Abstract
The LMAN1-MCFD2 (lectin, mannose binding 1/multiple coagulation factor deficiency protein 2) cargo receptor complex transports coagulation factors V (FV) and VIII (FVIII) from the endoplasmic reticulum (ER) to the ER-Golgi intermediate compartment (ERGIC). LMAN1 (ERGIC-53) is a hexameric transmembrane protein with a carbohydrate recognition domain (CRD) on the ER luminal side. Here, we show that mutations in the first beta sheet of the CRD abolish MCFD2 binding without affecting the mannose binding, suggesting that LMAN1 interacts with MCFD2 through its N-terminal beta sheet, consistent with recently reported crystal structures of the CRD-MCFD2 complex. Mutations in the Ca2+- and sugar-binding sites of the CRD disrupt FV and FVIII interactions, without affecting MCFD2 binding. This interaction is independent of MCFD2, as LMAN1 mutants defective in MCFD2 binding can still interact with FVIII. Thus, the CRD of LMAN1 contains distinct, separable binding sites for both its partner protein (MCFD2) and the cargo proteins (FV/FVIII). Monomeric LMAN1 mutants are defective in ER exit and unable to interact with MCFD2, suggesting that the oligomerization of LMAN1 is necessary for its cargo receptor function. These results point to a central role of LMAN1 in regulating the binding in the ER and the subsequent release in the ERGIC of FV and FVIII.
Introduction
Combined deficiency of factor V and factor VIII (F5F8D) is an autosomal recessive disorder1,2 caused by mutations in LMAN1 (lectin, mannose binding 1) or MCFD2 (multiple coagulation factor deficiency protein 2).3–5 LMAN1 is a mannose-selective lectin recycling from the endoplasmic reticulum (ER) to the ER-Golgi intermediate compartment (ERGIC), a vesicular tubular structure unique in higher eukaryotes. MCFD2 is a small, soluble protein with 2 EF hand domains at its C-terminus.4 LMAN1 forms a Ca2+-dependent complex with MCFD2 with a 1:1 stoichiometry.6 Both FV and FVIII interact with the LMAN1-MCFD2 complex.6,7 On the basis of these findings, the LMAN1-MCFD2 complex is proposed as a cargo receptor that ferries FV and FVIII from the ER to the Golgi. LMAN1 also potentially functions as a cargo receptor for cathepsin C, cathepsin Z, and α1-antitrypsin.8–10 Cargo receptors are transmembrane proteins that bind specific secretory proteins in the ER lumen. They also contain ER exit signals on the cytoplasmic side that are recognized by the SEC24 component of coat protein complex II (COPII).11–13 To date, most cargo receptors are identified in yeast.14 The LMAN1-MCFD2 complex is the only well-characterized cargo receptor in mammalian cells. Therefore, understanding the mechanism of the LMAN1-MCFD2–mediated transport will not only provide new insights into FV and FVIII biosynthesis and possible therapies for hemophilia A, but also help us to understand the general mechanisms of the receptor-mediated transport pathway.
The requirement of both LMAN1 and MCFD2 is a unique feature for this cargo receptor. The exact function of LMAN1 in the secretion of FV and FVIII remains to be determined. LMAN1 is a 53-kDa type I transmembrane protein that has all the characteristics of a cargo receptor.15 The cytosolic tail of LMAN1 contains a diphenylalanine ER exit motif, which interacts with the COPII, and a dilysine ER retrieval motif that mediates the recycling of the protein.15 The ER luminal part of the protein contains a carbohydrate recognition domain (CRD) and a series of 4 α-helixes that are predicted to form coiled-coil domains.16 The CRD exhibits mannose-specific carbohydrate binding activity and likely binds the highly glycosylated FV and FVIII. In addition, the CRD was also shown to bind MCFD2 in vitro.17 MCFD2 is dispensable for the secretion of cathepsins C and Z, suggesting that it may be specifically required for the transport of FV and FVIII.15 MCFD2 can interact with FV and FVIII independent of LMAN1 and the EF hand domains of MCFD2 are sufficient for binding both LMAN1 and FV/FVIII.6,7 During the preparation of this manuscript, crystal structures of the CRD-MCFD2 complex were published,18,19 which identify the binding surface consisting of the EF hand domain of MCFD2 and the N-terminal side of the CRD. Most MCFD2 missense mutations either disrupt the protein structure or change a crucial amino acid residue involved in binding the CRD.18,19
LMAN1 was first reported to exist as both homodimers and homohexamers in cells, maintained by 2 membrane proximal cysteines (C466 and C475).20,21 By using nondenaturing analysis techniques together with cross-linking studies, LMAN1 was later demonstrated to exclusively exist as homohexamers in cells.22 LMAN1 hexamers include 2 forms: one as a disulfide-linked and sodium dodecyl sulfate (SDS)–resistant complex and the other as a noncovalent, SDS-sensitive complex. The helical domain appears to be important for the formation of the latter type of complex.22 Whether oligomerization of LMAN1 is required for its ER exit is still controversial. Endo H resistance analysis demonstrated that the C466A/C475A mutant significantly decreased the ER exit rate.21 In contrast, immunofluorescence and cell fractionation analysis showed that the 2 cysteines are not essential for the intracellular distribution of LMAN1.22 Furthermore, although appearing to be a monomer on nonreducing SDS–polyacrylamide gel electrophoresis (PAGE), the C466A/C475A mutant actually forms a noncovalent hexamer when analyzed under native conditions.22 Therefore, the importance of oligomerization for the cargo receptor function of LMAN1 needs further investigation.
In this study, we performed systematic mutagenesis analysis of LMAN1 and tested the effects of mutations on the interactions with both MCFD2 and FV/FVIII in cells. We found that the CRD of LMAN1 contains separate binding sites for MCFD2 and FV/FVIII, and that the oligomerization of LMAN1 is required for MCFD2 binding and the ER exit, but dispensable for FV/FVIII binding.
Methods
Plasmid constructs
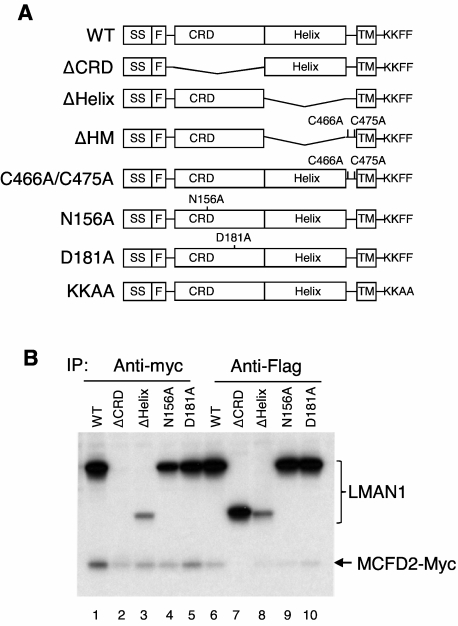
LMAN1 mutant constructs are shown schematically in Figures 1A and 2A. The pED-Flag-LMAN1 plasmid was derived from pED-LMAN123 by replacing the LMAN1 signal sequence with that of calreticulin24 and introducing a Flag tag immediately after the signal sequence. Other mutant constructs, including ΔCRD (R44-E269), ΔHelix (G271-N457), Y48A, K53A, W67S, N156A, D181A, as well as the Y48A/K53A and C466A/C475A double mutants were made by introducing the respective mutations into the pED-Flag-LMAN1 plasmid using the Stratagene mutagenesis II XL kit. The KKAA mutation and the LMAN1 β sheet deletion mutations, including Δβ1 (H43-Q59), Δβ2 (H43-N72), Δβ3 (H43-S76), and Δβ4 (H43-A83), were introduced into the pED-Flag-LMAN1 plasmid by polymerase chain reaction (PCR)-based methods. The ΔHM mutant was made by introducing 2 point mutations (C466A and C475A) into the ΔHelix mutant plasmid. All constructs were confirmed by DNA sequencing. The effects of these mutants are summarized in supplemental Table 1 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Figure 1.
