Abstract
Phagocytosis of the shed outer segment discs of photoreceptors is a major function of the retinal pigmented epithelium (RPE). We demonstrate for the first time that βA3/A1-crystallin, a major structural protein of the ocular lens, is expressed in RPE cells. Further, by utilizing the Nuc1 rat, in which the βA3/A1-crystallin gene is mutated, we show that this protein is required by RPE cells for proper degradation of outer segment discs that have been internalized in phagosomes. We also demonstrate that in wild-type RPE, βA3/A1-crystallin is localized to the lysosomes. However, in the Nuc1 RPE, βA3/A1-crystallin fails to translocate to the lysosomes, perhaps because misfolding of the mutant protein masks sorting signals required for proper trafficking. The digestion of phagocytized outer segments requires a high level of lysosomal enzyme activity, and cathepsin D, the major enzyme responsible for proteolysis of the outer segments, is decreased in mutant RPE cells. Interestingly, our results also indicate a defect in the autophagy process in the Nuc1 RPE, which is probably also linked to impaired lysosomal function, because phagocytosis and autophagy might share common mechanisms in degradation of their targets. βA3/A1-crystallin is a novel lysosomal protein in RPE, essential for degradation of phagocytosed material.
Keywords: βA3/A1-crystallin, Cathepsin D, Retinal pigmented epithelium (RPE), Photoreceptor outer segment discs, Lysosome, Phagocytosis, Autophagy
Introduction
The retinal pigmented epithelium (RPE) is a single layer of pigmented and polarized cells, with the apical surface facing the photoreceptors and the basal side facing Bruch's membrane. The RPE serves many crucial physiological roles within the eye; phagocytosis of the outer segments of photoreceptors is one of its most important functions (Strauss, 2005). Each photoreceptor maintains a constant length by generating new outer segment discs at its base while releasing mature spent discs to the subretinal space. Phagocytosis of the shed discs by the RPE is essential to the survival of photoreceptor cells (Kevany and Palczewski, 2010). Several lines of evidence link abnormal phagocytic function of the RPE to degenerative retinal diseases.
Elegant studies have shown that phagocytosis by the RPE uses many of the same molecular components involved in phagocytosis by macrophages (Kevany and Palczewski, 2010). RPE cells are polarized and, therefore, the phagocytosis of the outer segment discs in vivo occurs at the apical surface of the RPE cells, which face the photoreceptor layer of the neurosensory retina (Strauss, 2005). Once the discs are internalized, the phagosome moves from the apical surface of the RPE to its basal surface, where the contents of the phagosome are degraded (Hoppe et al., 2004). It has been estimated that 30,000 outer segment discs are phagocytized each day by RPE cells in a single rat eye (Bok and Young, 1979).
A number of molecules that are essential in outer segment recognition, binding and internalization have been identified (Boesze-Battaglia and Albert, 1992; Burnside and Bost-Usinger, 1998; Finnemann et al., 1997; Finnemann and Silverstein, 2001; Nandrot et al., 2004; Nandrot et al., 2007; Ryeom et al., 1996a; Ryeom et al., 1996b). A defect in the initial stages of RPE phagocytosis can lead to photoreceptor death, as seen in the Royal College of Surgeons (RCS) rat. This rat carries a mutation in the Mertk (a receptor tyrosine kinase) gene that impairs the phagocytic ability of the RPE (D'Cruz et al., 2000; Gal et al., 2000). Once internalized in the RPE cells, the phagosome proceeds through a maturation process, followed by fusion with lysosomes to form phagolysosomes (Bosch et al., 1993; Hoppe et al., 2004; Kevany and Palczewski, 2010). The process of phagosome maturation and final degradation of photoreceptor outer segments in the RPE cell remains obscure (Deguchi et al., 1994; Kevany and Palczewski, 2010). Incomplete digestion of outer segments in the RPE also results in the formation of lipofuscin, an autofluorescent cellular waste product, which accumulates inside the RPE (Wolf, 2003). Although lipofuscin is a heterogeneous mixture of complex molecules, the main component is N-retinylidene-N-retinylethanolamine (A2E) and its photoisomers. A2E accumulates and remains within intralysosomal storage bodies (Eldred and Lasky, 1993).
In this study, we demonstrate for the first time that βA3/A1-crystallin, a major structural protein of the ocular lens, is expressed in RPE cells. βA3- and βA1-crystallins are two separate polypeptides produced from a single βA3/A1-crystallin mRNA by leaky ribosomal scanning (Werten et al., 1999). These polypeptides, like other β-crystallins, comprise two domains, and in the lens form mixed dimers or higher oligomers with homologous β-crystallin polypeptides. Each β-crystallin domain comprises two β-stranded greek key motifs (Dolinska et al., 2009). By utilizing the Nuc1 mutant rat, in which the βA3/A1-crystallin gene has undergone mutation (Sinha et al., 2008), we show that this protein is required by RPE cells for the proper degradation of outer segment discs that have been internalized in phagosomes. In the normal RPE, βA3/A1-crystallin is localized to the lysosomes. In the RPE of Nuc1 rats, βA3/A1-crystallin fails to translocate into the lysosomes and is found in the cytosol. Electron microscopy clearly shows that the mutant RPE cells have an impaired ability to degrade rod outer segments, resulting in accumulation of autofluorescent cellular waste products. Although some proteins that are transported from the rough endoplasmic reticulum via the Golgi apparatus to the lysosomes for participation in the phagosomal degradation process have been identified (Braulke and Bonifacino, 2009; Lubke et al., 2009; Saftig and Klumperman, 2009; van Meel and Klumperman, 2009), it is clear that some are yet to be identified, especially in RPE. Our findings implicate βA3/A1-crystallin as a novel lysosomal component required by the RPE for the proper degradation of the shed outer segment discs of photoreceptors.
Results
βA3/A1-crystallin is expressed in RPE cells
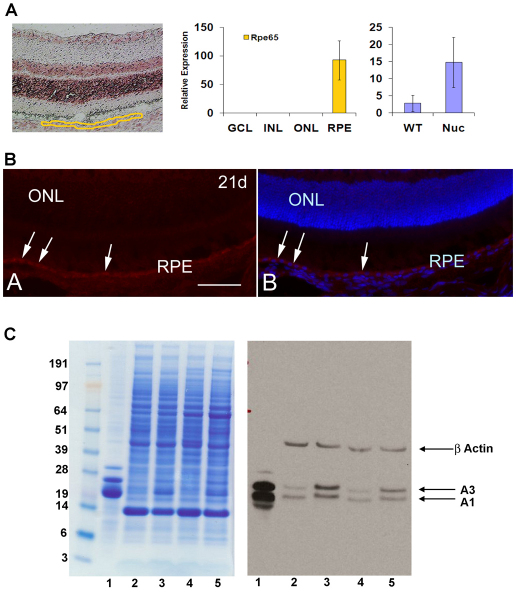
We have previously reported that βA3/A1-crystallin is expressed only in the ganglion cell layer in the neural retina. This observation was made by laser capture microdissection (Sinha et al., 2008), which we also used here to demonstrate that βA3/A1-crystallin is expressed in the RPE. Fig. 1A shows the region sampled for this analysis (left panel) and demonstrates the purity of the preparation (middle panel) as determined by the expression of the RPE-specific marker Rpe65 (Hamel et al., 1993). The data suggest, based on the mRNA levels, that expression of the βA3/A1-crystallin gene might be higher in the RPE of the Nuc1 mutant rat than in normal RPE (right panel). The presence of βA3/A1-crystallin protein in RPE was demonstrated by immunohistochemical analysis with an antibody specific for βA3/A1-crystallin on a 21-day-old wild-type retina (Fig. 1B). Further confirmation of this finding was obtained by western blotting of protein extracts from RPE plus choroid preparations from wild-type and Nuc1 rats (Fig. 1C). Samples from animals 3 weeks and 3 months of age are shown, as well as a normal rat lens protein sample that was used as a positive control. The left panel shows the staining patterns for each sample, whereas the right panel shows the western blot probed with the anti-βA3/A1-crystallin antibody. Bands for both βA3- and βA1-crystallin (produced from a single gene using two different ATG start sites) are evident in each sample. Note that both proteins are slightly larger (1083 Da) in the Nuc1 samples because of the nine additional amino acid residues resulting from the Nuc1 insertion mutation (Sinha et al., 2008) and that, consistent with the mRNA data, the protein level appears to be increased in the RPE of the mutant animals.
