Abstract
Stil (Sil, SCL/TAL1 interrupting locus) is a cytosolic and centrosomal protein expressed in proliferating cells that is required for mouse and zebrafish neural development and is mutated in familial microcephaly. Recently the Drosophila melanogaster ortholog of Stil was found to be important for centriole duplication. Consistent with this finding, we report here that mouse embryonic fibroblasts lacking Stil are characterized by slow growth, low mitotic index and absence of clear centrosomes. We hypothesized that Stil regulates mitosis through the tumor suppressor Chfr, an E3 ligase that blocks mitotic entry in response to mitotic stress. Mouse fibroblasts lacking Stil by genomic or RNA interference approaches, as well as E9.5 Stil−/− embryos, express high levels of the Chfr protein and reduced levels of the Chfr substrate Plk1. Exogenous expression of Stil, knockdown of Chfr or overexpression of Plk1 reverse the abnormal mitotic phenotypes of fibroblasts lacking Stil. We further demonstrate that Stil increases Chfr auto-ubiquitination and reduces its protein stability. Thus, Stil is required for centrosome organization, entry into mitosis and cell proliferation, and these functions are at least partially mediated by Chfr and its targets. This is the first identification of a negative regulator of the Chfr mitotic checkpoint.
Keywords: Centrosome, Chfr, Mitosis, Sil, Stil
Introduction
The STIL (SCL/TAL1 interrupting locus) gene was cloned from a common chromosomal rearrangement associated with T cell acute lymphoblastic leukemia (Aplan et al., 1991). It encodes a 150 kDa cytosolic protein with no homology to other known proteins and is expressed in proliferating cells (Izraeli et al., 1997). Mice lacking a functional Stil protein die at mid-gestation with marked growth retardation, defects in the developing neural fold and loss of left–right symmetry (Izraeli et al., 1999). Similarly, inactivating mutations in zebrafish Stil cause disseminated apoptosis in the developing nervous system associated with mitotic defects (Pfaff et al., 2007). The same study demonstrated localization of Stil to the centrosomes of HeLa cells during metaphase. Recently, several homozygous mutations in STIL were identified in autosomal-recessive primary microcephaly (MCPH7, OMIM # 612703) (Kumar et al., 2009). Interestingly, the other five genes known to be mutated in this disorder – MCPH1, CDK5RAP2, ASPM, CENPJ and CEP152 – also encode centrosomal proteins (Graser et al., 2007; Guernsey et al., 2010; Lin et al., 2005; Tibelius et al., 2009; Woods et al., 2005; Zhong et al., 2005). It has thus been proposed that primary microcephaly is caused by a reduced number of neurons owing to defective mitoses of fetal neural precursors (Bond et al., 2005; Thornton and Woods, 2009). Together, these observations suggest that STIL might affect neural development through a role in mitosis and centrosomal biology. A recent study has demonstrated that the Drosophila Ana2 protein, which is related to the Caenorhabditis elegans SAS-5 protein and to human STIL by sequence similarity of a C-terminal ‘STAN’ motif, is essential for centriole duplication (Stevens et al., 2010). This implies that STIL might regulate centrosome structure and integrity through its function in centriole duplication.
Our previous studies have demonstrated that STIL is a mitotic regulator. The STIL protein accumulates during the cell cycle, reaching peak levels in G2 phase, and then degrades upon exit from mitosis (Campaner et al., 2005; Izraeli et al., 1997). STIL is expressed in multiple cancers and its expression is correlated with an elevated mitotic index and cancer progression (Erez et al., 2004; Ramaswamy et al., 2003). STIL expression is regulated by E2F1, a transcription factor that also regulates the expression of mitotic checkpoint genes such as MAD2 that are coexpressed with STIL (Erez et al., 2008; Erez et al., 2004). Knockdown of STIL by RNA interference in cancer cells causes a marked delay in entry into mitosis associated with reduced activation of CDK1 (cyclin B kinase) (Erez et al., 2007).
The Chfr protein (checkpoint with FHA and RING finger domains) is a tumor suppressor that delays entry into mitosis in response to mitotic stress (Kang et al., 2002; Scolnick and Halazonetis, 2000; Summers et al., 2005; Yu et al., 2005). It is an E3 ubiquitin ligase that ubiquitinates its substrates through its RING finger domain (Kang et al., 2002). Ubiquitination by Chfr leads to degradation of mitotic kinases including Plk1, delaying the activation of Cdc25c phosphatase, and consequently delayed activation of Cdk1 and entry into mitosis (Kang et al., 2002; Shtivelman, 2003; Yu et al., 2005). Ubiquitination of target proteins is required for Chfr activity also in a proteosome-independent manner (Matsusaka and Pines, 2004). Its subcellular localization might be cell-cycle-dependent. It has been shown to localize in nuclear promyelocytic leukemia (PML) bodies and to regulate nuclear dynamics and genomic stability (Daniels et al., 2004; Kwon et al., 2009; Oh et al., 2009). Other studies have demonstrated localization at the spindle poles during metaphase and interaction with microtubule-associated proteins (Burgess et al., 2008; Maddika and Chen, 2009). Chfr might auto-regulate itself by auto-ubiquitination (Chaturvedi et al., 2002). However, the upstream negative regulators of Chfr are unknown. Thus, the mechanisms of Chfr activity and regulation are incompletely understood.
Because Stil is required for entry into mitosis and Chfr is known to regulate the G2–M transition, we hypothesized that Stil might be involved in the regulation of Chfr. The present study demonstrates that Stil has a role in centrosome organization, entry into mitosis and cell proliferation, and that these functions are at least partially mediated by Chfr. In this regard, we show that overexpression of Stil results in increased auto-ubiquitination and reduced protein stability of Chfr.
