During meiosis, the APC/C is activated by either Cdc20 or the meiosis-specific activator Ama1. Upon exit from meiosis II, APC/CAma1 mediates Cdc20 destruction using Db1 and GxEN degrons. The amino terminus of Ama1, which contains the Cdc20-binding domain, is sufficient for Cdc20 degradation but not spore formation.
Abstract
The execution of meiotic divisions in Saccharomyces cerevisiae is regulated by anaphase-promoting complex/cyclosome (APC/C)–mediated protein degradation. During meiosis, the APC/C is activated by association with Cdc20p or the meiosis-specific activator Ama1p. We present evidence that, as cells exit from meiosis II, APC/CAma1 mediates Cdc20p destruction. APC/CAma1 recognizes two degrons on Cdc20p, the destruction box and destruction degron, with either domain being sufficient to mediate Cdc20p destruction. Cdc20p does not need to associate with the APC/C to bind Ama1p or be destroyed. Coimmunoprecipitation analyses showed that the diverged amino-terminal region of Ama1p recognizes both Cdc20p and Clb1p, a previously identified substrate of APC/CAma1. Domain swap experiments revealed that the C-terminal WD region of Cdh1p, when fused to the N-terminal region of Ama1p, could direct most of Ama1p functions, although at a reduced level. In addition, this fusion protein cannot complement the spore wall defect in ama1Δ strains, indicating that substrate specificity is also derived from the WD repeat domain. These findings provide a mechanism to temporally down-regulate APC/CCdc20 activity as the cells complete meiosis II and form spores.
INTRODUCTION
The anaphase-promoting complex/cyclosome (APC/C) is a highly conserved ubiquitin ligase that directs the destruction of key regulatory proteins necessary for proper mitotic and meiotic progression (reviewed in Yu, 2007). During mitotic cell division in budding yeast, APC/C activation and substrate specificity are directed by two highly conserved Trp-Asp (WD40) repeat proteins: Cdc20p and Cdh1p (Dawson et al., 1995; Schwab et al., 1997; Sigrist and Lehner, 1997; Visintin et al., 1997). Ama1p is the meiosis-specific APC/C activator (Chu et al., 1998; Cooper et al., 2000) that directs the ubiquitylation of the B-type cyclin Clb1p (Cooper et al., 2000) plus other unknown substrates (Rabitsch et al., 2001). APC/CAma1 activates Smk1p, the meiotic mitogen-activated protein kinase (MAPK) required for spore wall morphogenesis through an unknown mechanism (McDonald et al., 2005). Ama1p also coordinates exit from meiosis II and is required for the early stages of spore wall assembly (Rabitsch et al., 2001; Coluccio et al., 2004; Diamond et al., 2009).
To associate with the APC/C, activators contain two short motifs called the C-box (CB) and IR motifs (Schwab et al., 2001; Passmore et al., 2003; Vodermaier et al., 2003). Although the precise mechanism has not been elucidated, it has been proposed that the IR motif targets the activators to the APC/C, while the CB motif promotes an activating change in APC/C conformation that is independent of the activators’ substrate-recruiting function (Vodermaier et al., 2003; Dube et al., 2005; Kimata et al., 2008). Interestingly, the IR box of Cdc20p is not required for function in Saccharomyces cerevisiae but contributes to APC/C-dependent turnover (Thornton et al., 2006).
