Abstract
Ciliopathies are caused by mutations in genes encoding proteins required for cilia organization or function. We show through colocalization with PCM-1, that OFD1 (the product of the gene mutated in oral-facial-digital syndrome 1) as well as BBS4 and CEP290 (proteins encoded by other ciliopathy genes) are primarily components of centriolar satellites, the particles surrounding centrosomes and basal bodies. RNA interference experiments reveal that satellite integrity is mutually dependent upon each of these proteins. Upon satellite dispersal, through mitosis or forced microtubule depolymerization, OFD1 and CEP290 remain centrosomal, whereas BBS4 and PCM-1 do not. OFD1 interacts via its fifth coiled-coil motif with the N-terminal coiled-coil domain of PCM-1, which itself interacts via its C-terminal non-coiled-coil region with BBS4. OFD1 localization to satellites requires its N-terminal region, encompassing the LisH motif, whereas expression of OFD1 C-terminal constructs causes PCM-1 and CEP290 mislocalization. Moreover, in embryonic zebrafish, OFD1 and BBS4 functionally synergize, determining morphogenesis. Our observation that satellites are assembly points for several mutually dependent ciliopathy proteins provides a further possible explanation as to why the clinical spectrum of OFD1, Bardet–Biedl and Joubert syndromes overlap. Furthermore, definition of how OFD1 and PCM-1 interact helps explain why different OFD1 mutations lead to clinically variable phenotypes.
Keywords: Basal body, BBS4, Centriole, CEP290, Cilia, Laterality, PCM-1, OFD1, Zebrafish
Introduction
Most mammalian cells possess a single hair-like projection called a primary cilium, which extends into the extracellular milieu. These organelles act like antennae, sensing mechanical and chemical signals and thus coordinating cell growth, survival, polarity and differentiation (Berbari et al., 2009; Marshall and Nonaka, 2006; Singla and Reiter, 2006). Primary cilia are generally non-motile and contain a 9+0 organization of microtubule pairs within the ciliary axoneme, distinct from the 9+2 organization found within flagella and the motile cilia of multiciliated epithelia. However, there appear to be subsets of primary cilia that, although having a 9+0 arrangement, have a motile function, e.g. within the embryonic node of mammals or the Kuppfer's vesicle of fish. Through rotating in a clockwise manner, these nodal cilia create a flow that is necessary for defining left–right asymmetry during body pattern determination (Marshall and Nonaka, 2006).
Much of our understanding regarding primary cilia has come from studies on genetic human disorders, collectively known as ciliopathies, many features of which are considered to result from defects in the assembly or function of these organelles. These features include laterality defects, digital and brain anomalies, retinal degeneration, obesity and kidney cysts (Badano et al., 2006; Gerdes et al., 2009; Hildebrandt and Otto, 2005). The oral-facial-digital (OFD) syndromes are linked by dysmorphology of the mouth, face and digits (Macca and Franco, 2009). OFD type 1 (OFD1), first reported in 1954 by Papillon-League and Psaume, is the commonest, occurring in 1:50,000 births. OFD1 is distinguished by its X-linked dominant inheritance pattern (Feather et al., 1997), with prenatal lethality in affected males. Affected females are born with OFD dysmorphology and develop polycystic kidneys (Feather et al., 1997; Saal et al., 2010). Mutations of OFD1, located on the X chromosome, account for most cases of OFD1 syndrome with most mutations found in the first half of the gene (Ferrante et al., 2001; Macca and Franco, 2009; Romio et al., 2003). Human OFD1 escapes X-inactivation and affected females are probably composed of cells with reduced levels of normal OFD1 protein (Ferrante et al., 2003). There remain, however, some OFD1 syndrome individuals for which OFD1 mutations cannot be detected (Thauvin-Robinet et al., 2009). In human embryos, OFD1 is expressed in many organs, including those that develop abnormally in the syndrome (Romio et al., 2004; Romio et al., 2003). Intriguingly, OFD1 mutations have recently been associated with other disease phenotypes, including the nephronophthisis (NPHP)-related ciliopathy, Joubert Syndrome (Budny et al., 2006; Coene et al., 2009; and see Discussion).
The classic OFD1 syndrome is typical of a ciliopathy. Moreover, studies support the hypothesis that the developmental abnormalities are, at least in part, due to abnormal cilia-dependent signalling events. In cultured cells, OFD1 is clearly required for primary cilia formation (Corbit et al., 2008; Graser et al., 2007; Singla et al., 2010). Furthermore, experimental downregulation of Ofd1 in both mice (Ferrante et al., 2006) and zebrafish (Ferrante et al., 2009) causes laterality defects of internal organs, including the heart, subsequent to embryonic node (Ferrante et al., 2006) and Kuppfer's vesicle (Ferrante et al., 2009) structural defects in cilia, the normal functions of which are crucial for the breaking of embryonic symmetry (Bakkers et al., 2009). In mice lacking Ofd1, altered expression of Sonic hedgehog (Shh) pathway genes has been noted (Ferrante et al., 2006). Although Ofd1 might not be involved in Shh signalling in zebrafish (Ferrante et al., 2009), Ofd1 functionally synergizes in this organism with Slb/Wnt11 and Tri/Vangl2 to direct convergent extension movements, suggesting a role in the non-canonical Wnt–planar cell polarity (PCP) signalling pathway. Notably, other experiments show that both Shh and PCP signalling can be initiated within cilia (Berbari et al., 2009).
Like many proteins encoded by ciliopathy disease genes, OFD1 localizes to the centrosome throughout the cell cycle (Romio et al., 2004). The centrosome is a cytoplasmic organelle composed of two barrel-shaped centrioles held within a proteinaceous matrix of pericentriolar material (PCM) that together act as the primary microtubule organizing centre (Nigg and Raff, 2009). During cell division, the duplicated centrosomes form the poles of the microtubule-based mitotic spindle. In post-mitotic cells, the unduplicated centrosome moves to the apical cell surface where the older, or mother, centriole, docks with the plasma membrane and subtends the axonemal microtubules of the primary cilium. When centrioles participate in ciliogenesis, they are called basal bodies and, in ciliated cells, OFD1 has been localized both to basal bodies and the stalk of the cilium (Romio et al., 2004). Importantly, OFD1 was recently shown to localize specifically to the distal ends of centrioles and, in mouse ES cells lacking OFD1, centriole distal ends were disturbed (Singla et al., 2010). Specifically, centrioles exhibited excessive elongation and failure to properly assemble distal appendages. These defects could potentially lead to problems in attachment of the mother centriole to the apical cell surface. It remains possible, however, that the actions of OFD1 are not confined to the generation of cilia because these appear to be present during tubular cystogenesis induced by renal epithelial-specific downregulation of Ofd1 in mice (Zullo et al., 2010). OFD1 protein has also been detected in nuclei as part of the TIP60 chromatin remodelling complex, and thus might play roles in regulating gene expression (Giorgio et al., 2007). Supporting this idea is the observation that downregulation of Ofd1 function leads to a striking increase in levels of Ofd1 transcripts (Ferrante et al., 2009).
OFD1 contains 23 exons and generates two main splice variants, OFD1a and OFD1b, the latter coding for an unstudied putative protein of 367 amino acids (aa) derived from exons 1–11 (Budny et al., 2006). More is known about OFD1a, the protein encoded by exons 1–23, itself with a variant lacking exon 10, with a predicted molecular weight of ~110 kDa (hereafter, for simplicity, we call OFD1a, ‘OFD1’). OFD1 contains an N-terminal Lis1 homology (LisH) motif and an extended C-terminal domain containing what have been alternatively indicated as either five or six putative coiled-coils (Budny et al., 2006; Coene et al., 2009; de Conciliis et al., 1998; Emes and Ponting, 2001). This C-terminal region, which we show as containing six coiled-coils based on SMART analysis (http://smart.embl-heidelberg.de/), is essential for localizing OFD1 to the centrosome (Romio et al., 2004). It is also essential for interaction with the LCA5-encoded ciliary protein, lebercilin (Coene et al., 2009), itself mutated in Leber congenital amaurosis. LisH motifs are present in many proteins, where they contribute to protein–protein interactions, dimerization, stability and/or localization (Gerlitz et al., 2005). Furthermore, the LisH motif might regulate microtubule dynamics directly or indirectly via cytoplasmic dynein (Emes and Ponting, 2001; Ferrante et al., 2003; Gerlitz et al., 2005). Interestingly, Miller–Dieker lissencephaly and Treacher Collins syndrome are caused by mutations in genes encoding LisH-containing proteins, and both disorders have been attributed to incorrect cell migration resulting from cytoskeletal defects (Emes and Ponting, 2001). Hence, certain neuronal components of the OFD1 syndrome might involve aberrant cell migration. In addition, missense mutation of the OFD1 LisH domain deregulates centriole elongation (Singla et al., 2010).
We set out to test the hypothesis that OFD1 contributes to the physical organization of basal bodies and, as such, has an architectural role in ciliogenesis. We unexpectedly found that, as well as localizing to centrosomes, OFD1 localizes to centriolar satellites, which are granular structures surrounding the centrosome that are implicated in trafficking of material required for centriole assembly (Kubo et al., 1999). Other ciliopathy disease proteins are known to localize to satellites. These include the Bardet–Biedl syndrome (BBS) protein BBS4, one component of a large complex of BBS subunits known as the BBSome, and CEP290, which is a protein mutated in NPHP and Joubert syndrome (Kim et al., 2008; Kim et al., 2004; Nachury, 2008). We therefore addressed the dependencies of these proteins on each other for localization to satellites by using RNA interference and expression of truncated OFD1 constructs. Our data support a model whereby centriolar satellites regulate the delivery of several ciliopathy disease proteins to the basal body.
