Abstract
Human γδ T-cell lymphoma is a rare clinicopathologic entity with aggressive course and poor prognosis. The etiology and pathogenesis of γδ T-cell lymphoma is unknown. We show here that mice with deficiency in inhibitory helix-loop-helix protein Id3 (Id3−/−) developed γδ T-cell lymphoma that resembled human γδ T-cell lymphoma. The Id3−/− mice with lymphoma showed splenomegaly, hepatomegaly, and lymphadenopathy with involvement of bone marrow, thymus, kidney, and lungs between 6 and 15 months of age. Phenotypic analysis revealed that lymphomatous cells were cluster of differentiation (CD)3+, γδ T-cell receptor (TCR)+, and αβ TCR−, and expressed CD8+CD4−, CD4+CD8−, or a mixture of the two. Id3−/− γδ T-cell lymphoma used predominantly Vγ1.1, some Vγ3, yet no Vγ2 TCR, and some showed increased levels of the oncogene c-Myc. Strikingly, adoptive transfer of the γδ T-cell lymphoma into syngeneic Rag1−/− mice resulted in aggressive γδ T-cell lymphoma, identical to the Id3−/− donor. Thus, our data demonstrate that Id3 regulates the development of γδ T-cell lymphoma in mice, raising a possibility of Id3 gene mutation in human γδ T-cell lymphoma. Our model will provide a tool for studying the molecular mechanisms and development of human γδ T-cell lymphoma.
Introduction
Human γδ T-cell lymphoma represents a subgroup of peripheral T-cell lymphoma expressing γδ T-cell receptors (TCRs).1 Hepatosplenic T-cell lymphoma (HSTCL) is the prototype peripheral T-cell lymphoma expressing the γδ TCR, although non-HSTCL γδ T-cell lymphoma also exist.1 In contrast to the majority of T-cell lymphoma that express the αβ TCR,2,3 human γδ T-cell lymphoma is a rare clinicopathologic entity with an aggressive course and poor prognosis.4–10 Patients usually have splenomegaly, hepatomegaly, and/or cutaneous lesions and, paradoxically, this lymphoma often accompanies autoimmune disease.4,6,7 The etiology and the genes that control the development of γδ T-cell neoplastic cells are unknown. In addition, there is a lack of animal models that resemble human γδ T-cell lymphoma.
The helix-loop-helix (HLH) proteins are a family of transcriptional regulatory proteins comprising 2 major subclasses; the basic and inhibitory HLH proteins.11–13 The basic HLH proteins include the mammalian E2A, E2-2, and Hela E-box-binding (HEB) proteins and the Drosophila gene product Daughterless. It has been reported that E2A inactivation in mice leads to the rapid development of a T-cell lymphoma expressing no or low levels of surface TCRs, and cluster of differentiation (CD)4 and CD8.14 The lymphoma are monoclonal and highly malignant and displayed a cell surface phenotype similar to that of immature thymocytes.15,16 The inhibitory HLH proteins include 4 mammalian members Id1, Id2, Id3, and Id4,17–21 which function as inhibitors of the basic HLH proteins, preventing their binding to target DNA. Id3 has been shown to be involved in thymocyte maturation20; playing a role in the restriction of the γδ lineage development during thymopoiesis.22 Interestingly, Id3 plays a role in controlling the development of autoimmune-like Sjogren Syndrome in mice.23,24
Here we reveal a novel role for Id3 in controlling the development of T-cell lymphoma expressing γ/δ TCRs. Id3−/− mice developed an aggressive γδ T-cell lymphoma between 6 and 15 months of age. The mice with lymphoma showed massive splenomegaly and hepatomegaly with involvement of bone marrow, thymus, kidney, and lungs, which share many pathological features with human HSTCL. We have established an animal model for the analysis of the pathogenesis of γδ T-cell lymphoma and propose that inactivation of Id3 gene might contribute to the development of γδ T-cell lymphoma in humans.
Methods
Mice
C57BL/6 (wild-type [WT]) mice were obtained from The Jackson Laboratory. Id3−/− mice on a C57BL/6 background (a gift from Dr Y. Zhuang, Duke University Medical Center, Chapel Hill, NC) were bred in our facilities under specific pathogen-free conditions. Id3−/− mice that developed symptoms such as dermatitis, hunched posture, weight loss, and/or listlessness were killed for analysis. Age-matched WT mice were used as controls. All animal studies were performed according to National Institutes of Health guidelines for use and care of live animals and approved by Animal Care and Use Committee of National Institute of Dental and Craniofacial Research.
Histopathological and immunohistochemical analysis
Immediately after the mice were killed, the tissues were fixed with 10% phosphate-buffered saline (PBS)–formalin, embedded in paraffin, and sectioned. Sections were stained with hematoxylin and eosin (H&E) for histological examination. The immunohistochemical staining was performed at the Diagnostic and Research Services Branch, Division of Veterinary Resources, National Institutes of Health.
Cell purification
Thymi, spleens, and lymph nodes were gently minced in complete Dulbecco Modified Eagle Medium containing 10% fetal bovine serum (BioWhittaker) and single cell suspensions were obtained. Lung and liver were excised, minced, and digested for 40 minutes at 37°C in a Hanks Buffered Salt Solution containing 2 mg/mL of collagenase (GIBCO BRL). The homogenate was passed through 40-μm pore size filters, and the cell suspensions were washed and either stained for flow cytometric analysis or stored at −80°C for mRNA isolation.