Results
Mitotic defects in mouse embryonic fibroblasts lacking Stil
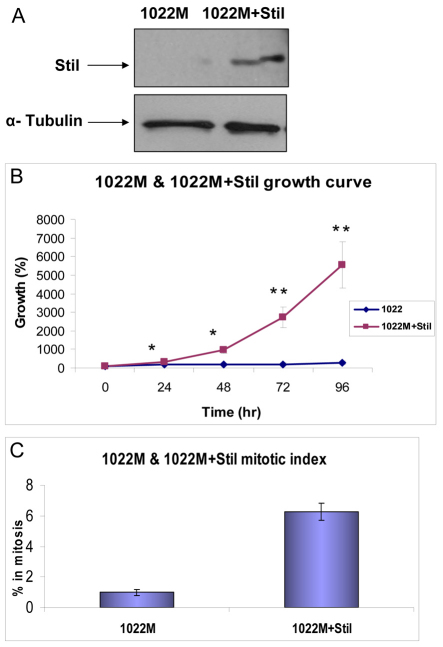
Mouse embryonic fibroblasts (MEFs) derived from Stil knockout embryos (1022M) are characterized by a low proliferation rate (doubling time of 72 hours) and a low mitotic index (1%) (Fig. 1; supplementary material Table S1). A similar phenotype was observed in 3T3 fibroblasts expressing a short hairpin RNA (shRNA) targeting Stil (supplementary material Table S1). Restoration of Stil expression in Stil knockout 1022M MEFs by retroviral transduction (1022M+Stil) resulted in increased cell growth (doubling time of 18 hours) and a higher mitotic index (6%) (Fig. 1; supplementary material Table S1).
Fig. 1.
Reduced proliferation and mitoses in mouse fibroblasts lacking Stil. (A) Immunoblotting for Stil in embryonic fibroblasts derived from Stil−/− embryos (1022M) compared with the same cells stably transduced with a retroviral vector encoding Stil (1022M+Stil). α-tubulin was used as a loading control. (B) Growth curve analysis of 1022M and 1022M+Stil. The doubling times of 1022M and 1022M+Stil are 72 hours and 18 hours, respectively. t-test: *, P<0.03, **, P<0.02. (C) Mitotic index of both cell lines. Error bars in B and C indicate s.e.
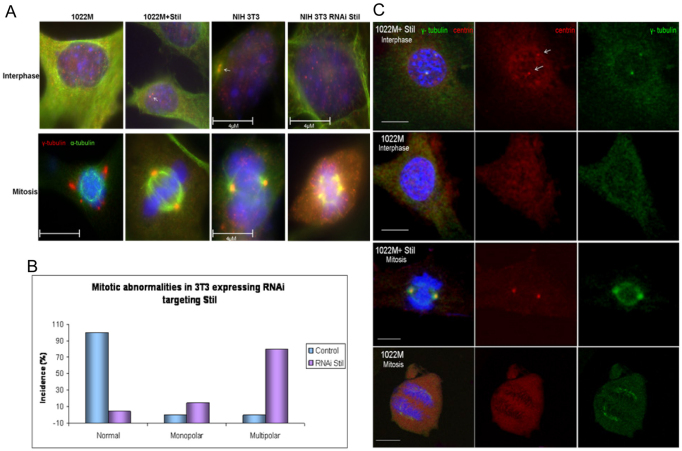
Because Stil is localized to the centrosomes during metaphase (Pfaff et al., 2007), we analyzed the status of centrosomes during interphase and mitosis in 1022M, 1022M+Stil and 3T3 fibroblasts with and without shRNA-mediated knockdown of Stil (Fig. 2A). The cells were immunostained for γ-tubulin (centrosomes) and α-tubulin (microtubules). Non-infected 3T3 and 1022M+Stil MEFs have two focused centrosomes during interphase and bipolar spindles with one centrosome per spindle pole during mitosis. By contrast, knockout or knockdown of Stil as in 1022M and 3T3 fibroblasts, respectively, was associated with an absence of identifiable centrosomes during interphase and multiple spindle poles with disrupted γ-tubulin signals during mitosis (Fig. 2B; supplementary material Table S1).
Fig. 2.
Impaired expression of Stil results in centrosomal and mitotic abnormalities. (A) Immunostaining for α-tubulin (green) and γ-tubulin (red) in interphase (upper panel) and mitosis (lower panel). Centrosomes are marked by white arrows. (B) Mitotic abnormalities in the absence of Stil. 3T3 cells were infected with virions (pLKO-Stil) encoding shRNA targeting Stil or a control sequence. The majority of the observed mitoses in Stil knockdown cells are multipolar compared with cells expressing normal levels of Stil. (C) Centrin 1 staining in 1022M cells demonstrates that lack of Stil expression results in an absence of centrioles in interphase and mitosis compared with 1022M+Stil cells (centrioles are marked by white arrows). DAPI shows chromosomes, γ-tubulin shows centrosomes and centrin 1 shows centrioles. The images were acquired using Zeiss LSM510 confocal microscope. Magnitude: ×63. Scale bar: 10 μM.
Because Ana2, a suggested Stil ortholog in Drosophila, was shown to be important for centriole duplication, we next analyzed whether the abnormal centrosomal phenotypes in Stil knockout and knockdown cells are associated with centriole abnormalities. To do this, 1022M and 1022M+Stil MEFs were co-immunostained with antibodies to centrin (which marks centrioles) and γ-tubulin following analysis by confocal microscopy. In contrast to 1022M+Stil MEFs, 1022M cells show no centrin signals that colocalize with γ-tubulin patches in mitosis, implying that the structures present in cells lacking Stil are clusters of γ-tubulin without associated centrioles rather than functional centrosomes. In addition, no detectible centrioles were found in 1022M cells during interphase, implying that Stil is crucial for the integrity of the centrosomes through its functions in the biology of the centrioles.
Stil negatively regulates Chfr protein levels in vitro and in vivo
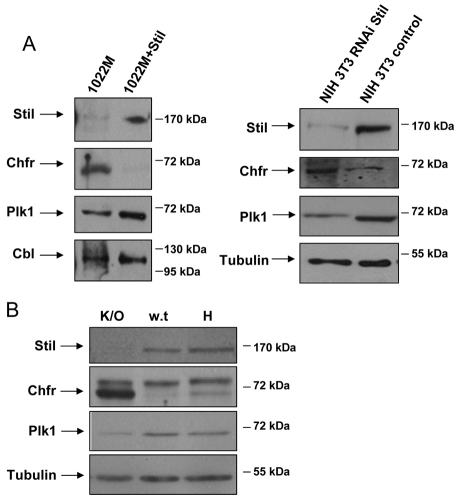
As Stil is crucial for entry to mitosis and Chfr inhibits this process, we examined Chfr levels in cells lacking Stil. Chfr protein levels were higher in Stil knockout MEFs (1022M) compared with 1022M+Stil cells (Fig. 3A). Knockdown of Stil in 3T3 fibroblasts also resulted in high levels of Chfr protein compared with control cells (Fig. 3A). This increase in Chfr protein levels was accompanied with a modest reduction in the protein levels of the mitotic kinase Plk1 (Fig. 3A). This reduction could be due to direct ubiquitination of Plk1 by Chfr (Kang et al., 2002), or due to a more general effect on the cell cycle.