Fig. 1.
βA3/A1-crystallin in the RPE. (A) Laser-capture microdissection. The left panel shows the region (circled in yellow) taken as the retinal pigmented epithelium (RPE); the center panel shows the specificity of expression of the RPE marker RPE65 to this sample. Markers specific for the other retinal layers were not detected in the RPE preparation (data not shown). The right panel shows relative mRNA levels for βA3/A1-crystallin in the wild-type and Nuc1 RPE samples. Error bars indicate s.e. (B) The presence of βA3/A1-crystallin protein in the RPE of a wild-type 21-day-old rat by immunostaining with an antibody specific for βA3/A1-crystallin. The moderately intense red fluorescence indicated by the arrows represents βA3/A1-crystallin. In the right panel, nuclei are labeled with DAPI for orientation, showing the outer nuclear layer (ONL) and the RPE cell nuclei. (C) Western blotting confirms the presence of βA3/A1-crystallin in RPE. The left panel shows the staining pattern and the right panel the western blot from an identical gel run in parallel and probed with antiserum specific for βA3/A1-crystallin. Lane 1 is a normal rat lens sample run as a positive control. Lanes 2 and 3 were loaded with RPE extracts from 3-week-old wild-type and Nuc1 rats, respectively. Lanes 4 and 5 were loaded with wild-type and Nuc1 RPE extracts from 3-month-old rats, respectively. Both the βA3 and βA1 polypeptides are detected in all samples. Note that the βA3 and βA1 polypeptides migrate slightly slower in the Nuc1 samples, consistent with the insertion mutation, which increases the molecular mass by about 1000 Da. Scale bar: 50 μm
Effects of the Nuc1 mutation on RPE cell ultrastructure
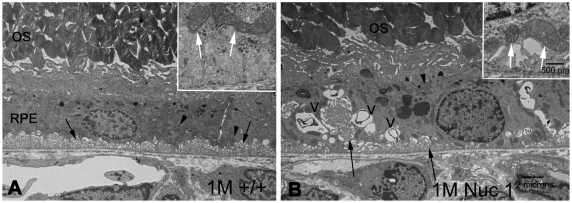
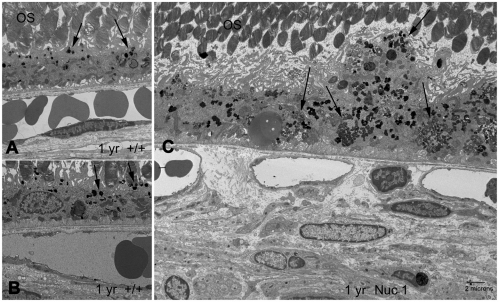
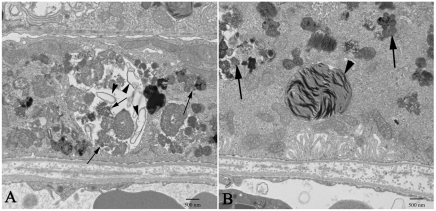
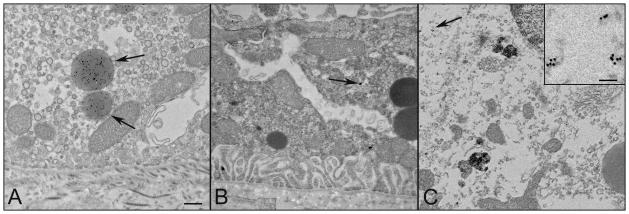
Transmission electron microscopy (TEM) was used to compare the cellular structure of the RPE in Nuc1 and wild-type rats. Fig. 2 shows representative images for 1-month-old animals (n=7 for wild type; n=7 for Nuc1). In Nuc1 RPE cells (Fig. 2B), basal infoldings appear disorganized (arrows), and more-numerous vacuole-like structures (V) were evident compared with wild-type RPE cells (Fig. 2A). No differences in mitochondria (arrowhead) were observed between wild-type and Nuc1 RPE. This is shown at higher magnification in insets (Fig. 2A,B, white arrows). The accumulation of undigested outer segment discs in Nuc1 RPE cells progressed with age (data not shown). At 1 year of age, wild-type RPE (Fig. 3A,B) shows some accumulation of lipofuscin-like particles (arrows) in the apical cytoplasm, whereas in Nuc1 animals, the RPE cells are filled with large aggregates of lipofuscin-like material (Fig. 3C, arrows) throughout the entire cytoplasm. Nuc1 photoreceptor outer segments (OS), however, look similar to wild-type outer segments (Figs 2, 3). Eyes from four wild-type animals and eight Nuc1 animals were analyzed. Fig. 4 shows in greater detail the abnormal structures present in the Nuc1 RPE at 1 year of age. Fig. 4A shows a large vacuole containing both partially degraded cellular organelles (arrowheads) and lipofuscin-like aggregates (arrows). Fig. 4B shows similar structures plus a phagosome containing undigested photoreceptor outer segment discs (arrowhead).
Fig. 2.
Transmission electron microscopy of RPE from 1-month-old rats. Photoreceptor outer segments (OS) are shown at the top and choriocapillaris at the bottom; both appear to be normal in wild-type and Nuc1 animals. (A) Wild type. Arrows indicate basal infoldings of RPE; arrowheads show normal-appearing mitochondria. Insert shows higher magnification (white arrows). (B) Nuc1. Basal infoldings appear disrupted (arrows) and mitochondria look normal (arrowheads). Insert shows higher magnification (white arrows). Many vacuole-like structures (V) are seen in the Nuc1 RPE cytoplasm. Degenerated membrane-bound cellular organelles are present inside the vacuoles. Many undigested outer segment structures are seen inside the RPE. n=7 wild-type animals and 7 Nuc1 animals. Scale bars: 2 μm in A,B, main panels; 500 nm in insets.
Fig. 3.
Transmission electron microscopy of RPE from 1-year-old rats. (A,B) In the 1-year-old wild-type rats, normal accumulation of lipofuscin-like particles (arrows) are seen in the apical cytoplasm of RPE cells. (C) In Nuc1 rats, the RPE cell is filled with large aggregates of lipofucsin-like material (arrows) that are present throughout the entire cytoplasm of the RPE cells. Some cells bulge beyond the limits of the monolayer into the subretinal space. n=4 wild-type animals and 8 Nuc1 animals. Scale bar: 2 μm.
Fig. 4.
At higher magnification, prominent features of RPE cells from 1-year-old Nuc1 animals are shown. (A) An RPE cell showing large aggregates of lipofuscin-like material. A large vacuole contains many degenerated cellular organelles (arrowheads) intermixing with lipofuscin-like aggregates (arrows). (B) Abundant lipofuscin-like aggregates (arrows) and a large phagosome containing undigested outer segment discs (arrowhead) are present in the cytoplasm of an RPE cell. Scale bars: 500 nm.