Flow cytometric analysis
For the staining of cell surface markers, cells were incubated with antibodies at 4°C for 30 minutes. Isotype-matched negative controls were used for all flow cytometric analysis. For intracellular staining for c-myc, after surface staining, the cells were resuspended in fixation/permeabilization solution (eBioscience), then blocked with purified anti-Fc gamma receptor (FcR)II/III (CD32/CD16) at 4°C for 15 minutes, and then incubated with antibodies for 30 minutes at 4°C. For intracellular interleukin-17 (IL-17) and interferon-γ (IFN-γ), cells were stimulated with 5 ng/mL phorbol 12-myristate 13-acetate (Sigma-Aldrich), 250 ng/mL Ionomycin (Sigma-Aldrich), in the presence of 1 μL/mL GolgiPlug (BD PharMingen) for 4-5 hours. Cells were then harvested, washed and stained with antibodies to surface markers followed by intracellular cytokine antibodies using the BD Cytofix/Cytoperm kit (BD Biosciences). Cells were analyzed using a FACSCalibur (BD Biosciences). The following anti-murine antibodies were purchased from BD Biosciences: phycoerythrin (PE)–conjugated anti-CD45RB, anti-CD44, anti-CD69, anti-CD62L, anti–IFN-g, anti–mouse Notch1, and fluorescein isothiocyanate (FITC)–, PE-, or allophycocyanin (APC)–conjugated anti-murine CD25, peridinin–chlorophyll–protein complex-, or fluorescein isothiocyanate-conjugated anti-CD4 or anti-CD8, anti-TCR Vγ2 and Vγ3, purified anti-FcRII/III (CD32/CD16), as well as their respective isotype control antibodies. APC-conjugated anti-Perforin and PE-conjugated anti-Granzyme B antibodies were purchased from eBioscience. APC-conjugated antimouse IL-17 was purchased from BioLegend. PE-conjugated antimouse c-Myc was purchased from Santa Cruz Biotechnology.
RT-PCR analysis
mRNA extractions were performed using RNeasy mini kit (QIAGEN) following the manufacturer's protocol. cDNA was synthesized from 200 ng to 1 μg of RNA from cells with a SuperScript first synthesis system for reverse transcription–polymerase chain reaction (RT-PCR; Invitrogen), and mRNA were quantified by real-time PCR using the ABI 7500 Real-time PCR system (Applied Biosystems). Tal1 (NM_213237.1), Pax5 (NM_008782.2), P21 (NM_007669.4), and hypoxanthine-guanine phosphoribosyltransferase (HPRT; NM_013556.2) were analyzed according to the protocol of TaqMan gene expression assay kit (Applied Biosystems). All primers were purchased from Applied Biosystems. PCR reaction system contained 0.5μM primers and 0.2μM TaqMan probe. PCR amplification was preceded by a 10-minute incubation at 95°C denaturation step and amplification step that consisted of 40 cycles of 15 seconds at 95°C and 60 seconds at 60°C.
c-Myc expression was analyzed using SYBR Green Supermix kit (Bio-Rad) and a quantitative real-time PCR iCycler iQ detection system (Bio-Rad). The PCR primers used to detect mouse c-Myc and glyceraldehyde 3-phosphate dehydrogenase (GAPDH; Internal control) were: c-Myc Fw: 5′-GCCCAGTGAGGATATCTGGA, c-Myc Rev: 3′-ATCGCAGATGAAGCTCTGGT, GAPDH Fw: 5′-ACTCCACTCACGGCAAATTC, GAPDH Rev: 3′-TCTCCATGGTGGTGAAGACA.
PCR amplification was preceded by a 10-minute incubation at 95°C denaturation step and amplification step that consisted of 40 cycles 30 seconds at 95°C, 30 seconds at 57°C, and 30 seconds at 72°C. The amount of target gene was normalized to the endogenous reference HPRT or GAPDH using the 2(-Delta C [T]) method (2−DCt), and presented as a relative expression. The γδ TCR-specific primers and PCR reaction conditions were performed as described elsewhere.25,26 The PCR products were visualized on a 2% agarose gel stained with ethidium bromide.
Adoptive transfer of lymphoma cells to Rag1−/− mice
Enlarged lymph nodes dissected from tumor mice were gently minced in complete Dulbecco Modified Eagle Medium containing 10% fetal bovine serum (BioWhittaker) and single cell suspensions were obtained. Red cells were lysed by ammonium chloride-potassium (K) lysing buffer (BioWhittaker) at room temperature for 5 minutes, followed by 2 washes of the cells in PBS. Cells were counted and then resuspended in PBS. Cells (1 × 107 or 1 × 105) were intraperitoneally injected per mouse into 6-8-week-old Rag1−/− mice. In some experiments, γδ T cells were purified from the spleens with MACS kits (Miltenyi Biotec) in Id3−/− mice (4 weeks old) and adoptively transferred into Rag1−/− mice (1 × 105 or 4 × 106 cells per recipient).