The CRD domain of LMAN1 is responsible for MCFD2 binding in vivo. (A) Diagram of LMAN1 mutants used in the experiments. SS, signal peptide; F, Flag; TM, transmembrane. KKFF represents the last 4 amino acids of wild-type LMAN1 important for ER exit and retrieval, while KKAA represents mutations of the last 2 amino acids. (B) Co-IP of LMAN1 mutants with MCFD2. COS1 cells were cotransfected with Flag-tagged LMAN1 mutants and myc-tagged wild-type MCFD2. Cell lysates were immunoprecipitated with anti-myc for MCFD2 and anti-Flag for LMAN1.
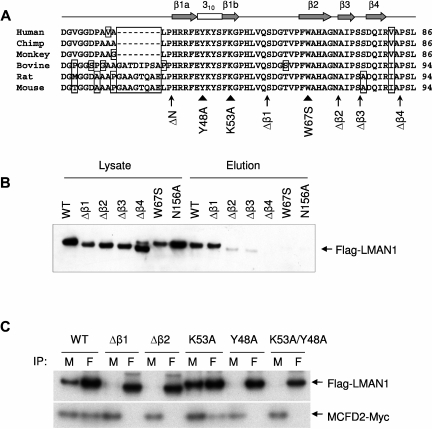
Figure 2.
The first β-sheet of the CRD is the binding motif for MCFD2. (A) Alignment of the first 56 amino acids of the mature human LMAN1 with the orthologs from chimp, monkey, bovine, rat, and mouse. Amino acid residues that differ from the consensus are boxed. The secondary structures are indicated on top of the sequences. The locations of deletions (arrows) and point mutations (arrow heads) used in the study are denoted under the sequences. (B) Mannose binding activities of different LMAN1 mutants. COS1 cells were transfected with the wild-type and the indicated LMAN1 mutants. LMAN1 proteins that are in the cell lysate and that are eluted from the mannose agarose beads are detected by Western blot analysis. Lysate lanes represent 20% of the input added to the mannose beads. (C) Co-IP of LMAN1 mutants with MCFD2. COS1 cells were cotransfected with Flag-tagged LMAN1 mutants and myc-tagged wild-type MCFD2. Cell lysates were immunoprecipitated with anti-myc for MCFD2 and anti-Flag for LMAN1.
Antibodies
We purchased monoclonal anti-myc from Santa Cruz Biotechnology, monoclonal anti-Flag from Sigma-Aldrich, and monoclonal antihuman FV from Hematologic Technologies. Monoclonal anti-LMAN1 was a gift from H.-P. Hauri (University of Basel). Monoclonal anti-human FVIII was a gift from D. Pittman (Pfizer). Monoclonal anti-MCFD2 was reported previously.4 Rabbit anti-SEC22B was a gift from J. C. Hay (University of Montana). Rabbit anti-ribophorin I was a gift from T. Rapoport (Harvard University). Rabbit anti–translocon-associated protein-alpha (TRAP-α) was a gift from R. Hegde (National Institute of Child Health and Human Development, National Institutes of Health).
Cell culture, transfection, and metabolic labeling
Cells were grown in Dulbecco modified Eagle medium (DMEM), supplemented with 10% fetal bovine serum (FBS), 100 IU/mL penicillin, and 100 IU/mL streptomycin, and transfected using FuGENE 6 (Roche). Twenty hours after transfection, cells were metabolically labeled with [35S]-methionine/cysteine (250 μCi/mL in methionine/cysteine-free DMEM) (MP Biomedicals) for 45 minutes, followed by a 30-minute incubation in complete medium.
Cross-linking and immunoprecipitation
Cross-linking using DSP [dithio-bis(succinimidyl propionate)] (Thermo Scientific) and immunoprecipitation was described previously.6 Each experiment was performed at least twice.
Immunofluorescence
Indirect immunofluorescence staining of HeLa cells transfected with different LMAN1 expression vectors was essentially done as previously described.4 Images were viewed on a Leica DMRXE confocal microscope using a 63× oil-immersion objective with a 1.40 numeric aperture. Confocal images were acquired using the Leica Confocal Software Version 2.61.
BiFC assay
Venus (a mutant of EYFP)-based BiFC cloning vectors pFlag-VN173N (for N-terminal fusion with amino acids 1-172 of Venus), pHA-VC155N (for N-terminal fusion with amino acids 155-238 of Venus), and pHA-VC155C (for C-terminal fusion with amino acids 155-238 of Venus) were gifts from Chang-Deng Hu (Purdue University).25,26 These vectors were modified to express ER-targeted fusion proteins by inserting the signal sequence of calreticulin after the initiation codon. Bimolecular fluorescence complementation (BiFC) constructs are schematically shown in supplemental Figure 4. Wild-type and LMAN1 mutants, including Δβ1, Y48A, K53A, N156A, D181A, and ΔHM, were cloned into the modified pFlag-VN173N to express Flag-tagged LMAN1 fusion proteins. In addition, wild-type LMAN1 and the ΔHM mutant were also cloned into the modified pHA-VC155N to express hemagglutinin (HA)–tagged LMAN1 fusion proteins. MCFD2 was cloned into the modified pHA-VC155C, expressing a C-terminally HA-tagged fusion protein.
HeLa cells were grown in 6-well plates and cotransfected with 0.25 μg of each plasmid. Ten hours after transfection, cells were washed once using phosphate-buffered saline (PBS) and observed under a LEICA DMI3000 inverted fluorescence microscope. Images were taken by the attached camera, and intensities of at least 50 fluorescent cells from each transfection were measured using ImageJ software Version 1.44f (National Institutes of Health). Cells were immediately lysed after pictures were taken and analyzed by immunoblotting.