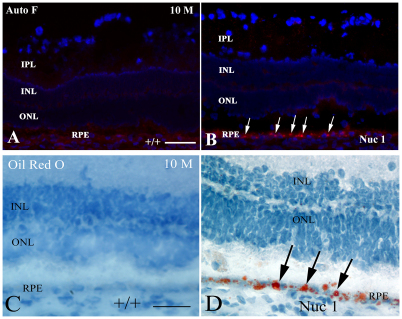
Impaired outer segment degradation in RPE of Nuc1 rats
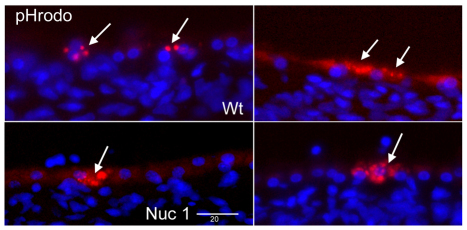
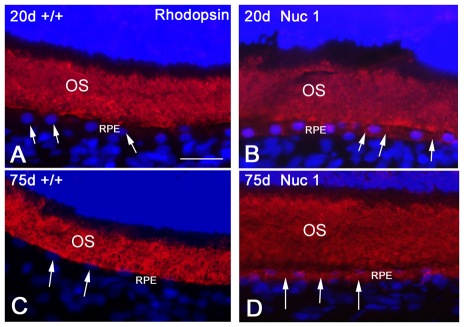
The TEM data indicate an accumulation of undigested outer segment discs and debris in the Nuc1 RPE. This is substantiated by an increased level of lipofuscin autofluorescence as seen in 10-month-old Nuc1 RPE (Fig. 5B) compared with wild type (Fig. 5A) at the same age. Oil Red O staining, indicative of neutral lipid accumulation, is barely detectable in the wild-type RPE (Fig. 5C). However, as shown in Fig. 5D, RPE cells of Nuc1 rats stain strongly with Oil Red O (arrows). Analysis of the ability of RPE cells, in situ, to phagocytize fluorescently labeled bovine rod outer segments revealed that wild-type and Nuc1 cells ingested the material equally well, as demonstrated by the fluorescent microscopy images in Fig. 6. In Fig. 7, an anti-rhodopsin antibody was used to compare, by immunohistochemistry, the amount of this protein in the RPE of wild-type and Nuc1 rats. Rhodopsin is not expressed by RPE but is the major protein in the photoreceptor outer segments, accounting for the heavy staining of this layer. The RPE, indicated by arrows, is the single layer of cells directly beneath the outer segments. It is clear that higher amounts of rhodopsin are present, both at 20 and 75 days, in the Nuc1 RPE (Fig. 7B,D, arrows) compared with wild type (Fig. 7A,C, arrows).
Fig. 5.
Demonstration of abnormal lipid accumulation in the Nuc1 RPE. (A,B) At 10 months of age, minimal autofluorescence is seen in the RPE cells of the wild-type animals (A). However, abundant autofluorescence (arrows) is observed in the RPE of 10-month-old Nuc1 animals, consistent with accumulation of lipofuscin (B). (C,D) Autofluorescence viewed with a red filter (560–620 nm). Oil Red O staining, indicating the presence of neutral lipids, shows abundant intense staining in the RPE cells of the 10-month-old Nuc 1 retinas (D, arrows), but not in the control wild-type animals (C). Scale bars: 50 μm.
Fig. 6.
Uptake of bovine outer segments by wild-type and Nuc1 RPE cells. Outer segments were labeled with the fluorescent dye pHrodo. In both wild-type (top two images) and Nuc 1 samples (bottom two images), the RPE from 6-week-old animals phagocytized the outer segments as indicated by the fluorescent particles within the cells (arrows). Sections are counterstained with DAPI. Scale bar: 20 μm.
Fig. 7.
Rhodopsin immunostaining in wild-type and Nuc1 RPE cells. In both 20-day-old and 75-day-old wild-type animals (A,C), weak staining is seen in the RPE cells (arrows) with anti-rhodopsin antibody. However, rhodopsin labeling is much stronger in the RPE layer in the Nuc1 animals (B,D), particularly in the older animal (D). OS, outer segment. Scale bar: 25 μm.
The primary enzyme involved in the proteolysis of rhodopsin in the RPE phagosome is cathepsin D (Bosch et al., 1993; Deguchi et al., 1994; Rakoczy et al., 1997). Fig. 8 demonstrates by western analysis that this enzyme is present at lower levels in the Nuc1 cells than in wild type at both 2 and 8 months of age; in particular, the mature active form of the enzyme is absent or greatly reduced in the mutant RPE preparation.
Fig. 8.
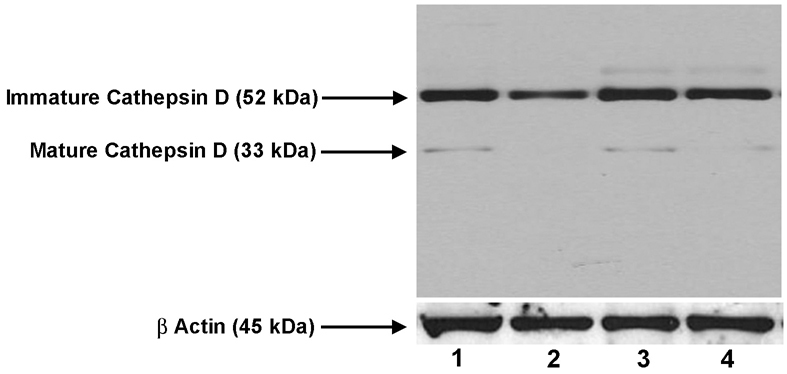
Cathepsin D in wild-type and Nuc1 RPE. Western blotting with cathepsin D antibody. RPE samples were as follows: Lane 1, 2-month-old wild type; lane 2, 2-month-old Nuc1; lane 3, 8-month-old wild type; lane 4, 8-month-old Nuc1. The immature (inactive) pro-enzyme form of cathepsin is present in all samples. The mature (active) form of the enzyme is present in the wild-type samples, but is barely detectable in the Nuc1 samples. Actin is shown as a loading control.
Intracellular localization of βA3/A1-crystallin in RPE cells
In order to localize βA3/A1-crystallin in RPE cells, immuno-electron microscopy was performed using immuno-gold labeling. Fig. 9 shows representative data from these studies. Panel A shows an image from a 5.5-month-old wild-type rat in which the gold particles are strongly localized to lysosomes (arrow). By contrast, few, if any, gold particles are present in the lysosomes of Nuc1 homozygous animals (Fig. 9B,C). Rather, the gold particles appear to be randomly scattered throughout the cytoplasm. Gold particles are less apparent in panel C because the silver enhancement step was shorter than in A or B; the inset shows the region indicated by the arrow at higher magnification to better demonstrate the gold particles. In general, there were more lysosomes present in the aged Nuc1 RPE compared with wild type. This is consistent with acid phosphatase staining (supplementary material Fig. S1), which shows stronger staining in the Nuc1 RPE relative to wild type.
Fig. 9.
Immuno-electron microscopy to determine the intracellular localization of βA3/A1-crystallin in RPE cells. (A) A representative image from a 5.5-month-old wild-type animal. The arrows indicate lysosomes, which contain nearly all the gold particles. (B) An image from an age-matched Nuc1 animal showing several lysosomes, which lack gold particles. In this sample, the gold is randomly distributed within the cytoplasm; the arrow indicates an example. (C) A 22-month-old Nuc1 animal. Part of a large lysosome is visible at the lower right and does not contain gold particles. As in B, gold particles are distributed throughout the cytoplasm. In this section, they are not as apparent because a shorter silver-enhancement step was used. To demonstrate, the particles the area indicated by the arrow are shown at higher magnification in the insert. Scale bars: 500 nm in A-C; 125 nm in C, inset.