Results
Id3 null mutation causes the development of γδ T-cell lymphoma in mice
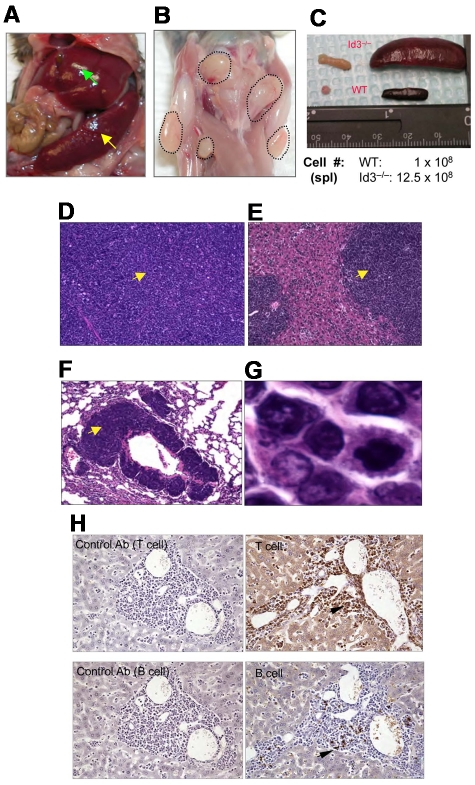
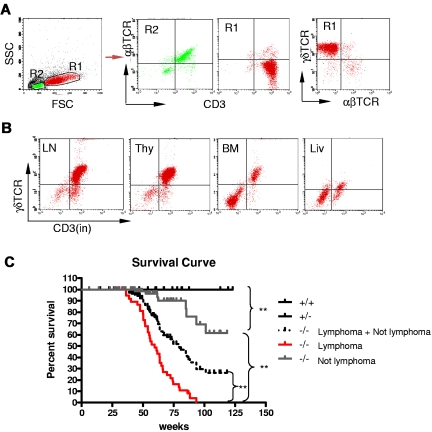
Because Id3−/− mice showed autoimmune-like Sjögren Syndrome at relatively late age,24 we were initially interested in studying the T-cell role in inflammation in the salivary glands in aged knock-out mice. Unexpectedly, we observed that some aged Id3−/− mice (> 6 months old) exhibited massive splenomegaly and hepatomegaly (Figure 1A, and data not shown), and peripheral and mesenteric lymphadenopathy (Figure 1B-C and data not shown). The size of the spleen in Id3−/− mice was almost 10-fold larger than that in the age-matched control mice (C57BL/6), and the total number of the cells was 10- to 20-fold higher than that in control mice (Figure 1C). Histopathology analysis with H&E staining showed that the architecture of the spleen in the Id3−/− mice was effaced by neoplastic cells (Figure 1D). The lymph nodes also showed similar morphology and cellular infiltrate (data not shown). Analysis of other vital organs revealed that liver, lungs, kidneys, and bone marrow showed massive cell infiltrations (Figure 1E-F and data not shown). Neoplastic cells have morphology consistent with lymphoblasts including large hyperbasophilic nuclei and scant basophilic cytoplasm (Figure 1D-G). Immunobiochemistry staining of the tissues revealed that the tumor cells were CD3+, suggesting T-cell lymphoma (Figure 1H). Flow cytometric analysis showed that T lymphomatous cells were CD3 positive on both the cell surface and in the cytoplasm (Figure 2A-B). Unexpectedly, the T lymphomatous cells expressed no αβTCRs on the cell surface (Figure 2A, R1) or intracellularly (data not shown), which was distinct from the remaining very minor population of non-lymphoma lymphocytes that were αβTCR+ (Figure 2A, R2). Further analysis with antibodies against γδTCRs revealed that the lymphoma cells were γδTCR+ (Figure 2A far right). Significantly, the same γδ T-cell lymphoma cells were detected in the lymph nodes, thymus, liver, bone marrow, kidneys, lungs, and blood (Figure 2B and data not shown). Of the Id3−/− mice with lymphoma examined by flow cytometry, most mice had a γδTCR T-cell lymphoma (73.3%, 11/15), and few were negative for γδTCR– (13.3%, 2/15) or expressed B220+CD19+ CD3– (13.3%, 2/15; supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). To study the prevalence of lymphoma, we collected a total of 142 Id3−/− mice of 6 to 18 months of age and found 38 mice developed lymphoma (26.7%), whereas none (0/50 mice) of the age-matched control mice (Id3+/+ or Id3+/−) showed lymphoma (Figure 2C). The median survival time for Id3−/− mice with lymphoma was approximately 60 weeks, which was significantly shorter than that of non-lymphoma Id3−/− mice (> 125 weeks, P < .01) and all Id3−/− mice (approximately 80 weeks, P < .01). None of the control mice died during the period of time monitored (Figure 2C). Thus, mutation of the Id3 gene endowed mice with an increased likelihood of developing γδ T-cell lymphoma.
Figure 1.
Mutation of Id3 results in lymphoma. (A) Splenomegaly (indicated by yellow arrow) and hepatomegaly (indicated by green arrow) in 1 representative Id3−/− mouse. (B) Massive lymphadenopathy in a representative Id3−/− mouse (highlighted with dotted lines). (C) Comparison of spleen and lymph nodes between Id3−/− (> 12 months) and age-matched WT control mice. The numbers indicate the total number of spleen cells. (D-G) H&E staining of the spleen (D), liver (E), or lungs (F) in a representative Id3−/− mouse with lymphoma. Magnification ×10. Arrows indicate neoplastic cell infiltration. (G) High magnification of tumor cells in liver (×10). (H) Immunohistochemical staining of tumor cells with CD3-specific antibody (T cell, top right) and B–cell–specific antibody (B220, B cell, bottom right). Arrows indicate positive cells. Data shown in left column are same tissues stained with isotype control antibodies (rat immunoglobulin G2a for T cell; rabbit immunoglobulin G for B cell) with no positive staining.