Results
Ciliopathy disease proteins are components of centriolar satellites
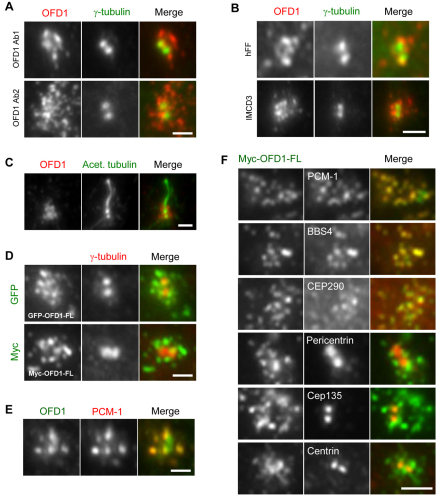
We set out to examine the regulation and function of the OFD1 protein at the cellular level by first revisiting its localization, primarily in telomerase-immortalized human retinal pigment epithelial (hTERT-RPE1) cells. These non-cancer-derived cells undergo normal proliferation and lack detectable cilia when subconfluent, but enter quiescence and produce primary cilia when grown to confluence and serum-starved. Here, we used two different polyclonal antibodies raised to distinct OFD1 antigens: Ab1 from Romio and colleagues (Romio et al., 2003) was raised against a peptide representing amino acids 867–891 (so would not detect OFD1b) and used for the studies shown in Figs 1 and 2, except where indicated. Ab2 from this study (supplementary material Fig. S1A,B) was raised against amino acids 145–1012 (so should detect OFD1a and OFD1b) and used in all other experiments. With both antibodies, we found that in proliferating hTERT-RPE1 cells the bulk of OFD1 was not primarily localized to the centrosome, but was distributed in small aggregates that mostly surrounded the centrosome (Fig. 1A and supplementary material Fig. S1D). A similar distribution was observed in proliferating human foreskin fibroblast (hFF) and renal inner medullary collecting duct (IMCD3) cells (Fig. 1B). Moreover, in contrast to what was observed in renal proximal tubule epithelial cells (RPTEC) (Romio et al., 2004), OFD1 was not detected along the stalk of the cilium in ciliated hTERT-RPE1 cells (Fig. 1C and supplementary material Fig. S1E). Localization to small pericentriolar aggregates was confirmed in hTERT-RPE1 cells overexpressing GFP- or Myc-tagged full-length OFD1 protein (Fig. 1D and supplementary material Fig. S1F), consistent with a recent study on cells expressing enhanced cyan fluorescent protein (eCFP)–OFD1 (Coene et al., 2009).
Fig. 1.
OFD1 localizes to centriolar satellites. (A) hTERT-RPE1 cells were co-stained with antibodies against γ-tubulin to detect the centrosome, and with two different antibodies against OFD1 (Ab1 and Ab2). (B) hFF and IMCD3 cells were co-stained with antibodies against γ-tubulin and OFD1. (C) Ciliated hTERT-RPE1 cells were co-stained with antibodies against acetylated tubulin to detect the axonemal microtubules and against OFD1. (D) hTERT-RPE1 cells were transfected with constructs expressing GFP- or Myc-tagged full-length (FL) OFD1 and stained with antibodies against GFP or Myc and γ-tubulin. (E) hTERT-RPE1 cells were co-stained with antibodies against OFD1 and PCM-1 to identify centriolar satellites. (F) hTERT-RPE1 cells were transfected with Myc-tagged full-length OFD1 and co-stained with antibodies against Myc to identify transfected cells and against PCM-1, BBS4, CEP290, pericentrin, Cep135 or centrin. Scale bars: 2 μm. Merged images are shown for all panels.
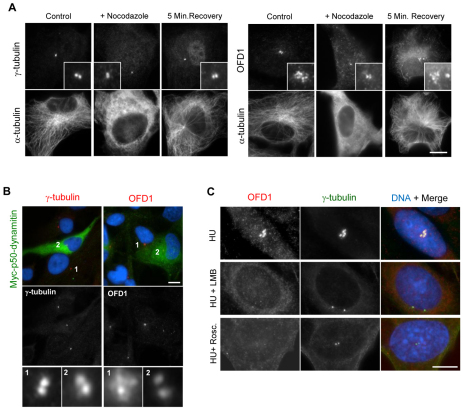
Fig. 2.
Localization of OFD1 is disturbed following centriolar satellite dispersal. (A) hTERT-RPE1 cells were either mock-treated (Control), treated with nocodazole (+ Nocodazole) to depolymerize the microtubule network, or treated with nocodazole and then allowed to recover for 5 minutes (5 Min. Recovery). Cells were then stained with antibodies against α-tubulin and either γ-tubulin or OFD1. Insets show enlargements of the centrosome. (B) hTERT-RPE1 cells were transfected with Myc-tagged p50-dynamitin to disrupt dynein–dynactin function and co-stained with antibodies against Myc and against either γ-tubulin or OFD1. Enlargements of γ-tubulin and OFD1 signals from untransfected (1) or transfected (2) cells are shown. (C) CHO cells were treated with hydroxyurea (HU) alone for 48 hours, HU for 18 hours followed by HU and leptomycin B (LMB) for 30 hours, or HU for 18 hours followed by HU and roscovitine for 30 hours. Cells were stained with antibodies against OFD1 and γ-tubulin. Merged images, including DNA stain (blue), are shown in B and C. Scale bars: 10 μm.
The size and distribution of OFD1 aggregates were highly reminiscent of centriolar satellites, non-membranous 70–100 nm cytoplasmic granules that concentrate around the centrosome and contain the protein PCM-1 (Dammermann and Merdes, 2002; Kubo et al., 1999; Kubo and Tsukita, 2003). Using antibodies to detect endogenous OFD1 or expression of recombinant Myc–OFD1, we found that the small aggregates that surround the centrosome colocalized precisely with PCM-1 (Fig. 1E,F). Furthermore, co-staining for two other ciliopathy disease proteins known to be satellite components, CEP290 and BBS4 (Kim et al., 2008; Kim et al., 2004), also revealed colocalization with OFD1. Note that all studies on BBS4 here were done with an antibody raised in our laboratory (supplementary material Fig. S1A,C). Interestingly, other reported satellite markers, notably centrin, pericentrin and Cep135 (Dammermann and Merdes, 2002; Ohta et al., 2002), did not form obvious pericentriolar aggregates in hTERT-RPE1 cells but were predominantly associated with the centrosome itself and showed no colocalization with OFD1 (Fig. 1F). Thus, we conclude that the bulk of the ciliopathy disease proteins, OFD1, CEP290 and BBS4, colocalize in interphase hTERT-RPE1 cells primarily within centriolar satellites.
Redistribution of centriolar satellites disturbs OFD1 localization
Centriolar satellites cluster around the centrosome in a manner that is dependent on microtubules and the microtubule-dependent motor protein, dynein (Dammermann and Merdes, 2002). Consistent with this, interfering with microtubule-dependent transport leads to dispersal not only of PCM-1, but also of BBS4 and CEP290 (Kim et al., 2008; Kim et al., 2004; May-Simera et al., 2009). We therefore analysed the localization pattern of OFD1 in hTERT-RPE1 cells following, first, depolymerization of the microtubule network with nocodazole and, second, disruption of dynein function by overexpression of the dynactin subunit, p50-dynamitin. Under both conditions, γ-tubulin remained on centrosomes and the pericentriolar aggregates of OFD1 dispersed, which was consistent with redistribution of centriolar satellites into the cytoplasm (Fig. 2A,B). A fraction of OFD1 specifically on the centrosome also became more obvious to detect. Microtubule regrowth following nocodazole washout led to re-accumulation of the pericentriolar OFD1 aggregates, which was consistent with focusing of centriolar satellites (Fig. 2A). We have previously shown that treatments that block centrosome overduplication in S-phase-arrested Chinese hamster ovary (CHO) cells, including inhibition of nuclear export or Cdk activity, lead to loss of detectable centriolar satellites (Prosser et al., 2009). As predicted, these same treatments led to loss of the pericentriolar aggregates of OFD1 (Fig. 2C). Hence, using a variety of approaches known to disturb centriolar satellites, we have confirmed that OFD1 behaves as a bona fide centriolar satellite component.
Redistribution of ciliopathy disease proteins during mitotic progression
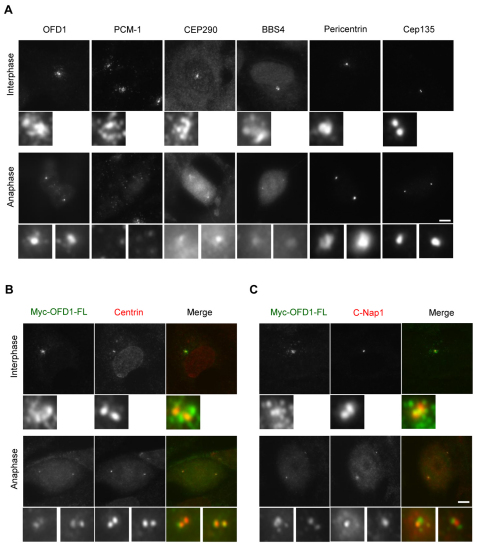
Through analysis of PCM-1, it has been previously reported that centriolar satellites disperse in mitosis, such that in metaphase and anaphase they show little point of focus (Balczon et al., 1994; Dammermann and Merdes, 2002). We therefore looked at the behaviour of OFD1 and other ciliopathy disease proteins in hTERT-RPE1 cells during mitotic progression (Fig. 3A). In the case of OFD1, although the satellites disappeared, the protein was very distinctly associated with spindle poles (Fig. 3A). CEP290 exhibited very similar behaviour and was retained on spindle poles in mitosis, although it could also be detected on spindle microtubules in anaphase (Fig. 3A). BBS4 behaved more like PCM-1 in being mainly dispersed into the cytoplasm, albeit with small amounts detectable at the centrosome (Kim et al., 2004). Pericentrin and Cep135, both proteins that are mainly present on centrosomes in interphase, remained associated with spindle poles in mitosis and showed no significant alteration during mitotic progression (Fig. 3A). Using recombinant Myc-tagged OFD1 and co-staining for centrin, a marker of centriole distal ends (Fig. 3B), and C-Nap1, a marker of centriole proximal ends (Fig. 3C), we confirmed that OFD1 is specifically associated with the distal ends of the parental centrioles in mitosis. Thus, although certain centriolar satellite proteins are completely dispersed in mitosis (PCM-1, BBS4), others redistribute onto spindle poles (OFD1, CEP290).