Possible effects of the Nuc1 mutation on autophagy in RPE cells
In view of the obvious abnormalities in the lysosomes of the mutant rat RPE and the defects in degradation and elimination of phagocytized photoreceptor outer segments in the mutant cells, we have investigated the status of autophagy in the RPE of wild-type and Nuc1 rats. The marker generally used to assess autophagy is microtubule-associated protein 1A/1B light chain 3 (LC3). When autophagosomes are formed, the soluble form of LC3 (LC3-I) is conjugated with phosphatidylethanolamine and localizes to the autophagosome membrane. This form of LC3, called LC3-II, is the best indicator of autophagy activity (Glick et al., 2010). Fig. 10 compares, by western blotting, the levels of LC3 in the mutant and wild-type RPE extracts. There is a definite decrease in the amount of LC3-II in the mutant cells, consistent with a lower level of autophagy in the Nuc1 RPE.
Fig. 10.
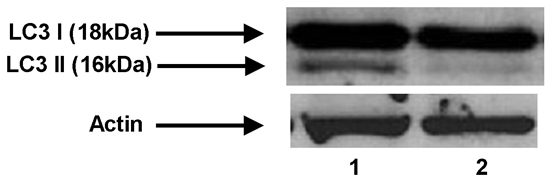
Western blotting for LC3. Lane 1 is loaded with wild-type and lane 2 with Nuc1 RPE extracts from rats 2 months of age. LC3-II, the active form of the protein, which indicates active autophagy, is markedly reduced in the Nuc1 sample. Actin immunostaining is shown as a loading control.
Molecular modeling of βA3/A1-crystallin and Nuc1 mutation
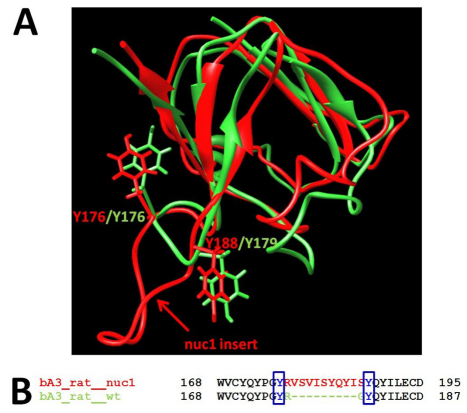
A possible explanation for the failure of the mutant protein to be properly trafficked to lysosomes is the loss of a sorting signal. One such signal important in trafficking to lysosomes is the GYxxϕ sequence motif, where ϕ is a bulky hydrophobic residue (Braulke and Bonifacino, 2009). It is of interest that residues 175–179 of βA3-crystallin (GYRGY) could potentially be such a sorting signal, and further that this sequence is the site of the Nuc1 mutation, which eliminates G178 and inserts ten new amino acid residues into the sequence. Thus, the insertion into the mutant protein would destroy this putative sorting signal. Molecular modeling of wild-type βA3/A1-crystallin and the mutant variant was performed in order to determine whether this site is exposed on the surface of the molecule where it could be recognized as a sorting signal (Fig. 11). In this figure, structures of the carboxy-terminal domains of βA3/A1-crystallin and the Nuc1 mutant variant are superimposed using the UCSF Chimera, version 1.4.1 (http://www.cgl.ucsf.edu/chimera) and shown in green and red, respectively. It is clear that the site in question is on the surface of the wild-type βA3/A1-crystallin. As we have shown previously, the ten-residue insertion in the mutant protein creates a highly hydrophobic surface loop, predicted to sterically affect dimer formation and proper protein folding of the carboxy-terminal domain (Sinha et al., 2008).
Fig. 11.
Superposition of C-terminal domains of wild-type rat βA3-crystallin and Nuc1 mutant protein. (A) Polypeptide ribbons for wild-type rat βA3-crystallin and the Nuc1 mutant are shown in green and red, respectively. The putative tyrosine-based sorting signal GYxxϕ is located at positions 175–179 on the protein surface. Tyrosines 176 and 179 are shown in green. The tyrosine-based sorting signal GYxxϕ is interrupted by the ten-residue insert located between Y176 and Y188 in the Nuc1 mutant protein (red). (B) The sequence alignment of wild-type rat βA3-crystallin and Nuc1 mutant protein. The positions of the two tyrosine residues from the putative tyrosine-based sorting signal GYxxϕ are shown in blue.
Discussion
The RPE performs several functions that are crucial for the homeostasis of the neural retina. In addition, the RPE and the photoreceptors are dependent upon each other for proper functioning. Phagocytosis of outer segment discs by the RPE is absolutely essential for maintaining a constant outer segment length (Travis et al., 2007). Although various studies have shown that phagocytosis by the RPE is different from that carried out by professional phagocytes such as macrophages and dendritic cells, several molecules involved in phagocytosis by the RPE have been shown to be identical to those known to function in general phagocytosis (Kevany and Palczewski, 2010). We provide evidence, for the first time, that βA3/A1-crystallin has a pivotal role in the degradation process following phagocytosis of outer segment discs by the RPE.
Crystallins are the very abundant soluble proteins that constitute as much as 90% of the total protein in the ocular lens (Piatigorsky, 2007) and are largely responsible for its refractivity and transparency (Delaye and Tardieu, 1983). Clearly, crystallins are specialized structural proteins in the lens but, contrary to prior belief, they are not lens-specific (Andley, 2007). It is now believed that all crystallins have been derived from pre-existing proteins that are expressed in other tissues and have other functions (Wistow, 1995). The α-crystallins are small heat-shock proteins that function, in the lens and elsewhere, as chaperones with anti-aggregative activity. We show here for the first time that βA3/A1-crystallin is expressed by RPE cells, appearing shortly after birth (Parthasarathy et al., 2011), during the time-period when the photoreceptor outer segments mature and when active shedding of outer segment discs begins (Fig. 1).
In order to understand the possible function of βA3/A1-crystallin in RPE cells, we took advantage of the Nuc1 rat model, which has a spontaneous insertion mutation in the βA3/A1-crystallin gene (Sinha et al., 2008). Molecular modeling studies indicate that the mutation creates a loop near the C-terminal end that prevents the mutant protein from folding properly and having normal protein-protein interactions. Nuc1 RPE cells appear to have an increased amount of βA3/A1-crystallin protein (Fig. 1), possibly because the mutant protein exists in a mis-folded or aggregated state and does not turnover as efficiently as the normal protein.
The process of photoreceptor disc shedding and renewal and the role for RPE in phagocytizing the shed discs have been known since the work of Young and colleagues (Young, 1967; Young and Bok, 1969; Young and Droz, 1968). Although some components involved in the process have been identified, the phagocytosis of outer segment discs by RPE cells remains poorly understood. The process of phagocytosis can be divided into four distinct stages: recognition and attachment of the phagosome, ingestion of the phagosomes, fusion with acidified vesicles containing lysosomal enzymes and, finally, digestion (Bosch et al., 1993). Our data with the Nuc1 spontaneous mutant rat indicate that βA3/A1-crystallin has no apparent role in recognition or ingestion of the phagosomes. This is based on two lines of evidence. First, engulfment of outer segments labeled with the dye pHrodo in an in situ assay clearly showed that both wild-type and Nuc1 RPE cells were able to internalize outer segments (Fig. 6). Second, there is abnormal accumulation of rhodopsin in the RPE of the Nuc1 animals (Fig. 7), which could not occur if the shed discs had not been internalized. In addition, our TEM studies show no build-up of photoreceptor outer segments between the retina and RPE in Nuc1 eyes (Figs 2, 3), as would be expected if there were, as in the RCS rat, a defect in the internalization process.