Figure 2.
Development γδ T-cell lymphoma in Id3−/− mice. (A-B) Flow cytometric analysis of the lymphomatous cells in a representative Id3−/− mouse (mouse no. 44). The lymphomatous cells are γδTCR+CD3+, but αβTCR− (A, R1); The gated R2 area represents the non-γδ T cells. γδ T-cell lymphoma cells (R1) were seen in the lymph nodes (LN), thymus (thy), bone marrow (BM), and liver (liv) of Id3−/− mice (B). (C) The survival curve of Id3−/− mice. Control mice (Id3+/+ and +/−, n = 50 mice); Id3−/− mice with lymphoma (−/− lymphoma, n = 38 mice); Id3−/− mice without lymphoma (−/− not lymphoma, n = 104 mice). ** P < .01.
Characterization of the γδ T-cell lymphoma
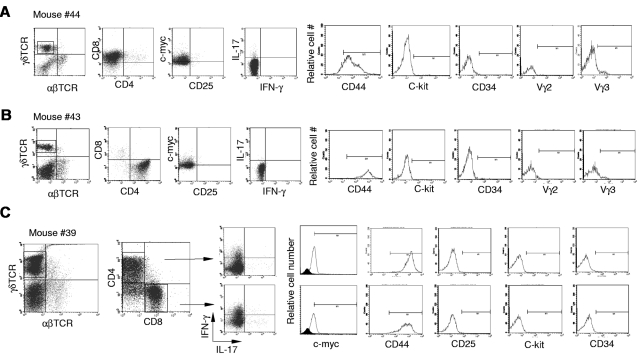
Next, we further characterized the γδ T-cell lymphoma cells in Id3−/− mice. The lymphomatous cells from spleen were considerably larger in size demonstrated by higher forward scatter (FSC) compared with cells derived from spleens of control mice or even the remaining non-lymphomatous T-cell (Figure 2A, R1 versus R2). We observed that the majority of γδ T-cell lymphoma cells were either CD4+CD8– (Figure 3A), CD8+CD4− (Figure 3B), or a mixture of CD4+CD8− and CD4−CD8+ (Figure 3C). This is distinct from normal γδ T cells, which are negative for both CD4 and CD825,27,28 (data not shown). Lymphomatous cells were positive for CD44 but negative for CD25 (Figure 3). The lymphomatous cells did not express stem cell-like marker CD34 or C-kit, suggesting the tumor cells were likely derived from mature γδ T cells (Figure 3). γδ T-cell lymphoma cells were negative for the T regulatory cell specific gene Foxp329,30 (data not shown). Staining of γδ T-cell lymphomatous cells for B-cell (B-220 and CD19), macrophages (f4/80), and dendritic cells (CD11c) was negative (data not shown). We then characterized the functional status of the γδ T-cell lymphoma cells in the Id3−/− mice. We directly examined the cytokine expression ex vivo by flow cytometry and observed that the γδ T-cell lymphomatous cells produced no or small amounts of IFN-γ (Figure 3) and IL-17 (Figure 3). No IL-4, IL-2, or tumor necrosis factor was observed in the γδ T-cell lymphoma cells (data not shown). Some γδ T-cell lymphomatous cells were positive for intracellular granzyme-B and perforin (data not shown). Thus, the data demonstrate that the γδ T-cell lymphoma in Id3−/− mice exhibit a mature T-cell phenotype.
Figure 3.
Characterization of the γδ T-cell lymphomatous cells by flow cytometry. Lymphoma cells were isolated from the spleen in indicated representative mice and stained with indicated antibodies. For intracellular staining of the nuclear molecule c-myc, cells were first stained with antibodies to surface markers followed by intracellular staining with the nuclear buffer from eBioscience. For intracellular cytokine determination, cells were treated with phorbol 12-myristate 13-acetate plus ionomycine for 5 hours before staining. The tumor cells in the spleens are stained with antibodies against αβTCRs and γδTCRs (far left dot plots in no. 44, no. 43, and no. 39), and the following dot plots or histograms in each mouse are from gated γδ T-cell lymphoma cells. (A) Mouse no. 44 (same mouse as in Figure 1D-G), a representative CD4−CD8+ γδ T-cell lymphoma. (B) Mouse no. 43, a representative CD4+CD8− γδ T-cell lymphoma. (C) Mouse no. 39, a mixture of CD4+CD8− and CD4−CD8+ γδ T-cell lymphoma. The isotype control for c-Myc staining is included as filled histogram.
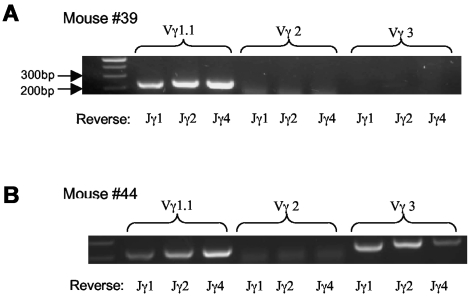
We then examined TCR usage in the γδ T-cell lymphoma. Staining with antibodies against Vγ2 and Vγ3 revealed that the tumor cells were negative for Vγ2 and marginally positive (5%-10%) for Vγ3 (Figure 3). Similar results were obtained with mRNA analysis (Figure 4A-B) As a Vγ1.1 specific antibody was not available to us, we determined Vγ1.1 mRNA expression in the γδ T-cell lymphoma with RT-PCR26 and observed that all tumor cells from mice examined were strongly positive for Vγ1.1 (Figure 4A-B and data not shown).