Fig. 3.
Redistribution of centriolar satellite proteins during mitosis. (A) hTERT-RPE1 cells were stained with antibodies against OFD1, PCM-1, CEP290, BBS4, pericentrin or Cep135. (B,C) hTERT-RPE1 cells transfected with a Myc-tagged OFD1 full-length (FL) construct were co-stained with antibodies against Myc and either centrin2 (B) or C-Nap1 (C). Cells from interphase and anaphase are shown together with enlargements of the centrosomes and spindle poles, respectively. The C-Nap1 images in anaphase are overexposed compared to interphase due to the reduction in C-Nap1 on spindle poles. Scale bars: 5 μm.
Ciliopathy disease proteins are mutually dependent for centriolar satellite localization
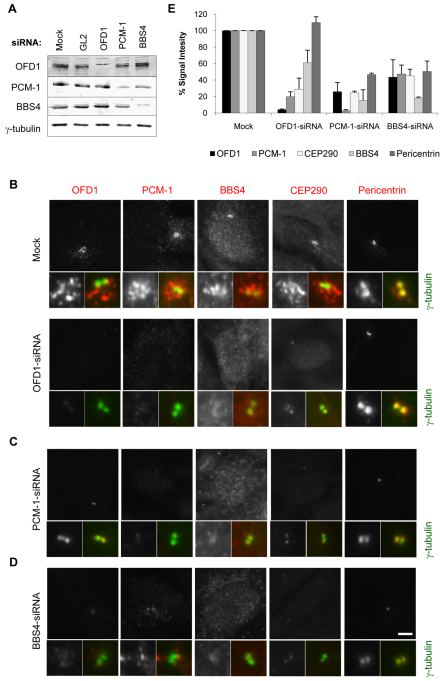
To address the function of OFD1, we used siRNA oligonucleotides to deplete the protein in hTERT-RPE1 cells (Fig. 4A). OFD1 depletion did not affect either the anchoring or nucleation of the microtubule network in these cells, nor did it alter the abundance of γ-tubulin (data not shown). However, as expected, OFD1 depletion did interfere with ciliogenesis without affecting cell cycle progression (supplementary material Fig. S2). We then addressed the consequence of depleting OFD1 on centriolar satellite organization. It has been reported that depletion of PCM-1 leads to reduced centrin, pericentrin and ninein, but not γ-tubulin, at the centrosome (Dammermann and Merdes, 2002), whereas depletion of BBS4 leads to dispersal of PCM-1 (Kim et al., 2004). Interestingly, depletion of CEP290 causes PCM-1 to become concentrated around the centrosome, whereas depletion of PCM-1 causes loss of CEP290 from centriolar satellites but not the centrosome (Kim et al., 2008). Strikingly, we found that depletion of OFD1 led to complete loss of PCM-1 and BBS4 from the vicinity of the centrosome (Fig. 4B). In the case of CEP290, the satellites disappeared but weak staining of the centrosome was apparent. We interpret this to mean that OFD1 is essential to the integrity of centriolar satellites but, in their absence, CEP290 can be retained at the centrosome. Similarly, when PCM-1 was depleted, the pericentriolar aggregates of OFD1, CEP290 and BBS4 disappeared but OFD1 and CEP290 could be detected on centrosomes (Fig. 4C). Depletion of BBS4 led to loss of all proteins from centriolar satellites; however, CEP290 could again be weakly detected on centrosomes, but OFD1 could not (Fig. 4D). Localization of γ-tubulin was not altered in response to depletion of OFD1, PCM-1 or BBS4. Interestingly, the intensity of pericentrin at the centrosome was reduced by ~50% upon depletion of PCM-1, as previously reported (Dammermann and Merdes, 2002), and upon depletion of BBS4 but not OFD1 (Fig. 4E). These data, obtained with pooled siRNA duplexes, were verified using two distinct single duplexes for each protein (supplementary material Fig. S3). Hence, taken together with data published for PCM-1, BBS4 and CEP290 depletions (Dammermann and Merdes, 2002; Kim et al., 2008; Kim et al., 2004), it is clear that the localization of the centriolar satellite proteins OFD1, PCM-1, BBS4 and CEP290 are all mutually dependent with respect to the satellites. However, once satellites are disturbed then the individual proteins behave in a more complex manner with respect to their localization to centrosomes.
Fig. 4.
Ciliopathy disease proteins are mutually dependent for their centriolar satellite localization. (A) hTERT-RPE1 cells were either mock-depleted or depleted with pooled siRNA oligonucleotides against luciferase (GL2), OFD1, PCM-1 or BBS4 for 72 hours and depletion assessed by western blot with the antibodies indicated. (B–D) hTERT-RPE1 cells were either mock-depleted or depleted of OFD1 (B), PCM-1 (C) and BBS4 (D) and co-stained with antibodies against γ-tubulin (green in merge) and against OFD1, PCM-1, BBS4, CEP290 or pericentrin (greyscale images and red in merge). Enlargements of centrosome are shown. Scale bar: 5 μm. (E) Total fluorescence intensities for the different antibodies were determined within a 2 μm square centred on the centrosome in cells treated with different siRNAs. The histogram presents the mean total intensities relative to that in the mock-treated sample (set at 100%); ten cells were counted in two different experiments. Error bars show s.d.
OFD1 interacts with PCM-1 via their coiled-coil domains
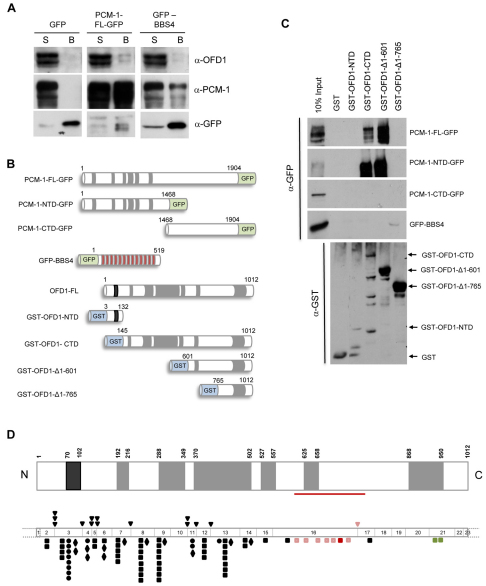
OFD1 is composed of a short N-terminal domain that contains a LisH motif and a much longer C-terminal domain containing six predicted coiled-coils. Given the colocalization of OFD1 and PCM-1 to centriolar satellites and their mutual dependency for this association, we investigated whether these two proteins were capable of interaction. Following transfection of hTERT-RPE1 cells, we found that endogenous OFD1 could co-precipitate with GFP-tagged full-length PCM-1 but not GFP alone (Fig. 5A). Interestingly, two bands were observed for OFD1 and are likely to reflect the full-length OFD1 and the version lacking exon 10, indicating that interaction with PCM-1 does not require the sequence encoded by exon 10. When a co-precipitation experiment was performed with GFP-tagged full-length BBS4, endogenous OFD1 was only very weakly detected, despite good interaction of GFP–BBS4 with PCM-1 (Fig. 5A), as previously reported (Kim et al., 2004). Hence, OFD1 exhibits a much more robust interaction with PCM-1 than with BBS4.
Fig. 5.
OFD1 and PCM-1 interact via their respective coiled-coil domains. (A) HEK293 cells were transfected with GFP alone, GFP-tagged full-length (FL) PCM-1 or GFP-tagged full-length BBS4. Cell lysates were immunoprecipitated with an anti-GFP antibody, and both the unbound supernatant (S) and immunoprecipitates (bound, B) separated by SDS-PAGE and immunoblotted with antibodies against OFD1, PCM-1 and GFP. (B) Schematic representation of GFP-tagged PCM-1 and BBS4 and GST-tagged OFD1 constructs used for interaction studies shown in C. Coiled-coil (grey), LisH (black) and TPR (red) motifs, as well as amino acid numbers, are indicated. (C) Purified GST alone or GST-tagged OFD1 constructs, as indicated, were incubated with lysates prepared from HEK293 cells transfected with PCM-1-GFP or GFP-BBS4 constructs. 10% of the input, together with the GST-bound material, were immunoblotted with an anti-GFP antibody. An anti-GST immunoblot shows the purified proteins in each fraction. (D) Schematic representation of OFD1 protein (upper diagram) is shown as in B, indicating the region (residues 601–765) essential for association with PCM-1 (red line). The lower diagram depicts the 23 exons of OFD1 gene drawn to scale so that the protein domains are aligned with their coding sequence. All point mutations identified to date are shown: frameshift (square), splice-site (triangle), nonsense (diamond) and missense (circle). Those found within the PCM-1-interacting region are shown in red and pink. The frameshift mutation in exon 16 depicted in red has been described in X-linked Simpson–Golabi–Behmel-like syndrome (Budny et al., 2006) and the frameshift mutations in exon 21 (green) have been described in an X-linked mental retardation syndrome with Joubert-like features (Coene et al., 2009). Full descriptions of mutations have been published (Macca and Franco, 2009).