Phagolysosomes, formed by the fusion of a phagosome with a lysosome, are formed through a number of fusion stages known as phagosomal maturation (Kinchen and Ravichandran, 2008). Proteins that are involved in the phagosome maturation process in macrophages have been identified (Elliott and Ravichandran, 2010), but have not been studied in the context of RPE phagocytosis. Interestingly, we find that three such proteins, engulfment and cell motility protein 1 (Elmo1), dedicator of cytokinesis (Dock180) and Ras-related C3 botulinum toxin substrate 1 (Rac1) are also expressed in the RPE cells of the rat, indicating that a similar signaling pathway might be involved in phagosome maturation in RPE. We see no difference in the expression of these proteins between wild-type and Nuc1 RPE cells (supplementary material Fig. S2). Moreover, Rab5 GTPase, which has recently been shown to be crucial in macrophages in the final maturation process before degradation by the lysosomal enzymes occurs (Zhou and Yu, 2008), is expressed at normal levels in the Nuc1 RPE (supplementary material Fig. S2). These studies, in conjunction with the phagocytosis assay (Fig. 6) and TEM data (Figs 2, 3, 4), suggest that phagosomes form normally in the Nuc1 RPE, but that degradation of the engulfed outer segments does not occur normally (Fig. 7), perhaps as a result of a defective lysosomal digestion process.
Several proteolytic enzymes participate in the lysosomal digestion of photoreceptor outer segments but cathepsin D has been shown to be the major enzyme responsible for the proteolysis of rhodopsin (Hoppe et al., 2004; Lai et al., 2000; Kevany and Palczewski, 2010). A decrease in the activity of cathepsin D leads to the accumulation of photoreceptor-outer-segment-derived debris in RPE cells (Bosch et al., 1993; Deguchi et al., 1994; Rakoczy et al., 1997; Lai et al., 2000). Our studies clearly show that the active (34 kDa) form of cathepsin D is markedly decreased in Nuc1 RPE compared with wild type (Fig. 8). It is known that the inactive form of cathepsin D undergoes a series of activation steps in the lysosomal compartments to produce the active form, and βA3/A1-crystallin, an acidic protein, could be involved in the activation process of cathepsin D or even in the stabilization of the cathepsin D hetero-tetrameric native structure by maintaining the optimal pH necessary for the cathepsin D aspartyl-protease activity in the acidic environment of lysosomes.
The localization of βA3/A1-crystallin in the lysosomes in wild-type RPE (Fig. 9A) suggests that this protein might function as a lysosomal enzyme. When the protein fails to translocate to the lysosomes, as shown by our immuno-electron microscopy studies (Fig. 9B,C), Nuc1 RPE cells fail to clear the shed outer segment discs. We have shown previously, by molecular modeling and gel chromatography studies, that the mutant protein folds abnormally and forms aggregates in the lens (Sinha et al., 2008). Abnormal conformation is also consistent with our studies comparing the wild-type and Nuc1 mutant recombinant proteins, where the mutant protein is found in inclusion bodies rather than in the soluble fraction where the wild-type protein is found (supplementary material Fig. S3).
The failure of βA3/A1-crystallin to localize to the lysosome in the Nuc1 RPE could result from loss of a sorting signal(s) necessary for proper trafficking. Tyrosine-based sorting signals with a GYxxϕ motif (where ϕ is a bulky hydrophobic residue) have been described (Braulke and Bonifacino, 2009). βA3/A1-crystallin has such a motif at residues 175–179 (GYRGY). The codon for G178 is the site of the insertion mutation in Nuc1, which destroys this putative signaling motif. Molecular modeling demonstrates that, in the wild-type protein, this motif is on the surface of the protein (Fig. 11), where it could be recognized as a sorting signal. Further studies are required to determine whether this particular sequence is a lysosomal sorting signal for βA3/A1-crystallin.
These studies lead us to speculate that βA3/A1-crystallin functions as a lysosomal enzyme in RPE cells and that it could be essential for the digestion of engulfed photoreceptor outer segment discs. It should be noted that proteolytic activity has been reported previously for βA3/A1-crystallin (Srivastava and Srivastava, 1999; Srivastava et al., 2008). Although this activity is not fully characterized and, as described, would not be expected to function at the acidic pH of the lysosome, it does lend support to the concept that βA3/A1-crystallin might have protease activity.
It has been proposed earlier that phagocytosis and autophagy might share a common mechanism because they share key molecular regulators in the maturation process of the phagosomes and that the fusion with lysosomes is required for ultimate degradation (Kinchen and Ravichandran, 2008). In autophagy, an autophagosome fuses with a lysosome for a lysosomal-mediated destruction during cell remodeling (Glick et al., 2010; Hamasaki and Yoshimori, 2010). Interestingly, our results also show a defect in the autophagy process in Nuc1 RPE cells. Beclin 1, a protein that is required for the initiation of autophagosome formation in autophagy, is upregulated in the Nuc1 RPE cells, possibly to facilitate clearance of the debris accumulating inside the cells (data not shown). However, as the cells age and the normal clearance mechanisms fail, the loss of membrane-associated autophagy protein LC3-II in Nuc1 becomes obvious (Fig. 10), and there is significant deposition of lipid and lipofuscin-like granules in Nuc1 RPE cells (Fig. 5).
Finally, because abnormalities in the phagocytic function of RPE can result in degenerative eye diseases, it is tempting to speculate that βA3/A1-crystallin might be a relevant therapeutic target in conditions where RPE phagocytosis is impaired. Before such speculation could become practical, further investigations are needed to elucidate fully the mechanisms whereby βA3/A1-crystallin participates in the digestion of phagosomes in the RPE cell.
Materials and Methods
Animals and sample preparation
Animal experiments were performed using Nuc1 (Sinha et al., 2005) and wild-type Sprague Dawley rats in accordance with the Guide for the Care and Use of Laboratory Animals (National Academy Press) and were approved by the Animal Care and Use Committee of Johns Hopkins University. RPE plus choroid was prepared from freshly enucleated eyes by first cleaning all muscle and other external tissue from the globe then removing the anterior chamber by a circumferential cut just posterior to the limbus. The lens and vitreous were removed and then the retina was carefully teased free from the RPE and removed. Protein was extracted from the remaining tissue as described in the next section.
SDS-PAGE and western blot analysis
RPE plus choroid preparations from freshly dissected rats were rinsed in PBS and homogenized in lysis buffer (150 mM NaCl, 1% NP-40, 5 mM EDTA, 1 mM DTT, 25% glycerol, 50 mM Tris, pH 8.0) with 1% protease inhibitor cocktail (Sigma-Aldrich, St Louis, MO). Samples were incubated on a rotator at 4°C for 30 minutes followed by centrifugation at 13,000 g for 15 minutes. Approximately 25 μg of protein from the supernatant was mixed with 2× LDS sample buffer (Invitrogen, Carlsbad, CA) and then heated in a boiling water bath for 2 minutes. Each sample was loaded onto a 4–12% Bis-Tris Nu-PAGE gel and run with MES Buffer (Invitrogen). The gels were stained with Colloidal Coomassie Brilliant Blue. For western blotting, proteins were transferred to nitrocellulose membranes (Invitrogen) for 60 minutes and then blocked with 3% BSA in TTBS (Tris-buffered saline, 0.1% Tween-20) overnight at 4°C. Blots were incubated with primary antibodies to βA3/A1-crystallin (1:8000), cathepsin D (1:500, Santa Cruz Biotechnology, Santa Cruz, CA), LC3 (1:500, MBL International, Woburn, MA) or actin (1:1000, Sigma-Aldrich, St Louis, MO) for 1 hour at room temperature followed by four washes of 10 minutes each. Blots were incubated with HRP-conjugated secondary antibodies (Kirkegaard and Perry Laboratories, Gaithersburg, MD) for 1 hour at room temperature at a dilution of 1:20,000 followed by four washes of 10 minutes each. ECL western blotting detection reagents (GE Healthcare, Piscataway, NJ) were used for detection with varying exposure times. The βA3/A1-crystallin antibody was raised against a synthetic peptide sequence common to both βA3- and βA1-crystallin and was produced at Spring Valley Laboratories, Woodbine, MD. The peptide sequence is from the N-terminal region of the protein and is not affected by the mutation in the C-terminal region; the antibody therefore reacts with both normal and mutant forms of both βA3- and βA1-crystallin.