Figure 4.
Vγ TCR repertoire of lymphoma cells. (A-B) mRNA analysis of Vγ TCR usage with RT-PCR; 2 mice shown are representative of the total of 5 mice examined.
To study the underlying mechanism responsible for the transformation of γδ T cells in the absence of Id3, we examined the mRNA expression of a number of oncogenes including Lyl1 (NM_013556.2), Tal1 (NM_213237.1), Pax5 (NM_008782.2), and P21 (NM_007669.4) in the Id3−/− γδ T-cell lymphoma by real-time RT-PCR analysis. The expression of these oncogenes has been reported to be associated with αβTCR lymphoma; however, we found no significant changes in their expression compared with normal cells (data not shown). We observed that some γδ T-cell lymphomatous cells had increased c-myc mRNA (data not shown) and protein expression (Figure 3 and supplemental Table 1). Taken together, the data indicate that the γδ T-cell lymphoma cells in Id3−/− mice mainly express CD4+ or CD8+ molecules, which is distinct from the normal peripheral γδ T cells that are CD4−CD8−. The malignant γδ T-cell transformation in the absence of Id3 might be associated with the abnormal expression of the oncogene c-myc, although a causative relationship remains to be elucidated.
Transfer of γδ T-cell lymphoma to immunodeficient Rag1−/− mice
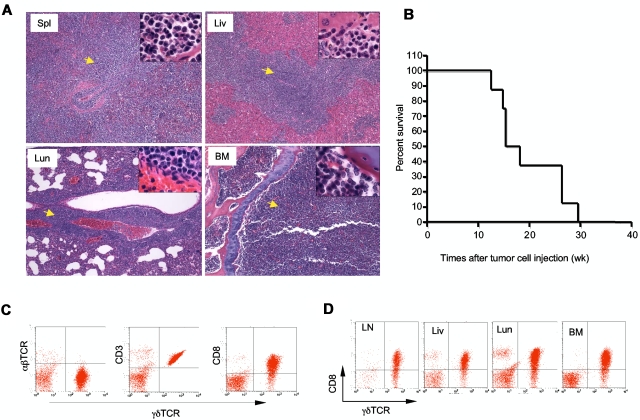
To determine whether the γδ T-cell lymphoma from Id3−/− mice were able to invade a WT host, we injected γδ T-cell lymphomatous cells (1 × 107 per mouse) into the Rag1 deficient (Rag1−/−) mice. Fifteen weeks later, we found all the recipients developed aggressive T-cell lymphoma that invaded all vital organs, including spleen, lymph nodes, liver, bone marrow, and lungs (Figure 5A and data not shown). We then monitored the life-span of the Rag1−/− mice receiving the γδ T-cell lymphoma and observed that the recipients developed an aggressive tumor that caused death of transfer recipients; beginning at approximately 11 to 12 weeks after tumor cell transplantation and causing all mice to die by 30 weeks posttransfer (Figure 5B). The lymphomatous cells isolated from the recipients were identical to that observed to the original Id3−/− mice (Figure 5C-D). Strikingly, even 1 × 105 of γδ T-cell lymphoma cells could result in the development of lymphoma in the Rag1−/− recipients (supplemental Figure 1). In contrast, transfer of non-lymphoma γδ T cells (1 × 105 or 4 × 106 cells per recipient) isolated from the young Id3−/− mice into Rag1−/− mice failed to cause development of lymphoma in the recipients, although some Rag1−/− recipients showed some mild inflammatory infiltration in the liver and lungs (supplemental Figure 1 and data not shown). The data demonstrate the malignant and metastatic characteristics of the γδ T-cell lymphoma cells from Id3−/− mice. These studies also established a more consistent and rapid animal model of γδ T-cell lymphoma, which may be used to test potential therapeutic interventions.
Figure 5.
Adoptive transfer of γδ T-cell lymphoma to Rag1−/− recipients. γδ T-cell lymphomatous cells were isolated from the lymph nodes of Id3−/− mice and injected into Rag1−/− mice (1 × 107 per mouse). (A) Histopathological analysis of the indicated tissues from a representative mouse that received T-cell lymphoma 15 weeks ago (H&E staining, 5×; the inset in each slide was an enlarged area, 100×, n = 4 mice). Spl: spleen; Liv: liver; Lun: lungs; BM: bone marrow. Arrows indicate lymphoma cells. (B) Survival curve for recipient Rag1−/− mice (n = 8 mice) that received γδ T-cell lymphomatous cells from Id3−/− donors. Lymphomatous cells from 2 separate Id3−/− mice were transferred; all 8 Rag1−/− recipient mice died before 30 weeks. The data represent 2 independent experiments. (C-D). Flow cytometric analysis of γδ T-cell lymphoma isolated from the spleen (C) or other tissues (D) of Rag1−/− mice receiving γδ T-cell lymphoma cells 15 weeks ago. LN: lymph nodes; Liv: liver; Lun: lungs; BM: bone marrow. Data shown are dot plots of indicated markers on the lymphoma cells in a representative mouse of a total of 4 mice.
