Recent studies have revealed a role for the Ub/proteasome system in the regulation and turnover of OMM-associated proteins. The data presented show that an AAA-ATPase, p97, is required for the proteasomal degradation of Mcl1 and Mfn1, two unrelated OMM proteins, and establishes p97 as a novel and essential part of the OMM-protein degradation pathway.
Abstract
Recent studies have revealed a role for the ubiquitin/proteasome system in the regulation and turnover of outer mitochondrial membrane (OMM)-associated proteins. Although several molecular components required for this process have been identified, the mechanism of proteasome-dependent degradation of OMM-associated proteins is currently unclear. We show that an AAA-ATPase, p97, is required for the proteasomal degradation of Mcl1 and Mfn1, two unrelated OMM proteins with short half-lives. A number of biochemical assays, as well as imaging of changes in localization of photoactivable GFP-fused Mcl1, revealed that p97 regulates the retrotranslocation of Mcl1 from mitochondria to the cytosol, prior to, or concurrent with, proteasomal degradation. Mcl1 retrotranslocation from the OMM depends on the activity of the ATPase domain of p97. Furthermore, p97-mediated retrotranslocation of Mcl1 can be recapitulated in vitro, confirming a direct mitochondrial role for p97. Our results establish p97 as a novel and essential component of the OMM-associated protein degradation pathway.
INTRODUCTION
Mitochondria are the primary site of energy production in animal cells. To eliminate surplus or dysfunctional mitochondrial proteins, or entire damaged organelles that could negatively influence cellular homeostasis, regulation of mitochondrial biogenesis and clearance is required. Within the mitochondrial matrix, the remnants of bacterial ATP-stimulated mitochondrial proteases, including Lon protease, play a role in the degradation of misfolded oxidized proteins (reviewed in Bulteau et al., 2006; Ngo and Davies, 2007). In contrast to the inner mitochondrial compartments (the inner mitochondrial membrane and matrix), information about the proteostasis of the outer mitochondrial membrane (OMM) is very limited. The OMM functions as a barrier separating mitochondria from the cytosol and plays vital roles in mitochondrial function, including the regulation of metabolism, apoptosis, and other signaling events, such as mitochondrial membrane dynamics. Therefore, quality control of OMM-associated proteins is likely of highest importance for maintaining cellular function. In addition, the OMM is a platform-integrating mitochondria with the cytosol, with other membrane-bound organelles and likely with cytosol-localized degradation pathways, including the proteasome and autophagic machinery.
In eukaryotes, short-lived proteins are degraded by the ubiquitin (Ub)/proteasome system. In addition to a variety of previously identified proteasome substrates, it is now known that certain OMM-associated proteins are under the control of the Ub/proteasome system (Yonashiro et al., 2006; Karbowski et al., 2007; Neutzner et al., 2008; Ziviani et al., 2010). For example, the degradation of OMM-associated anti-apoptotic proteins, such as Bcl-2 and Mcl1, involves their polyubiquitination and requires the activity of the 26S proteasome (Zhong et al., 2005; Azad et al., 2006). We have shown that, in yeast, the mitochondrial fusion protein Fzo1p is modified at Lys-48, targeting it to the proteasome, and that the proteasome inhibitor MG132, as well as proteasome mutations, can suppress the degradation of Fzo1p (Neutzner et al., 2007). These results indicate that poly-Ub-dependent proteasomal degradation is involved in Fzo1p turnover. In further support of this notion, it has recently been shown that the degradation of dMfn, a Drosophila melanogaster homologue of Fzo1p, also depends on the proteasome (Ziviani et al., 2010). In addition, a number of E3 Ub ligases associated with mitochondria, including MARCH5 (Nakamura et al., 2006; Yonashiro et al., 2006; Karbowski et al., 2007), IBRDC2 (Benard et al., 2010), and Parkin (Narendra et al., 2008), as well as a deubiquitinating protein, USP30 (Nakamura and Hirose, 2008), have recently been identified.
Despite this recent progress, which establishes a significant role for the Ub/proteasome system in the regulation of mitochondria, the molecular steps of Ub-dependent mitochondrial protein degradation are largely unknown (Neutzner et al., 2008). One can assume that the cytosolic localization of essential components of the Ub/proteasome system, including E1s, E2s, and the proteasome itself, would make it necessary for proteins integral to the OMM to be extracted prior to degradation. In support of this hypothesis, we show here that p97 (valosin-containing protein, p97/VCP; hereafter referred to as p97) provides the main driving force for extraction of the Mfn1 and Mcl1 proteins from the OMM, thereby regulating their degradation by the proteasome in the cytosol. p97 is ubiquitous and a member of the highly conserved AAA (ATPases associated with diverse cellular activities) family of proteins that are known for their chaperone-like activities in various cellular locations. p97 is an essential biochemical component of a wide range of Ub-associated biological pathways, including Ub/proteasome system-mediated protein degradation (Ye et al., 2004; Bar-Nun, 2005; Halawani and Latterich, 2006; Jentsch and Rumpf, 2007), Golgi and endoplasmic reticulum (ER) membrane fusion (Acharya et al., 1995), and transcription factor activation (Rape et al., 2001). p97 is known to mediate the movement of polypeptides from the ER membrane to the cytosol (Bays et al., 2001; Ye et al., 2001, 2004). This extraction step, known as dislocation or retrotranslocation, is a hallmark of the ER-associated degradation (ERAD) pathway. Although the mechanisms that operate during dislocation and the nature of the channel through which ERAD substrates egress from the ER are only beginning to be elucidated, numerous studies have firmly established the critical role of p97 in this process (Bays et al., 2001; Ye et al., 2004; Bar-Nun, 2005; Ballar et al., 2007; Jentsch and Rumpf, 2007). The data shown here indicate that, in addition to extracting proteins from the ER, p97 serves as a membrane protein-extracting factor in mitochondria. We show that the inhibition of p97 impairs the proteasomal degradation of Mcl1 and Mfn1, two unrelated, short-lived OMM proteins. Furthermore, imaging and biochemical assays revealed that p97 regulates the removal of Mcl1 from the mitochondrial membrane, suggesting that, as in the case of ERAD substrates, p97 regulates Mcl1 retrotranslocation from the OMM to the cytosol.
RESULTS
Proteasome and p97-dependent regulation of Mfn1 and Mcl1 stability
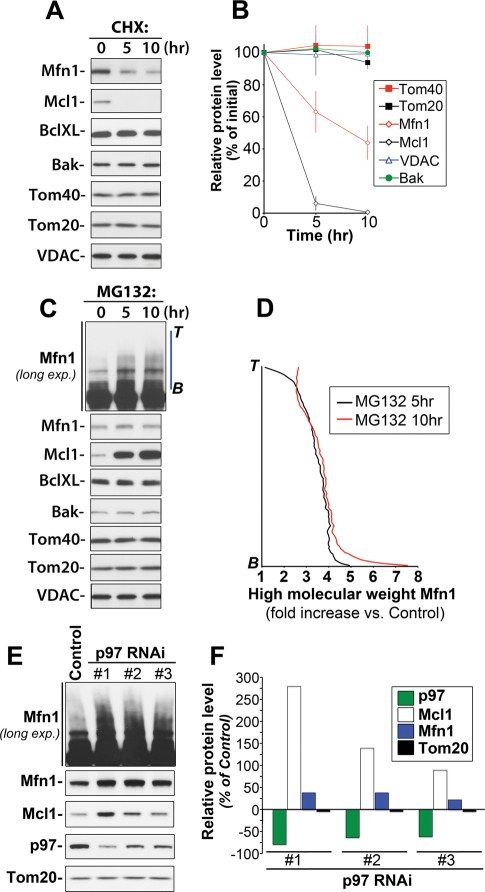
We analyzed the stability and Ub/proteasome dependence of an array of OMM-associated proteins (Figure 1,A–C). We found that, in addition to Mcl1, an OMM-associated anti-apoptotic member of the Bcl2 protein family with an estimated half-life of 40–60 min (Yang et al., 1995; Nijhawan et al., 2003), Mfn1, a mitochondrial fusion factor, was also stabilized by proteasome inhibition (Figure 1C) and degraded in the presence of the protein synthesis inhibitor cycloheximide (CHX; Figure 1A). Under these experimental conditions, the estimated half-life of Mfn1 is approximately 6 h (Figure 1B). Therefore, Mfn1 displays a relatively short half-life when compared with the half-life of the entire mitochondrion, which, depending on cell type, is estimated to be between 8.65 and 23.3 d (Beattie et al., 1967). In contrast to Mcl1 and Mfn1, levels of other tested proteins (Figure 1, A–C) did not exhibit noticeable changes within the time frame of CHX or MG132 treatment (Figure 1, A–C), further confirming the instability of Mcl1 and Mfn1.