Fig. 3.
Inverse correlation between Stil and Chfr protein levels in vitro and in vivo. (A) Stil, Chfr and Plk1 protein levels in 1022M and 1022M+Stil cells (left panel), and in 3T3 cells expressing shRNA targeting Stil (right panel). (B) Immunoblot analysis of protein lysates from E9.5 embryos derived from mating between Stil+/− mice. K/O=Stil−/−, WT=Stil+/+, H=Stil+/−. α-Tubulin was used as loading control.
To test whether this upregulation of Chfr levels occurs also in vivo, we examined Chfr protein levels in Stil−/− embryos. These embryos die by embryonic day (E) 10.5, exhibiting marked growth retardation and significantly fewer cells compared with wild-type (WT) embryos (Izraeli et al., 1999). Embryos from timed matings of Stil+/− mice were harvested on day E9.5. Subsequently, protein was extracted and analyzed by western blot for Chfr and Plk1 (Fig. 3B). Compared with WT and Stil heterozygous embryos, Stil knockout embryos expressed markedly higher levels of Chfr protein and lower levels of Plk1. Thus, the upregulation of Chfr caused by the absence of Stil is also observed in vivo.
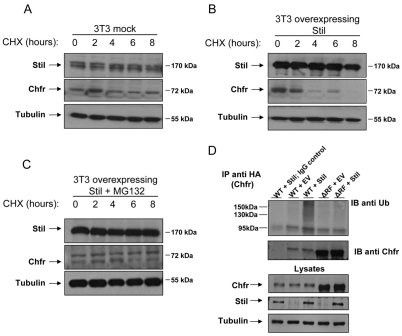
The changes in the levels of Chfr in the Stil knockout and knockdown cells were not correlated with changes in Chfr mRNA levels (supplementary material Fig. S1), suggesting that Stil might affect Chfr protein stability rather than its transcription. We therefore examined the stability of the Chfr protein in cells with different levels of Stil. 3T3 cells were transfected with Stil or control vectors and were exposed to cycloheximide (CHX) two days after transfection for the indicated time-points and analyzed for Stil and Chfr protein levels (Fig. 4A,B; supplementary material Fig. S2 for mRNA levels). We found that overexpression of Stil was associated with a significantly decreased stability of Chfr. Addition of a proteasome inhibitor (MG132) blocked the reduction of Chfr in the presence of overexpressed Stil (Fig. 4C), suggesting that Stil destabilizes Chfr via proteasome-mediated degradation.
Fig. 4.
Stil overexpression destabilizes the Chfr protein in a proteasome-dependent manner. (A,B) 3T3 cells were transfected with control (A) or Stil (B) encoding vectors. Forty-eight hours post-transfection, the cells were treated with cyclohexamide (CHX, 1 μg/ml) for the indicated time-points (in hours). Overexpression of Stil decreases Chfr levels at 4 hours, whereas at 8 hours no Chfr protein is detectable compared with its stability in the control. (C) 3T3 cells transfected with Stil and treated with CHX and proteasome inhibitor (MG132) show relatively stable Chfr protein compared with CHX alone. α-tubulin was used as a loading control. (D) HEK293T cells were transiently transfected with vectors encoding HA–Chfr wild-type (WT) or an HA–Chfr ΔRF mutant (ΔRF) and Myc-tagged ubiquitin, with either empty vector (EV) or with Stil. The cells were treated with MG132 (10 μM) and lysates were immunoprecipitated for Chfr (IP anti-HA). HA immunoprecipitates were immunoblotted for ubiquitin (IB anti-Ub) and Chfr (IB anti-Chfr). Ubiquitinated Chfr is evident by the smear of higher-molecular-weight bands. Whole-cell lysates (lysates, lower panel) were immunoblotted for Chfr, Stil and α-tubulin (loading control). For the IP negative control, we used lysate from cells expressing HA–Chfr WT, Myc–Ub and Stil that was precipitated only with the anti-mouse IgG-conjugated beads (WT+Stil; IgG control).
Proteasomal degradation of Chfr is regulated by its auto-ubiquitination (Chaturvedi et al., 2002). To examine the effect of Stil on the ubiquitination levels of Chfr, we examined HEK293T cells expressing Chfr with or without overexpression of Stil (Fig. 4D). In cells cotransfected with Chfr and Stil, the ubiquitination of Chfr was elevated compared with cells cotransfected with Chfr and empty vector. Because auto-ubiquitination is the only known mechanism by which Chfr is ubiquitinated (Chaturvedi et al., 2002), we tested whether mutant Chfr lacking its RING finger domain (ΔRF), which is crucial for its E3 ligase activity (Fukuda et al., 2008), is still ubiquitinated in the presence of high Stil levels. We found that the ΔRF mutant is not ubiquitinated, although it was expressed at much higher levels than the WT Chfr protein (Fig. 4D). This finding was further corroborated by demonstrating that a Chfr construct with a single point mutation in its leucine 306 residue, known to be well conserved within a subgroup of RING finger domains and required for its ubiquitin ligase activity (Kang et al., 2002; Kwon et al., 2009), led to a similar pattern of ubiquitination (supplementary material Fig. S3). Together, these experiments demonstrate that Stil expression is associated with increased auto-ubiquitination and proteasomal degradation of Chfr.
The mitotic phenotypes of cells lacking Stil depend on Chfr
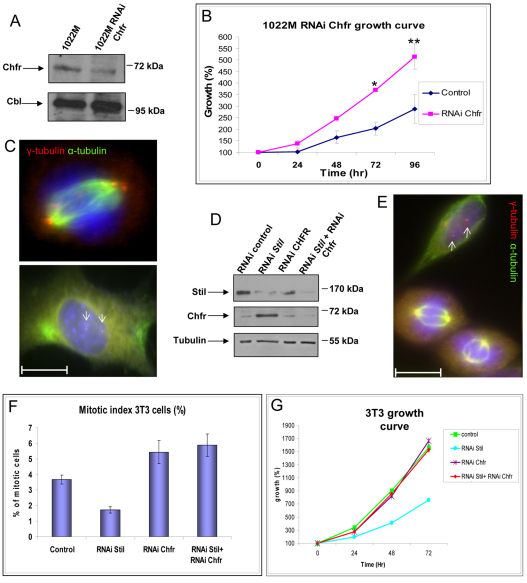
To explore whether the stabilization of Chfr observed in the cells lacking Stil (Fig. 3A) accounts for the abnormal mitotic phenotypes, we transduced 1022M cells with lentiviruses encoding shRNA targeting Chfr and generated stable polyclonal populations of 1022M expressing RNAi to Chfr. (Fig. 5A-C; Table 1). These cells demonstrated a higher mitotic index and a higher proliferation rate compared with 1022M cells transduced with a control vector (72 hours and 30 hours doubling time for 1022M and 1022M-RNAi Chfr cells, respectively). Downregulation of Chfr in 1022M cells also partially restored the integrity of centrosomes in interphase cells and the bipolarity of mitotic spindle poles. Similarly, double knockdown of Chfr and Stil in 3T3 cells corrected the growth and mitotic phenotypes compared with Stil knockdown alone (Fig. 5D–G; Table 1 for statistics). Chfr localizes to the spindle poles during mitosis (Burgess et al., 2008), but no evidence of its direct localization or function at centrosomes during interphase has been previously reported. We now demonstrate colocalization of endogenous Chfr and γ-tubulin by immunostaining of 3T3 cells (supplementary material Fig. S4), confirming the centrosomal localization of Chfr during interphase. Together, these observations suggest that the mitotic arrest and centrosomal abnormalities caused by the absence of Stil are at least partially mediated by Chfr.