Discussion
Here we have shown that deletion of the inhibitory HLH protein Id3 results in the development of an aggressive γδ T-cell lymphoma, which exhibited similar clinical symptoms to the HSTCL in humans. The massive splenomegaly and hepatomegaly with widespread organ invasion, especially of the bone marrow, resembles the main clinical and pathological features of patients with HSTCL. Of particular interest are the observations that Id3−/− mice develop spontaneous autoimmune-like diseases such as Sjogren Syndrome.23,24 As HSTCL is often accompanied by autoimmune conditions, such as autoimmune hemolytic anemia,6 the γδ T-cell lymphoma seen in Id3−/− mice shares further characteristics with that seen in HSTCL patients. In addition, a few lymphoma with αβ TCRs or B-cell receptors were also observed in the Id3−/− mice, which appeared also the case in human HSTCL.1 Although the HSTCL is a rare subtype of peripheral T-cell lymphoma in humans, the γδ T-cell lymphoma is an aggressive lymphoma with poor prognosis. The etiology and genes that control and regulate the development of HSTCL in human are unknown. The development of γδ T-cell lymphoma after inactivation of the Id3 gene in mice has clearly raised an intriguing possibility to explore whether this gene is also linked and associated with HSTCL in humans.
Examination of the lymphoma in Id3−/− mice showed the malignant cells were a γδ T-cell lymphoma with predominant CD8+CD4− or CD4+CD8− phenotype, which is in marked contrast to the phenotype of normal γδ T cells (CD4−CD8−). All the γδ T-cell lymphoma cells expressed surface and cytoplasmic CD3, suggesting the lymphoma had a mature lymphocyte phenotype rather than immature thymocytes. Intriguingly, some γδ T-cell lymphoma cells expressed a mixed phenotype with CD4+CD8− and CD4−CD8+. The underlying mechanisms responsible for this mixed phenotype remain unknown. We speculate that these lymphoma cells lack monoclonality, or alternatively, the tumor cells acquired either CD4 or CD8 during the tumorigenesis. Moreover, we noted that in the spleens of mice with γδ T-cell lymphoma there were populations of cells located in the area of tumor cells (gated on the fluorescence-activated cell sorting analysis) showing negative for γδ TCR−, αβTCR− and background levels of B220/CD19. Because this population of cells only appears in the spleen, not in lymph nodes or the thymus, it remains unknown whether these cells represent other types of tumor cells or some other non-tumor cells in the spleen. This γδ T-cell lymphoma is different from the premature T-cell lymphoma caused by the mutation of the basic HLH protein E2A,15,16 which belongs to the αβ T-cell lineage and causes a CD4−CD8– double negative preTCR lymphoma. In addition, the γδ T-cell lymphoma is negative for CD34 and C-kit stem cell-like marker, further indicating their mature feature. Despite the difference in phenotype between the malignant entities seen after mutation of E2A and loss of Id3, these data highlight a previously unexplored role for HLH proteins in the transformation and/or expansion of malignant T cells.
In addition to the dominant phenotype of γδ T-cell lymphoma, there are also few lymphoma that express negative for γδTCRs. Specifically, some lymphoma cells express markers for B cells including B220+CD19+ and are negative for T-cell markers such as CD3 and αβTCRs. Although these lymphoma cells represent as B–cell–like lymphoma, it remains to be known whether they are truly B cell lymphoma or if they acquire B cell markers such as B220 and down-regulate TCR. Morphological analysis of the tumor cells between the B cell–like lymphomas and γδ T-cell lymphoma revealed no significant difference. In addition, very few lymphoma cells expressed no γδTCRs (gd TCR−), but were CD3+, suggesting a T-cell lineage. Whether this type of lymphoma belongs to αβ T-cell lymphoma remains unknown. Nevertheless, the data raise an exciting question of a possible association between γδ T-cell lymphoma and B-cell lymphoma or even αβ T-cell lymphoma, especially in the context of autoimmune diseases such as Sjogren Syndrome, which merits further investigation.
The underlying mechanisms that are responsible for the development and/or transformation to malignant γδ T tumor cells in the absence of Id3 remain unknown. The increased TCR γ gene rearrangement in the Id3−/− mice22 may provide some clue to further delineate the impact for the malignant transformation. Id3−/−E2Aflox/floxLCKCretg mice do not have γδ T-cell production ability, suggesting that E2A somehow controlls γδ T-cell generation and Vγ rearrangement in Id3−/− mice.22 The lack of increase in some lymphoma associated genes, including Lyl1, Tal1, P21, and Pax5, suggests these genes are not involved in the γδ T-cell lymphoma seen in the Id3−/− mice. The up-regulation of the oncogene c-myc in some γδ T-cell lymphoma suggests an association between c-myc dysregulation and the development of γδ T-cell lymphoma in Id3−/− mice. However, it is unclear at present why only a subset of lymphoma increases c-myc and how the inactivation of Id3 leads to activation and increase in c-myc expression in γδ T cells. Alternatively, the development of γδ T-cell lymphoma in the aged Id3−/− mice also raises an important question whether it is associated with or even caused by chronic inflammation in these Id3 knockout mice. It has been gradually recognized that chronic inflammation responses play an important role at different stages of tumor development, including initiation, promotion, and metastasis. Thus, the possibility that γδ T-cell lymphoma is caused by chronic inflammation cannot be excluded in the Id3−/− mice and merits careful studies in the future.