FIGURE 1:
Proteasome- and p97-dependent regulation of Mcl1 and Mfn1 stability. HeLa cells were treated with the protein synthesis inhibitor CHX (20 μg/ml) (A) or a proteasome inhibitor, MG132 (50 μM) (C), for 0, 5, or 10 h followed by Western blotting as indicated in the figure. In (B), relative protein levels in cells treated as described in (A) were quantified and plotted against the time of treatment with CHX [data represent the mean ± SD of three (VDAC, Bak, BclXL) or four (Mfn1, Mcl1, Tom20, Tom40) independent experiments]. In (D), the levels of high-molecular-weight species of Mfn1 were quantified in control cells and cells treated with MG132 for 5 or 10 h (along blue lines as shown in C; Mfn1-long exp.; B, bottom; T, top). The data shown represent fold increases of Mfn1 levels in different points of intensity plots versus the respective values in control samples. The data were normalized with control values at the respective points of intensity plots taken as 1. In panel E, total cell lysates obtained from cells transfected with 3 different p97 shRNAi constructs (#1, #2, and #3) or with a GFP shRNAi construct (Control) were analyzed by Western blot as indicated in the figure. In panel F, changes in the protein levels of p97, Mcl1, Mfn1, and Tom20 in p97 RNAi cells (clones #1, #2, and #3) were quantified and plotted as the percentage of the protein levels in control RNAi cells.
Others and we have shown that stress-induced degradation of Mfn1 homologues in Saccharomyces cerevisiae and D. melanogaster is also ubiquitin- and proteasome-dependent (Neutzner et al., 2007, 2008; Ziviani et al., 2010). The data shown here (Figure 1, C and D) indicate that, in mammalian cells, proteasome might also be required for Mfn1 turnover under normal growth conditions. In contrast to Mcl1, for which a monomeric form accumulated in the presence of proteasome inhibitor (Figure 1C), MG132 treatment led to the accumulation of high-molecular-weight forms of Mfn1, with little effect on the monomeric form of Mfn1 (Figure 1C). Immunoprecipitation of Mfn1 under denaturing conditions revealed that these high-molecular-weight forms represent ubiquitinated forms of Mfn1 (data not shown). The quantification of the levels of high-molecular-weight degradation intermediates of Mfn1 revealed marked ∼3.5-fold increases in the detectable levels of these intermediates in MG132-treated cells (Figure 1D), as compared with the untreated cells; this further suggests proteasomal dependence of Mfn1 degradation. The lack of ubiquitinated forms of Mcl1 in MG132-treated cell lysates is consistent with the recently described, rapid deubiquitination of this protein by deubiqutinase USP9x (Schwickart et al., 2010).
Because Mfn1 and Mcl1 are OMM-associated proteins, with Mfn1 having two membrane-spanning domains and Mcl1 being anchored to the membrane through its hydrophobic C-terminal tail, these proteins likely need to be extracted from the OMM before their degradation by the proteasome. It has been shown that in S. cerevisiae, a mutation in CDC48, an AAA ATPase with established function in the retrotranslocation of ubiquitinated proteins from the ER membrane to the cytosol (Ye et al., 2001, 2004; Braun et al., 2002), resulted in mitochondrial impairment and alteration of the mitochondrial proteome (Braun et al., 2006). Furthermore, a number of mitochondria-localized AAA ATPases, including YME1L1 (Griparic et al., 2007) and paraplegin (Ishihara et al., 2006), regulate the stability of inner mitochondrial membrane- and matrix-localized proteins. A number of proteomic studies revealed that p97 also associates with mitochondria in mammalian cells (Taylor et al., 2003; Reifschneider et al., 2006; Zhang et al., 2008). These results suggest a conserved role for AAA-ATPase protein family members in the regulation of mitochondrial proteostasis. Therefore, we asked whether p97, a mammalian homologue of CDC48, regulates the turnover of OMM-associated proteins. To address this question, the expression of p97 protein was knocked down using an shRNAi-based method (Figure 1, E and F), followed by Western blotting to detect Mcl1 and Mfn1 (Figure 1D). The data show that the effects of p97 downregulation closely resemble those observed in MG132-treated cells (Figure 1E). Namely, accumulation of both the monomeric form of Mcl1 and the high-molecular-weight forms of Mfn1 was apparent (Figure 1E). Densitometric evaluation of Western blots revealed that p97 knockdown achieved with three independent shRNAi constructs increased the protein level of both Mcl1 and Mfn1, suggesting a specific role for p97 in regulating the stability of Mcl1 and Mfn1 (Figure 1F). Thus, our results agree well with a previous report showing that mutation of CDC48 induced distinct changes in the mitochondrial proteome (Braun et al., 2006). Clearly, the role of CDC48/p97 in the regulation of mitochondrial protein content is evolutionally conserved in mammals and yeast.
Mutation of the p97 ATPase domain inhibits the proteasomal degradation of Mcl1
Given the short half-life and the well-established proteasome dependence of Mcl1 turnover (Yang et al., 1995; Nijhawan et al., 2003; Zhong et al., 2005), we analyzed Mcl1 as a representative OMM-localized, p97-dependent proteasome substrate in most of the following experiments aimed at defining the mechanisms underlying the mitochondrial role of p97.
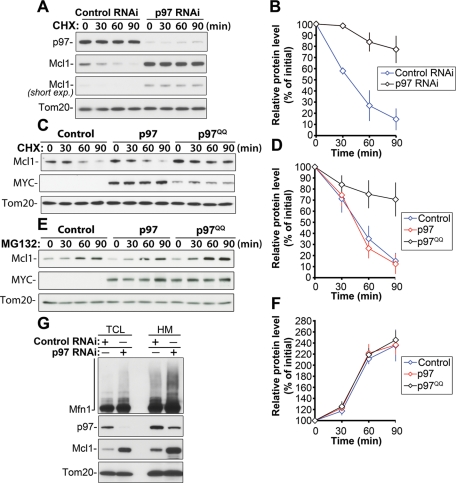
If p97 is involved in the regulation of Mcl1 turnover, then downregulation of p97 would affect the rate of Mcl1 degradation, and this change would be readily detectable upon inhibition of protein synthesis. Control RNAi and p97 RNAi cells were treated with CHX for 30, 60, or 90 min. Total cell lysates were analyzed by Western blot for the level of Mcl1 (Figure 2A). The data show almost complete inhibition of p97 degradation in CHX-treated p97 RNAi cells compared with CHX-treated control RNAi cells (Figure 2, A and B), validating the notion that p97 acts in the regulation of Mcl1 turnover.