Fig. 5.
Knockdown of Chfr in Stil-depleted cells restores growth rate and spindle bipolarity. (A) Immunoblot confirming Chfr knockdown in 1022M cells expressing shRNA targeting Chfr. Cbl was used as a loading control. (B) Increased growth rate of 1022M cells after shRNA knockdown of Chfr (from a doubling time of 72 hours to 30 hours). t-test: *, P<0.003, **, P<0.05. Error bars indicate s.e. (C) Immunostaining for α-tubulin (green) and γ-tubulin (red) in 1022M cells expressing shRNA targeting Chfr demonstrates partial correction of the abnormal centrosomal phenotypes caused by absence of Stil (centrosomes are marked by white arrows). (D) Immunoblotting of 3T3 cells (polyclonal populations) stably expressing shRNA targeting Stil, Chfr or both. (E) Immunostaining for α-tubulin (green) and γ-tubulin (red) shows restoration of bipolar centrosomal phenotypes in 3T3 cells with knockdown of Stil and Chfr. (F,G) Analysis of mitotic index (F) and growth rate (G) demonstrates that knockdown of Chfr partially corrects the defects caused by knockdown of Stil in these 3T3 cells. Error bars indicate s.e. Scale bars: 11 μM.
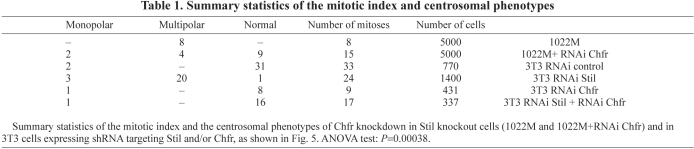
Table 1.
Summary statistics of the mitotic index and centrosomal phenotypes
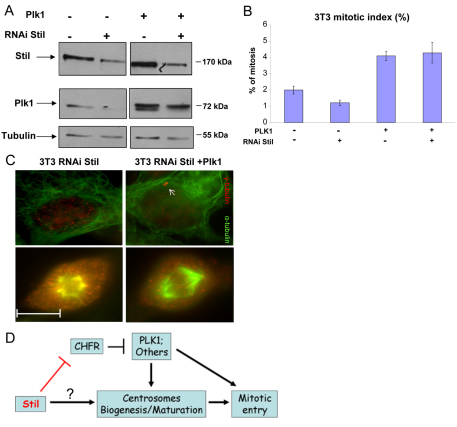
Because the elevated levels of Chfr in the absence of Stil are associated with lower levels of Plk1, we next examined if forced expression of Plk1 can rescue the Stil knockdown phenotype as well. 3T3 fibroblasts were cotransfected with a vector encoding shRNA targeting Stil and an expression vector encoding Plk1. After verification of Stil downregulation and Plk1 protein overexpression (Fig. 6A), the cells were stained for α-tubulin and γ-tubulin to examine the mitotic index and the centrosomal phenotypes. Exogenous expression of Plk1 increased the mitotic index in Stil-knockdown 3T3 cells from 1.25% without Plk1 to 4.3% with Plk1 (Fig. 6B). Importantly, the centrosomal defects decreased as a consequence of Plk1 overexpression. All observed mitoses were bipolar, with properly organized spindles (Fig. 6C). Also, centrosomes in these cells were clearly visible during interphase. Thus, the centrosomal phenotype in the absence of Stil can be corrected by overexpression of Plk1.
Fig. 6.
Partial correction of the mitotic phenotype of Stil knockdown in fibroblasts by ectopic expression of Plk1. 3T3 cells were transfected with vectors encoding shRNA targeting Stil (RNAi Stil) or control sequence, with or without a vector encoding Plk1. (A) Immunoblot for Stil and Plk1 levels. The endogenous Plk1 is the lower band at the transfected lanes, and the transfected Plk1 is the upper band. Note that the levels of the endogenous Plk1 decrease when Stil is downregulated. (B) Mitotic index of the transfected 3T3 cells. (C) Immunostaining for α-tubulin (green) and γ-tubulin (red) of the transfected 3T3 cells shows that transfection of Plk1 partially corrects the centrosomal aberrations caused by the absence of Stil (centrosomes are marked by white arrows). Upper panel, interphase; lower panel, mitosis. Scale bar: 11 μM. (D) A diagram depicting the pathway discovered in this manuscript. The mechanisms by which Chfr regulates centrosomal biology and other downstream components of Stil need to be elucidated by additional research.
Discussion
Stil, Chfr and mitosis
Mitosis is a highly regulated process that ensures faithful segregation of chromosomes into two daughter cells. This regulation is maintained by careful coordination of the protein levels of many mitotic regulators. Too much or too little of these proteins impairs mitosis (De Luca et al., 2006; Lu et al., 2008; Sotillo et al., 2007; Sumara et al., 2004; Toyoda and Yanagida, 2006). Chfr is an E3 ligase that delays entry into mitosis in response to mitotic stress and might also have further roles in spindle formation, centrosome separation and chromosome segregation. Little is known about how Chfr is regulated. It is probably degraded through auto-ubiquitination and stabilized by the de-ubiquitination enzyme USP7 (HAUSP) (Oh et al., 2007).