The recognition of substrates by the APC/C is more complex. Initially, it was proposed that the APC/C activators played an equivalent role to F-box proteins in the SCF ubiquitin ligase, serving as a bridge between the APC/C catalytic domain and the substrate (Deshaies, 1999). In support of this model, APC/C activators were shown to recognize specific degrons (destruction box, KEN box, A-box, and the destruction degron [GxEN]) on their target protein (Burton and Solomon, 2001; Burton et al., 2005; Hilioti et al., 2001; Pfleger et al., 2001a; Littlepage and Ruderman, 2002; Meyn et al., 2002; Castro et al., 2003; Kraft et al., 2003). However, more recent studies have reported that the core APC/C also recognizes substrates, although the presence of activators is almost always required for this binding (Carroll and Morgan, 2002; Carroll et al., 2005; Passmore et al., 2003; Passmore and Barford, 2005; Yamano et al., 2004; Eytan et al., 2006). Taken together, a current model proposes that substrate selection is aided by direct binding to the core APC/C itself. Although the exact mechanism for how this promotes substrate ubiquitylation remains unknown, many models have been proposed (reviewed in Yu, 2007). Recent evidence has favored a “multivalency” model in which activator binding to the APC/C (through its CB and IR motifs) creates a bipartite binding site for the substrate through the activator and the APC/C core (Matyskiela and Morgan, 2009). Activator binding in turn may also create a conformational change to the APC/C, promoting ubiquitylation of the substrate.
In vegetative cells, Cdc20p is transcribed from S phase through G2 phase and subjected to proteolysis in G1 (Fang et al., 1998a; Prinz et al., 1998; Shirayama et al., 1998; Huang et al., 2001; Morris et al., 2003). This proteolysis is mediated by APC/CCdh1 in late mitosis/early G1 and a Cdh1p-independent mechanism during G1/S (Goh et al., 2000; Robbins and Cross, 2010). In addition, APC/CCdc20 is negatively regulated by the binding of the spindle checkpoint protein Mad2p (reviewed in Bharadwaj and Yu, 2004) or by protein kinase A phosphorylation (Searle et al., 2004; Mallory et al., 2007). In contrast, Cdh1p is controlled by posttranslational mechanisms including cell cycle–dependent nuclear export (Jaquenoud et al., 2002), G1 cyclin–Cdc28p phosphorylation (Zachariae et al., 1998; Jaspersen et al., 1999), and specific inhibitors (Martinez et al., 2006; Dial et al., 2007; Choi et al., 2008; Crasta et al., 2008; Enquist-Newman et al., 2008; Hall et al., 2008; Ostapenko et al., 2008). Cdh1p is activated at anaphase when the Cdc28p-antagonizing phosphatase Cdc14p is released from the nucleolus (Anghileri et al., 1999). The meiosis-specific activator Ama1p is controlled at the level of transcription (Chu et al., 1998) and splicing (Cooper et al., 2000; Spingola and Ares, 2000). APC/CAma1 activity is inhibited by the APC/C subunit Mnd2p and B cyclin–Cdc28p kinase activity until late in the meiotic program (Oelschlaegel et al., 2005; Penkner et al., 2005; Carlile and Amon, 2008).
In this study, we demonstrate that APC/CAma1, and not APC/CCdh1, mediates the degradation of Cdc20p at the end of the second meiotic division. Genetic studies and coimmunoprecipitation assays identified two conserved degrons on Cdc20p (destruction box, GxEN) that are recognized by the divergent amino-terminus of Ama1p. Interestingly, replacing the C-terminal WD40 of Ama1p with that of Cdh1p provided partial APC/CAma1 function. These findings indicate that unique information is provided by both the N-terminal and carboxyl terminal domains of Ama1p.