FIGURE 2:
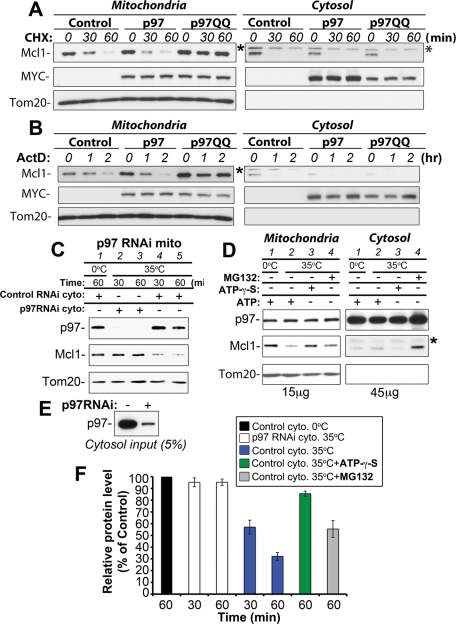
p97 is required for the degradation of Mcl1 in an ATPase domain activity-dependent manner. Control and p97 RNAi cells (clone #1) were treated with CHX for 0, 30, 60, or 90 min, followed by Western blot analyses to detect protein levels of Mcl1 (two exposures of the same Mcl1 blot are shown) and p97. Tom20 served as a loading control (A). In panel B, relative Mcl1 protein levels in control RNAi and p97 RNAi cells were quantified and plotted as a function of the length of CHX treatment. Mcl1 levels detected in untreated samples (time, 0 min) were set at 100%. Control and p97- and p97QQ-expressing cells were treated with CHX (C) or MG132 (E) for 0, 30, 60, or 90 min, followed by Western blot analysis to detect protein levels of Mcl1. Expression levels of p97 and p97QQ were detected with anti-MYC antibody (MYC). Tom20 was used as a loading control. Relative protein levels in cells treated with CHX (D) and MG132 (F) as described in panels C and E were quantified (data represent the mean ± SD of at least three independent experiments). Mcl1 levels detected in untreated samples (time, 0 min) were set at 100%. In panel G, the total cell lysates (TCL) and mitochondria-enriched heavy membrane fractions (HM) obtained from control RNAi and p97 RNAi cells were analyzed by Western blot for the levels of Mcl1, Mfn1, and p97. Tom20 was used as a loading control. Note the mitochondrial accumulation of Mcl1 and Mfn1 in p97 RNAi cells.
To test whether p97 ATPase activity is required for Mcl1 degradation, we used a dominant negative p97 ATPase mutant (p97E305Q, E578Q; p97QQ [Ye et al., 2001]). It has been shown that expression of this mutant inhibits p97-mediated retrotranslocation of various ER proteins from the ER membrane to the cytosol and, therefore, leads to their stabilization (Ye et al., 2001; Halawani and Latterich, 2006). Cells were transfected with MYC-tagged wild-type p97 (p97), MYC-tagged p97QQ (p97QQ), or pEYFP vector as a control (Control), followed by treatment with CHX for 30, 60, or 90 min approximately 24 h after transfection. Total cell lysates were analyzed by Western blot for the level of Mcl1 (Figure 2C). The data show that the expression of p97QQ results in an increase in Mcl1 levels (155.6 ± 23.3%, n = 6) compared with control cells. Furthermore, upon CHX-induced inhibition of protein synthesis, p97QQ significantly delayed Mcl1 degradation (after 90 min of CHX treatment, 70.6 ± 15.3% of the initial protein level of Mcl1 remained in p97QQ-expressing cells compared with 15.0 ± 7.3% in control cells and 12.3 ± 8.9% in p97-expressing cells; n = 4; see Figure 2, C and D). Thus, it appears that the ATPase domain of p97 is required for the regulation of Mcl1 degradation.
We also investigated whether the p97QQ-dependent delay in Mcl1 degradation is due to an effect on the overall proteasomal degradation pathway or on proteasomal degradation of Mcl1 in particular. To test this, control and p97- and p97QQ-expressing cells were treated with the proteasome inhibitor MG132, followed by Western blot analysis, as shown in Figure 2E. The data show that the rate of proteasome inhibition-induced accumulation of Mcl1 was not noticeably altered by p97QQ expression (Figure 2, E and F), indicating that overall proteasome function was not affected by the inhibition of p97. Therefore, we conclude that in Mcl1 degradation pathway, p97 acts upstream of the proteasome and may regulate mitochondrial steps of this process. Consistent with this conclusion, we found that the accumulation of Mcl1 and high-molecular-weight species of Mfn1 was apparent in mitochondrial fractions purified from p97 RNAi cells (Figure 2G), as compared with control RNAi cells. These results indicate that the inhibition of p97 not only stabilizes Mcl1 and Mfn1 but also hinders their movement from the OMM to the cytosol.
p97 regulates apoptosis-induced Mcl1 degradation
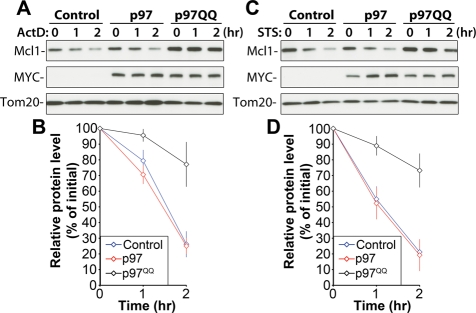
The data described above strongly support a role for p97 in the control of steady state levels of Mcl1 and Mfn1, two unrelated, short-lived, and proteasome-dependent OMM-associated proteins, and indicate a housekeeping role for p97 in the regulation of OMM protein turnover. We also sought to determine whether p97 participates in Mcl1 degradation under conditions of stress. Upon the activation of apoptosis, Mcl1 is rapidly degraded through the Ub/proteasome pathway. Disappearance of Mcl1, achieved by a combination of degradation and blocked synthesis, is associated with the onset of apoptosis (Yang et al., 1995; Cuconati et al., 2003; Nijhawan et al., 2003; Zhong et al., 2005). To determine whether apoptosis-induced degradation of Mcl1 is also regulated by p97, we applied two unrelated inducers of mitochondria-dependent apoptosis: actinomycin D (ActD), a DNA replication and transcription inhibitor, and staurosporine (STS), a general kinase inhibitor. Control and p97- and p97QQ-expressing cells were treated with ActD (Figure 3, A and B) or STS (Figure 3, C and D) for 1 or 2 h and then analyzed for levels of Mcl1 expression. The data show significant inhibition of Mcl1 degradation in both ActD-treated cells (after 2 h of ActD treatment, 77.11 ± 14.27% of the initial protein level of Mcl1 remained in p97QQ-expressing cells compared with 26.16 ± 8.26% in control cells and 25.03 ± 5.09% in p97-expressing cells, n = 3) and STS-treated cells (after 2 h of STS treatment, 73.21 ± 10.82% of the initial protein level of Mcl1 remained in p97QQ-expressing cells compared with 21.20 ± 8.09% in control and 19.22 ± 10.15% in p97QQ-expressing cells, n = 3) when p97QQ is expressed (Figure 3, B and D). These data indicate that p97 regulates the degradation of Mcl1 not only under normal growth conditions but also during stress.