Here, we show that the Stil protein is a negative regulator of Chfr. Genomic or RNAi-mediated elimination of Stil in mouse fibroblasts did not affect Chfr mRNA levels but was associated with increased levels of Chfr protein. This Chfr accumulation was reversed upon reintroduction of Stil. This marked accumulation of Chfr was also observed in Stil knockout embryos in vivo. Conversely, overexpression of Stil induced Chfr auto-ubiquitination and destabilized the endogenous protein in a proteasome-dependent manner. The mechanism by which Stil negatively regulates Chfr is not clear. One possibility is that Stil acts as a scaffold protein that facilitates Chfr degradation similarly to the recently described role of Dyrk2 in ubiquitination and degradation of the mitotic regulator katanin (Maddika and Chen, 2009). This kind of regulation requires interaction between the two proteins. As we now confirm a previous report (Burgess et al., 2008) that Chfr is, like Stil, a centrosomal protein, such a direct interaction is probable. However, we have been unable so far to convincingly demonstrate direct interaction between the two proteins and hence an indirect mechanism by which Stil regulates Chfr stability is also possible.
Stil has previously been shown to be involved in mitosis but the mechanism of its activity has remained unclear. Stil protein accumulates in a cell-cycle-dependent manner, reaching peak levels at the G2–M boundary (Campaner et al., 2005; Izraeli et al., 1997). Earlier, we have shown that knockdown of STIL inhibits CDK1 activation and blocks mitotic entry before any evident nuclear membrane breakdown (Erez et al., 2007). Interestingly, at this cell cycle stage, Chfr inhibits progression into mitosis. Consistently, we show here that loss of Stil leads to a marked reduction in the proliferation and mitotic index of mouse fibroblasts. These mitotic and growth phenotypes were partially corrected by knockdown of Chfr. Together, this suggests that the block at mitosis entry in the absence of Stil is mediated by stabilization of the Chfr protein and activation of its checkpoint.
Stil, Chfr and centrosomes
In addition to reduced entry into mitosis, we observed marked centrosomal abnormalities in fibroblasts lacking Stil. Stil was recently demonstrated to localize to the centrosome during mitosis (Pfaff et al., 2007) (our unpublished data). A role for Stil in centrosomal biology is consistent with the recent identification of Stil as one of the genes mutated in autosomal-recessive microcephaly (Kumar et al., 2009), a disorder caused by mutations in centrosomal proteins. All other identified genes involved in this disorder are centrosomal proteins as well. Although a loss-of-function mutation in zebrafish Stil was reported to cause centrosome amplification (Pfaff et al., 2007), we have not seen any centrosomes in mouse fibroblasts lacking Stil. However, Pfaff et al. used γ-tubulin for centrosomal staining and their findings are similar to our observations of the multipolar mitoses (Pfaff et al., 2007) (Fig. 2A). Staining with antibodies to centrin (Fig. 2C) confirmed the absence of matured centrioles in fibroblasts lacking Stil. Thus, we suggest that Stil might be required for centriolar biogenesis and, in its absence, aggregates of pericentriolar material containing γ-tubulin but lacking centrioles provide alternative spindle poles. Although the absence of centrioles in mammalian cells lacking Stil needs to be verified by electron microscopy, this observation is consistent with the recent report describing Ana2 as a functional ortholog of Stil in Drosophila (Stevens et al., 2010). This protein was found to localize to centrioles and regulate their duplication and formation, functions that are related to its Caenorhabditis elegans ortholog SAS-5. Overexpression of Ana2 can drive de novo formation of centriole-like structures, whereas its knockdown abolishes centriole duplication (Dobbelaere et al., 2008), similar to what we see in cells depleted of Stil. Thus, Stil seems to be important for centrosome biogenesis and integrity. As centrosomes are involved through their roles in primary cilia formation, the functions of Stil in centrosome biology might explain the other phenotypes in Stil-knockout mice, including the randomized left–right asymmetry and blocked hedgehog signaling (Izrael et al., 1999).
Initial activation of CDK1 and subsequent entry into mitosis is known to occur at centrosomes in the late G2 phase of the cell cycle (Jackman et al., 2003; Krämer et al., 2004). Therefore, the loss of functional centrosomes in cells lacking Stil might explain why Stil depletion causes a marked delay in entry into mitosis associated with reduced activation of CDK1 (cyclin B kinase) (Erez et al., 2007).
The centrosomal abnormalities observed in the absence of Stil were partially corrected with either knockdown of Chfr or exogenous expression of Plk1. Chfr was found to be localized to the spindle poles during mitosis (Burgess et al., 2008), but no evidence of its direct localization or function at centrosomes during interphase was reported. The centrosomal localization of endogenous Chfr during interphase (supplementary material Fig. S4) and the partial correction of the centrosomal phenotype in cells lacking Stil by knockdown of Chfr suggest, for the first time, a role for Chfr in centrosomal biogenesis and maturation. Plk1, a major regulator of mitosis and a substrate of Chfr, is known to localize to centrosomes and regulates their maturation and organization (Lane and Nigg, 1996; Sumara et al., 2004; van Vugt et al., 2004). It is unclear, however, whether Plk1 mediates all the mitotic defects caused by Chfr accumulation in cells lacking Stil. The reduction in Plk1 protein levels in these cells is relatively small and could be secondary to the cell cycle arrest of these cells. Although exogenous overexpression of Plk1 partially restored the centrosomal defects and the mitotic index, these effects might also have been caused by phosphorylation of targets of other mitotic kinases such as Plk4, the ortholog of Drosophila SAK, that is also crucial for centrosomal biogenesis (Bettencourt-Dias et al., 2005). Additional research is needed to elucidate how Chfr regulates centrosomal biology.
Our studies reveal, for the first time, functional interactions between Stil and Chfr in the regulation of centrosomal biology and entry to mitosis. We suggest a model by which Stil regulates centrosmal biogenesis and maturation and mitotic entry through destabilization of the Chfr protein (Fig. 6D). However, because the correction of the centrosomal defects in cells lacking Stil by elimination of Chfr were not complete, there might be additional functions for Stil in the regulation of centrosomes and mitosis. It will be interesting to learn which of the marked developmental abnormalities observed in mice, zebrafish and humans lacking a fully functional Stil protein are caused by the stabilization of the Chfr protein.
Materials and Methods
Cell culture
All cell lines were grown in Dulbecco's Modified Eagle's Medium (DMEM) supplemented with 10% fetal calf serum, penicillin (100 U/ml) and streptomycin (100 μg/ml) at 37°C and 5% CO2.
Transfections
HEK293T cells were transfected by calcium phosphate with plasmid DNA, and expression of the encoded proteins was analyzed 2 days after transfection. All other cell lines were transfected with Lipofectamine 2000 (Invitrogen) or jetPEI (Polyplus) transfection reagents using standard protocols.
