. Autophagy 3, 295-299 [DOI] [PubMed] [Google Scholar]
- Eskelinen E. L., Cuervo A. M., Taylor M. R., Nishino I., Blum J. S., Dice J. F., Sandoval I. V., Lippincott-Schwartz J., August J. T., Saftig P. (2005). Unifying nomenclature for the isoforms of the lysosomal membrane protein LAMP-2. Traffic 6, 1058-1061 [DOI] [PubMed] [Google Scholar]
- Finn P. F., Dice J. F. (2005). Ketone bodies stimulate chaperone-mediated autophagy. J. Biol. Chem. 280, 25864-25870 [DOI] [PubMed] [Google Scholar]
- Jadot M., Wattiaux R., Mainferme F., Dubois F., Claessens A., Wattiaux-De Coninck S. (1996). Soluble form of Lamp II in purified rat liver lysosomes. Biochem. Biophys. Res. Comm. 223, 353-359 [DOI] [PubMed] [Google Scholar]
- Kabuta T., Furuta A., Aoki S., Furuta K., Wada K. (2008). Aberrant interaction between Parkinson disease-associated mutant UCH-L1 and the lysosomal receptor for chaperone-mediated autophagy. J. Biol. Chem. 283, 23731-23738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushik S., Cuervo A. M. (2009). Methods to monitor chaperone-mediated autophagy. Methods Enzymol. 452, 297-324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushik S., Massey A. C., Cuervo A. M. (2006). Lysosome membrane lipid microdomains: novel regulators of chaperone-mediated autophagy. EMBO J. 25, 3921-3933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiffin R., Christian C., Knecht E., Cuervo A. (2004). Activation of chaperone-mediated autophagy during oxidative stress. Mol. Biol. Cell 15, 4829-4840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiffin R., Kaushik S., Zeng M., Bandyopadhyay U., Zhang C., Massey A. C., Martinez-Vicente M., Cuervo A. M. (2007). Altered dynamics of the lysosomal receptor for chaperone-mediated autophagy with age. J. Cell Sci. 120, 782-791 [DOI] [PubMed] [Google Scholar]
- Kon M., Cuervo A. M. (2010). Chaperone-mediated autophagy in health and disease. FEBS Lett. 584, 1399-1404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Wang P., Song W., Sun X. (2009). Degradation of regulator of calcineurin 1 (RCAN1) is mediated by both chaperone-mediated autophagy and ubiquitin proteasome pathways. FASEB J. 23, 3383-3392 [DOI] [PubMed] [Google Scholar]
- Mak S. K., McCormack A. L., Manning-Bog A. B., Cuervo A. M., Di Monte D. A. (2010). Lysosomal degradation of alpha-synuclein in vivo. J. Biol. Chem. 285, 13621-13629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Vicente M., Talloczy Z., Kaushik S., Massey A., Mazzulli J., Mosharov E., Hodara R., Fredenburg R., Wu D., Follenzi A., et al. (2008). Dopamine-modified alpha-synuclein blocks chaperone-mediated autophagy. J. Clin. Invest. 118, 777-788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massey A. C., Kaushik S., Sovak G., Kiffin R., Cuervo A. M. (2006). Consequences of the selective blockage of chaperone-mediated autophagy. Proc. Nat. Acad. Sci. USA 103, 5905-5910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N., Levine B. (2010). Autophagy in mammalian development and differentiation. Nat. Cell Biol. 12, 823-830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N., Levine B., Cuervo A. M., Klionsky D. J. (2008). Autophagy fights disease through cellular self-digestion. Nature 451, 1069-1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orenstein S. J., Cuervo A. M. (2010). Chaperone-mediated autophagy: Molecular mechanisms and physiological relevance. Semin. Cell Dev. Biol. 21, 719-726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothenberg C., Srinivasan D., Mah L., Kaushik S., Peterhoff C. M., Ugolino J., Fang S., Cuervo A. M., Nixon R. A., Monteiro M. J. (2010). Ubiquilin functions in autophagy and is degraded by chaperone-mediated autophagy. Hum. Mol. Genet. 19, 3219-3232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvador N., Aguado C., Horst M., Knecht E. (2000). Import of a cytosolic protein into lysosomes by chaperone-mediated autophagy depends on its folding state. J. Biol. Chem. 275, 27447-27456 [DOI] [PubMed] [Google Scholar]
- Sooparb S., Price S. R., Shaoguang J., Franch H. A. (2004). Suppression of chaperone-mediated autophagy in the renal cortex during acute diabetes mellitus. Kidney Int. 65, 2135-2144 [DOI] [PubMed] [Google Scholar]
- Venugopal B., Mesires N., Kennedy J., Curcio-Morelli C., Laplante J. M., Dice J. F., Slaugenhaupt S. A. (2009). Chaperone-mediated autophagy is defective in mucolipidosis type IV. J. Cell. Physiol. 219, 344-353 [DOI] [PubMed] [Google Scholar]
- Wang Y., Martinez-Vicente M., Kruger U., Kaushik S., Wong E., Mandelkow E. M., Cuervo A. M., Mandelkow E. (2009). Tau fragmentation, aggregation and clearance: the dual role of lysosomal processing. Hum. Mol. Genet. 18, 4153-4170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsch T., Younsi A., Disanza A., Rodriguez J. A., Cuervo A. M., Scita G., Schmidt J. (2010). Eps8 is recruited to lysosomes and subjected to chaperone-mediated autophagy in cancer cells. Exp. Cell Res. 316, 1914-1924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wing S., Chiang H. L., Goldberg A. L., Dice J. F. (1991). Proteins containing peptide sequences related to KFERQ are selectively depleted in liver and heart, but not skeletal muscle, of fasted rats. Biochem. J. 275, 165-169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xilouri M., Vogiatzi T., Vekrellis K., Park D., Stefanis L. (2009). Abberant alpha-synuclein confers toxicity to neurons in part through inhibition of chaperone-mediated autophagy. PLoS ONE 4, e5515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q., She H., Gearing M., Colla E., Lee M., Shacka J. J., Mao Z. (2009). Regulation of neuronal survival factor MEF2D by chaperone-mediated autophagy. Science 323, 124-127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Klionsky D. J. (2010). Mammalian autophagy: core molecular machinery and signaling regulation. Curr. Opin. Cell Biol. 22, 124-131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Cuervo A. M. (2008). Restoration of chaperone-mediated autophagy in aging liver improves cellular maintenance and hepatic function. Nat. Med. 14, 959-965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D., Li P., Lin Y., Lott J. M., Hislop A. D., Canaday D. H., Brutkiewicz R. R., Blum J. S. (2005). Lamp-2a facilitates MHC class II presentation of cytoplasmic antigens. Immunity 22, 571-581 [DOI] [PubMed] [Google Scholar]