FIGURE 3:
p97 inhibits apoptosis-induced degradation of Mcl1. Control HeLa cells or cells transfected with p97 or p97QQ were treated with ActD (20 μM; A) or STS (1 μM; C), two unrelated inducers of mitochondria-dependent apoptosis, for 0, 1, or 2 h, followed by Western blotting, as indicated. In panels B and D, relative protein levels in cells treated as described in panels A and C were quantified and plotted against the length of treatment with ActD (B) or STS (D). Data represent the mean ± SD of three independent experiments. Mcl1 levels detected in untreated cells (time, 0 min) were set at 100%.
p97 regulates the movement of Mcl1 from the mitochondria to the cytosol
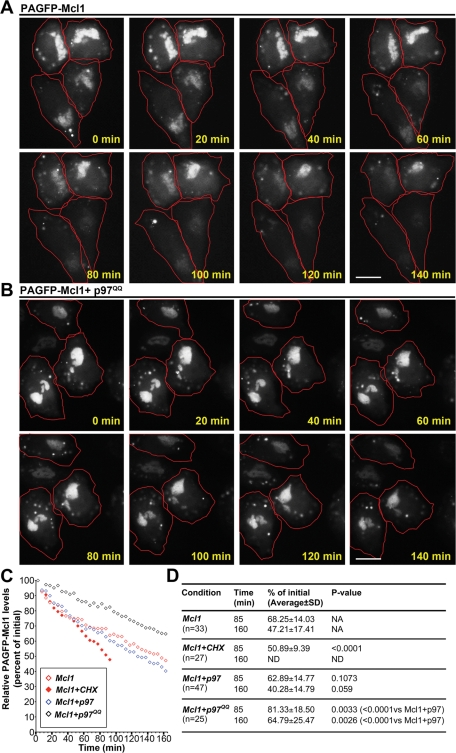
Considering the data described above showing mitochondrial accumulation of Mcl1 and Mfn1 in p97 RNAi cells (see Figure 2G), as well as the importance of p97 for the retrotranslocation of proteins from the ER membrane to the cytosol prior to proteasomal degradation (Ye et al., 2004; Bar-Nun, 2005; Jentsch and Rumpf, 2007), we tested whether the retrotranslocation of Mcl1 from the OMM to the cytosol is also mediated by p97. To do so, we constructed a mammalian expression vector containing Mcl1 fused with a photoactivable green fluorescent protein (PAGFP-Mcl1). PAGFP is a variant of the Aequorea victoria GFP that, after irradiation with 413-nm light, displays fluorescence increased 100-fold when excited with 488-nm light (Patterson and Lippincott-Schwartz, 2002). Because activated PAGFP is extremely stable (up to several days under aerobic conditions; Patterson and Lippincott-Schwartz, 2002) and the fluorescence produced reflects the level of PAGFP-fusion protein in specific subcellular locations (Karbowski et al., 2004), we used PAGFP-Mcl1 to assess the effects of p97 and p97QQ on the movement of Mcl1 from the mitochondria to the cytosol. In contrast to biochemical methods, this approach enables the detection of mitochondrial changes in Mcl1 levels in cells not treated with any compound (e.g., CHX or MG132) and, therefore, the estimation of steady-state rates of Mcl1 retrotranslocation from mitochondria.
Photoactivation of whole image fields in cells labeled with the vital mitochondrial probe Mitotracker CMXROS, followed by imaging with 488-nm light, revealed that PAGFP-Mcl1 correctly localized to the mitochondria (data not shown). To test the effect of p97 on the retrotranslocation of Mcl1 from the OMM, changes in the fluorescence of mitochondria-associated PAGFP-Mcl1 were quantified over time in cells cotransfected with PAGFP-Mcl1 and p97 or Mcl1 and p97QQ (transfection ratio 1:9), as well as in cells expressing only PAGFP-Mcl1 (Control) (Figure 4, A and B; data not shown). The data show that in control cells, mitochondrial levels of photoactivated PAGFP-Mcl1 (Figure 4A) gradually decreased, with a rate similar to that detected in p97-expressing cells (47.21 ± 17.41% vs. 40.28 ± 14.79% of the initial value at 160 min in control and p97-expressing cells, respectively; see Figure 4, A, C, and D). Importantly, the expression of p97QQ significantly delayed the decline in the levels of mitochondria-associated photoactivated PAGFP-Mcl1 (64.79 ± 17.41% of initial value at 160 min; Figure 4,B–D), indicating that p97QQ hinders the retrotranslocation of photoactivated PAGFP-Mcl1 from the mitochondria to the cytosol. Control experiments revealed that photobleaching rates of photoactivated PAGFP-Mcl1 were much lower than the rate of fluorescence decrease observed in the long-term imaging experiments described above (approximately 7% of the initial value after 60 image acquisitions with light intensity comparable to the experiments shown in Figure 4, A and B; data not shown). Thus, the decreases detected in the mitochondrial pool of PAGFP-Mcl1 are unlikely to be due to nonspecific photobleaching. This notion is further supported by results found after the inhibition of protein synthesis and therefore inhibition of the mitochondrial import of newly synthesized endogenous Mcl1 and nonactivated PAGFP-Mcl1. Protein synthesis inhibition significantly increased the elimination of the mitochondrial pool of PAGFP-Mcl1 (after 85 min, 68.25 ± 14.03% of the initial level remained vs. 50.89 ± 9.39% of the initial value in untreated and CHX-treated control cells, respectively; see Figure 4, C and D). However, due to the high background noise, we were not able to detect the accumulation of PAGFP-Mcl1 in the cytosol, even when cells were treated with proteasome inhibitor MG132 (data not shown). Thus, based on these data, one can only conclude that the changes in the fluorescence intensity of PAGFP-Mcl1 reflect the role of p97 in the PAGFP-Mcl1 degradation. However, these data also suggest that p97 might also regulate Mcl1 retrotranslocation from the mitochondria to the cytosol, with subsequent dilution of PAGFP-Mcl1 fluorescence in the cytosol.
FIGURE 4:
p97 regulates the retrotranslocation of Mcl1 from the mitochondria to the cytosol. HeLa cells transfected with PAGFP-Mcl1 (A) or cotransfected with PAGFP-Mcl1 and p97QQ (B) were imaged over time, after whole imaging fields were photoactivated with a short pulse of UV light. Note the gradual decrease in mitochondrial PAGFP-Mcl1 fluorescence in cells expressing only PAGFP-Mcl1 (A) and the relatively stable mitochondrial PAGFP-Mcl1 fluorescence in cells expressing p97QQ (B). Cells are outlined in red. Bars in panels A and B: 20 μm. In panel C, the averages of mitochondrial PAGFP-Mcl1 fluorescence changes in numerous imaging experiments (see panel D) were plotted as a function of time. In panel D, mitochondrial PAGFP-Mcl1 fluorescence changes were quantified, and the mean ± SD and P values obtained with unpaired, two-tailed t-tests are shown (see figure for details).
We tested the hypothesis that p97 mediates retrotranslocation of Mcl1 from the OMM further by analyzing endogenous Mcl1 in mitochondrial and cytosolic fractions isolated from control and p97- and p97QQ-expressing cells treated with CHX to inhibit protein synthesis (Figure 5A). The data show a gradual decrease in the mitochondrial pool of Mcl1 in CHX-treated control and p97-expressing cells (Figure 5A, left panels). In contrast, mitochondria-associated Mcl1 was stabilized by p97QQ expression (Figure 5A, left panels). Consistent with an OMM-specific function of p97, p97QQ expression did not noticeably affect the degradation of the Mcl1 already present in the cytosol (Figure 5A, right panels). We have also found that, in ActD-treated cells, p97QQ expression stabilized mitochondria-associated, but not cytosolic, Mcl1 (Figure 5B), suggesting a conserved p97-dependent mechanism underlying steady state and stress-induced degradation of Mcl1. On the basis of these data, we conclude that p97 acts specifically on the mitochondria and controls the retrotranslocation of Mcl1 from the OMM for its subsequent degradation by the proteasome.