Laser-capture microdissection, RNA isolation and cDNA synthesis
Enucleated eyes were cryoprotected with an increasing concentration gradient of sucrose at 4°C. Eyes were embedded in Tissue-Tek O.C.T. compound (Sakura Finetek, Inc., Torrance, CA) with snap-freezing on dry ice then stored at −80°C. Sections (7 μm) were cut with a Microm cryostat and placed on PEN-membrane LCM slides (Leica Microsystems, Bannockburn, IL). Sections were dehydrated and stained with Mayer's Hematoxylin Solution (Sigma-Aldrich) for visualization of cell nuclei. Cells from three layers of the neural retina (ganglion cell layer: GCL, inner nuclear layer: INL, outer nuclear layer: ONL) and from the RPE were cut by a Leica LMD6000 instrument. Samples were collected in 0.5 ml tube caps in 45 μl RLT lysis buffer (Qiagen, Valencia, CA) then stored at −80°C until processed. Total RNA was purified with the RNeasy Micro kit (Qiagen) according to the manufacturer's protocol. The purified total RNA was reverse-transcribed in a 20 μl reaction containing 100 ng random hexamer (Invitrogen), 0.4 μl 25 mM dNTP Mix, 4 μl 5× First Strand Buffer (Invitrogen), 2 μl 0.1 M DTT, 1 μl RNaseOut ribonuclease inhibitor (40 U/μl; Invitrogen) and 1 μl SuperScriptIII (200 U/μl; Invitrogen). The reaction mix was incubated at 50°C for 1 hour then inactivated at 70°C for 15 minutes. The reaction mix was diluted 10-fold and used as template in quantitative real-time PCR (qRT-PCR) reactions.
qRT-PCR
A Bio-Rad IQ5 Multicolor Real-Time PCR Detection System was used for analysis. Reactions (20 μl) contained 6 pmol of each primer, 10 μl of 2× iQ SYBR Green Supermix (Bio-Rad, Hercules, CA) and 5 μl cDNA template. Cycling conditions were: 95°C for 10 seconds, 59°C for 30 seconds, 72°C for 30 seconds. Detection was carried out at 72°C after the extension step. The integrity of PCR products was verified with melting curve analysis. Relative expression was calculated using the expression of HPRT as a reference gene (Pfaffl, 2001).
Immunohistochemistry
Specimens for immunohistochemistry and histochemical analysis were obtained from freshly euthanized Nuc1 and wild-type Sprague Dawley rats. Eyes were removed immediately after the animals were euthanized, and fixed in freshly prepared 2% paraformaldehyde in phosphate-buffered saline (PF-PBS) for 3 hours, transferred to 20% sucrose for cryoprotection and then embedded in Tissue-Tek O.C.T. (Sakura, Tokyo) in chilled methyl-butane in dry ice. The blocks were kept at −80°C until sectioning. The eyeballs were serially sectioned longitudinally at 10 μm thickness. Sections were air-dried at room temperature for 10 minutes and successively incubated with 5% blocking serum in PBS for 30 minutes, followed by a primary antibody overnight at 4°C and then a secondary fluorescent antibody conjugated with either Cy2 or Cy3 (Jackson ImmunoResearch, West Grove, PA) for 1 hour at room temperature. The primary antibodies used for immunofluorescence in this study were the βA3/A1-crystallin antibody (1:400) and anti-rhodopsin monoclonal antibody (1:400, Thermo Scientific, Fremont, CA). The sections were finally counterstained with 4′,6-diamidino-2-phenylindole (DAPI; Invitrogen) and mounted with DAKO Fluorescent Mounting Medium (DAKO, Carpinteria, CA). Fluorescent digital images were taken with a Leica 6000 fluorescence microscope (Leica, Germany). Confocal microscopy was done on a Zeiss LSM 510 (Zeiss North America, Thornwell, NY).
Transmission electron microscopy
Eyes were enucleated immediately after euthanasia; a small slit was made at the limbus and then fixative was injected into the globe. Each eye was then immersed in the same fixative (2.5% glutaraldehyde with 2% paraformaldehyde in 0.1 mol/l cacodylate buffer, pH 7.4). After removal of the anterior segment, the eyecups were washed in 0.05 mol/l cacodylate buffer and post-fixed in 1% OsO4 in 0.05 mol/l cacodylate buffer for 90 minutes. The eyecups were then washed in 0.05% cacodylate buffer and dehydrated in a graded series of ethyl alcohol and stained en bloc with 1% uranyl acetate in 100% ethanol. The tissue was placed in propylene oxide twice for 15 minutes each time and then was kept overnight in a 1:1 propylene oxide and resin mixture. The tissue was infiltrated in 100% LX112 resin (Ladd Research Industry, Burlington, VT) for 4–6 hours under vacuum, embedded in a final change of 100% LX112 resin and then polymerized at 60°C for 36–48 hours. Ultrathin sections were cut with a Leica Ultramicrotome UCT (Leica Microsystems, Wetzlar, Germany), stained with uranyl acetate and lead citrate and visualized with an H7600 transmission electron microscope (Hitachi, Tokyo, Japan). Images analyzed were from the central retina, 1–2 disc widths from the optic nerve.
Immuno-electron microscopy
Immuno-gold labeling was performed using a pre-embedding incubation protocol. Briefly, the animals were deeply anesthetized and perfused for 10 minutes with 1X PBS with 0.1% heparin followed by 0.1% glutaraldehyde with 4% paraformaldehyde in 0.1 M phosphate buffer. The eyes were enucleated and immediately immersed in the same fixative for 50 minutes at 4°C followed by several washes in 0.1 M phosphate buffer. The anterior chambers were removed and the eyecups cut in half. The tissues were then incubated in 0.1% sodium borohydride in 0.1 M phosphate buffer for 10 minutes to inactivate aldehydes. After permeabilization with 0.05% Triton X-100, tissues were blocked in 5% normal goat serum in 0.1 M phosphate buffer with BSAc (Aurion, The Netherlands) and cold water fish gelatin (Aurion). Incubation in primary antibody (rabbit anti-βA3/A1-crystallin, 1:100) was performed overnight at 4°C. Negative controls were incubated without primary antibody. After washing in PBS, eyecups were incubated overnight at 4°C in goat anti-rabbit IgG labeled with gold (Aurion Ultra Small gold, diluted 1:100), then washed with incubation buffer followed by PBS before fixing in 3% glutaraldehyde for 30–60 minutes at 4°C. Tissues were washed in 0.1 M phosphate buffer followed by distilled water, then floated in Silver Enhancement (Aurion) mixture for 30–40 minutes. After washing in distilled water, tissues were immersed in 0.5% osmium for 30 minutes, washed with distilled water, dehydrated through an alcohol series and then put into 1% uranyl acetate in 100% ethanol for 60 minutes in the dark. Tissues were embedded, ultrathin-sectioned and visualized with an H7600 transmission electron microscopy as described above. Sections were not stained with additional uranyl acetate or lead nitrate.
Oil Red O staining
One percent Oil Red O was made in 70% alcohol and acetone. The frozen sections were washed with distilled water and rinsed in 70% alcohol. The sections were then placed in Oil Red O solution for 10 minutes, counterstained in Harris Hematoxylin for 1 minute and mounted in aqueous mounting media.
Acid phosphatase staining
Acid phosphatase activity was visualized on frozen sections of wild-type and Nuc1 eyes using Naphthol AS-MX phosphoric acid, as follows. Naphthol AS-MX phosphoric acid (4 mg) was dissolved in 0.1 ml DMSO and then added to 25 ml 0.2 M acetate buffer, pH 5.2. Fast Blue RR salt (30 mg) was then added along with two drops of 10% manganese chloride. The solution was mixed and filtered through a 0.2 μm filter. Sections were incubated at 37°C for 30–60 minutes, rinsed in distilled H2O and mounted with aqueous mounting medium.