The fact that γδ T-cell lymphomatous cells develop a rapid and aggressive lymphoma in the Rag1−/− recipients provides a model to further study the pathogenesis of γδ T-cell lymphoma. Intriguingly, transfer of splenic γδ T cells from non-lymphomatous Id3−/− mice into Rag1−/− mice failed to develop γδ T-cell lymphoma in the recipients for 12 weeks after transplantation (supplemental Figure 1). These data suggest that the γδ T-cell lymphoma might develop in the early stage of γδ T-cell development in the thymus. However, the possibility that low numbers of tumor cells in the spleen in Id3−/− mice without lymphoma fail to form lymphoma in the recipients cannot be completely excluded and remains to be investigated. More importantly, in light of the poor response to the current therapeutic regimens of human HSTCL, this model of transferred γδ T-cell lymphoma will facilitate the exploration and development of a range of therapeutic interventions that could ultimately be beneficial for patients with HSTCL.
Supplementary Material
Acknowledgments
We thank Dr Y. Zhuang (Duke University) for the gift of Id3−/− mice.
This research was supported by the Intramural Research Program of the National Institutes of Health, National Institute of Dental and Craniofacial Research.
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: J.L. and T.M. designed and performed experiments, analyzed data, and contributed to the writing of the manuscript; P.Z., J.E.K., and B.Z. performed experiments and contributed to the writing of the manuscript; V.H. performed diagnostic analysis for the tumor; and W.J.C. initiated and directed the whole study, designed experiments, analyzed data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: WanJun Chen, MIU, NIDCR, Bldg 30, Rm 304, 30 Convent Dr, Bethesda, MD 20892; e-mail: wchen@mail.nih.gov.
References
- 1.Vega F, Medeiros LJ, Gaulard P. Hepatosplenic and other gammadelta T-cell lymphomas. Am J Clin Pathol. 2007;127(6):869–880. doi: 10.1309/LRKX8CE7GVPCR1FT. [DOI] [PubMed] [Google Scholar]
- 2.Grabher C, von Boehmer H, Look AT. Notch 1 activation in the molecular pathogenesis of T-cell acute lymphoblastic leukaemia. Nat Rev Cancer. 2006;6(5):347–359. doi: 10.1038/nrc1880. [DOI] [PubMed] [Google Scholar]
- 3.Li X, Gounari F, Protopopov A, Khazaie K, von Boehmer H. Oncogenesis of T-ALL and nonmalignant consequences of overexpressing intracellular NOTCH1. J Exp Med. 2008;205(12):2851–2861. doi: 10.1084/jem.20081561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Belhadj K, Reyes F, Farcet JP, et al. Hepatosplenic gammadelta T-cell lymphoma is a rare clinicopathologic entity with poor outcome: report on a series of 21 patients. Blood. 2003;102(13):4261–4269. doi: 10.1182/blood-2003-05-1675. [DOI] [PubMed] [Google Scholar]
- 5.Kao GF, Resh B, McMahon C, et al. Fatal subcutaneous panniculitis-like T-cell lymphoma gamma/delta subtype (cutaneous gamma/delta T-cell lymphoma): report of a case and review of the literature. Am J Dermatopathol. 2008;30(6):593–599. doi: 10.1097/DAD.0b013e318182c7bf. [DOI] [PubMed] [Google Scholar]
- 6.Minauchi K, Nishio M, Itoh T, et al. Hepatosplenic alpha/beta T cell lymphoma presenting with cold agglutinin disease. Ann Hematol. 2007;86(2):155–157. doi: 10.1007/s00277-006-0212-3. [DOI] [PubMed] [Google Scholar]
- 7.Saenz AD, Amador A, Ruiz BM, Davis J, Ruiz P. Cytofluorographic and molecular identification of a CD8-positive, TCR-alpha/beta-negative intraocular T cell lymphoma: a case report and review of the literature. J Med Case Reports. 2007;1:114. doi: 10.1186/1752-1947-1-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sallah S, Smith SV, Lony LC, Woodard P, Schmitz JL, Folds JD. Gamma/delta T-cell hepatosplenic lymphoma: review of the literature, diagnosis by flow cytometry and concomitant autoimmune hemolytic anemia. Ann Hematol. 1997;74(3):139–142. doi: 10.1007/s002770050272. [DOI] [PubMed] [Google Scholar]
- 9.Taguchi A, Miyazaki M, Sakuragi S, Shinohara K, Kamei T, Inoue Y. Gamma/delta T cell lymphoma. Intern Med. 2004;43(2):120–125. doi: 10.2169/internalmedicine.43.120. [DOI] [PubMed] [Google Scholar]
- 10.Willemze R, Meijer CJ. Classification of cutaneous T-cell lymphoma: from Alibert to WHO-EORTC. J Cutan Pathol. 2006;33(suppl 1):18–26. doi: 10.1111/j.0303-6987.2006.00494.x. [DOI] [PubMed] [Google Scholar]
- 11.Kee BL. E and ID proteins branch out. Nat Rev Immunol. 2009;9(3):175–184. doi: 10.1038/nri2507. [DOI] [PubMed] [Google Scholar]