RESULTS
Ama1p is required for Cdc20p destruction during meiosis
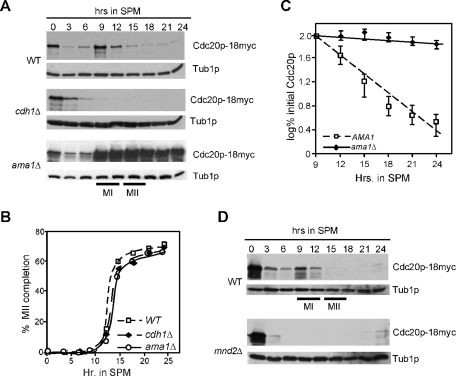
CDC20 mRNA levels decline as cells exit the mitotic cell cycle, followed by a transient induction during the first meiotic prophase, which peaked at the meiotic divisions (Chu et al., 1998). To investigate Cdc20p regulation during meiosis, a strain harboring a chromosomally amino-terminal tagged CDC20 allele (CDC20-18myc) was induced to enter meiosis, and time points were taken before the shift (0 h) until the completion of spore formation. Similar to its mRNA profile, Western blot analysis of protein samples prepared from each time point revealed a transient accumulation of Cdc20p peaking during the meiotic nuclear divisions (Figure 1A, top). In vegetative cells, Cdc20p is subjected to proteolysis in G1 mediated by APC/CCdh1 as well as an APC/C-independent mechanism (Prinz et al., 1998; Fang et al., 1998a; Shirayama et al., 1998; Huang et al., 2001; Morris et al., 2003). To determine whether Ama1p or Cdh1p plays a role in Cdc20p proteolysis during meiosis, the meiotic time course described above was repeated in homozygous diploid cells deleted for these genes. Despite progressing though meiosis with the same kinetics as wild type (Figure 1B), Cdc20p levels are at the limits of detection in the cdh1Δ strain during the meiotic divisions (Figure 1A, middle). Longer exposure of this blot revealed that Cdc20p is present but, unlike wild-type cells, does not seem to be up-regulated as the cells execute meiosis I and II. In addition, CDC20 mRNA is expressed normally in cdh1Δ strains (Supplemental Figure S1A). However, in ama1Δ cells, Cdc20p was not down-regulated as the cells exited from meiosis II (monitored by the appearance of tetra-nucleated cells; Figure 1B, quantitated in Figure 1C). These results suggest that APC/CAma1 is required for Cdc20p proteolysis as cells exit from meiosis II. They also suggest that, different from mitotic cell divisions, this degradation is not assisted by an APC/C-independent mechanism.
FIGURE 1:
APC/CAma1 is required for of Cdc20p destruction during meiosis. (A) Wild-type (RSY695), cdh1Δ (RSY1210), and ama1Δ (RSY853) strains harboring integrated Cdc20p-18myc were induced to enter meiosis and time points taken as indicated. Cdc20p-18myc levels were monitored by Western blot analysis. (B) The percent population of tetranucleated cells in the strains described in (A) are plotted. (C) Quantitation of the Cdc20p-18myc signal from (A) is plotted from the 9-h time point. The results shown are the averages from three separate experiments with error bars included. (D) Wild-type (RSY695) or mnd2Δ (KCY440) strains harboring integrated Cdc20p-18myc were induced to enter meiosis and time points taken as indicated. Western blot analysis of protein extracts was conducted to detect Cdc20p. The mnd2Δ strain remained mononucleated as previously described (Penkner et al., 2005). The blots were stripped and reprobed for Tub1p as a loading control. MI and MII indicate the approximate times of meiosis I (MI) and meiosis II (MII) as determined by DAPI analysis.
Loss of Ama1p or Cdc16p function leads to defects in meiotic progression (Cooper et al., 2000). Therefore one possible explanation is that the maintenance of Cdc20p levels in these mutants is an indirect consequence of this meiotic arrest. However, we and others (Diamond et al., 2009) have shown that the abundance of proteins that are normally degraded at the completion of meiosis II (Cooper et al., 2000; Carlile and Amon, 2008) are comparable in wild-type and ama1Δ strains. This suggests that the failure to destroy Cdc20p at the end of meiosis II in the ama1Δ mutant is directly due to lack of APC/CAma1 activity.
Mnd2p prevents premature meiotic Cdc20p degradation
The above results suggest a model in which Ama1p regulates Cdc20 degradation as the cells exit from meiosis II. Although Ama1p is synthesized early in the meiotic program (Cooper et al., 2000), it is kept inactive until Mnd2p is degraded (Oelschlaegel et al., 2005). Therefore it would be predicted that Cdc20p would be prematurely degraded during meiosis in mnd2Δ cells. To test this possibility, Cdc20p-18myc levels were followed in wild-type and mnd2Δ cultures during meiosis. The results show that Cdc20p fails to accumulate in mnd2Δ cells compared with the wild type (Figure 1D). Northern analysis confirmed that CDC20 mRNA was expressed in mnd2Δ cells (Supplemental Figure S1B). This result is consistent with the model that APC/CAma1 regulates Cdc20p degradation as cells exit from the meiotic divisions.