In situ phagocytosis assay
The ability of wild-type and Nuc1 RPE to phagocytize fluorescently labeled bovine photoreceptor outer segments was assessed using intact RPE layers in freshly dissected posterior eyecup preparations. The bovine photoreceptor outer segments were labeled with the pH-sensitive rhodamine-based dye pHrodo (Invitrogen) and were the kind gift of R. N. Fariss and N. V. Gordiyenko of the National Eye Institute, NIH. The isolation method for the outer segments and the labeling procedure has been described (Gordiyenko et al., 2010).
For our studies, eyes were obtained from freshly euthanized wild-type and Nuc1 rats, 6 weeks of age. The anterior segments were carefully excised and the neural retina was then teased away from the RPE and carefully removed from the eyecup. The eyecups were immersed in 500 μl medium (DMEM/F12, SAFC Biosciences, Lenexa, Kansas) in wells of a 48-well tissue culture plate and were allowed to equilibrate for 30–60 minutes in a standard incubator (37°C, 5% CO2). Fifty microliters of a suspension of outer segments in BSS (~1 mg/ml) was added to each well and incubation was continued for 90 minutes. The eyecups were then fixed in 2% paraformaldehyde and cryosectioned as described above (see Immunohistochemistry). Sections were analyzed by fluorescent microscopy as described above.
Cloning and expression of βA3-crystallin
The full-length cDNA sequences of wild-type and Nuc1 βA3/A1-crystallin were directionally cloned into pET17b vector using NdeI and XhoI sites. Recombinant vectors were initially transformed into chemically competent NEB-5α and subsequently into BL21 (DE3) for protein expression. To obtain soluble and insoluble fractions, single-colony BL21 (DE3) transformants were grown overnight in MagicMedia for T7 expression systems (Invitrogen) and aliquots subjected to sonication. The supernatants containing soluble protein, if any, were separated from the insoluble fraction by centrifugation at 12,350 g for 30 minutes at 4°C. The insoluble pellets were dissolved in 20 μl of 4% sodium dodecyl sulfate. SDS-PAGE and western blot analysis were performed as described above.
Molecular modeling
The model of the rat βA3/A1-crystallin was built by homology modeling based on crystal coordinates of the bovine βB2-crystallin (RCSB Protein Data Bank ID: 1blb.pdb) as the structural template. The primary protein sequences were aligned (Needleman and Wunch, 1970) and GENE-MINE Look software, version 3.5.2 (Lee and Subbiah, 1991) was used for the three-dimensional structure prediction. The dimeric βA3-crystallin was built using the automatic segment matching method in Look (Levitt, 1992), followed by 500 cycles of energy minimization. The same program was used to generate the conformation for the βA3-Nuc1 mutant protein, refined by self-consistent ensemble optimization (Lee, 1994), which applies the statistical mechanical mean-force approximation iteratively to achieve the global energy minimum structure. Finally, structures were minimized in the presence of water molecules by 100 cycles of steepest descent, followed by 200 cycles of conjugate gradient.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Institutes of Health: EY018636 (D.S.), EY019037 (D.S.), EY019037-S (D.S.), EY09357 (G.A.L.), EY016151-5 (G.A.L.), EY14005 (J.T.H.), EY01765 (Wilmer Imaging Core), Intramural Research Program, National Eye Institute (J.S.Z. and Y.S.), Helena Rubinstein Foundation (D.S.) and Research to Prevent Blindness (an unrestricted grant to The Wilmer Eye Institute). We thank Robert N. Fariss and Nataliya Goriyenko of the National Eye Institute for help with the in situ phagocytosis assay and the staff members at Spring Valley Laboratories, Woodbine, MD, for taking care of the experimental animals. We thank Noriko Esumi and Morton Goldberg for critical reading and discussion regarding this manuscript. Deposited in PMC for release after 12 months.
Footnotes
Supplementary material available online at http://jcs.biologists.org/cgi/content/full/124/4/523/DC1
References
- Andley U. P. (2007). Crystallins in the eye: function and pathology. Prog. Retin. Eye Res. 26, 78-98 [DOI] [PubMed] [Google Scholar]
- Boesze-Battaglia K., Albert A. D. (1992). Phospholipid distribution among bovine rod outer segment plasma membrane and disk membrane. Exp. Eye Res. 54, 821-823 [DOI] [PubMed] [Google Scholar]
- Bok D., Young R. W. (1979). Phagocytic properties of the retinal pigment epithelium. In The Retinal Pigment Epithelium (ed. Marmor M. F., Zinn K. M.), pp. 148-174 Cambridge, MA: Harvard University Press; [Google Scholar]
- Bosch E., Horwitz J., Bok D. (1993). Phagocytosis of outer segments by retinal pigment epithelium: phagosome-lysosome interaction. J. Histochem. Cytochem. 41, 253-263 [DOI] [PubMed] [Google Scholar]
- Braulke T., Bonifacino J. S. (2009). Sorting of lysosomal proteins. Biochim. Biophys. Acta 1793, 605-614 [DOI] [PubMed] [Google Scholar]
- Burnside B., Bost-Usinger L. (1998). The retinal pigment epithelium cytoskeleton. In The Retinal Pigment Epithelium: Function and Disease (ed. Marmor M. F., Wolfensberger T. J.), pp. 41-67 New York: Oxford University Press; [Google Scholar]
- D'Cruz P. M., Yasumura D., Weir J., Matthes M. T., Abderrahim H., LaVail M. M., Vollrath D. (2000). Mutation of the receptor tyrosine kinase gene Mertk in the retinal dystrophic RCS rat. Hum. Mol. Genet. 9, 645-651 [DOI] [PubMed] [Google Scholar]
- Deguchi J., Yamamoto A., Yoshimori T., Sugasawa K., Moriyama Y., Futai M., Suzuki T., Kato K., Uyama M., Tashiro Y. (1994). Acidification of phagosome and degradation of rod outer segments in rat retinal pigment epithelium. Invest. Ophthalmol. Vis. Sci. 35, 568-579 [PubMed] [Google Scholar]
- Delaye M., Tardieu A. (1983). Short range order of crystallin proteins accounts for eye lens transparency. Nature 302, 415 [DOI] [PubMed] [Google Scholar]
- Dolinska M. B., Sergeev Y. V., Chan M. P., Palmer I., Wingfield P. T. (2009). N-terminal extension of βB1-crystallin: identification of a critical region that modulates protein interaction with βA3-crystallin. Biochemistry 48, 9684-9695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eldred G. E., Lasky M. R. (1993). Retinal age pigments generated by self-assembling lysosomotropic detergents. Nature 361, 724-726 [DOI] [PubMed] [Google Scholar]
- Elliott M. R., Ravichandran K. S. (2010). Clearance of apoptotic cells: implications in health and disease. J. Cell Biol. 189, 1059-1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnemann S. C., Silverstein R. L. (2001). Differential roles of CD36 and αvβ5 integrin in photoreceptor phagocytosis by the retinal pigment epithelium. J. Exp. Med. 194, 1289-1298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnemann S. C., Bonilha V. L., Marmorstein A. D., Rodriguez-Boulan E. (1997). Phagocytosis of rod outer segments by retinal pigment epithelial cells requires αvβ5 integrin for binding binding but not for internalization. Proc. Natl. Acad. Sci. USA 94, 12932-12937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gal A., Li Y., Thompson D. A., Weir J., Orth U., Jacobson S. G., Apfelstedt-Sylla E., Vollrath D. (2000). Mutations in MERTK, the human orthologue of the RCS rat retinal dystrophy gene, cause retinitis pigmentosa. Nat. Genet. 26, 270-271 [DOI] [PubMed] [Google Scholar]