Mannose binding assay
This assay was performed as previously described,27 with some modifications. Briefly, COS1 cells were harvested on ice in homogenate buffer (10mM Tris [tris(hydroxymethyl)aminomethane]-HCl, pH 7.4, 150mM NaCl, lmM CaCl2, and 1mM MgCl2) 48 hours after transfection with LMAN1 expression constructs. Cells were passed through a ball-bearing homogenizer at 18-μm clearance 20 times and cleared by centrifugation at 500g for 10 minutes. Membrane fraction from the postnuclear supernatant was pelleted at 100 000g for 1 hour. The pellet was solubilized for 1 hour in lysis buffer (10mM Tris-HCl, pH 7.4, 150mM NaCl, 10mM CaCl2, 1mM MgCl2 containing 1% Triton X-100, and protease inhibitors), followed by a centrifugation at 100 000g for 1 hour and dialysis of the supernatant overnight against binding buffer (10mM Tris-HCl, pH 7.4, 150mM NaCl, 10mM CaCl2, 1 mM MgCl2, and 0.15% Triton X-100). The dialysate was incubated with D-mannose agarose beads (Sigma-Aldrich) overnight, and bound LMAN1 was eluted using 0.2M mannose in binding buffer. Eluted LMAN1 was immunoprecipitated using a monoclonal anti-Flag antibody and detected by Western blot analysis using a rabbit anti-Flag antibody.
In vitro vesicle formation assay
The in vitro vesicle formation assay protocol was adapted from the Schekman Lab.28,29 COS1 cells were transfected with various LMAN1 expression constructs. Twenty-eight hours later, cells were treated with 0.1% digitonin to generate semipermeabilized (SP) cells. Rat liver cytosol was prepared as described previously.28 Vesicle-budding reactions were performed in KHM buffer (110mM KOAc, 20mM HEPES [N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid] at pH 7.2, and 2mM Mg(OAC)2) with 0.1mM guanine nucleotides, an adenosine triphosphate (ATP) regeneration system, SP cells, and cytosol (4 mg/mL) as a resource of COPII proteins. After 1 hour of incubation at 30°C, the reactions were placed on ice for 3 minutes, and the transport vesicle fractions were separated from donor membranes by centrifugation at 16 000g for 20 minutes at 4°C. Vesicles were collected by centrifugation at 55 000 rpm for 25 minutes at 4°C in a Beckman TLA100 rotor. The membrane fraction and the vesicle fraction were solubilized with 60 and 16 μL of buffer C (10mM Tris-HCl at pH 7.6, 100mM NaCl, 1% Triton X-100, and protease inhibitor), respectively. The resulting samples were mixed with the lithium dodecyl sulfate (LDS) sample buffer (Invitrogen) and heated at 42°C for 15 minutes. Twenty percent of the total membrane fraction (ie, input) and 100% of the vesicle fraction were separated by SDS-PAGE, transferred to the nitrocellulose membrane, and analyzed by immunoblotting.
Results
The CRD domain of LMAN1 is responsible for MCFD2 binding in vivo
To define the structural features in LMAN1 required for MCFD2 binding in vivo, we engineered a series of LMAN1 mutant constructs (Figure 1A), including deletion of the CRD (ΔCRD), deletion of the helical region (ΔHelix) of the stalk domain, and 2 missense mutations. The N156A mutation changes a critical amino acid in the carbohydrate binding pocket and has been reported to abolish mannose binding.27 The D181A mutation changes a critical residue involved in chelating both calcium cations in the Ca2+ binding site of LMAN1 (supplemental Figure 1B).30 This mutation is expected to result in localized conformational changes that also disrupt mannose binding. We confirmed that the D181A mutant is defective in mannose binding (data no shown). As shown in Figure 1B, the ΔHelix mutant can still coimmunoprecipitate with MCFD2 (lanes 3 and 8), whereas the ΔCRD mutation abolishes the coimmunoprecipitation (co-IP) with MCFD2 (lanes 2 and 7). These results indicate that the CRD of LMAN1 is responsible for MCFD2 binding in vivo. This is in agreement with the in vitro observation that purified CRD can form a stable complex with MCFD2.16,18,19, In addition, both N156A and D181A mutants can still bind MCFD2 efficiently, suggesting that the binding of mannose or Ca2+ is not a prerequisite for LMAN1 to interact with MCFD2.
MCFD2 interacts with the N-terminus of LMAN1
The CRD structure is primarily composed of 15 β-strands, along with a small α-helix and one turn of 310 helix.31 To further narrow down the MCFD2-binding motif in LMAN1, we generated sequential deletions (ΔN, Δβ1, Δβ2, Δβ3, and Δβ4) from the N-terminus of CRD (Figure 2A). The sequence preceding the first β-sheet is not well conserved and is shorter in primates (Figure 2A). Deletion of this sequence (ΔN) has no effect on the co-IP of LMAN1 with MCFD2 (data not shown). We used mannose binding assay to distinguish mutations that exhibit localized effect and mutations that disrupt the correct folding of the protein, because the mannose binding site is located at the opposite side of the N-terminal β-sheets.30,31 In this assay, proteins in the cell lysate that can bind mannose are retained in, and subsequently eluted from, a mannose column.27 The mannose binding–deficient N156A mutant served as a negative control. As shown in Figure 2B, only the Δβ1 mutant maintained mannose binding activity comparable with the wild-type protein (Figure 2B). Further deletions (Δβ2, Δβ3, and Δβ4) markedly decreased the mannose binding, suggesting that these deletion mutations interfere with the correct folding of LMAN1 in the ER. A LMAN1 missense mutation (W67S) recently identified in a F5F8D patient32 was located in the second β-sheet. This mutation also abolished the mannose binding, consistent with the previous report.32
Next, we used a co-IP assay to test the LMAN1-MCFD2 interaction. The Δβ1 and Δβ2 mutants were cotransfected into COS1 cells with myc-tagged MCFD2, metabolically labeled, and immunoprecipitated with anti-myc and -Flag antibodies. Interestingly, both the Δβ1 and the Δβ2 mutants failed to coimmunoprecipitate with MCFD2 (Figure 2C). Unlike the Δβ2 mutant, the Δβ1 mutant retains the mannose binding ability, which suggests that it does not cause global conformational changes in the CRD (Figure 2B). Therefore, these results imply that β1 is directly involved in MCFD2 binding, consistent with the crystal structures of the CRD-MCFD2 complex.18,19 To identify critical amino acid residues in the β1 region that are involved in MCFD2 binding, we developed a model of the LMAN1-MCFD2 complex using molecular docking methods (detailed methods and results in supplemental Document 1) from the nuclear magnetic resonance structure of MCFD2 (PDB code: 2VRG)33 and a homology model of the human CRD (supplemental Figure 1A), based on the crystal structure of the rat CRD (PDB code: 1R1Z).30,31 In this model, the N-terminus of the CRD (primarily β1) directly contacts MCFD2 (supplemental Figure 2). It also predicts potential LMAN1-MCFD2 contacting residues (supplemental Table 2). We chose to mutate 2 potential interaction residues on the first β-sheet (Y48A and K53A) individually and in combination. As shown in Figure 2C, the K53A mutation had no effect on the interaction of LMAN1 with MCFD2. The Y48A mutation disrupted MCFD2 binding (Figure 2C), so did the Y48A/K53A double mutations. These results suggest that Y48 is a key amino acid involved in the interaction with MCFD2 and further support our conclusion that the first β-sheet of the CRD is the binding site for MCFD2.