To define the regions of OFD1 and PCM-1 required for these interactions, a series of GST-tagged OFD1 truncation constructs were expressed in bacteria and mixed with extracts from hTERT-RPE1 cells transfected with GFP-tagged full-length PCM-1 or, separately, the PCM-1 N-terminal domain encompassing its coiled-coils (residues 1–1468) or the C-terminal non-coiled-coil domain (residues 1468–1904) (Dammermann and Merdes, 2002) (Fig. 5B). Both full-length PCM-1 and the PCM-1 N-terminal domain interacted strongly with the C-terminal domain of OFD1 (residues 145–1012), but not the N-terminal region containing the LisH motif (residues 3–132) (Fig. 5C). Although the GST–OFD1 N-terminal domain protein was present at lower amounts in this experiment, when analysed at equivalent amounts to the GST–OFD1 C-terminal domain protein, there was still no sign of interaction with the N- or C-terminal domains of PCM-1-GFP (supplementary material Fig. S4). Hence, these proteins interact via their respective coiled-coil regions. There was no binding of GFP–BBS4 with the GST-tagged N- or C-terminal OFD1 proteins in this experiment, supporting the notion that there is unlikely to be direct association between BBS4 and OFD1. Further truncation of the OFD1 coiled-coil region indicated that removal of the first four coiled-coils had no significant effect on the interaction with PCM-1, but the additional removal of the fifth coiled-coil led to its complete loss (Fig. 5C). We therefore conclude that the N-terminal coiled-coil domain of PCM-1 associates with residues 601–765 of OFD1, and that although OFD1 and BBS4 might weakly associate in cells, they are unlikely to interact directly. The importance of these data are highlighted by the fact that several mutations reported in OFD1 patients lie within the region shown to be required for PCM-1 interaction (Fig. 5D).
Expression of truncated OFD1 constructs disrupts centriolar satellites
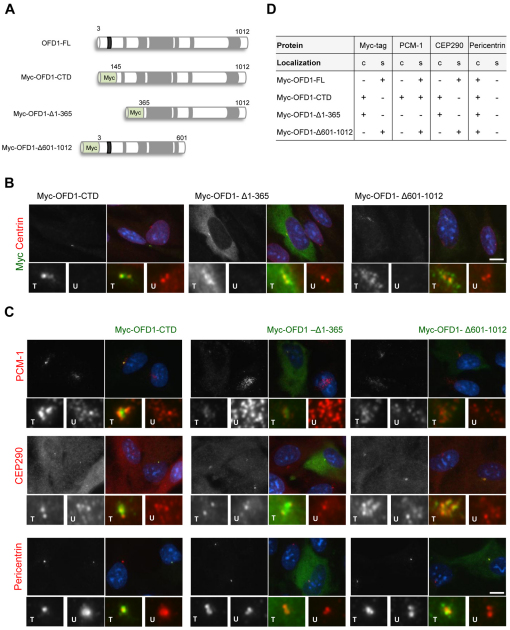
Previous work had reported that the C-terminal domain of OFD1 was necessary for localization to the centrosome, but without distinguishing between centriolar satellites and centrioles (Romio et al., 2004). We therefore decided to test the localization of different Myc-tagged OFD1 truncation constructs following expression in hTERT-RPE1 cells (Fig. 6A). As indicated earlier, full-length OFD1 is primarily associated with centriolar satellites and only weakly present at the centrosome itself (Fig. 1D). Moreover, in agreement with Romio and colleagues (Romio et al., 2004), the C-terminal domain encompassing the coiled-coil motifs does indeed localize to the centrosome; strikingly, though, it did not localize to centriolar satellites (Fig. 6B). Deleting the first two coiled-coils in additional to the N-terminal domain from this construct (OFD1-Δ1-365) led to the protein localizing less specifically to the centrosome, with a cytoplasmic distribution becoming obvious; however, this was diffuse with no apparent staining of satellites (Fig. 6B). By contrast, a protein starting at the N-terminus and encompassing both the LisH motif and the first four coiled-coils, hence only lacking the last two coiled-coils (OFD1-Δ601-1012), specifically localized to centriolar satellites suggesting that the LisH-containing N-terminal domain is crucial for this localization (Fig. 6B). Unfortunately, a construct containing only the LisH-containing N-terminal domain could not be expressed in hTERT-RPE1 cells, most probably as a result of unstable mRNA or protein.
Fig. 6.
Ciliopathy protein localizations upon expression of truncated OFD1 constructs. (A) Schematic representation of Myc-tagged OFD1 deletion constructs. Coiled-coil (grey) and the LisH (black) motif, as well as amino acids numbers, are indicated. (B) hTERT-RPE1 cells were transfected with Myc-tagged OFD1 constructs as indicated and co-stained with antibodies against the Myc tag (greyscale and green in merge) to determine the localization of recombinant OFD1 protein and centrin2 (red in merge). Enlargements show centrosomal regions in transfected (T) and untransfected (U) cells. (C) hTERT-RPE1 cells were transfected with Myc-tagged OFD1 constructs as indicated and co-stained with antibodies against the Myc tag (green) and against PCM-1, CEP290 or pericentrin (greyscale and red in merge). Enlargements show centrosomal regions in transfected and untransfected cells. Merge images including DNA (blue) are shown in B and C. Scale bars: 10 μm. (D) The primary localization (+ present; − absent) of the Myc–OFD1 protein, PCM-1, CEP290 and pericentrin in the presence of the different OFD1 constructs is indicated. Results for each stain are based on at least 100 transfected cells for Myc-OFD1 full-length (FL), C-terminal domain (CTD) and Δ1-365, and 25 cells for Myc-OFD1 Δ601-1012, which transfected with much lower efficiency. c, centrosomes; s, centriolar satellites.
We then examined the effect of expressing these truncated OFD1 constructs on the distribution of other satellite proteins. The OFD1 C-terminal domain protein that localized to centrosomes but not satellites, led to the redistribution of PCM-1 and CEP290 onto the centrosome at the expense of their localization to satellites (Fig. 6C). Even pericentrin became more intensively concentrated on centrosomes in cells expressing this construct. This suggests that, even though pericentrin was not easily detected on satellites in these cells, a fraction might be satellite-associated and becomes redistributed by this OFD1 construct. By contrast, in cells expressing the OFD1-Δ1-365 protein that is only weakly centrosome-associated, satellites containing PCM-1 and CEP290 were dispersed, although CEP290 was still weakly detected on centrosomes (Fig. 6C). We interpret this to mean that both these C-terminal OFD1 constructs can bind to PCM-1 and CEP290 and disturb their localization to satellites, but the one lacking the first two coiled-coils is less efficient at centrosome targeting. Finally, expression of the OFD1-Δ601-1012 protein, which is still capable of localizing to satellites, did not obviously affect the localization of other satellite proteins (Fig. 6C). Hence, OFD1 constructs that themselves mislocalize cause mislocalization of other centriolar satellite components, consistent with their interaction in cells (Fig. 6D).
Functional interaction of Ofd1 and Bbs4 in embryogenesis
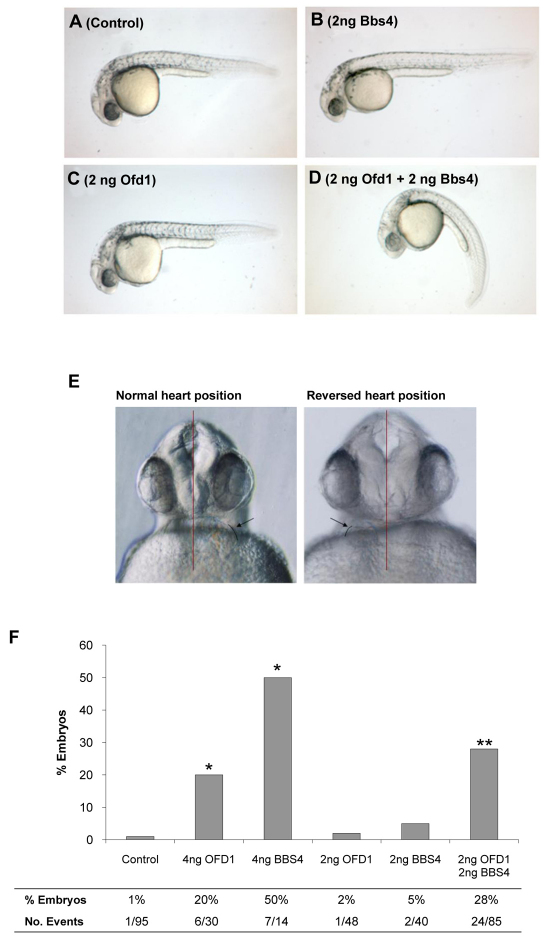
Given the striking colocalization of OFD1, BBS4 and PCM-1 proteins in pericentriolar satellites, we elected to use a zebrafish model to test biological effects of downregulation of these genes using antisense morpholinos. This model also allows the assessment of potential functional synergy between molecules by observing effects of administration of low doses of more than one type of morpholino. Abnormal body curvature of embryos, characteristic of ciliopathies, was elicited by co-administration of these low dose morpholinos, whereas alone neither produced this effect (Fig. 7A–D). Using heart primordium lateralization as a readout of ciliary function, administration of 4 ng of either Ofd1 or Bbs4 morpholino produced defects in 20% and 50% of embryos, respectively, whereas only 1% of controls had these defects (Fig. 7E,F). When a lower dose (2 ng) of either morpholino was administered alone, laterality defect frequencies were similar to controls. Strikingly, co-administration of 2 ng of Ofd1 and 2 ng Bbs4 morpholinos led to laterality defects in 28% of embryos (Fig. 7F). Unfortunately, Pcm-1 morpholinos caused unspecific delays in early embryogenesis and thus could not be used to test effects on heart laterality and later phenotypes.
Fig. 7.