- Glick D., Barth S., Macleod K. F. (2010). Autophagy: cellular and molecular mechanisms. J. Pathol. 221, 3-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordiyenko N. V., Fariss R. N., Zhi C., MacDonald I. M. (2010). Silencing of the CHM gene alters phagocytic and secretory pathways in the retinal pigment epithelium. Invest. Ophthalmol. Vis. Sci. 51, 1143-1150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamasaki M., Yoshimori T. (2010). Where do they come from? Insights into autophagosome formation. FEBS Lett. 584, 1296-1301 [DOI] [PubMed] [Google Scholar]
- Hamel C. P., Tsilou E., Pfeffer B. A., Hooks J. J., Detrick B., Redmond T. M. (1993). Molecular cloning and expression of RPE65, a novel retinal pigment epithelium-specific microsomal protein that is post-transcriptionally regulated in vitro. J. Biol. Chem. 268, 15751-15757 [PubMed] [Google Scholar]
- Hoppe G., O'Neil J., Hoff H. F., Sears J. (2004). Accumulation of oxidized lipid-protein complexes alters phagosome maturation in retinal pigment epithelium. Cell. Mol. Life Sci. 61, 1664-1674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kevany B. M., Palczewski K. (2010). Phagocytosis of retinal rod and cone photoreceptors. Physiology 25, 8-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinchen J. M., Ravichandran K. S. (2008). Phagosome maturation: going through the acid test. Nat. Rev. Mol. Cell Biol. 9, 781-795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai C. M., Robertson T., Papadimitriou J., Shen W. Y., Daw N., Constable I. J., Rakoczy P. E. (2000). Controlled production of active Cathepsin D in Retinal Pigment Epithelial cells following adenovirus-mediated gene delivery. Mol. Ther. 2, 476-484 [DOI] [PubMed] [Google Scholar]
- Lee C. (1994). Predicting protein mutant energetic by self-consistent ensemble optimization. J. Mol. Biol. 236, 918-939 [DOI] [PubMed] [Google Scholar]
- Lee C., Subbiah S. (1991). Prediction of protein side chain conformation by packing optimization. J. Mol. Biol. 217, 373-388 [DOI] [PubMed] [Google Scholar]
- Levitt M. (1992). Accurate modeling of protein conformation by automatic segment matching J. Mol. Biol. 226, 507-533 [DOI] [PubMed] [Google Scholar]
- Lubke T., Lobel P., Sleat D. (2009). Proteomics of the lysosome. Biochim. Biophys. Acta 179, 625-635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandrot E. F., Kim Y., Brodie S. E., Huang X., Sheppard D., Finnemann S. C. (2004). Loss of synchronized retinal phagocytosis and age-related blindness in mice lacking αvβ5 integrin. J. Exp. Med. 200, 1539-1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandrot E. F., Anand M., Almeida D., Atabai K., Sheppard D., Finnemann S. C. (2007). Essential role for MFG-E8 as ligand for αvβ5 integrin in diurnal retinal phagocytosis. Proc. Natl. Acad. Sci. USA 104, 12005-12010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Needleman S. B., Wunch C. D. (1970). A general method applicable to the search for similarities in the amino acid sequence of two proteins. J. Mol. Biol. 48, 443-453 [DOI] [PubMed] [Google Scholar]
- Parthasarathy G., Ma B., Zhang C., Gongora C., Samuel Zigler J., Jr, Duncan M.K., Sinha D. (2011) Expression of βA3/A1-crystallin in the developing and adult rat eye. J. Mol. Histol. [Epub ahead of print] PMID: 21203897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl M. W. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29, 45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piatigorsky J. (2007). Gene Sharing and Evolution: The Diversity of Protein Function. Cambridge, MA: Harvard University Press; [Google Scholar]
- Rakoczy P. E., Lai C. M., Baines M., Di Grandi S., Fitton J. H., Constable I. J. (1997). Modulation of cathepsin D activity in retinal pigment epithelial cells. Biochem. J. 324, 935-940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryeom S. W., Silverstein R. L., Scotto A., Sparrow J. R. (1996a). Binding of anionic phospholipids to retinal pigment epithelium may be mediated by the scavenger receptor CD36. J. Biol. Chem. 271, 20536-20539 [DOI] [PubMed] [Google Scholar]
- Ryeom S. W., Sparrow J. R., Silverstein R. L. (1996b). CD36 participates in the phagocytosis of rod outer segments by retinal pigment epithelium. J. Cell Sci. 109, 387-395 [DOI] [PubMed] [Google Scholar]
- Saftig P., Klumperman J. (2009). Lysosome biogenesis and lysosomal membrane proteins: trafficking meets function. Nat. Rev. Mol. Cell Biol. 10, 623-635 [DOI] [PubMed] [Google Scholar]
- Sinha D., Hose S., Zhang C., Neal R., Ghosh M., O'Brien T. P., Sundin O., Goldberg M. F., Robison G. W., Russell P., et al. (2005). Rat spontaneous mutation affects programmed cell death during the early development of the eye. Exp. Eye Res. 80, 323-335 [DOI] [PubMed] [Google Scholar]
- Sinha D., Klise A., Sergeev Y., Hose S., Bhutto I. A., Hackler L., Jr, Malpic-llanos T., Samtani S., Grebe R., Goldberg M. F., et al. (2008). βA3/A1-Crystallin in astroglial cells regulates retinal vascular remodeling during development. Mol. Cell. Neurosci. 37, 85-95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava O. P., Srivastava K. (1999). Characterization of a sodium-activatable proteinase activity associated with βA3/A1-crystallin of human lenses. Biochem. Biophys. Acta 1434, 331-346 [DOI] [PubMed] [Google Scholar]
- Srivastava O. P., Srivastava K., Chaves J. M. (2008). Isolation and characterization of βA3-crystallin associated proteinase from α-crystallin fraction of human lenses. Mol. Vis. 14, 1872-1885 [PMC free article] [PubMed] [Google Scholar]
- Strauss O. (2005). The retinal pigment epithelium in visual function. Phys. Rev. 85, 841-881 [DOI] [PubMed] [Google Scholar]
- Travis G. H., Golczak M., Moise A. R., Palczewski K. (2007). Diseases caused by defects in the visual cycle: retinoids as potential therapeutic agents. Annu. Rev. Pharmacol. Toxicol. 47, 469-512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Meel E., Klumperman J. (2008). Imaging and imagination: understanding the endo-lysosomal system. Histochem. Cell Biol. 129, 253-266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werten P. J. L., Stege G. J. J., de Jong W. W. (1999). The short 5′ untranslated region of the βA3/A1-crystallin mRNA is responsible for leaky ribosomal scanning. Mol. Biol. 26, 201-205 [DOI] [PubMed] [Google Scholar]
- Wistow G. (1995). Molecular Biology and Evolution of Crystallins: Gene Recruitment and Multifunctional Proteins in the Eye Lens. New York: Springer; [Google Scholar]
- Wolf G. (2003). Lipofuscin and macular degeneration. Nutr. Rev. 61, 342-346 [DOI] [PubMed] [Google Scholar]
- Young R. W. (1967). The renewal of photoreceptor cell outer segments. J. Cell Biol. 33, 61-72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young R. W., Bok D. (1969). Participation of the retinal pigment epithelium in the rod outer segment renewal process. J. Cell Biol. 42, 392-403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young R. W., Droz B. (1968). The renewal of protein in retinal rods and cones. J. Cell Biol. 39, 169-184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z., Yu X. (2008). Phagosomematuration during the removal of apoptotic cells: receptors lead the way. Trends Cell Biol. 18, 474-485 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.