FIGURE 5:
In vitro recapitulation of p97-dependent retrotranslocation of Mcl1. (A, B) Mitochondria-enriched heavy membrane (left panels) and cytosolic fractions (right panels) obtained from control and p97- and p97QQ-expressing cells treated with CHX for 0, 30, 60, or 90 min (A) or ActD for 0, 60, or 120 min were analyzed for levels of endogenous Mcl1 protein. Expression of exogenous p97 and p97QQ was detected with anti-MYC tag monoclonal antibody (MYC). Tom20 was used as a loading control. (C) Mitochondrial fractions isolated from p97 RNAi cells (lines 1–5) were combined with cytosolic fractions isolated from control RNAi (lines 1, 4, 5) or p97 RNAi cells (lines 2 and 3) and incubated at 35°C for 30 min (lines 2 and 4) or for 60 min (lines 3 and 5) or left on ice for 60 min (line 1). Following the incubation, samples were centrifuged (20,000 × g for 30 min), supernatants were removed, and pellets (15 μg of protein) were analyzed by Western blotting for levels of Mcl1 and p97. Tom20 was used as a loading control. (D) Cytosolic fractions isolated from control RNAi cells were incubated with ATP (lines 1 and 2), ATP-γ-S (lines 3), or MG132 (lines 4), and then combined with mitochondrial fractions obtained from p97 RNAi cells and incubated at 35°C for 60 min (lines 2–4) or left on ice for 60 min (line 1). Following incubation, samples were centrifuged (20,000 × g for 30 min), and supernatants (cytosolic fractions; 45 μg) and pellets (mitochondrial fractions; 15 μg) were analyzed by Western blotting for levels of Mcl1 and p97. Tom20 was used as a loading control. In panel E, the amount of p97 in cytosolic fractions obtained from control RNAi and p97 RNAi cells, which was used in experiments described in panel C, is shown. In panel F, quantification of Mcl1 levels in two independent experiments described in panels C and D is shown (data represent the mean ± SD). *, unknown/cross-reactive band detectable in cytosolic fractions incubated with anti-Mcl1 antibody.
Recent studies have shown that p97-dependent retrotranslocation of proteins from the ER membrane to the cytosol can be recapitulated using isolated microsomes and p97-containing cytosol (Wahlman et al., 2007; Garza et al., 2009). We tested whether retrotranslocation of Mcl1 from the OMM could also be recapitulated using an in vitro reconstituted system and whether this process is p97-dependent. To this end, Mcl1-enriched and p97-depleted mitochondrial fractions isolated from p97 RNAi cells (as shown in Figure 2G) were combined with cytosolic fractions obtained from either control RNAi cells (with normal levels of p97; Figure 5C) or p97 RNAi cells (p97-depleted; Figure 5C). Samples consisting of p97 RNAi mitochondria mixed with p97 RNAi or control RNAi cytosol were incubated for 30 or 60 min at 35°C or for 60 min on ice, followed by centrifugation and Western blot analysis of p97, Mcl1, and Tom20 protein levels in the pellets (Figure 5C). The data show that membrane-associated Mcl1 levels were unaltered in p97 RNAi cytosol-treated mitochondrial fractions incubated at 35°C for 30 or 60 min (Figure 5C, lines 2 and 3). However, the levels of membrane-associated Mcl1 in control RNAi cytosol-treated mitochondrial fractions were notably reduced at 30 and 60 min of incubation at 35°C (Figure 5C, lines 4 and 5). Importantly, incubation of control RNAi cytosol-treated mitochondrial fractions for 60 min on ice resulted in Mcl1 recovery similar to that detected in mitochondria treated with p97 RNAi cytosol and incubated at 35°C (Figure 5C, compare line 1 with lines 2 and 3). On the other hand, we detected increased membrane association of p97 in all samples containing cytosol from control RNAi cells (Figure 5C, lines 1, 4, and 5) compared with samples containing p97 RNAi cytosol (Figure 5C, lines 2 and 3). We also tested the effects of ATP-γ-S, a substrate and inhibitor of ATP-dependent enzymes that is hydrolyzed very slowly by and affects activities of most ATPases, including p97 (Briggs et al., 2008; Tang et al., 2010). Cytosolic fractions obtained from control RNAi cells were incubated with ATP-γ-S for 30 min on ice, prior to addition to the mitochondrial fraction obtained from p97 RNAi cells and incubation at 35°C for 60 min. The data show that ATP-γ-S noticeably inhibited p97-mediated retrotranslocation of Mcl1 from the mitochondria to the cytosol (Figure 5D), further supporting the notion that p97 ATPase activity is essential for p97 function in the retrotranslocation of Mcl1. Furthermore, we found that proteasome inhibition does slightly inhibit p97-mediated retrotranslocation of Mcl1 (Figure 5D), indicating that Mcl1 retrotranslocation might be, to some degree, synchronized with proteasomal degradation of this protein. However, the degree of this inhibition is much less pronounced than the effect of ATP-γ-S (Figure 5D, compare lines 3 and 4; left panels). In addition, Western blot analysis of the Mcl1 levels in cytosolic fractions (Figure 5D, right panels) revealed an accumulation of this protein in the MG132-incubated samples, as compared with ATP- or ATP-γ-S-treated samples (Figure 5D). These data indicate that p97 regulates movement of Mcl1 from the OMM to the cytosol and that cytosolic Mcl1 is rapidly degraded by the proteasome. Quantification of the changes in mitochondrial Mcl1 levels in all analyzed samples is shown in Figure 5F.
In summary, these data indicate that the p97-dependent retrotranslocation of Mcl1 can be reconstituted outside an intact cell and therefore support a direct mitochondrial mechanism of p97-dependent regulation of Mcl1 retrotranslocation. Furthermore, because the incubation temperature affected the p97-dependent retrotranslocation of Mcl1, but not the association of p97 with the membrane fraction, it appears that the retrotranslocation of Mcl1, but not the mitochondrial membrane association of p97, depends on the enzymatic activity of p97 (see Figure 5C).
Indeed, studies using similar approaches have shown that p97 acts on the ER membrane and mediates the retrotranslocation of misfolded substrates of the ERAD pathway or the turnover of ER resident proteins (Wahlman et al., 2007; Garza et al., 2009; Soetandyo and Ye, 2010). Therefore, one can conclude that p97 acts on the mitochondria in a manner resembling its well-established mechanism of action in the ER.
Spatial relationship between p97 and mitochondria
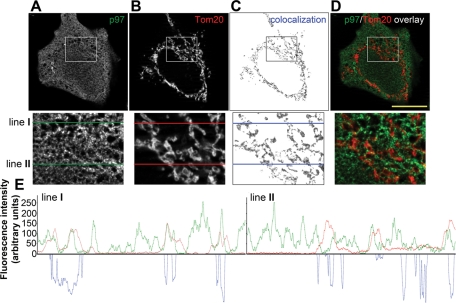
The data described above clearly show that p97 is required for the proteasomal degradation of OMM-localized Mcl1 and Mfn1. Furthermore, the data shown in Figure 5C suggest that the association of p97 with the OMM is required for Mcl1 retrotranslocation, suggesting that, under normal growth conditions, a subset of p97 might be associated with mitochondria. Published reports indicate that p97 is a ubiquitous protein, distributed in the nucleus, cytosol, and membrane compartments of the cell, including the ER and the Golgi complex (Chi et al., 1995; Kobayashi et al., 2002). Using structured illumination fluorescence microscopy, we analyzed the spatial relationship between endogenous p97 (Figure 6A) and Tom20, an OMM marker (Figure 6B). Consistent with published data, our results show that p97 localizes to the nucleus, cytosol, and cellular membranes (Figure 6A). Importantly, we found that a subset of p97 was also colocalized or in close association with the mitochondria (Figure 6D). In addition, “colocalizing” pixels extracted from images of p97 and Tom20 immunolabeled cells (image areas positive for green-p97-derived and red-Tom20-derived pixels; Figure 6, C) closely resemble the staining pattern of Tom20, suggesting that the association between p97 and Tom20. Furthermore, the line scans performed using the lines shown in magnified insets in Figure 6, A–C (lines I and II) further confirm a close spatial association between p97 and the OMM (Figure 6E). Although these descriptive data do not prove that p97 acts at the OMM, we believe that the partial mitochondrial localization of p97, in combination with the data shown in earlier sections of this work, provides evidence supporting a direct mitochondrial role for p97. In addition, we found that p97 interacts with USP30, an OMM-associated deubiquitinating protein (Nakamura and Hirose, 2008), as well as Mfn1 (a substrate of p97-dependent degradation, as detailed above) (data not shown), further connecting p97 with the OMM. These data have been also supported by the proteomic studies showing mitochondrial association of p97 in highly purified mitochondrial fractions (Taylor et al., 2003; Reifschneider et al., 2006; Zhang et al., 2008).