Oligomerization of LMAN1 is required for MCFD2 binding in vivo
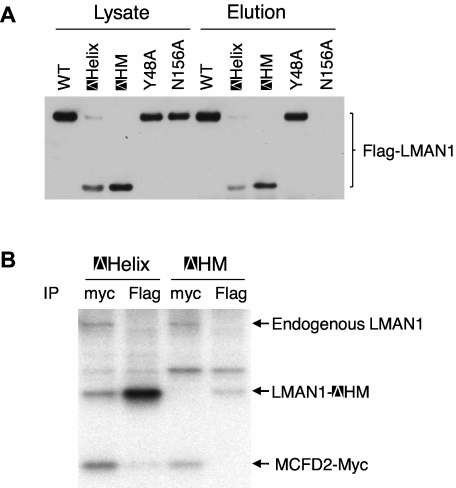
It was reported that LMAN1 exists exclusively as homohexamers in cells.22 Two membrane-proximal cysteine residues (C466 and C475) mediate the disulfide-linked hexamer formation, and the helical region likely mediates the noncovalent hexamer formation.22 To investigate if oligomerization of LMAN1 is necessary for MCFD2 binding, we engineered a mutant LMAN1 with both deletion of the helical region and mutations of the 2 cysteines (C466A and C475A), which we termed ΔHM. Deletion of both elements required for the oligomerization is expected to result in a monomeric protein. We first assessed the oligomerization state of this mutant by nonreducing SDS-PAGE, which detects the wild-type LMAN1 as both dimers and hexamers.20,21 As shown in supplemental Figure 3, a majority of the ΔHM mutant exists as a monomer, as well as the C466A/C475A mutant, as expected.20,21 Although the C466A/C475A mutant runs as a monomer on SDS-PAGE, it still forms a hexamer in cells.22 As a comparison, deletion of the first 4 β-sheets, deletion of the helix domain alone (ΔHelix), and mutation of the ER exit signal (KKAA) have no effect on the oligomerization of the protein. Next, we asked whether the ΔHM mutant can fold correctly by examining whether it can bind mannose. The results showed that the ΔHM mutant can still bind mannose as efficiently as the ΔHelix mutant (Figure 3A), indicating that the monomeric LMAN1 mutant contains a functional CRD domain. To test whether monomeric LMAN1 can bind MCFD2, we cotransfected the Flag-tagged ΔHM with the myc-tagged MCFD2 into COS1 cells and immunoprecipitated the metabolic-labeled cell lysates with anti-myc and -Flag antibodies. As shown in Figure 3B, while the ΔHelix mutant and MCFD2 pulled down each other, the ΔHM mutant failed to coimmunoprecipitate with MCFD2, suggesting that oligomerization is required for MCFD2 binding in vivo.
Figure 3.
Oligomerization of LMAN1 is required for MCFD2 binding. (A) Mannose binding activities of different LMAN1 mutants. COS1 cells were transfected with the wild-type and the indicated LMAN1 mutants. LMAN1 proteins that are in the cell lysate and that are eluted from the mannose agarose beads are detected by Western blot analysis. Lysate lanes represent 20% of the input added to the mannose beads. (B) Co-IP of LMAN1 mutants with MCFD2. COS1 cells were cotransfected with Flag-tagged ΔHelix or ΔHM mutant and myc-tagged wild-type MCFD2. Cell lysates were immunoprecipitated with anti-myc for MCFD2 and anti-Flag for LMAN1.
Monomeric LMAN1 is defective in ER exit
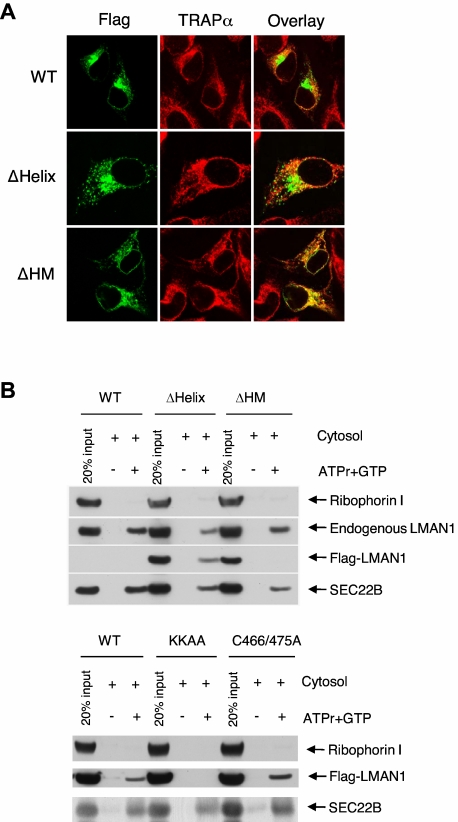
To further investigate why monomeric LMAN1 cannot coimmunoprecipitate with MCFD2, we examined the steady-state intracellular localization of the ΔHM mutant by indirect immunofluorescence staining. As shown in Figure 4A, the wild-type LMAN1 and the ΔHelix mutant showed typical ERGIC localization, characterized by a juxtanuclear and peripheral punctate staining pattern. In contrast, the ΔHM mutant exhibited a very different localization pattern, and it largely colocalized with an ER marker, TRAP-α. These results suggest that the ΔHM mutant is retained in the ER after synthesis. To test directly whether monomeric LMAN1 is defective in ER exit, we performed an in vitro vesicle formation assay. The Flag-tagged wild-type LMAN1, the ΔHelix mutant, the ΔHM mutant, the KKAA mutant, and the C466A/C475A mutant were transfected into HeLa cells separately. Transfected HeLa cells were semipermeabilzed by digitonin and incubated with COPII proteins supplied in rat liver cytosol.29 Vesicle-budding reaction was initiated by the addition of GTP and the ATP regeneration mix. As shown in Figure 4B, the wild-type LMAN1, the ΔHelix mutant, and the C466A/C475A mutant were all efficiently packaged into the COPII vesicles. However, the ΔHM mutant was nearly undetectable in the budded vesicles, similar to the ER-exit–deficient KKAA mutant. As internal controls, the endogenous LMAN1 still efficiently packaged into the vesicles in the ΔHM transfected cells, compared with the wild-type and the ΔHelix-transfected cells. Another COPII-dependent transmembrane cargo protein, SEC22B, was packaged into the vesicles in all transfected cells. The absence of an ER resident protein, ribophorin I, indicates minimal contamination of the ER membrane in the isolated COPII vesicles. Thus, we conclude that monomeric LMAN1 is defective in ER exit as a result of inefficient packaging into the COPII vesicles.
Figure 4.