Bbs4 and Ofd1 morpholinos synergize in disrupting body axis formation and laterality in zebrafish. (A–D) Whole mount images of zebrafish embryos 28 hours after fertilization. Shown are untreated control (A) and embryos treated with 2 ng Bbs4 morpholino (B), 2 ng Ofd1 morpholino (C) and 2 ng Bbs4 + 2 ng Ofd1 morpholino (D). Only the embryo administered both Ofd1 and Bbs4 morpholinos exhibits a bent body axis. (E) Frontal view of two zebrafish embryos (28 hours post-fertilization) injected with 4 ng of Ofd1 morpholino. Red vertical lines indicate the embryonic midline, and the arrow and black curved lines indicate the position of the heart. In the left image, the heart is positioned normally on the left side of the embryo, whereas in the right image the position of the heart is reversed. (F) Histogram indicates frequency of heart laterality defects in one-day-old embryos treated with single or combined morpholinos. *P<0.001 versus control, **P<0.001 versus effects from low dose morpholinos administered individually.
OFD1 associates with amplified basal bodies in cells with motile cilia
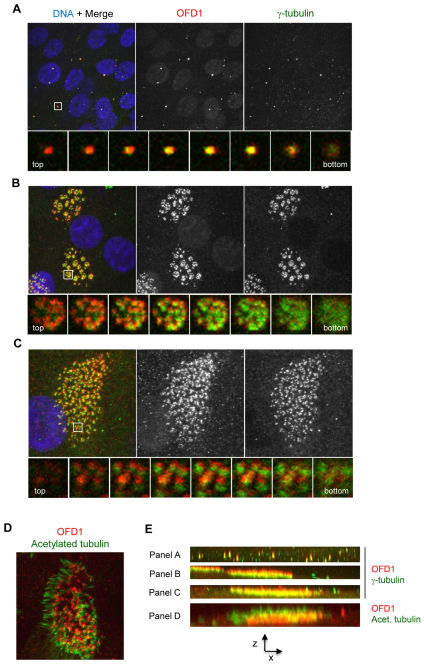
The laterality defects seen upon depletion of Ofd1 in zebrafish are suggestive of loss of function of nodal cilia. Because these are specialized types of primary cilia with motile function, we decided to examine the localization of OFD1 in cultured human multiciliated cells with motile cilia. For this purpose, primary human nasal epithelial cells were cultured at an air–liquid interface (ALI) to induce differentiation (Hirst et al., 2010). As cells differentiate, they assemble around 200 basal bodies, via a combination of centriolar and non-centriolar duplication pathways, which each then generate a motile cilium (Hagiwara et al., 2004). OFD1 localization was examined prior to (10 days at ALI) and after (28 days at ALI) differentiation, and cells co-stained for γ-tubulin. In contrast to results in hTERT-RPE1 cells, prior to differentiation, OFD1 was found to concentrate on centrosomes and no distinct centriolar satellites were observed (Fig. 8A). Very similar results were found for PCM-1 (supplementary material Fig. S5A). In differentiated cells, OFD1 was closely associated with each of the multiple basal bodies, both when the basal bodies were arranged in distinct rosette-like clusters (presumably representing a midway point in the differentiation process), and when cells were full of basal bodies (fully differentiated) (Fig. 8B,C). As in cells with primary cilia, co-staining of OFD1 with acetylated tubulin showed no evidence of OFD1 along the axonemal microtubules (Fig. 8D). It was notable that both in x–y and z-sections the OFD1 and γ-tubulin signals did not precisely align, suggesting that they are part of different structures although they are both associated with basal bodies (Fig. 8B,C,E). Analysis of z-stack images revealed that OFD1 sits towards the distal ends of basal bodies between the focus of the γ-tubulin signal and the axonemal microtubules (Fig. 8E). PCM-1 also localized at the sites of basal bodies in multiciliated cells but not within the ciliary axonemes (supplementary material Fig. S5B). Thus, in multiciliated cells, OFD1 and PCM-1 concentrate on basal bodies at the foot of the ciliary axoneme.
Fig. 8.
OFD1 localizes to basal bodies in multiciliated epithelia. (A–C) Primary human nasal epithelial cells were grown to different stages of differentiation by transfer to an air–liquid interface before fixation and co-staining with antibodies against OFD1 (red) and γ-tubulin (green) (plus DNA, blue). Maximum intensity projections are shown in the upper panels. The lower panels show enlargements of the boxed areas through sequential z-sections. (D) Cells as in A–C were fixed co-stained with antibodies against OFD1 (red) and acetylated tubulin (green). (E) z-axis projections of the large panels in A–D. Cells in A are at day 10 post-transfer. Cells in B–D are at day 28 post-transfer.
Discussion
We show that the protein encoded by the OFD1 disease gene localizes not only to centrosomes and basal bodies, but also to centriolar satellites. Here, it colocalizes with other ciliopathy disease proteins, BBS4 and CEP290, and with a key centriolar satellite component, PCM-1. Our experiments further demonstrate that localization of these proteins to centriolar satellites is mutually dependent. Indeed, proteins implicated in several ciliopathies rely on each other for this particular subcellular association. Thus, centriolar satellites are crucial assembly points for proteins implicated in human ciliopathies, including OFD1. Additionally, on the basis of our zebrafish experiments, OFD1 and BBS4 functionally synergize in the whole animal (e.g. determining laterality), even if the proteins do not directly interact. Together, these observations provide some explanation as to why the clinical spectrum of OFD1, BBS and Joubert syndromes shows considerable overlap, for example featuring polydactyly and renal cysts.
With BBS4 and CEP290, OFD1 joins a growing list of ciliopathy proteins localizing to centriolar satellites. Intriguingly, OFD1, BBS4 and CEP290 all physically interact with PCM-1. PCM-1 mutations have not yet been associated with ciliopathies, although we suggest that it might be fruitful to seek such mutations in patients with apparent ciliopathies who do not have defined mutation of other genes. PCM-1 was the first component of centriolar satellites to be described and was suggested to act as the core structural element of satellites (Kubo et al., 1999; Kubo and Tsukita, 2003). Consistent with this, localization of OFD1, CEP290 and BBS4 to centriolar satellites are all dependent on PCM-1. However, depletion of OFD1, BBS4 or CEP290 also leads to mislocalization of PCM-1 and other ciliopathy disease proteins (see also Kim et al., 2008; Kim et al., 2004), which suggests that removal of any satellite component destabilises this structure. Clearly, centriolar satellites are large macromolecular assemblies that rely on multiple protein–protein interactions for their integrity.
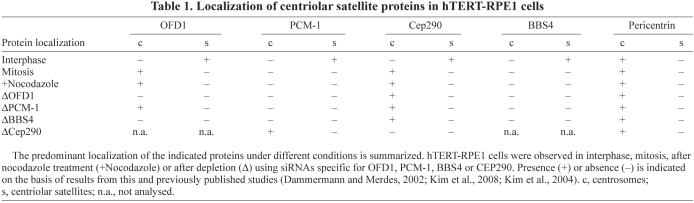
Our discovery that PCM-1 binds OFD1, and that the region encompassing the fifth coil-coil of OFD1 is crucial for this interaction, might help explain why different OFD1 mutations lead to clinically variable phenotypes. There exist rare males with OFD1 mutations who do not die in early gestation nor have typical female-associated OFD1 features (Budny et al., 2006; Coene et al., 2009). Instead, their disorders are dominated by mental retardation, sometimes associated with Joubert syndrome-like brain lesions, with variable digital anomalies (Coene et al., 2009). Again contrasting with classical OFD1 symptomatology, females carrying the same mutations are healthy (Budny et al., 2006). Males affected by the atypical disease have mutations towards the end of OFD1 (frameshifts in either exon 16 or 21) and, should these mutant transcipts be translated, the resulting proteins would retain the fifth coiled-coil domain (Fig. 5D) (see also figure 7 in Coene et al., 2009). By striking contrast, frameshift and nonsense mutations described in females with the typical OFD1 syndrome (Macca and Franco, 2009; Romio et al., 2003) (and presumably males in the same families with similar mutations and who die in early gestation) would, if mutant transcripts were stable enough to be translated, overwhelmingly often generate proteins lacking the fifth coiled-coil (Fig. 5D) (see also figure 7 in Coene et al., 2009). Given the pivotal biochemical role of PCM-1 in binding not only OFD1 but also BBS4 and CEP290 within satellites, we speculate that OFD1 mutations that abolish interaction with PCM-1 would result in destabilization of PCM-1, CEP290 and BBS4 in vivo, as demonstrated by our in vitro experiments summarized in Table 1. This would lead to catastrophic biological effects in males, who carry only one OFD1 allele on their single X chromosome, leading to early embryonic lethality. By contrast, males with mutations predicted to leave the fifth coil-coil domain intact, thus allowing potential interaction with PCM-1, are viable (Budny et al., 2006; Coene et al., 2009). This, in turn, suggests that the C-terminus of OFD1, beyond the last two coiled-coil domains, has yet-to-be-defined roles, especially in brain maturation.
Table 1.
Localization of centriolar satellite proteins in hTERT-RPE1 cells
Localization of these ciliopathy disease proteins to centrosomes themselves is harder to pin down. In untreated interphase hTERT-RPE1 cells, OFD1 does not strongly colocalize with core centrosomal markers such as centrin, Cep135 or γ-tubulin. Similar results were reported for CEP290 and BBS4 (Kim et al., 2008; Kim et al., 2004). These data suggest that, if there is a centrosomal component to these proteins in cycling cells, it represents a much smaller fraction of the protein than is present at centriolar satellites. PCM-1 is exclusively localized to centriolar satellites and, interestingly, upon progression through mitosis when centriolar satellites disperse, both OFD1 and CEP290 become obviously associated with centrosomes whereas PCM-1 and BBS4 do not. Although we and others (Singla et al., 2010) have not observed defects in mitosis in cells depleted of OFD1, the fact that the protein is present on spindle poles could indicate an as-yet-unidentified role in mitosis. In this respect, it is worth noting that OFD1 was identified as essential for mitosis in a recent genome-wide study by the MitoCheck consortium (Neumann et al., 2010).