FIGURE 6:
Spatial relationship between p97 and the OMM. HeLa cells were immunostained to detect p97 (A and green fluorescence on overlay images shown in panel D) and Tom20, a marker of the OMM (B and red fluorescence on overlay images shown in panel D), followed by image acquisition and analysis using Zeiss AxioVision Colocalization software (Zeiss MicroImaging). The colocalizing pixels, representing the subcellular areas within a representative z-section containing both p97 and Tom20, are shown in panel C. The bottom images in panels A–D show magnified areas (bounded by squares). In panel E, line scans along the lines marked with I and II in the magnified areas taken from panels A–C are plotted (red lines represent Tom20 fluorescence pattern, green lines represent p97 fluorescence patterns, and blue represents colocalization of the green and red channels in pixels along lines I and II). Bar in D: 20 μm.
DISCUSSION
The findings described here extend the emerging understanding of the mechanism and role of the Ub/proteasome system in the regulation of mitochondrial homeostasis. Several recently published reports indicate that Ub-dependent degradation or regulation of mitochondrial proteins is vital for the maintenance of mitochondrial function (Yonashiro et al., 2006; Karbowski et al., 2007; Narendra et al., 2008; Neutzner et al., 2008; Benard et al., 2010; Ziviani et al., 2010). For example, Parkin, an IBR-type E3 Ub ligase, is recruited to mitochondria and participates in the autophagy of dysfunctional mitochondria (Narendra et al., 2008). In D. melanogaster, Parkin appears to regulate the stress-induced degradation of dMfn (Ziviani et al., 2010), a fly homologue of mammalian Mfn proteins. Moreover, we and others have also shown that a number of critical mitochondrial events, including mitochondrial membrane remodeling, mitochondrial communication with other organelles, and mitochondrial steps involved in apoptosis, are also regulated by E3 Ub ligases (Yonashiro et al., 2006; Karbowski et al., 2007; Margineantu et al., 2007; Neutzner et al., 2007, 2008; Neuspiel et al., 2008). These reports clearly indicate a critical role for the Ub/proteasome system in the regulation of mitochondrial homeostasis. However, the molecular steps coordinating mitochondrial and cytosolic events leading to the proteasomal degradation of OMM-associated proteins are not clear.
In this work, we show that p97, an AAA-ATPase with an established role in the retrotranslocation of ER proteins from the ER membrane to the cytosol (Ye et al., 2001, 2004), is also required for the retrotranslocation of OMM proteins. Impairing the function of p97 leads to the stabilization and mitochondrial accumulation of Mcl1 and Mfn1, mitochondrial substrates of p97. These data significantly increase our current understanding of the outer mitochondrial membrane–associated degradation (OMMAD) pathway. Although we have previously proposed that the proteasomal degradation of OMM-associated proteins (a yeast homologue of Mfn1, Fzo1p, in particular) might require a membrane extraction step before Fzo1p is degraded in the cytosol (Neutzner et al., 2007, 2008), the mechanism and factors required for this process were, until now, unknown. Therefore, the data shown here that link p97 with the retrotranslocation of OMM-proteins establish a new part of the OMMAD pathway. Exactly how p97-driven substrate release from the OMM is linked to mitochondrial substrate ubiquitination, the cofactors needed for the mitochondrial function of p97, and the extent of p97’s role in the regulation of mitochondrial proteostasis need to be clarified. The current work in our lab focuses on these important issues.
Nonetheless, the exceptional importance of the OMM to the integration of mitochondria into cell signaling pathways as well as to mitochondrial function has been underscored by findings that this compartment hosts proteins central to the regulation of apoptosis, mitochondrial membrane dynamics, and other important pathways. Therefore, we believe that the identification of p97 as a critical factor in OMM protein homeostasis, reported here, is an important discovery.
MATERIALS AND METHODS
Cell culture and transfection
HeLa cells were cultured in DMEM medium supplemented with 10% heat-inactivated fetal bovine serum, 2 mM Glutamax, 1 mM sodium pyruvate, MEM nonessential amino acids (Invitrogen, Carlsbad, CA), 100 U/ml penicillin, and 100 mg/ml streptomycin in 5% CO2 at 37°C. Cells were transfected with FuGeneHD transfection reagent (Roche, Indianapolis, IN), according to the manufacturer’s instructions. The fine-tuned transfection conditions resulted in at least 80% of cells being transfected.
Cloning and shRNAi
PCR fragments containing the gene encoding Mcl1 were generated using the proofreading Pfx DNA polymerase (Invitrogen, Carlsbad, CA) and a commercially available cDNA clone (OriGene, Rockville, MD) as the template. The primers used were 5′-GGCGGCGGAATTCAATGTTTGGCCTCAAAAG-3′and 5′-ACTTACAGGATCCCTATCTTATTAGATATG-3′. PCR fragments were purified, digested with the appropriate restriction enzymes, and cloned into a PAGFP encoding mammalian expression vector (Patterson and Lippincott-Schwartz, 2002; Karbowski et al., 2004) between EcoRI and BamHI restriction sites to generate PAGFP-Mcl1. MYC-tagged wild-type p97 and p97QQ mammalian expression vectors were described previously (Yang et al., 2010). GFP shRNAi and p97 shRNAi constructs and GFP control shRNAi were purchased from Sigma-Aldrich (St. Louis, MO). Cells were transfected with respective constructs, and then at ∼24 h after transfection, to select transfected cells, they were incubated with 3 μg/ml puromycine for an additional 48–60 h.
Immunofluorescence, fluorescence microscopy, and image analysis
Immunofluorescence and fluorescence microscopy analyses were performed as previously described (Benard et al., 2010). The primary antibodies used for immunofluorescence studies were anti-Tom20 polyclonal antibody (Santa Cruz Biotechnology, Santa Cruz, CA) and anti-p97 (BD Biosciences, Franklin Lakes, NJ). Immunofluorescence labeling was performed as previously described (Benard et al., 2010), with one modification. Prior to blocking with bovine serum albumin (BSA), nonspecific sites were blocked with Image-iTFX signal enhancer (Invitrogen). We found that using this reagent prior to BSA blocking dramatically increased the signal-to-noise ratio in fluorescence images. For live cell imaging experiments, images were acquired using a Zeiss AxioObserver Z1 fluorescence microscope, equipped with a 100/1.45 a-Plan-FLUAR objective lens (Zeiss MicroImaging, Thornwood, NJ), an ApoTome unit (enabling high-resolution structured illumination image acquisition), environment control units (for temperature and pH control), a Definitive Focus module, and a high-sensitivity CCD camera (QuantEM 512SC; Photometrics, Tucson, AZ). Using this system, image acquisition took approximately 50–100 ms per channel, with low illumination levels enabling the acquisition of several hundred images without considerable photobleaching or cytotoxicity. Colocalization analyses were performed using the colocalization module of AxioVision 4 software (Zeiss MicroImaging).