LMAN1 monomer is defective in ER exit. (A) Immunofluorescence staining of the wild-type LMAN1 (WT), ΔHelix, and ΔHM mutants. LMAN1 proteins were detected with a monoclonal anti-Flag antibody. TRAP-α was used as an ER marker. (B) ER exit of wild-type LMAN1, ΔHelix, ΔHM, KKAA, and C466A/C475A mutants. HeLa cells were transfected with the indicated LMAN1 constructs. Membrane from transfected cells were incubated with rat liver cytosol. COPII vesicles were isolated and analyzed for the presence of a resident ER protein, ribophorin I, the endogenous LMAN1, the transfected LMAN1 mutants, and a control cargo protein, SEC22B.
Interactions of LMAN1 mutants with MCFD2 in living cells
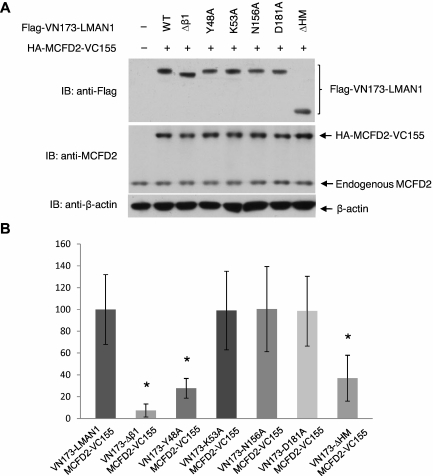
The bimolecular fluorescence complementation (BiFC) assay is based on the observation that 2 fragments of a fluorescent protein can complement each other and form a fluorescent complex when the 2 fragments are fused separately to 2 interacting proteins.25 It is a semiquantitative method that evaluates protein-protein interactions in living cells at expression levels comparable with the endogenous proteins.26 A similar assay was previously used to visualize the interaction of LMAN1 with MCFD2.15,24 Here, LMAN1 mutants were fused with the N-terminal half of Venus (VN173) with a Flag tag, and MCFD2 was fused with the C-terminal half of Venus (VC155) with a HA tag (supplemental Figure 4). We cotransfected 0.25 μg of each plasmid into Hela cells in 6-well plates, and BiFC signals were observed 10 hours after transfection. Immunoblotting revealed equal expression of fusion proteins, and the MCFD2 fusion protein was expressed at approximately twice the level of the endogenous MCFD2 (Figure 5A). Strong BiFC signals were observed in cells cotransfected with VN173-wtLMAN1 and MCFD2-VC155, as well as in cells cotransfected with VN173-N156A and MCFD2-VC155 or VN173-D181A and MCFD2-VC155 (Figure 5B and supplemental Figure 5). This is in agreement with our co-IP results (Figure 1B), and indicates that Ca2+ and sugar binding site mutations in LMAN1 have no significant effect on its interaction with MCFD2. Similarly, the K53A mutation has no significant effect on MCFD2 binding (Figure 2C), and BiFC signals of this mutant fusion (VN173-K53A) with MCFD2-VC155 were comparable with those of the wild-type fusion proteins. In contrast, very weak signals were seen in cells cotransfected with fusions of the 2 β1 mutants that failed to bind MCFD2 (VN173-Δβ1 and VN173-K48A) and MCFD2-VC155.
Figure 5.
BiFC analysis of the interactions of LMAN1 mutants with MCFD2 in living cells. (A) Fusion protein levels in cotransfection experiments were detected by immunoblotting (IB) using the indicated antibodies. Cells were lysed immediately after microscopic observations. (B) BiFC signals relative to the wild-type LMAN1 and MCFD2 fusion proteins. Fifty cotransfected cells from each set of transfection were counted, with the background subtracted for each image. Bars represent means ± SD from 3 replicates. Statistical analysis was performed using the Student t test, and asterisks indicate significant differences (P < .05).
The oligomerization deficiency of the ΔHM mutant was confirmed in living cells using BiFC assay (supplemental Figure 6). To this end, we generated 2 additional constructs fusing the wild-type LMAN1 and the ΔHM mutant with the C-terminal half of the Venus with a HA tag (supplemental Figure 4). BiFC signals from VN173-ΔHM and VC155-ΔHM were reduced to ∼ 15% of the level of the signals from VN173-wtLMAN1 and VC155-wtLMAN1 (supplemental Figure 6). These observations confirm that the ΔHM primarily exists as a monomer in cells. Consequently, BiFC signals from VN173-ΔHM and MCFD2-VC155 were significantly reduced (Figure 5B), consistent with the observation that the ΔHM failed to coimmunoprecipitate MCFD2 (Figure 3B).
The Ca2+ and sugar binding sites of LMAN1 mediate interactions with FV and FVIII
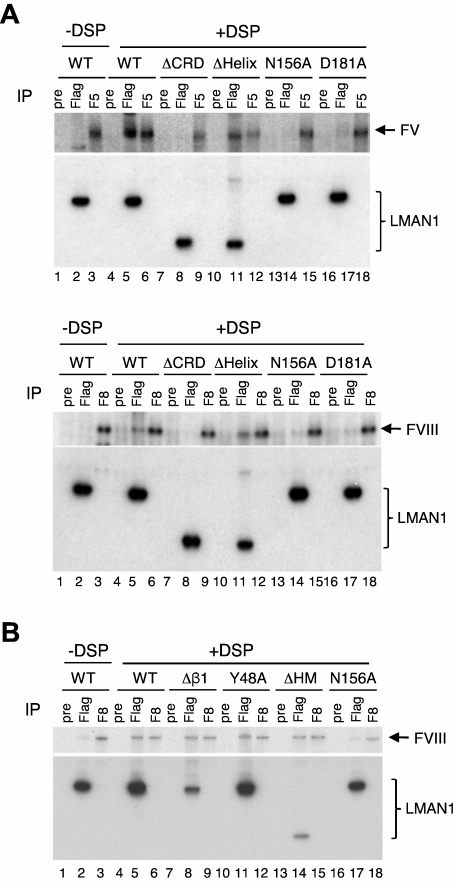
We have shown that functional Ca2+ and sugar binding sites in LMAN1 are not required for MCFD2 binding (Figures 1,5). Next, we asked whether these binding sites are required for FV and FVIII interactions. LMAN1 mutants depicted in Figure 1A were cotransfected with FV or FVIII into COS1 cells, metabolically labeled, cross-linked using DSP, and immunprecipitated with anti-Flag and -FV or -FVIII antibodies, as well as with preimmune serum as a negative control. We have previously demonstrated specificity of cross-linking of FVIII to the LMAN1-MCFD2 complex.6,7 As expected, both the wild-type LMAN1 and the ΔHelix mutant can be cross-linked to FV and FVIII (Figure 6A). In contrast, the ΔCRD mutant cannot be cross-linked to FV or FVIII (Figure 6A). These results indicate that the CRD is responsible for the interaction with FV/FVIII, and that the helix domain is dispensable for this interaction. LMAN1 with either N156A or D181A mutation cannot be cross-linked to FV and FVIII (Figure 6A), although both mutants can still bind MCFD2 (Figure 1B). These results suggest that the sugar and Ca2+ binding sites in the CRD are critical for the interaction of LMAN1 with FV and FVIII.