The ciliopathy disease proteins also display distinct patterns in terms of centrosome localization following treatments that either disrupt centriolar satellites or disturb their pericentriolar distribution. First, we found that CEP290 can be weakly detected at centrosomes upon depletion of OFD1, PCM-1 or BBS4, despite apparent loss of satellites. However, OFD1 was detected at centrosomes upon depletion of PCM-1 but not depletion of BBS4. It has been reported that PCM-1 moves to centrosomes in the absence of CEP290 (Kim et al., 2008), although when CEP290 localization was disturbed in our hands by either OFD1 or BBS4 depletion, there was no accumulation of PCM-1 at the centrosome. Depolymerization of the microtubule network leads to dispersal of satellites into the cytoplasm. These appear to retain OFD1 because cytoplasmic aggregates of similar size to satellites could be detected. Similar results have recently been described for another LisH-containing centriolar satellite component, FOR20 (Sedjai et al., 2010). However, under these conditions, OFD1 and CEP290 were also clearly detected on centrosomes, whereas BBS4 was not (Kim et al., 2008; Kim et al., 2004). Although these rather confusing results indicate that ciliopathy disease proteins exhibit a much more complex dependency in terms of centrosome localization, they do suggest that in many respects OFD1 and CEP290 display remarkably similar behaviour (summarised in Table 1). This is intriguing in light of the recent finding that mutations in both proteins can lead to Joubert syndrome (Coene et al., 2009; Kim et al., 2008).
OFD1 and PCM-1 interaction required the regions encompassing the N-terminal coiled-coil domain of PCM-1 and at least the fifth coiled-coil motif of OFD1, which implies the possibility of a direct coiled-coil interaction. PCM-1 is also capable of oligomerization via its coiled-coil region (Kubo and Tsukita, 2003); whether this competes with binding to OFD1 is not clear. By contrast, PCM-1 interacts with BBS4 via its C-terminal non-coiled-coil region (Kim et al., 2004). Hence, PCM-1 could bridge binding of OFD1 and BBS4, although we only detected very weak interaction between OFD1 and BBS4. Our interaction data is consistent with the consequences of overexpressing a C-terminal fragment of OFD1 containing the third to sixth coiled-coil, leading to PCM-1 mislocalization. This fragment also caused CEP290 mislocalization, suggesting either that OFD1 directly interacts with CEP290 through this region or it disturbs CEP290 via PCM-1. Indeed, CEP290 co-precipitates with PCM-1, although the region of interaction is unknown (Kim et al., 2008). Intriguingly, a C-terminal fragment of OFD1 that includes the fifth and sixth coiled-coils was recently found to promote interaction with SDCCAG8, the product of a novel NPHP-related ciliopathy gene (Otto et al., 2010). Meanwhile, the section of OFD1 protein encompassing the second to fourth coiled-coils is important for recruitment of IFT88 and association with lebercilin, the gene for which is mutated in Leber congenital amaurosis (Coene et al., 2009; Singla et al., 2010). Importantly, an N-terminal OFD1 fragment lacking the region required for PCM-1 interaction is localized to satellites, indicating that association with PCM-1 is unlikely to be the primary mechanism for this localization and raising the prospect that the LisH motif could contribute to this targeting. In this regard, it is intriguing that satellite localization of FOR20 is also dependent on its LisH motif (Sedjai et al., 2010).
Unsurprisingly, many ciliopathy disease proteins are essential for primary cilia formation, and not simply because of a requirement for cell cycle progression. However, a specific function for centriolar satellites in ciliogenesis remains elusive. Certainly, satellites mediate protein transport along microtubules to the centrosome or basal body. For example, the incorporation of centrin, pericentrin and ninein to the centrosome depends on PCM-1 (Dammermann and Merdes, 2002). Moreover, both BBS4 and CEP290 interact with microtubule-based motor protein subunits (Chang et al., 2006; Kim et al., 2004; McEwen et al., 2007). This trafficking might provide the machinery for basal body biogenesis, cilia extension or intraflagellar transport. Indeed, both CEP290 and BBSome components are required for transport of Rab8 to the cilia, where this GTPase enables vesicle docking and fusion events essential to assembly of the ciliary membrane (Kim et al., 2008; Nachury, 2008). Interestingly, in our experiments, mislocalization of PCM-1, either by depletion of OFD1 or expression of truncated OFD1 constructs, did not obviously affect pericentrin levels at the centrosome. This suggests that there could be important differences in the way that trafficking responds to the mislocalization or absence of PCM-1.
Regarding basal body biogenesis, centriolar satellites (probably the same as structures previously termed fibrous granules) do accumulate in highly ciliated tissues (Hagiwara et al., 2004). Moreover, we found intense staining of OFD1 and PCM-1 at the basal bodies of multiciliated nasal epithelia, although discrete satellites were not apparent. However, neither we, nor others (Singla et al., 2010), observed defects in centriole duplication in cells depleted of OFD1, and PCM-1 was reported not to be essential for ciliogenesis in multiciliated cells (Vladar and Stearns, 2007). Nevertheless, tantalizing evidence exists for a role for OFD1 in multiciliated cells, based on the report of a family carrying an OFD1 mutation. This mutation would lead to an OFD1a protein truncated beyond the fifth coil-coil domain, but with normal expression of the little-studied OFD1b (Budny et al., 2006). Affected individuals had severe recurrent respiratory infections, and ciliary beating in respiratory epithelia was perturbed. However, respiratory infections are not a symptom seen in classic OFD1 and thus further work is required to define the possible roles of OFD1 and other ciliopathy proteins in multiciliated cells.
Materials and Methods
Plasmid constructions
OFD1 fragments were amplified by polymerase chain reaction (PCR) using full-length human OFD1 cDNA as the template (Romio et al., 2004). Amplified fragments were subcloned into the mammalian expression vector pCMV-Tag3C (Stratagene) and bacterial expression vectors pGEX-4T-1 (GE Healthcare) and pETM-11 (EMBL) providing N-terminal Myc, GST or His tags, respectively. A GST-tagged C-terminal fragment of BBS4 was also generated in pGEX-4T-1 by amplifying the BBS4 fragment by PCR from a GFP–BBS4 construct. All constructs were verified by DNA sequencing within the Protein and Nucleic Acid Laboratory, Leicester, UK.
Antibody generation
For production of antibodies against OFD1, rabbits were immunised with a His-tagged C-terminal fragment of OFD1 that spanned amino acids 145–1012. This fragment was expressed in Escherichia coli strain Rosetta 2 (DE3) (Novagen) and purified under denaturing conditions according to standard protocols. For production of antibodies against BBS4, rabbits were immunized with the GST-tagged BBS4 C-terminal fragment (residues 235–519) following expression in E. coli strain Rosetta 2 (DE3) (Novagen) and purification according to standard protocols. Immunizations and affinity purification of antibodies was performed by Cambridge Research Biochemicals, Billingham, UK.
Cell culture and transfections
Cells were grown in the following media: hTERT-RPE1 cells in Dulbecco's modified Eagle's medium (DMEM)/Ham's F12 (1:1) supplemented with 0.348% sodium bicarbonate solution; IMCD3 cells in DMEM/Ham's F12 (1:1); hFF and HEK293 cells in DMEM; and CHO cells in Ham's F12. All cells were supplemented with 10% fetal calf serum, 100 IU/ml penicillin, and 100 μg/ml streptomycin. Cells were grown at 37°C in a 5% CO2 atmosphere. Primary cilia formation was induced by culturing cells in serum-free medium for 48 hours. Human nasal epithelial cells were obtained with ethical approval from healthy volunteers and cultured and induced to differentiate as described (Hirst et al., 2010). Transient transfections were performed with Lipofectamine 2000 (Invitrogen), Fugene HD (Roche) or JetPrime (Polyplus Transfection) according to the manufacturer's instructions and analysed after 24 hours.
Fluorescence microscopy
Immunofluorescence microscopy was carried out as previously described (Fry et al., 1998). Primary antibodies were against OFD1 Ab1 (1:100) (Romio et al., 2003), OFD1 Ab2 (1:100; this study), PCM-1 (1:1000) (Dammermann and Merdes, 2002), BBS4 (1:250), centrin2 (2 μg/ml; N-17 Santa Cruz Biotechnology), pericentrin (2 μg/ml; Abcam), acetylated tubulin (0.5 μg/ml; Sigma), γ-tubulin (0.15 μg/ml; Sigma), α-tubulin (0.3 μg/ml; Sigma), Cep135 (1 μg/ml) (Kleylein-Sohn et al., 2007), C-Nap1 (1 μg/ml) (Fry et al., 1998), CEP290 (1:200; Abcam ab84870), GFP (0.5 μg/ml; Sigma) and Myc (1:1000; Cell Signaling). Secondary antibodies were Alexa-Fluor-488- and Alexa-Fluor-594-conjugated goat anti-mouse and goat anti-rabbit IgGs (1 μg/ml; Invitrogen). Images were captured using a TE300 inverted microscope (Nikon) or Leica TCS SP5 confocal microscope equipped with a Leica DMI 6000B inverted microscope using a 63× oil objective (numerical aperture, 1.4). For quantification of immunofluorescence intensity, integrated pixel density was measured using Openlab software (Improvision).