Ama1p is required for normal meiotic nuclear divisions
We had previously reported that ama1Δ cells arrest before meiosis I (Cooper et al., 2000). However, other studies using the SK1 background reported that Ama1p is dispensable for meiosis I or meiosis II (Coluccio et al., 2004; Oelschlaegel et al., 2005; Penkner et al., 2005; Diamond et al., 2009). Reexamination of our strain revealed that it does duplicate its spindle pole bodies (Figure 2A) indicative of nuclear divisions taking place. This is consistent with our previous finding that early and middle genes are expressed in ama1Δ cells but not loci required for spore wall formation (Cooper et al., 2000). However, 70% of the ama1Δ mutants exhibited uneven nuclear divisions (Figure 2A and “other” category in Figure 2B). Closer examination revealed that cells with aberrant nuclear divisions also possessed defective spindle formation (Figure 2C). To conclude, we concur with other published reports that ama1 mutants can execute meiotic divisions. However, the data presented here show that ∼20% ama1Δ mutants arrest before meiosis I and that the segregation of nuclei in cells that do undergo nuclear divisions is abnormal.
FIGURE 2:
Ama1p is required for normal meiotic divisions. (A) Wild-type (WT) (KCY224) and ama1Δ mutant (KCY225) harboring integrated Tub4p-GFP were induced to enter meiosis. Samples were fixed and analyzed by fluorescence microscopy at the time points indicated. Nuclear, spindle, and spindle pole body morphologies were determined by DAPI staining (top), indirect immunofluorescence of tubulin, and direct immunofluorescence of Tub4p-GFP, respectively. Magnification is 1000×. (B) Terminal meiotic arrest phenotype of ama1Δ mutant. WT and ama1Δ strains were induced to enter meiosis, and samples were taken after 24 h. Cells were scored as described in Materials and Methods. “Other” represents cells exhibiting irregular DAPI and tubulin staining. For all morphology quantitations presented, the SDs were ≤6% for all values. (C) Indirect immunofluorescence of tubulin and DAPI analysis of WT (RSY335) and ama1Δ (RSY562) cells after 24 h in sporulation medium. 1200× magnification.
APC/CAma1 activity is dependent on the conserved APC/C-binding motifs
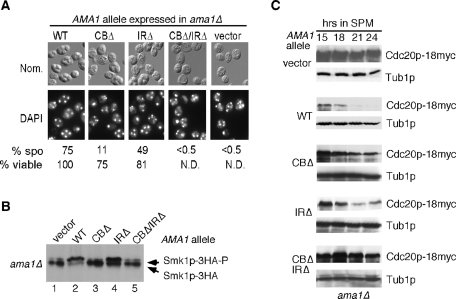
To further confirm a role for Ama1p in the meiotic destruction of Cdc20p, we examined the degradation of Cdc20p in nonfunctional ama1 mutants in which the conserved APC/C interaction motifs (CB and IR motifs) were deleted. Deletion of both of these motifs inactivated Ama1p, as indicated by the absence of viable spores (Figure 3A). Interestingly, the strain expressing the CBΔ mutant exhibited a sixfold reduction in spore formation with unevenly separated nuclei whereas the IRΔ mutant displayed a more modest defect in spore formation and nuclear spacing. Similar results were obtained when the ability of these mutants to activate the meiosis-specific MAPK Smk1p was tested (Figure 3B). As previously reported (McDonald et al., 2005), the ratio of active (phosphorylated) to inactive (unphosphorylated) forms of Smk1p is reduced in the ama1Δ background (compare lanes 1 and 2, Figure 3B).