Figure 6.
The Ca2+- and sugar-binding sites in the CRD interacts with FV and FVIII. COS-1 cells were cotransfected with the wild-type or indicated LMAN1 mutants, along with either FV (A) or FVIII (A-B). Cells were incubated with or without DSP before lysis. Cell lysates were immunoprecipitated with preimmune serum (pre) as a negative control, anti-Flag (Flag) for LMAN1, and anti-FV (F5) or -FVIII (F8) for the indicated coagulation factor.
MCFD2 has been shown to interact with FV/FVIII independent of LMAN1.7 The availability of LMAN1 mutants that disrupt MCFD2 binding provides an opportunity to test whether LMAN1 can interact with FV/FVIII independent of MCFD2. The wild-type LMAN1 and the Δβ1, Y48A, ΔHM, and N156A mutants were cotransfected with FVIII and analyzed by the cross-linking and co-IP assay (Figure 6B). The wild-type LMAN1, but not the N156A mutant, can be cross-linked to FVIII. All 3 LMAN1 mutants (Δβ1, Y48A, and ΔHM) that fail to bind MCFD2 can still be efficiently cross-linked to FVIII. These results suggest that the interaction of LMAN1 with FVIII, which requires the Ca2+- and sugar-binding sites, is independent of MCFD2 binding.
Discussion
Cargo receptors are thought to be required for the efficient ER to Golgi transport of many soluble secretory proteins.12 However, the LMAN1-MCFD2 complex is, so far, the only known cargo receptor in mammalian cells, required for the efficient secretion of FV and FVIII. Recently, we demonstrate that the EF hand domains of MCFD2 mediate interactions with both LMAN1 and FV/FVIII.7 In this study, we demonstrated that the first β-sheet (β1) in the CRD of LMAN1 is primarily responsible for the interaction with MCFD2, while Ca2+- and mannose-binding sites are required for FV/FVIII binding. The β1 is located on the convex side of the molecule and far away from the calcium/mannose binding sites, which are located on the concave side.30,31 We showed that mutations that disturb the MCFD2 binding site have no effect on the function of the FV/FVIII binding site and vice versa. Therefore, the CRD of LMAN1 contains distinct, separable binding sites for MCFD2 and FV/FVIII. In this study, we used 2 methods to study the interactions of MCFD2 with various mutants of LMAN1, namely, co-IP and BiFC assay, both of which produced results that are in agreement with each other. Because these assays are based on an overexpression system, we were careful in including proper positive and negative controls in each experiment.
Crystal structures of the LMAN1-MCFD2 complex recently reported by 2 groups18,19 are in agreement with our observation that the N-terminal β1 region is involved in MCFD2 interaction. We further demonstrated that deletion of β1 does not have a significant impact on the overall structural integrity of LMAN1. However, deletion into the second β-sheet disrupts the structure of the protein (Figure 2). Although both are in β1 and predicted to directly interact with MCFD2, mutation of K53 has no effect on the LMAN1-MCFD2 interaction, while mutation of Y48 abolishes MCFD2 binding, suggesting a more critical role of Y48 in the complex formation. Of note, a missense mutation of the Y48 interaction residue (D122V) in MCFD2 causes F5F8D.33,34 To date, only 2 missense mutations have been reported in LMAN1. The C475R mutation changes a membrane-proximal cysteine involved in oligomerization20,21 and destabilizes the protein. W67S is a recently reported missense mutation32 that is located in the second β-sheet. We showed that this mutation likely causes global destruction of the CRD structure. Our results predict that a missense mutation in Y48 would disrupt the MCFD2 binding without significantly affecting the protein structure and cause F5F8D.
Our observation that the Ca2+ and mannose binding activity of the CRD is required for LMAN1 to bind FV and FVIII (Figure 6) suggests that sugar residues in FV and FVIII are involved in the interaction with LMAN1. Previously, we showed that FVIII with a deletion of the heavily glycosylated B domain markedly decreased the binding to the LMAN1-MCFD2 complex.6 On the other hand, protein-protein interaction is also involved in the cargo to cargo-receptor interaction as the deglycosylated FVIII can still be cross-linked to the LMAN1-MCFD2 complex.6 Because our cross-linking assay can potentially capture transient interactions, we cannot distinguish whether carbohydrate binding is the primary interaction or is secondary to the protein-protein interaction. The LMAN1-FVIII interaction is independent of MCFD2 binding or the oligomeric state of LMAN1 (Figure 6B), suggesting that the mannose binding function is active in monomeric LMAN1 and is independent of MCFD2 binding. Indeed, the structure of the CRD-MCFD2 complex indicates that there is no major perturbation in the mannose binding site upon complex formation.18,19 Although our results and the structural studies18,19 suggest it to be unlikely, we cannot rule out the possibility that MCFD2 binding enhances the binding of FV/FVIII to LMAN1, as reported in a recent study.16
The CRD can form a stable complex with MCFD2 with a 1-to-1 stoichiometry in vitro and with no evidence of oligomerization.16,18,19, We provide evidence that the monomeric LMAN1 is not sufficient for MCFD2 binding in vivo. The requirement of oligomerization for ER exit is not unprecedented, as Emp47p, a yeast cargo receptor and a homolog of LMAN1, requires oligomeraization for ER exit.35 Although MCFD2 is also synthesized in the ER, it apparently fails to bind to the monomeric LMAN1 that is localized to the ER membrane (Figure 4A). These results suggest an intriguing possibility that the LMAN1-MCFD2 interaction normally occurs in specific locations of the ER, potentially near the ER exit sites.36 In this hypothesis, the oligomerization is required for LMAN1 to move to the ER exit sites, where it binds MCFD2. Alternatively, the MCFD2-binding site in monomeric LMAN1 may be sequestered by other associated proteins, such as chaperones. FV and FVIII loading may be initiated by an interaction with MCFD2 and stabilized by the follow-up interaction of oligosaccharide side chains of FV/FVIII with the sugar binding site of LMAN1. Supporting a more direct role of MCFD2 in cargo recognition is the observation that mean FV and FVIII levels in patients with MCFD2 mutations are significantly lower than patients with LMAN1 mutations.37 In this model, FV/FVIII molecules must engage in interactions with both MCFD2 and LMAN1 to form a sufficiently stable ternary complex for packaging into COPII vesicles.