Plasmids and infections
HEK293T cells (approximately 80% confluent in 100 mm plates) were cotransfected with pLKO (lentiviral vector expressing shRNA targeting STIL, CHFR and control non-targeting sequence; Open Biosystems) or pQCXIP (retroviral vector for the expression of Flag-Stil protein, Clontech), psSAX or pCGP (Gag-Pol of lentivirus and retrovirus, respectively) and pMD2.G (encodes envelope protein of VSV). After 8 hours, the supernatants were replaced with 10 ml of fresh medium. Two days post-transfection, supernatants of transfected cells were supplemented with HEPES (pH 7.0; 50 mM final concentration) filtered through a 0.45 μm pore size filter, and Polybrene (hexadimethrine bromide; Sigma) was added to a final concentration of 8 μg/ml. These viral preparations were used to infect naive cells for 2 hours, after which fresh medium was added. One day post-infection, the cells were selected for puromycin resistance and stable clones were generated (single and polyclonal populations).
Additional plasmids
For the expression of Chfr, we used pIRES-2HA-Chfr or HA–Chfr and the HA–Chfr ΔRF mutant in pcDNA3.1 vector. I306A Chfr was cloned by standard site-directed mutagenesis technique using the following primers: 5′-ACGCTGACATGCATCGCCTGCCAGGACCTGCTG-3′; 5′-CAGCAGGTCCTGGCAGGCGATGCATGTCAGCGT-3′. For expression of Plk1, we used the myc-plk1-CMV vector.
Protein extraction and western analysis
Cultured cells were washed twice in cold PBS and resuspended in lysis buffer (1% Triton-X100, 20 mM Tris-HCl, 120 mM NaCl) and a tablet of protease inhibitors (Complete, Mini, Roche), incubated for 1 hour on ice and centrifuged at 16,100 g for 20 minutes. Cell supernatant was collected, concentration was determined using the Bradford method (Pierce), then the supernatant was resuspended in loading buffer and denatured in 95°C for 5 minutes. SDS-PAGE analysis was carried out on Tris-glycine polyacrylamide gels and electro-transferred onto nitrocellulose membrane. For western blot analysis, membranes were blocked with 5% skim milk, incubated with the relevant antibody, then followed by incubation with a secondary, peroxidase-conjugated antibody (1:10,000; Jackson ImmunoResearch, #115-035-146 and #115-035-144). Peroxidase activity was detected by exposure of the membrane to chemiluminescence solution containing 150 mM Tris-HCl (pH 8.9), 0.22 mg/ml Luminol (Sigma), 0.033 mg/ml paracoumaric acid (Sigma) and 0.015% H2O2.
For the analysis of Chfr levels in Stil knockout embryos, we harvested embryos at E9.5 after time-mating of Stil+/− mice. The embryos were spun down and resuspended in lysis buffer (1% Triton-X100, 20 mM Tris-HCl, 120 mM NaCl) and a tablet of protease inhibitors (Complete, Mini, Roche) for 30 minutes on ice. The samples were spun down and subjected to SDS-PAGE analysis.
Antibodies and reagents
The antibodies we used for western blotting were against: Stil (243, affinity-purified; 0.4 μg/ml for 2 hours) (Izraeli et al., 1997), Chfr (Santa Cruz Biotechnologies, sc-28263), Plk1 (Abcam, ab14210), ubiquitin (Abcam, ab7254; Santa-Cruz Biotechnologies sc-8017 P4D1), HA (Santa-Cruz Biotechnologies, sc-7392), CBL (Santa Cruz Biotechnologies, sc-170) and α-tubulin (Sigma, T9026). The antibodies we used for immunoflouresence were against: Stil (2 μg/ml) (Izraeli et al., 1997), centrin-1 (Abcam, ab11257), α-tubulin (Sigma, T9026) and γ-tubulin (Abcam, ab11316; Sigma, T5192). The secondary antibodies used were conjugated to: Rhodamine Red-X (RRX, Jackson ImmunoResearch, 711-296-152) and FITC (Jackson ImmunoResearch, 715-096-150).
Immunofluorescence
Cells were grown on glass coverslips (15 mm diameter) in the appropriate growth medium. The cells were fixed with 4% paraformaldehyde (PFA) for 10 minutes and permeabilized with 0.25% Triton-X100 and 2% PFA in PBS for 10 minutes following blocking in 5% skimmed milk powder in PBS for 1 hour. For centrioles and Chfr staining, the cells were fixed using ice-cold methanol:acetone (1:1) for 10 minutes. The cells were then stained using primary antibody as indicated, in 5% skim milk in PBS for 40 minutes, washed 6 times in PBS with 0.1% Tween and incubated with secondary antibody as indicated, for 40 minutes in 5% skim milk in PBS. The coverslips were mounted using ProLong Gold antifade reagent with DAPI (Invitrogen, p36935) and images were acquired with an Olympus IX81 fluorescence microscope. Confocal images were acquired using a Zeiss LSM510 confocal scanning laser microscope coupled to a multi-photon laser and a dispersion compensation unit (MaiTai DeepSee, Spectraphysics).
Mitotic index
Mitotic index was calculated as the percentage of cells from the total population showing nuclear envelope breakdown and condensed chromosomes. The cells were stained with DAPI and analyzed by fluorescence microscopy.
Protein stability by CHX
3T3 cells were transfected with control and Stil-encoding vectors. Forty-eight hours post-transfection, the cells were exposed to cyclohexamide (CHX, 1 μg/ml) for the indicated time-points, with or without proteasome inhibitor (MG132, Sigma), and prepared for western blot analysis for detection of Stil and Chfr levels.
Ubiquitination assay
HEK293T cells were cotransfected with vectors encoding HA–Chfr WT or HA–Chfr ΔRF (Fukuda et al., 2008) and Myc–ubiquitin, with pQCXIP-Stil or pQCXIP (empty vector control). Forty-eight hours post-transfection, the cells were treated with proteasome inhibitor (MG132, 10 μM) for 3 hours following harvesting and lysis of the cells for immunoprecipitation using anti-HA antibody (to pull down Chfr). The samples were incubated at 4°C overnight, followed by addition of a secondary anti-mouse antibody conjugated to agarose beads (Sigma, A6531) for 1 hour. The samples were washed with washing buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.05% Triton-X100), resuspended in loading buffer and denatured at 95°C for 5 minutes and then analyzed by western blot for the detection of Chfr and Chfr–ubiquitin conjugates using an anti-ubiquitin antibody.