- 12.Murre C. Intertwining proteins in thymocyte development and cancer. Nat Immunol. 2000;1(2):97–98. doi: 10.1038/77881. [DOI] [PubMed] [Google Scholar]
- 13.Murre C. Helix-loop-helix proteins and lymphocyte development. Nat Immunol. 2005;6(11):1079–1086. doi: 10.1038/ni1260. [DOI] [PubMed] [Google Scholar]
- 14.Bacchetta R, Sartirana C, Levings MK, Bordignon C, Narula S, Roncarolo MG. Growth and expansion of human T regulatory type 1 cells are independent from TCR activation but require exogenous cytokines. Eur J Immunol. 2002;32(8):2237–2245. doi: 10.1002/1521-4141(200208)32:8<2237::AID-IMMU2237>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 15.Bain G, Engel I, Robanus Maandag EC, et al. E2A deficiency leads to abnormalities in alphabeta T-cell development and to rapid development of T-cell lymphomas. Mol Cell Biol. 1997;17(8):4782–4791. doi: 10.1128/mcb.17.8.4782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yan W, Young AZ, Soares VC, Kelley R, Benezra R, Zhuang Y. High incidence of T-cell tumors in E2A-null mice and E2A/Id1 double-knockout mice. Mol Cell Biol. 1997;17(12):7317–7327. doi: 10.1128/mcb.17.12.7317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Benezra R, Davis RL, Lassar A, et al. Id: a negative regulator of helix-loop-helix DNA binding proteins. Control of terminal myogenic differentiation. Ann N Y Acad Sci. 1990;599:1–11. doi: 10.1111/j.1749-6632.1990.tb42359.x. [DOI] [PubMed] [Google Scholar]
- 18.Christy BA, Sanders LK, Lau LF, Copeland NG, Jenkins NA, Nathans D. An Id-related helix-loop-helix protein encoded by a growth factor-inducible gene. Proc Natl Acad Sci U S A. 1991;88(5):1815–1819. doi: 10.1073/pnas.88.5.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Riechmann V, van Cruchten I, Sablitzky F. The expression pattern of Id4, a novel dominant negative helix-loop-helix protein, is distinct from Id1, Id2 and Id3. Nucleic Acids Res. 1994;22(5):749–755. doi: 10.1093/nar/22.5.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rivera RR, Johns CP, Quan J, Johnson RS, Murre C. Thymocyte selection is regulated by the helix-loop-helix inhibitor protein, Id3. Immunity. 2000;12(1):17–26. doi: 10.1016/s1074-7613(00)80155-7. [DOI] [PubMed] [Google Scholar]
- 21.Sun XH, Copeland NG, Jenkins NA, Baltimore D. Id proteins Id1 and Id2 selectively inhibit DNA binding by one class of helix-loop-helix proteins. Mol Cell Biol. 1991;11(11):5603–5611. doi: 10.1128/mcb.11.11.5603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ueda-Hayakawa I, Mahlios J, Zhuang Y. Id3 restricts the developmental potential of gamma delta lineage during thymopoiesis. J Immunol. 1. 2009;182(9):5306–5316. doi: 10.4049/jimmunol.0804249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hayakawa I, Tedder TF, Zhuang Y. B-lymphocyte depletion ameliorates Sjogren's syndrome in Id3 knockout mice. Immunology. 2007;122(1):73–79. doi: 10.1111/j.1365-2567.2007.02614.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li H, Dai M, Zhuang Y. A T cell intrinsic role of Id3 in a mouse model for primary Sjogren's syndrome. Immunity. 2004;21(4):551–560. doi: 10.1016/j.immuni.2004.08.013. [DOI] [PubMed] [Google Scholar]
- 25.Raulet DH. The structure, function, and molecular genetics of the gamma/delta T cell receptor. Annu Rev Immunol. 1989;7:175–207. doi: 10.1146/annurev.iy.07.040189.001135. [DOI] [PubMed] [Google Scholar]
- 26.Sandor M, Sperling AI, Cook GA, Weinstock JV, Lynch RG, Bluestone JA. Two waves of gamma delta T cells expressing different V delta genes are recruited into schistosome-induced liver granulomas. J Immunol. 1995;155(1):275–284. [PubMed] [Google Scholar]
- 27.Chien YH, Jores R, Crowley MP. Recognition by gamma/delta T cells. Annu Rev Immunol. 1996;14:511–532. doi: 10.1146/annurev.immunol.14.1.511. [DOI] [PubMed] [Google Scholar]
- 28.Haas W, Pereira P, Tonegawa S. Gamma/delta cells. Annu Rev Immunol. 1993;11:637–685. doi: 10.1146/annurev.iy.11.040193.003225. [DOI] [PubMed] [Google Scholar]
- 29.Chen W, Jin W, Hardegen N, et al. Conversion of peripheral CD4+CD25- naive T cells to CD4+CD25+ regulatory T cells by TGF-beta induction of transcription factor Foxp3. J Exp Med. 2003;198(12):1875–1886. doi: 10.1084/jem.20030152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fontenot JD, Rasmussen JP, Williams LM, Dooley JL, Farr AG, Rudensky AY. Regulatory T-cell lineage specification by the forkhead transcription factor foxp3. Immunity. 2005;22(3):329–341. doi: 10.1016/j.immuni.2005.01.016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.