How cargo proteins are released from the cargo receptor in the ERGIC is not clear. One possibility is that MCFD2 and FV/FVIII dissociate together from LMAN1 in the ERGIC. However, the endogenous LMAN1 and MCFD2 have similar half-lives,6 suggesting that MCFD2 bound to LMAN1 is not dissociated during the recycling between the ER and the ERGIC. Ca2+ concentrations are high in both the ER (0.4mM) and the Golgi (0.3mM), but it seems to be very low in the ERGIC.38 Although the pH value in the ERGIC is not clear, the organelle pH is known to drop from the ER to the Golgi.39 Previous studies demonstrate that acidification inhibits the binding of LMAN1 with another glycosylated cargo protein, cathepsin Z, and that organelle neutralization impairs the dissociation of this cargo protein.40 Although both LMAN1 and MCFD2 are Ca2+-binding proteins, the role of Ca2+ seems distinct between the 2 proteins. The Ca2+-induced folding of MCFD2 is important for LMAN1 interaction, but is not essential for FV/FVIII interaction.7 Mutations in the Ca2+-binding site of CRD disrupt FV and FVIII interaction, without affecting MCFD2 binding, suggesting that the Ca2+-binding site in LMAN1 is primarily required for the recognition of sugar residues in FV and FVIII. In vitro studies show that the LMAN1-MCFD2 interaction is insensitive to the pH.16 If the LMAN1-FV/FVIII interaction is more sensitive to the drop in Ca2+ concentration and pH than the LMAN1-MCFD2 interaction, the combination of lower Ca2+ and pH in the ERGIC may trigger the release of FV and FVIII from the LMAN1-MCFD2 receptor without dissociating the receptor complex itself. The empty receptor complex can then be recycled back to the ER for the next round of cargo loading.
Supplementary Material
Acknowledgments
We thank Min Zhu for help with the in vitro vesicle formation assay. We also thank Molegro Virtual Docker for providing a free license for the MVD docking work.
This work was supported by a Basil O'Connor Starter Scholar Research Award grant from the March of Dimes Foundation (#5-FY08-06) and by a grant from the National Institutes of Health (R01 HL094505; to B.Z.). C.Z. was supported, in part, by a postdoctoral fellowship from the American Heart Association.
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: C.Z. H.L., and B.Z. designed research; C.Z. and H.L. performed experiments; S.Y. and J.Z. performed the molecular docking analysis; and C.Z. and B.Z. wrote the manuscript, which was commented on by all the authors.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Bin Zhang, Genomic Medicine Institute, Lerner Research Institute, Cleveland Clinic Foundation, 9500 Euclid Ave, Cleveland, OH 44195; e-mail: zhangb@ccf.org.
References
- 1.Zhang B, Ginsburg D. Familial multiple coagulation factor deficiencies: new biologic insight from rare genetic bleeding disorders. J Thromb Haemost. 2004;2(9):1564–1572. doi: 10.1111/j.1538-7836.2004.00857.x. [DOI] [PubMed] [Google Scholar]
- 2.Zhang B. Recent developments in the understanding of the combined deficiency of FV and FVIII. Br J Haematol. 2009;145(1):15–23. doi: 10.1111/j.1365-2141.2008.07559.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nichols WC, Seligsohn U, Zivelin A, et al. Mutations in the ER-Golgi intermediate compartment protein ERGIC-53 cause combined deficiency of coagulation factors V and VIII. Cell. 1998;93(1):61–70. doi: 10.1016/s0092-8674(00)81146-0. [DOI] [PubMed] [Google Scholar]
- 4.Zhang B, Cunningham MA, Nichols WC, et al. Bleeding due to disruption of a cargo-specific ER-to-Golgi transport complex. Nat Genet. 2003;34(2):220–225. doi: 10.1038/ng1153. [DOI] [PubMed] [Google Scholar]
- 5.Zhang B, McGee B, Yamaoka JS, et al. Combined deficiency of factor V and factor VIII is due to mutations in either LMAN1 or MCFD2. Blood. 2006;107:1903–1907. doi: 10.1182/blood-2005-09-3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang B, Kaufman RJ, Ginsburg D. LMAN1 and MCFD2 form a cargo receptor complex and interact with coagulation factor VIII in the early secretory pathway. J Biol Chem. 2005;280(27):25881–25886. doi: 10.1074/jbc.M502160200. [DOI] [PubMed] [Google Scholar]
- 7.Zheng C, Liu HH, Zhou J, Zhang B. EF hand domains of MCFD2 mediate interactions with both LMAN1 and coagulation factor V or VIII. Blood. 2010;115(5):1081–1087. doi: 10.1182/blood-2009-09-241877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Appenzeller C, Andersson H, Kappeler F, Hauri HP. The lectin, ERGIC-53, is a cargo transport receptor for glycoproteins. Nat Cell Biol. 1999;1(6):330–334. doi: 10.1038/14020. [DOI] [PubMed] [Google Scholar]
- 9.Nyfeler B, Reiterer V, Wendeler MW, et al. Identification of ERGIC-53 as an intracellular transport receptor of a1-antitrypsin. J Cell Biol. 2008;180(4):705–712. doi: 10.1083/jcb.200709100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vollenweider F, Kappeler F, Itin C, Hauri HP. Mistargeting of the lectin ERGIC-53 to the endoplasmic reticulum of HeLa cells impairs the secretion of a lysosomal enzyme. J Cell Biol. 1998;142(2):377–389. doi: 10.1083/jcb.142.2.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee MC, Miller EA, Goldberg J, Orci L, Schekman R. Bi-directional protein transport between the ER and Golgi. Annu Rev Cell Dev Biol. 2004;20:87–123. doi: 10.1146/annurev.cellbio.20.010403.105307. [DOI] [PubMed] [Google Scholar]
- 12.Miller EA, Beilharz TH, Malkus PN, et al. Multiple cargo-binding sites on the COPII subunit, Sec24p, ensure capture of diverse membrane proteins into transport vesicles. Cell. 2003;114(4):497–509. doi: 10.1016/s0092-8674(03)00609-3. [DOI] [PubMed] [Google Scholar]
- 13.Mossessova E, Bickford LC, Goldberg J. SNARE selectivity of the COPII coat. Cell. 2003;114(4):483–495. doi: 10.1016/s0092-8674(03)00608-1. [DOI] [PubMed] [Google Scholar]
- 14.Baines AC, Zhang B. Receptor-mediated protein transport in the early secretory pathway. Trends Biochem Sci. 2007;32(8):381–388. doi: 10.1016/j.tibs.2007.06.006. [DOI] [PubMed] [Google Scholar]
- 15.Hauri HP, Kappeler F, Andersson H, Appenzeller C. ERGIC-53 and traffic in the secretory pathway. J Cell Sci. 2000;113(Pt 4):587–596. doi: 10.1242/jcs.113.4.587. [DOI] [PubMed] [Google Scholar]
- 16.Kawasaki N, Ichikawa Y, Matsuo I, et al. The sugar-binding ability of ERGIC-53 is enhanced by its interaction with MCFD2. Blood. 2008;111(4):1972–1979. doi: 10.1182/blood-2007-06-097022. [DOI] [PubMed] [Google Scholar]