RNA interference experiments
OFD1 was depleted using two siRNA oligonucleotides (5′-GCUCAUAGCUAUUAAUUCA-3′ and 5′-GAUCGAUCGUUCUGUCAAU-3′) (Dharmacon). Two PCM-1-specific siRNA oligonucleotides were obtained from QIAGEN (Dammermann and Merdes, 2002). BBS4 was depleted using a pool of four oligonucleotides (5′-GGAUAAGUGUAACCCUUUA-3′, 5′-CUGCAAGGCUGUUAUCAAA-3′, 5′-CAUUAUAUCCGGAAAGAUU-3′ and 5′-GCAAUGCACUGACUUAUGA-3′) (Dharmacon). Control depletions were performed with luciferase (GL2) siRNAs (Dharmacon). Cells were transfected with 100 nM siRNAs for 72 hours using Oligofectamine (Invitrogen).
Preparation of cell extracts, SDS-PAGE and western blotting
Cells were washed once in PBS and lysed in lysis buffer (50 mM HEPES–KOH pH 8.0, 100 mM KCl, 100 mM EDTA, 0.1% NP-40, 10 mM NaF, 50 mM β-glycerophosphate, 2 mM DTT, 1× Protease Inhibitor Cocktail, 10% glycerol) for 30 minutess on ice. Lysates were centrifuged for 10 minutes at 15,700 g, at 4°C, and protein concentrations of the cleared supernatant determined by Bradford assay. SDS-PAGE and western blotting were performed as previously described (Hames et al., 2005). Primary antibodies were against OFD1 (1:100), PCM-1 (1:1000), BBS4 (1:250), γ-tubulin (0.15 μg/ml), GFP (1:2000; Abcam) and GST (1:5000; Santa Cruz Biotechnology). Secondary antibodies were alkaline-phosphatase-conjugated anti-rabbit or anti-mouse IgGs (1:7500; Promega) or horseradish peroxidase (HRP)-labelled IgGs (Amersham).
Immunoprecipitations
Transfected cell extracts (4 mg) were pre-cleared for 30 minutess at 4°C with protein G agarose beads (Sigma) that had been pre-washed in lysis buffer. Beads were then removed and supernatants incubated with GFP antibody (1:500; Sigma) for 1 hour on ice. The complexes were then captured with protein G agarose beads (again pre-washed in lysis buffer) for 1 hour at 4°C. Beads were washed four times with lysis buffer, resuspended with protein sample buffer and analysed by SDS-PAGE and western blotting.
GST pull-downs
Purified GST-tagged protein (100 μg) was incubated for 1 hour at 4°C with glutathione Sepharose 4B beads (GE Healthcare) that had been pre-washed with NETN buffer (200 mM Tris-HCl pH 8.0, 200 mM NaCl, 0.5% NP-40, 1 mM EDTA) containing 0.5% milk. Beads were then washed four times with NETN buffer, resuspended in NETN buffer plus inhibitors (1 mM PMSF, 1 mM DTT, 1× Protease Inhibitor Cocktail) and incubated with 1 mg of cell extract overnight at 4°C. In this case, extracts of transfected cells were prepared by lysis with NETN buffer and processed as above. The next day, beads were washed four times with NETN buffer and resuspended in protein sample buffer for analysis by SDS-PAGE and western blotting.
Zebrafish morpholino experiments
Zebrafish were maintained at 28°C on a cycle of 14 hours light and 10 hours dark. Morpholino antisense oligonucleotides for Ofd1 (Spl6) and Bbs4 were injected in one- to two-cell-stage embryos using needles pulled from glass capillary tubes, as previously described (Ferrante et al., 2009; Gerdes et al., 2007). In whole embryos, the heart rudiment was visualized by direct inspection and the frequency of deviation from normal leftward displacement of the heart cone, or cardiac ‘jogging’, was recorded as a laterality readout (Ferrante et al., 2009). Comparison between groups was undertaken using Fisher's Exact Test. In addition, it was noted whether the bodies of embryos were straight or displayed abnormal curvature characteristic of ciliary defects (Ferrante et al., 2009).
Miscellaneous techniques
To depolymerize the microtubule cytoskeleton, cells were treated with 6 μg/ml nocodazole for 1 hour at 37°C and then kept on ice for 30 minutes. To allow microtubule regrowth, cells were subsequently washed 3× with 1× PBS and pre-warmed media added. To monitor centrosome duplication during S-phase arrest upon inhibition of nuclear export or Cdk2 activity, CHO cells were treated as previously described (Prosser et al., 2009). For flow cytometry, cells were stained with propidium iodide and analysed by flow cytometry (BD FACSCantoTM II) according to standard protocols.
Acknowledgments
We thank all members of our laboratory for useful discussion and Stephen W. Wilson (University College London, London, UK) for discussions and provision of fish facilities. We are indebted to Andreas Merdes (Institut des Sciences et Techniques du Médicament, Toulouse, France) for PCM-1 antibodies and constructs, Erich Nigg (Biozentrum, University of Basel, Basel, Switzerland) for Cep135 antibodies, Philip Beales (University College London, London, UK) for GFP-BBS4 andRichard Vallee (Columbia University, New York, NY) for Myc-dynamitin. This work was funded by Wellcome Trust grants to A.M.F. and A.S.W. C.A.M.L. is funded by Fundação para a Ciência e a Tecnologia (Portugal) and POPH/FSE (EU). A.S.W. acknowledges support from the Manchester NIHR Biomedical Research Centre. Deposited in PMC for release after 6 months.
Footnotes
Supplementary material available online at http://jcs.biologists.org/cgi/content/full/124/4/600/DC1
References
- Badano J. L., Mitsuma N., Beales P. L., Katsanis N. (2006). The ciliopathies: an emerging class of human genetic disorders. Annu. Rev. Genomics Hum. Genet. 7, 125-148 [DOI] [PubMed] [Google Scholar]
- Bakkers J., Verhoeven M. C., Abdelilah-Seyfried S. (2009). Shaping the zebrafish heart: from left-right axis specification to epithelial tissue morphogenesis. Dev. Biol. 330, 213-220 [DOI] [PubMed] [Google Scholar]
- Balczon R., Bao L., Zimmer W. E. (1994). PCM-1, A 228-kD centrosome autoantigen with a distinct cell cycle distribution. J. Cell Biol. 124, 783-793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berbari N. F., O'Connor A. K., Haycraft C. J., Yoder B. K. (2009). The primary cilium as a complex signaling center. Curr. Biol. 19, R526-R535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budny B., Chen W., Omran H., Fliegauf M., Tzschach A., Wisniewska M., Jensen L. R., Raynaud M., Shoichet S. A., Badura M., et al. (2006). A novel X-linked recessive mental retardation syndrome comprising macrocephaly and ciliary dysfunction is allelic to oral-facial-digital type I syndrome. Hum. Genet. 120, 171-178 [DOI] [PubMed] [Google Scholar]
- Chang B., Khanna H., Hawes N., Jimeno D., He S., Lillo C., Parapuram S. K., Cheng H., Scott A., Hurd R. E., et al. (2006). In-frame deletion in a novel centrosomal/ciliary protein CEP290/NPHP6 perturbs its interaction with RPGR and results in early-onset retinal degeneration in the rd16 mouse. Hum. Mol. Genet. 15, 1847-1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coene K. L., Roepman R., Doherty D., Afroze B., Kroes H. Y., Letteboer S. J., Ngu L. H., Budny B., van Wijk E., Gorden N. T., et al. (2009). OFD1 is mutated in X-linked Joubert syndrome and interacts with LCA5-encoded lebercilin. Am. J. Hum. Genet. 85, 465-481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbit K. C., Shyer A. E., Dowdle W. E., Gaulden J., Singla V., Chen M. H., Chuang P. T., Reiter J. F. (2008). Kif3a constrains beta-catenin-dependent Wnt signalling through dual ciliary and non-ciliary mechanisms. Nat. Cell Biol. 10, 70-76 [DOI] [PubMed] [Google Scholar]
- Dammermann A., Merdes A. (2002). Assembly of centrosomal proteins and microtubule organization depends on PCM-1. J. Cell Biol. 159, 255-266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Conciliis L., Marchitiello A., Wapenaar M. C., Borsani G., Giglio S., Mariani M., Consalez G. G., Zuffardi O., Franco B., Ballabio A., et al. (1998). Characterization of Cxorf5 (71-7A), a novel human cDNA mapping to Xp22 and encoding a protein containing coiled-coil alpha-helical domains. Genomics 51, 243-250 [DOI] [PubMed] [Google Scholar]
- Emes R. D., Ponting C. P. (2001). A new sequence motif linking lissencephaly, Treacher Collins and oral-facial-digital type 1 syndromes, microtubule dynamics and cell migration. Hum. Mol. Genet. 10, 2813-2820 [DOI] [PubMed] [Google Scholar]
- Feather S. A., Woolf A. S., Donnai D., Malcolm S., Winter R. M. (1997). The oral-facial-digital syndrome type 1 (OFD1), a cause of polycystic kidney disease and associated malformations, maps to Xp22.2-Xp22.3. Hum. Mol. Genet. 6, 1163-1167 [DOI] [PubMed] [Google Scholar]
- Ferrante M. I., Giorgio G., Feather S. A., Bulfone A., Wright V., Ghiani M., Selicorni A., Gammaro L., Scolari F., Woolf A. S., et al. (2001). Identification of the gene for oral-facial-digital type I syndrome. Am. J. Hum. Genet. 68, 569-576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrante M. I., Barra A., Truong J. P., Banfi S., Disteche C. M., Franco B. (2003). Characterization of the OFD1/Ofd1 genes on the human and mouse sex chromosomes and exclusion of Ofd1 for the Xpl mouse mutant. Genomics 81, 560-569 [DOI] [PubMed] [Google Scholar]
- Ferrante M. I., Zullo A., Barra A., Bimonte S., Messaddeq N., Studer M., Dolle P., Franco B. (2006). Oral-facial-digital type I protein is required for primary cilia formation and left-right axis specification. Nat. Genet. 38, 112-117 [DOI] [PubMed] [Google Scholar]