Supplementary Material
Acknowledgments
We thank Hitoshi Nakagama for providing the HA–Chfr and HA–Chfr ΔRF vectors, Thanos Halazonetis for providing the pIRES-2HA-Chfr vector and Hannah Tamary for the Myc–ubiquitin vector. We appreciate the advice of Aharon Ciechanover and the assistance from all members of the Izraeli laboratory. This research was partially funded by research grants from the US-Israel Binational Science Foundation (to S.I., P.D.A. and I.R.K.), the Israel Cancer Research Foundation (M.M.D. and S.I.), the Israel Science Foundation (S.I. and M.B.), the DKFZ-MOST grant (S.I. and A.K.) and the intramural funding of the NCI/NIH. This research was done as partial fulfillment of the requirement for a PhD degree of A.C. and A.D. at the Sackler Faculty of Medicine, Tel Aviv University. The authors declare they have no conflicts of interest. Deposited in PMC for release after 12 months.
Footnotes
Supplementary material available online at http://jcs.biologists.org/cgi/content/full/124/4/532/DC1
References
- Aplan P. D., Lombardi D. P., Kirsch I. R. (1991). Structural characterization of SIL, a gene frequently disrupted in T-cell acute lymphoblastic leukemia. Mol. Cell. Biol. 11, 5462-5469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettencourt-Dias M., Rodrigues-Martins A., Carpenter L., Riparbelli M., Lehmann L., Gatt M. K., Carmo N., Balloux F., Callaini G., Glover D. M. (2005). SAK/PLK4 is required for centriole duplication and flagella development. Curr. Biol. 15, 2199-2207 [DOI] [PubMed] [Google Scholar]
- Bond J., Roberts E., Springell K., Lizarraga S. B., Scott S., Higgins J., Hampshire D. J., Morrison E. E., Leal G. F., Silva E. O., et al. (2005). A centrosomal mechanism involving CDK5RAP2 and CENPJ controls brain size. Nat. Genet. 37, 353-355 [DOI] [PubMed] [Google Scholar]
- Burgess A., Labbe J. C., Vigneron S., Bonneaud N., Strub J. M., Van Dorsselaer A., Lorca T., Castro A. (2008). Chfr interacts and colocalizes with TCTP to the mitotic spindle. Oncogene 27, 5554-5566 [DOI] [PubMed] [Google Scholar]
- Campaner S., Kaldis P., Izraeli S., Kirsch I. R. (2005). Sil phosphorylation in a pin1 binding domain affects the duration of the spindle checkpoint. Mol. Cell. Biol. 25, 6660-6672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaturvedi P., Sudakin V., Bobiak M. L., Fisher P. W., Mattern M. R., Jablonski S. A., Hurle M. R., Zhu Y., Yen T. J., Zhou B. B. (2002). Chfr regulates a mitotic stress pathway through its RING-finger domain with ubiquitin ligase activity. Cancer Res. 62, 1797-1801 [PubMed] [Google Scholar]
- Daniels M. J., Marson A., Venkitaraman A. R. (2004). PML bodies control the nuclear dynamics and function of the CHFR mitotic checkpoint protein. Nat. Struct. Mol. Biol. 11, 1114-1121 [DOI] [PubMed] [Google Scholar]
- De Luca M., Lavia P., Guarguaglini G. (2006). A functional interplay between Aurora-A, Plk1 and TPX2 at spindle poles: Plk1 controls centrosomal localization of Aurora-A and TPX2 spindle association. Cell Cycle 5, 296-303 [DOI] [PubMed] [Google Scholar]
- Dobbelaere J., Josue F., Suijkerbuijk S., Baum B., Tapon N., Raff J. (2008). A genome-wide RNAi screen to dissect centriole duplication and centrosome maturation in Drosophila. PLoS Biol. 6, e224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erez A., Perelman M., Hewitt S. M., Cojacaru G., Goldberg I., Shahar I., Yaron P., Muler I., Campaner S., Amariglio N., et al. (2004). Sil overexpression in lung cancer characterizes tumors with increased mitotic activity. Oncogene 23, 5371-5377 [DOI] [PubMed] [Google Scholar]
- Erez A., Castiel A., Trakhtenbrot L., Perelman M., Rosenthal E., Goldstein I., Stettner N., Harmelin A., Eldar-Finkelman H., Campaner S., et al. (2007). The SIL gene is essential for mitotic entry and survival of cancer cells. Cancer Res. 67, 4022-4027 [DOI] [PubMed] [Google Scholar]
- Erez A., Chaussepied M., Castiel A., Colaizzo-Anas T., Aplan P. D., Ginsberg D., Izraeli S. (2008). The mitotic checkpoint gene, SIL is regulated by E2F1. Int. J. Cancer 123, 1721-1725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda T., Kondo Y., Nakagama H. (2008). The anti-proliferative effects of the CHFR depend on the forkhead associated domain, but not E3 ligase activity mediated by ring finger domain. PLoS ONE 3, e1776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graser S., Stierhof Y. D., Nigg E. A. (2007). Cep68 and Cep215 (Cdk5rap2) are required for centrosome cohesion. J. Cell Sci. 120, 4321-4331 [DOI] [PubMed] [Google Scholar]
- Guernsey D. L., Jiang H., Hussin J., Arnold M., Bouyakdan K., Perry S., Babineau-Sturk T., Beis J., Dumas N., Evans S. C., et al. (2010). Mutations in centrosomal protein CEP152 in primary microcephaly families linked to MCPH4. Am. J. Hum. Genet. 87, 40-51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izraeli S., Colaizzo-Anas T., Bertness V. L., Mani K., Aplan P. D., Kirsch I. R. (1997). Expression of the SIL gene is correlated with growth induction and cellular proliferation. Cell Growth Differ. 8, 1171-1179 [PubMed] [Google Scholar]
- Izraeli S., Lowe L. A., Bertness V. L., Good D. J., Dorward D. W., Kirsch I. R., Kuehn M. R. (1999). The SIL gene is required for mouse embryonic axial development and left-right specification. Nature 399, 691-694 [DOI] [PubMed] [Google Scholar]
- Jackman M., Lindon C., Nigg E. A., Pines J. (2003). Active cyclin B1-Cdk1 first appears on centrosomes in prophase. Nat. Cell Biol. 5, 143-148 [DOI] [PubMed] [Google Scholar]
- Kang D., Chen J., Wong J., Fang G. (2002). The checkpoint protein Chfr is a ligase that ubiquitinates Plk1 and inhibits Cdc2 at the G2 to M transition. J. Cell Biol. 156, 249-259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krämer A., Mailand N., Lukas C., Syljuåsen R. G., Wilkinson C. J., Nigg E. A., Bartek J., Lukas J. (2004). Centrosome-associated Chk1 prevents premature activation of cyclin B/Cdk1 kinase. Nat. Cell Biol. 6, 884-891 [DOI] [PubMed] [Google Scholar]