Western blot, in vitro retrotranslocation assay, and immunoprecipitation
Cells were harvested, and total cell protein lysates and subcellular fractions were prepared as previously described (Karbowski et al., 2007). Protein lysates were analyzed by Western blot using anti-p97 polyclonal antibody (Cell Signaling, Denvers, MA), anti-Mcl1 polyclonal antibody (Cell Signalling), anti-Tom20 polyclonal antibody (Santa Cruz Biotechnology), anti-MYC tag monoclonal antibody (Roche), anti-BclXL rabbit monoclonal antibody, anti-Bak polyclonal antibody (Upstate), and anti-VDAC1 polyclonal antibody (Cell Signaling). For in vitro retrotranslocation assay, mitochondrial fractions were isolated from p97 RNAi cells, as previously described (Karbowski et al., 2007). Cytosolic fractions (20,000 × g supernatants) were isolated from either control RNAi or p97 RNAi cells. To initiate the reactions, mitochondrial fractions (300 μg) were combined with cytosolic fractions (600 μg) preincubated with ATP (2 mM), ATP-γ-S (2 mM), or MG132 (20 μM) for 30 min on ice (for details, see Figure 5D). Samples were then incubated at 35°C for 30 or 60 min or left on ice for 60 min. Following the incubations, samples were centrifuged (20,000 × g for 30 min at 4°C), and after centrifugation, supernatants were carefully removed. Supernatants and pellets were immediately solubilized in SDS–PAGE sample buffer and then analyzed by Western blotting.
Immunoprecipitation under denaturing conditions was performed as previously described (Benard et al., 2010). For quantification of protein levels, Western blots were scanned and intensities of specific proteins were quantified using ImageJ software, as described (Benard et al., 2010).
Protein concentration was measured directly in the samples using NanoDrop 1000 spectrophotometer (Thermo Scientific, West Palm Beach, FL).
Acknowledgments
We thank Pamela Wright for insightful comments on the manuscript. We also gratefully acknowledge financial support from the National Institute of General Medical Science (grant RO1 GM083131, to M.K.).
Abbreviations used:
- OMM
outer mitochondrial membrane
- OMMAD
outer mitochondrial membrane–associated degradation
- Ub
ubiquitin
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E10-09-0748) on November 30, 2010.
REFERENCES
- Acharya U, Jacobs R, Peters JM, Watson N, Farquhar MG, Malhotra V. The formation of Golgi stacks from vesiculated Golgi membranes requires two distinct fusion events. Cell. 1995;82:895–904. doi: 10.1016/0092-8674(95)90269-4. [DOI] [PubMed] [Google Scholar]
- Azad N, Vallyathan V, Wang L, Tantishaiyakul V, Stehlik C, Leonard SS, Rojanasakul Y. S-nitrosylation of Bcl-2 inhibits its ubiquitin-proteasomal degradation. A novel antiapoptotic mechanism that suppresses apoptosis. J Biol Chem. 2006;281:34124–34134. doi: 10.1074/jbc.M602551200. [DOI] [PubMed] [Google Scholar]
- Ballar P, Zhong Y, Nagahama M, Tagaya M, Shen Y, Fang S. Identification of SVIP as an endogenous inhibitor of endoplasmic reticulum-associated degradation. J Biol Chem. 2007;282:33908–33914. doi: 10.1074/jbc.M704446200. [DOI] [PubMed] [Google Scholar]
- Bar-Nun S. The role of p97/Cdc48p in endoplasmic reticulum-associated degradation: from the immune system to yeast. Curr Top Microbiol Immunol. 2005;300:95–125. doi: 10.1007/3-540-28007-3_5. [DOI] [PubMed] [Google Scholar]
- Bays NW, Wilhovsky SK, Goradia A, Hodgkiss-Harlow K, Hampton RY. HRD4/NPL4 is required for the proteasomal processing of ubiquitinated ER proteins. Mol Biol Cell. 2001;12:4114–4128. doi: 10.1091/mbc.12.12.4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beattie DS, Basford RE, Koritz SB. The turnover of the protein components of mitochondria from rat liver, kidney, and brain. J Biol Chem. 1967;242:4584–4586. [PubMed] [Google Scholar]
- Benard G, Neutzner A, Peng G, Wang C, Livak F, Youle RJ, Karbowski M. IBRDC2, an IBR-type E3 ubiquitin ligase, is a regulatory factor for Bax and apoptosis activation. EMBO J. 2010;29:1458–1471. doi: 10.1038/emboj.2010.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun RJ, Zischka H, Madeo F, Eisenberg T, Wissing S, Buttner S, Engelhardt SM, Buringer D, Ueffing M. Crucial mitochondrial impairment upon CDC48 mutation in apoptotic yeast. J Biol Chem. 2006;281:25757–25767. doi: 10.1074/jbc.M513699200. [DOI] [PubMed] [Google Scholar]
- Braun S, Matuschewski K, Rape M, Thoms S, Jentsch S. Role of the ubiquitin-selective CDC48(UFD1/NPL4)chaperone (segregase) in ERAD of OLE1 and other substrates. EMBO J. 2002;21:615–621. doi: 10.1093/emboj/21.4.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs LC, Baldwin GS, Miyata N, Kondo H, Zhang X, Freemont PS. Analysis of nucleotide binding to P97 reveals the properties of a tandem AAA hexameric ATPase. J Biol Chem. 2008;283:13745–13752. doi: 10.1074/jbc.M709632200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulteau AL, Szweda LI, Friguet B. Mitochondrial protein oxidation and degradation in response to oxidative stress and aging. Exp Gerontol. 2006;41:653–657. doi: 10.1016/j.exger.2006.03.013. [DOI] [PubMed] [Google Scholar]
- Chi NC, Adam EJ, Adam SA. Sequence and characterization of cytoplasmic nuclear protein import factor p97. J Cell Biol. 1995;130:265–274. doi: 10.1083/jcb.130.2.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuconati A, Mukherjee C, Perez D, White E. DNA damage response and MCL-1 destruction initiate apoptosis in adenovirus-infected cells. Genes Dev. 2003;17:2922–2932. doi: 10.1101/gad.1156903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garza RM, Sato BK, Hampton RY. In vitro analysis of Hrd1p-mediated retrotranslocation of its multispanning membrane substrate 3-hydroxy-3-methylglutaryl (HMG)-CoA reductase. J Biol Chem. 2009;284:14710–14722. doi: 10.1074/jbc.M809607200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griparic L, Kanazawa T, Van Der Bliek AM. Regulation of the mitochondrial dynamin-like protein Opa1 by proteolytic cleavage. J Cell Biol. 2007;178:757–764. doi: 10.1083/jcb.200704112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halawani D, Latterich M. p97: The cell’s molecular purgatory? Mol Cell. 2006;22:713–717. doi: 10.1016/j.molcel.2006.06.003. [DOI] [PubMed] [Google Scholar]
- Ishihara N, Fujita Y, Oka T, Mihara K. Regulation of mitochondrial morphology through proteolytic cleavage of OPA1. EMBO J. 2006;25:2966–2977. doi: 10.1038/sj.emboj.7601184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jentsch S, Rumpf S. Cdc48 (p97): a “molecular gearbox” in the ubiquitin pathway? Trends Biochem Sci. 2007;32:6–11. doi: 10.1016/j.tibs.2006.11.005. [DOI] [PubMed] [Google Scholar]