- Ferrante M. I., Romio L., Castro S., Collins J. E., Goulding D. A., Stemple D. L., Woolf A. S., Wilson S. W. (2009). Convergent extension movements and ciliary function are mediated by ofd1, a zebrafish orthologue of the human oral-facial-digital type 1 syndrome gene. Hum. Mol. Genet. 18, 289-303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry A. M., Mayor T., Meraldi P., Stierhof Y. D., Tanaka K., Nigg E. A. (1998). C-Nap1, a novel centrosomal coiled-coil protein and candidate substrate of the cell cycle-regulated protein kinase Nek2. J. Cell Biol. 141, 1563-1574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes J. M., Liu Y., Zaghloul N. A., Leitch C. C., Lawson S. S., Kato M., Beachy P. A., Beales P. L., DeMartino G. N., Fisher S., et al. (2007). Disruption of the basal body compromises proteasomal function and perturbs intracellular Wnt response. Nat. Genet. 39, 1350-1360 [DOI] [PubMed] [Google Scholar]
- Gerdes J. M., Davis E. E., Katsanis N. (2009). The vertebrate primary cilium in development, homeostasis, and disease. Cell 137, 32-45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlitz G., Darhin E., Giorgio G., Franco B., Reiner O. (2005). Novel functional features of the Lis-H domain: role in protein dimerization, half-life and cellular localization. Cell Cycle 4, 1632-1640 [DOI] [PubMed] [Google Scholar]
- Giorgio G., Alfieri M., Prattichizzo C., Zullo A., Cairo S., Franco B. (2007). Functional characterization of the OFD1 protein reveals a nuclear localization and physical interaction with subunits of a chromatin remodeling complex. Mol. Biol. Cell 18, 4397-4404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graser S., Stierhof Y. D., Lavoie S. B., Gassner O. S., Lamla S., Le Clech M., Nigg E. A. (2007). Cep164, a novel centriole appendage protein required for primary cilium formation. J. Cell Biol. 179, 321-330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagiwara H., Ohwada N., Takata K. (2004). Cell biology of normal and abnormal ciliogenesis in the ciliated epithelium. Int. Rev. Cytol. 234, 101-141 [DOI] [PubMed] [Google Scholar]
- Hames R. S., Crookes R. E., Straatman K. R., Merdes A., Hayes M. J., Faragher A. J., Fry A. M. (2005). Dynamic recruitment of Nek2 kinase to the centrosome involves microtubules, PCM-1, and localized proteasomal degradation. Mol. Biol. Cell 16, 1711-1724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrandt F., Otto E. (2005). Cilia and centrosomes: a unifying pathogenic concept for cystic kidney disease? Nat. Rev. Genet. 6, 928-940 [DOI] [PubMed] [Google Scholar]
- Hirst R. A., Rutman A., Williams G., O'Callaghan C. (2010). Ciliated air-liquid cultures as an aid to diagnostic testing of primary ciliary dyskinesia (PCD). Chest 138, 1441-1447 [DOI] [PubMed] [Google Scholar]
- Kim J., Krishnaswami S. R., Gleeson J. G. (2008). CEP290 interacts with the centriolar satellite component PCM-1 and is required for Rab8 localization to the primary cilium. Hum. Mol. Genet. 17, 3796-3805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. C., Badano J. L., Sibold S., Esmail M. A., Hill J., Hoskins B. E., Leitch C. C., Venner K., Ansley S. J., Ross A. J., et al. (2004). The Bardet-Biedl protein BBS4 targets cargo to the pericentriolar region and is required for microtubule anchoring and cell cycle progression. Nat. Genet. 36, 462-470 [DOI] [PubMed] [Google Scholar]
- Kleylein-Sohn J., Westendorf J., Le Clech M., Habedanck R., Stierhof Y. D., Nigg E. A. (2007). Plk4-induced centriole biogenesis in human cells. Dev. Cell 13, 190-202 [DOI] [PubMed] [Google Scholar]
- Kubo A., Tsukita S. (2003). Non-membranous granular organelle consisting of PCM-1: subcellular distribution and cell-cycle-dependent assembly/disassembly. J. Cell Sci. 116, 919-928 [DOI] [PubMed] [Google Scholar]
- Kubo A., Sasaki H., Yuba-Kubo A., Tsukita S., Shiina N. (1999). Centriolar satellites: molecular characterization, ATP-dependent movement toward centrioles and possible involvement in ciliogenesis. J. Cell Biol. 147, 969-980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macca M., Franco B. (2009). The molecular basis of oral-facial-digital syndrome, type 1. Am. J. Med. Genet. C Semin. Med. Genet. 151, 318-325 [DOI] [PubMed] [Google Scholar]
- Marshall W. F., Nonaka S. (2006). Cilia: tuning in to the cell's antenna. Curr. Biol. 16, R604-R614 [DOI] [PubMed] [Google Scholar]
- May-Simera H. L., Ross A., Rix S., Forge A., Beales P. L., Jagger D. J. (2009). Patterns of expression of Bardet-Biedl syndrome proteins in the mammalian cochlea suggest noncentrosomal functions. J. Comp. Neurol. 514, 174-188 [DOI] [PubMed] [Google Scholar]
- McEwen D. P., Koenekoop R. K., Khanna H., Jenkins P. M., Lopez I., Swaroop A., Martens J. R. (2007). Hypomorphic CEP290/NPHP6 mutations result in anosmia caused by the selective loss of G proteins in cilia of olfactory sensory neurons. Proc. Natl. Acad. Sci. USA 104, 15917-15922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachury M. V. (2008). Tandem affinity purification of the BBSome, a critical regulator of Rab8 in ciliogenesis. Methods Enzymol. 439, 501-513 [DOI] [PubMed] [Google Scholar]
- Neumann B., Walter T., Heriche J. K., Bulkescher J., Erfle H., Conrad C., Rogers P., Poser I., Held M., Liebel U., et al. (2010). Phenotypic profiling of the human genome by time-lapse microscopy reveals cell division genes. Nature 464, 721-727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigg E. A., Raff J. W. (2009). Centrioles, centrosomes, and cilia in health and disease. Cell 139, 663-678 [DOI] [PubMed] [Google Scholar]
- Ohta T., Essner R., Ryu J. H., Palazzo R. E., Uetake Y., Kuriyama R. (2002). Characterization of Cep135, a novel coiled-coil centrosomal protein involved in microtubule organization in mammalian cells. J. Cell Biol. 156, 87-99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto E. A., Hurd T. W., Airik R., Chaki M., Zhou W., Stoetzel C., Patil S. B., Levy S., Ghosh A. K., Murga-Zamalloa C. A., et al. (2010). Candidate exome capture identifies mutation of SDCCAG8 as the cause of a retinal-renal ciliopathy. Nat. Genet. 42, 840-850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prosser S. L., Straatman K. R., Fry A. M. (2009). Molecular dissection of the centrosome overduplication pathway in S-phase-arrested cells. Mol. Cell. Biol. 29, 1760-1773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romio L., Wright V., Price K., Winyard P. J., Donnai D., Porteous M. E., Franco B., Giorgio G., Malcolm S., Woolf A. S., et al. (2003). OFD1, the gene mutated in oral-facial-digital syndrome type 1, is expressed in the metanephros and in human embryonic renal mesenchymal cells. J. Am. Soc. Nephrol. 14, 680-689 [DOI] [PubMed] [Google Scholar]
- Romio L., Fry A. M., Winyard P. J., Malcolm S., Woolf A. S., Feather S. A. (2004). OFD1 is a centrosomal/basal body protein expressed during mesenchymal-epithelial transition in human nephrogenesis. J. Am. Soc. Nephrol. 15, 2556-2568 [DOI] [PubMed] [Google Scholar]
- Saal S., Faivre L., Aral B., Gigot N., Toutain A., Van Maldergem L., Destree A., Maystadt I., Cosyns J. P., Jouk P. S., et al. (2010). Renal insufficiency, a frequent complication with age in oral-facial-digital syndrome type I. Clin. Genet. 77, 258-265 [DOI] [PubMed] [Google Scholar]
- Sedjai F., Acquaviva C., Chevrier V., Chauvin J. P., Coppin E., Aouane A., Coulier F., Tolun A., Pierres M., Birnbaum D., et al. (2010). Control of ciliogenesis by FOR20, a novel centrosome and pericentriolar satellite protein. J. Cell Sci. 123, 2391-2401 [DOI] [PubMed] [Google Scholar]
- Singla V., Reiter J. F. (2006). The primary cilium as the cell's antenna: signaling at a sensory organelle. Science 313, 629-633 [DOI] [PubMed] [Google Scholar]
- Singla V., Romaguera-Ros M., Garcia-Verdugo J. M., Reiter J. F. (2010). Ofd1, a human disease gene, regulates the length and distal structure of centrioles. Dev. Cell 18, 410-424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thauvin-Robinet C., Callier P., Franco B., Zuffardi O., Payet M., Aral B., Gigot N., Donzel A., Mosca-Boidron A. L., Masurel-Paulet A., et al. (2009). Search for genomic imbalances in a cohort of 20 patients with oral-facial-digital syndromes negative for mutations and large rearrangements in the OFD1 gene. Am. J. Med. Genet. A 149, 1846-1849 [DOI] [PubMed] [Google Scholar]
- Vladar E. K., Stearns T. (2007). Molecular characterization of centriole assembly in ciliated epithelial cells. J. Cell Biol. 178, 31-42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zullo A., Iaconis D., Barra A., Cantone A., Messaddeq N., Capasso G., Dolle P., Igarashi P., Franco B. (2010). Kidney-specific inactivation of Ofd1 leads to renal cystic disease associated with upregulation of the mTOR pathway. Hum. Mol. Genet. 19, 2792-2803 [DOI] [PMC free article] [PubMed] [Google Scholar]