- Kumar A., Girimaji S. C., Duvvari M. R., Blanton S. H. (2009). Mutations in STIL, encoding a pericentriolar and centrosomal protein, cause primary microcephaly. Am. J. Hum. Genet. 84, 286-290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon Y. E., Kim Y. S., Oh Y. M., Seol J. H. (2009). Nuclear localization of Chfr is crucial for its checkpoint function. Mol. Cells 27, 359-363 [DOI] [PubMed] [Google Scholar]
- Lane H. A., Nigg E. A. (1996). Antibody microinjection reveals an essential role for human polo-like kinase 1 (Plk1) in the functional maturation of mitotic centrosomes. J. Cell Biol. 135, 1701-1713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S. Y., Rai R., Li K., Xu Z. X., Elledge S. J. (2005). BRIT1/MCPH1 is a DNA damage responsive protein that regulates the Brca1-Chk1 pathway, implicating checkpoint dysfunction in microcephaly. Proc. Natl. Acad. Sci. USA, 102, 15105-15109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L. Y., Wood J. L., Minter-Dykhouse K., Ye L., Saunders T. L., Yu X., Chen J. (2008). Polo-like kinase 1 is essential for early embryonic development and tumor suppression. Mol. Cell. Biol. 28, 6870-6876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddika S., Chen J. (2009). Protein kinase DYRK2 is a scaffold that facilitates assembly of an E3 ligase. Nat. Cell Biol. 11, 409-419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsusaka T., Pines J. (2004). Chfr acts with the p38 stress kinases to block entry to mitosis in mammalian cells. J. Cell Biol. 166, 507-516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh Y. M., Yoo S. J., Seol J. H. (2007). Deubiquitination of Chfr, a checkpoint protein, by USP7/HAUSP regulates its stability and activity. Biochem. Biophys. Res. Commun. 357, 615-619 [DOI] [PubMed] [Google Scholar]
- Oh Y. M., Kwon Y. E., Kim J. M., Bae S. J., Lee B. K., Yoo S. J., Chung C. H., Deshaies R. J., Seol J. H. (2009). Chfr is linked to tumour metastasis through the downregulation of HDAC1. Nat. Cell Biol, 11, 295-302 [DOI] [PubMed] [Google Scholar]
- Pfaff K. L., Straub C. T., Chiang K., Bear D. M., Zhou Y., Zon L. I. (2007). The zebra fish cassiopeia mutant reveals that SIL is required for mitotic spindle organization. Mol. Cell. Biol. 27, 5887-5897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaswamy S., Ross K. N., Lander E. S., Golub T. R. (2003). A molecular signature of metastasis in primary solid tumors. Nat. Genet. 33, 49-54 [DOI] [PubMed] [Google Scholar]
- Scolnick D. M., Halazonetis T. D. (2000). Chfr defines a mitotic stress checkpoint that delays entry into metaphase. Nature 406, 430-435 [DOI] [PubMed] [Google Scholar]
- Shtivelman E. (2003). Promotion of mitosis by activated protein kinase B after DNA damage involves polo-like kinase 1 and checkpoint protein CHFR. Mol. Cancer Res. 1, 959-969 [PubMed] [Google Scholar]
- Sotillo R., Hernando E., Diaz-Rodriguez E., Teruya-Feldstein J., Cordon-Cardo C., Lowe S. W., Benezra R. (2007). Mad2 overexpression promotes aneuploidy and tumorigenesis in mice. Cancer Cell 11, 9-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens N. R., Dobbelaere J., Brunk K., Franz A., Raff J. W. (2010). Drosophila Ana2 is a conserved centriole duplication factor. J. Cell Biol, 188, 313-323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumara I., Gimenez-Abian J. F., Gerlich D., Hirota T., Kraft C., de la Torre C., Ellenberg J., Peters J. M. (2004). Roles of polo-like kinase 1 in the assembly of functional mitotic spindles. Curr. Biol. 14, 1712-1722 [DOI] [PubMed] [Google Scholar]
- Summers M. K., Bothos J., Halazonetis T. D. (2005). The CHFR mitotic checkpoint protein delays cell cycle progression by excluding Cyclin B1 from the nucleus. Oncogene 24, 2589-2598 [DOI] [PubMed] [Google Scholar]
- Thornton G. K., Woods C. G. (2009). Primary microcephaly: do all roads lead to Rome? Trends Genet. 25, 501-510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibelius A., Marhold J., Zentgraf H., Heilig C. E., Neitzel H., Ducommun B., Rauch A., Ho A. D., Bartek J., Kramer A. (2009). Microcephalin and pericentrin regulate mitotic entry via centrosome-associated Chk1. J. Cell Biol. 185, 1149-1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyoda Y., Yanagida M. (2006). Coordinated requirements of human topo II and cohesin for metaphase centromere alignment under Mad2-dependent spindle checkpoint surveillance. Mol. Biol. Cell 17, 2287-2302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Vugt M. A., van de Weerdt B. C., Vader G., Janssen H., Calafat J., Klompmaker R., Wolthuis R. M., Medema R. H. (2004). Polo-like kinase-1 is required for bipolar spindle formation but is dispensable for anaphase promoting complex/Cdc20 activation and initiation of cytokinesis. J. Biol. Chem. 279, 36841-36854 [DOI] [PubMed] [Google Scholar]
- Woods C. G., Bond J., Enard W. (2005). Autosomal recessive primary microcephaly (MCPH): a review of clinical, molecular, and evolutionary findings. Am. J. Hum. Genet. 76, 717-728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X., Minter-Dykhouse K., Malureanu L., Zhao W. M., Zhang D., Merkle C. J., Ward I. M., Saya H., Fang G., van Deursen J., et al. (2005). Chfr is required for tumor suppression and Aurora A regulation. Nat. Genet. 37, 401-406 [DOI] [PubMed] [Google Scholar]
- Zhong X., Liu L., Zhao A., Pfeifer G. P., Xu X. (2005). The abnormal spindle-like, microcephaly-associated (ASPM) gene encodes a centrosomal protein. Cell Cycle, 4, 1227-1229 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.