- 17.Nyfeler B, Zhang B, Ginsburg D, Kaufman RJ, Hauri HP. Cargo selectivity of the ERGIC-53/MCFD2 transport receptor complex. Traffic. 2006;7(11):1473–1481. doi: 10.1111/j.1600-0854.2006.00483.x. [DOI] [PubMed] [Google Scholar]
- 18.Nishio M, Kamiya Y, Mizushima T, et al. Structural basis for the cooperative interplay between the two causative gene products of combined factor V and factor VIII deficiency. Proc Natl Acad Sci U S A. 2010;107(9):4034–4039. doi: 10.1073/pnas.0908526107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wigren E, Bourhis JM, Kursula I, Guy JE, Lindqvist Y. Crystal structure of the LMAN1-CRD/MCFD2 transport receptor complex provides insight into combined deficiency of factor V and factor VIII. FEBS Lett. 2010;584(5):878–882. doi: 10.1016/j.febslet.2010.02.009. [DOI] [PubMed] [Google Scholar]
- 20.Lahtinen U, Svensson K, Pettersson RF. Mapping of structural determinants for the oligomerization of p58, a lectin-like protein of the intermediate compartment and cis-Golgi. Eur J Biochem. 1999;260(2):392–397. doi: 10.1046/j.1432-1327.1999.00158.x. [DOI] [PubMed] [Google Scholar]
- 21.Nufer O, Kappeler F, Guldbrandsen S, Hauri HP. ER export of ERGIC-53 is controlled by cooperation of targeting determinants in all three of its domains. J Cell Sci. 2003;116(Pt 21):4429–4440. doi: 10.1242/jcs.00759. [DOI] [PubMed] [Google Scholar]
- 22.Neve EP, Lahtinen U, Pettersson RF. Oligomerization and interacellular localization of the glycoprotein receptor, ERGIC-53, is independent of disulfide bonds. J Mol Biol. 2005;354(3):556–568. doi: 10.1016/j.jmb.2005.09.077. [DOI] [PubMed] [Google Scholar]
- 23.Moussalli M, Pipe SW, Hauri HP, Nichols WC, Ginsburg D, Kaufman RJ. Mannose-dependent endoplasmic reticulum (ER)-Golgi intermediate compartment-53-mediated ER to Golgi trafficking of coagulation factors V and VIII. J Biol Chem. 1999;274(46):32539–32542. doi: 10.1074/jbc.274.46.32539. [DOI] [PubMed] [Google Scholar]
- 24.Nyfeler B, Michnick SW, Hauri HP. Capturing protein interactions in the secretory pathway of living cells. Proc Natl Acad Sci U S A. 2005;102(18):6350–6355. doi: 10.1073/pnas.0501976102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu CD, Chinenov Y, Kerppola TK. Visualization of interactions among bZip and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol Cell. 2002;9(4):789–798. doi: 10.1016/s1097-2765(02)00496-3. [DOI] [PubMed] [Google Scholar]
- 26.Shyu YJ, Liu H, Deng X, Hu CD. Identification of new fluorescent protein fragments for bimolecular fluorescence complementation analysis under physiological conditions. Biotechniques. 2006;40(1):61–66. doi: 10.2144/000112036. [DOI] [PubMed] [Google Scholar]
- 27.Itin C, Roche AC, Monsigny M, Hauri HP. ERGIC-53 is a functional mannose-selective and calcium-dependent human homologue of leguminous lectins. Mol Biol Cell. 1996;7(3):483–493. doi: 10.1091/mbc.7.3.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim J, Hamamoto S, Ravazzola M, Orci L, Schekman R. Uncoupled packaging of amyloid precursor protein and presenilin 1 into coat protein complex II vesicles. J Biol Chem. 2005;280(9):7758–7768. doi: 10.1074/jbc.M411091200. [DOI] [PubMed] [Google Scholar]
- 29.Merte J, Jensen D, Wright K, et al. Sec24b selectively sorts Vangl2 to regulate planar cell polarity during neural tube closure. Nat Cell Biol. 2010;12(1):41–46. doi: 10.1038/ncb2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Velloso LM, Svensson K, Pettersson RF, Lindqvist Y. The crystal structure of the carbohydrate-recognition domain of the glycoprotein sorting receptor, p58/ERGIC-53, reveals an unpredicted metal-binding site and conformational changes associated with calcium ion binding. J Mol Biol. 2003;334(5):845–851. doi: 10.1016/j.jmb.2003.10.031. [DOI] [PubMed] [Google Scholar]
- 31.Velloso LM, Svensson K, Schneider G, Pettersson RF, Lindqvist Y. Crystal structure of the carbohydrate recognition domain of p58/ERGIC-53, a protein involved in glycoprotein export from the endoplasmic reticulum. J Biol Chem. 2002;277(18):15979–15984. doi: 10.1074/jbc.M112098200. [DOI] [PubMed] [Google Scholar]
- 32.Yamada T, Fujimori Y, Suzuki A, et al. A novel missense mutation causing abnormal LMAN1 in a Japanese patient with combined deficiency of factor V and factor VIII. Am J Hematol. 2009;84(11):738–742. doi: 10.1002/ajh.21532. [DOI] [PubMed] [Google Scholar]
- 33.Guy JE, Wigren E, Svard M, Hard T, Lindqvist Y. New insights into multiple coagulation factor deficiency from the solution structure of human MCFD2. J Mol Biol. 2008;381(4):941–955. doi: 10.1016/j.jmb.2008.06.042. [DOI] [PubMed] [Google Scholar]
- 34.Jayandharan G, Spreafico M, Viswabandya A, Chandy M, Srivastava A, Peyvandi F. Mutations in the MCFD2 gene are predominant among patients with hereditary combined FV and FVIII deficiency (F5F8D) in India. Haemophilia. 2007;13(4):413–419. doi: 10.1111/j.1365-2516.2007.01477.x. [DOI] [PubMed] [Google Scholar]
- 35.Sato K, Nakano A. Oligomerization of a cargo receptor directs protein sorting into COPII-coated transport vesicles. Mol Biol Cell. 2003;14(7):3055–3063. doi: 10.1091/mbc.E03-02-0115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Budnik A, Stephens DJ. ER exit sites—localization and control of COPII vesicle formation. FEBS Lett. 2009;583(23):3796–3803. doi: 10.1016/j.febslet.2009.10.038. [DOI] [PubMed] [Google Scholar]
- 37.Zhang B, Spreafico M, Yang A, et al. Genotype-phenotype correlation in combined deficiency of factor V and factor VIII. Blood. 2008;111(12):5592–5600. doi: 10.1182/blood-2007-10-113951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pezzati R, Bossi M, Podini P, Meldolesi J, Grohovaz F. High-resolution calcium mapping of the endoplasmic reticulum-Golgi-exocytic membrane system. Electron energy loss imaging analysis of quick frozen-freeze dried PC12 cells. Mol Biol Cell. 1997;8(8):1501–1512. doi: 10.1091/mbc.8.8.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu MM, Grabe M, Adams S, Tsien RY, Moore HP, Machen TE. Mechanisms of pH regulation in the regulated secretory pathway. J Biol Chem. 2001;276(35):33027–33035. doi: 10.1074/jbc.M103917200. [DOI] [PubMed] [Google Scholar]
- 40.Appenzeller-Herzog C, Roche AC, Nufer O, Hauri HP. pH-induced conversion of the transport lectin, ERGIC-53, triggers glycoprotein release. J Biol Chem. 2004;279(13):12943–12950. doi: 10.1074/jbc.M313245200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.