- Karbowski M, Arnoult D, Chen H, Chan DC, Smith CL, Youle RJ. Quantitation of mitochondrial dynamics by photolabeling of individual organelles shows that mitochondrial fusion is blocked during the Bax activation phase of apoptosis. J Cell Biol. 2004;164:493–499. doi: 10.1083/jcb.200309082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karbowski M, Neutzner A, Youle RJ. The mitochondrial E3 ubiquitin ligase MARCH5 is required for Drp1 dependent mitochondrial division. J Cell Biol. 2007;178:71–84. doi: 10.1083/jcb.200611064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T, Tanaka K, Inoue K, Kakizuka A. Functional ATPase activity of p97/valosin-containing protein (VCP) is required for the quality control of endoplasmic reticulum in neuronally differentiated mammalian PC12 cells. J Biol Chem. 2002;277:47358–47365. doi: 10.1074/jbc.M207783200. [DOI] [PubMed] [Google Scholar]
- Margineantu DH, Emerson CB, Diaz D, Hockenbery DM. Hsp90 inhibition decreases mitochondrial protein turnover. PLoS ONE. 2007;2:e1066. doi: 10.1371/journal.pone.0001066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura N, Hirose S. Regulation of mitochondrial morphology by USP30, a deubiquitinating enzyme present in the mitochondrial outer membrane. Mol Biol Cell. 2008;19:1903–1911. doi: 10.1091/mbc.E07-11-1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura N, Kimura Y, Tokuda M, Honda S, Hirose S. MARCH-V is a novel mitofusin 2- and Drp1-binding protein able to change mitochondrial morphology. EMBO Rep. 2006;7:1019–1022. doi: 10.1038/sj.embor.7400790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra D, Tanaka A, Suen DF, Youle RJ. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuspiel M, Schauss AC, Braschi E, Zunino R, Rippstein P, Rachubinski RA, Andrade-Navarro MA, McBride HM. Cargo-selected transport from the mitochondria to peroxisomes is mediated by vesicular carriers. Curr Biol. 2008;18:102–108. doi: 10.1016/j.cub.2007.12.038. [DOI] [PubMed] [Google Scholar]
- Neutzner A, Benard G, Youle RJ, Karbowski M. Role of the ubiquitin conjugation system in the maintenance of mitochondrial homeostasis. Ann N Y Acad Sci. 2008;1147:242–253. doi: 10.1196/annals.1427.012. [DOI] [PubMed] [Google Scholar]
- Neutzner A, Youle RJ, Karbowski M. Outer mitochondrial membrane protein degradation by the proteasome. Novartis Found Symp. 2007;287:4–14. [PubMed] [Google Scholar]
- Ngo JK, Davies KJ. Importance of the lon protease in mitochondrial maintenance and the significance of declining lon in aging. Ann NY Acad Sci. 2007;1119:78–87. doi: 10.1196/annals.1404.015. [DOI] [PubMed] [Google Scholar]
- Nijhawan D, Fang M, Traer E, Zhong Q, Gao W, Du F, Wang X. Elimination of Mcl-1 is required for the initiation of apoptosis following ultraviolet irradiation. Genes Dev. 2003;17:1475–1486. doi: 10.1101/gad.1093903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson GH, Lippincott-Schwartz J. A photoactivatable GFP for selective photolabeling of proteins and cells. Science. 2002;297:1873–1877. doi: 10.1126/science.1074952. [DOI] [PubMed] [Google Scholar]
- Rape M, Hoppe T, Gorr I, Kalocay M, Richly H, Jentsch S. Mobilization of processed, membrane-tethered SPT23 transcription factor by CDC48(UFD1/NPL4), a ubiquitin-selective chaperone. Cell. 2001;107:667–677. doi: 10.1016/s0092-8674(01)00595-5. [DOI] [PubMed] [Google Scholar]
- Reifschneider NH, Goto S, Nakamoto H, Takahashi R, Sugawa M, Dencher NA, Krause F. Defining the mitochondrial proteomes from five rat organs in a physiologically significant context using 2D blue-native/SDS-PAGE. J Proteome Res. 2006;5:1117–1132. doi: 10.1021/pr0504440. [DOI] [PubMed] [Google Scholar]
- Schwickart M et al. Deubiquitinase USP9X stabilizes MCL1 and promotes tumour cell survival. Nature. 2010;463:103–107. doi: 10.1038/nature08646. [DOI] [PubMed] [Google Scholar]
- Soetandyo N, Ye Y. The p97 ATPase dislocates MHC class I heavy chain in US2 expressing cells via an Ufd1-Npl4 independent mechanism. J Biol Chem. 2010;285:32352–32359. doi: 10.1074/jbc.M110.131649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang WK, Li D, Li CC, Esser L, Dai R, Guo L, Xia D. A novel ATP-dependent conformation in p97 N-D1 fragment revealed by crystal structures of disease-related mutants. EMBO J. 2010;29:2217–2229. doi: 10.1038/emboj.2010.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor SW et al. Characterization of the human heart mitochondrial proteome. Nat Biotechnol. 2003;21:281–286. doi: 10.1038/nbt793. [DOI] [PubMed] [Google Scholar]
- Wahlman J, DeMartino GN, Skach WR, Bulleid NJ, Brodsky JL, Johnson AE. Real-time fluorescence detection of ERAD substrate retrotranslocation in a mammalian in vitro system. Cell. 2007;129:943–955. doi: 10.1016/j.cell.2007.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Liu C, Zhong Y, Luo S, Monteiro MJ, Fang S. Huntingtin interacts with the cue domain of gp78 and inhibits gp78 binding to ubiquitin and p97/VCP. PLoS ONE. 2010;5:e8905. doi: 10.1371/journal.pone.0008905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T, Kozopas KM, Craig RW. The intracellular distribution and pattern of expression of Mcl-1 overlap with, but are not identical to, those of Bcl-2. J Cell Biol. 1995;128:1173–1184. doi: 10.1083/jcb.128.6.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y, Meyer HH, Rapoport TA. The AAA ATPase Cdc48/p97 and its partners transport proteins from the ER into the cytosol. Nature. 2001;414:652–656. doi: 10.1038/414652a. [DOI] [PubMed] [Google Scholar]
- Ye Y, Shibata Y, Yun C, Ron D, Rapoport TA. A membrane protein complex mediates retro-translocation from the ER lumen into the cytosol. Nature. 2004;429:841–847. doi: 10.1038/nature02656. [DOI] [PubMed] [Google Scholar]
- Yonashiro R et al. A novel mitochondrial ubiquitin ligase plays a critical role in mitochondrial dynamics. EMBO J. 2006;25:3618–3626. doi: 10.1038/sj.emboj.7601249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J et al. Systematic characterization of the murine mitochondrial proteome using functionally validated cardiac mitochondria. Proteomics. 2008;8:1564–1575. doi: 10.1002/pmic.200700851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Q, Gao W, Du F, Wang X. Mule/ARF-BP1, a BH3-only E3 ubiquitin ligase, catalyzes the polyubiquitination of Mcl-1 and regulates apoptosis. Cell. 2005;121:1085–1095. doi: 10.1016/j.cell.2005.06.009. [DOI] [PubMed] [Google Scholar]
- Ziviani E, Tao RN, Whitworth AJ. Drosophila parkin requires PINK1 for mitochondrial translocation and ubiquitinates mitofusin. Proc Natl Acad Sci USA. 2010;107:5018–5023. doi: 10.1073/pnas.0913485107. [DOI] [PMC free article] [PubMed] [Google Scholar]