FIGURE 3:
The C-box (CB) and IR motifs are required for normal meiotic nuclear divisions. (A) An ama1Δ culture (RSY562) harboring the AMA1 expression plasmids indicated above each panel was transferred to sporulation medium for 18 h and then analyzed by DAPI staining (bottom) or Nomarski imaging (top) to monitor nuclear segregation and spore formation, respectively. The percentage of sporulation was determined as described in Materials and Methods. The relative viability was scored with the wild-type (WT) value set at 100%. (B) The CB domain is required for Smk1p activation. SK1 diploid CMY15 (ama1Δ with an integrated SMK1-HA allele) harboring plasmids expressing either no Ama1p (vector), the WT T7-tagged Ama1p (pKC3036), mutant Ama1pCBΔ (pKC3045), Ama1pIRΔ (pKC3087), or Ama1pCBΔ/IRΔ (pKC3048) was induced to enter meiosis. After 10 h, samples were analyzed by Western blot using an HA antibody. The upper and lower immunoreactive species (arrows) correspond to the hyperphosphorylated (active) and hypophosphorylated (inactive) forms of Smk1p, respectively. (C) CB and IR motifs are required for Cdc20p destruction. Yeast strain RSY853 (ama1Δ, CDC20-18myc) harboring the same plasmids as (B) was induced to enter meiosis and time points were taken as indicated. Western blot analysis of protein extracts was conducted to detect Cdc20p. The blots were stripped and reprobed for Tub1p, which served as a loading control.
We next tested the requirement of the CB and IR motifs for Cdc20p degradation. Compared with the wild type, cultures expressing Ama1pCBΔ or Ama1pIRΔ displayed a modest defect in Cdc20p destruction (Figure 3C). However, Cdc20p was substantially protected from degradation in the Ama1pCBΔ/IRΔ double mutant (Figure 3C, bottom), suggesting that the phenotypes of these two mutations are additive. These results indicate that Ama1p activity is dependent on its conserved APC/C-binding motifs. In addition, our findings suggest that these domains are performing separate roles in mediating APC/CAma1 activity.
Cdc20p association with the APC/C is not required for its destruction
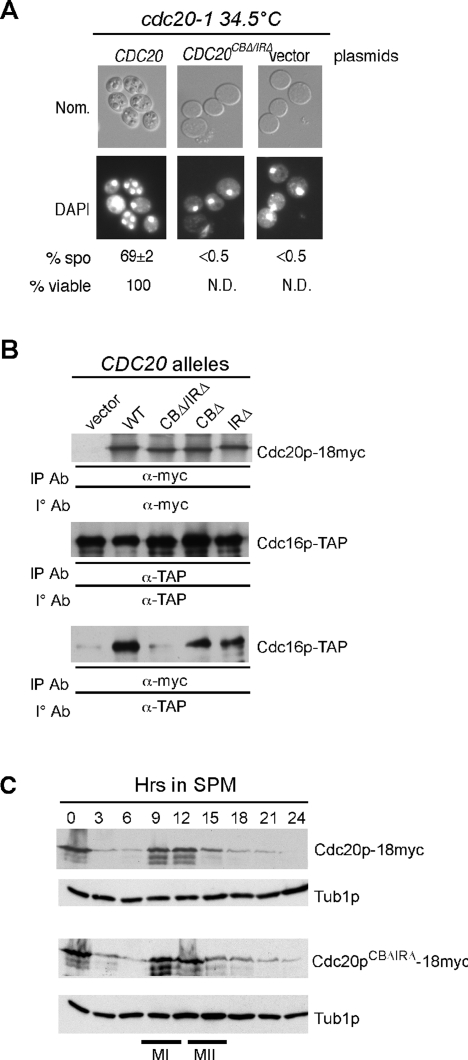
Cdc20p is both an activator and substrate of the APC/C. If these processes were separate, we would predict that APC/C association through its CB or IR domain would not be required for Cdc20p destruction. To test this model, both the CB and IR domains were mutated in a Cdc20p derivative and introduced into a cdc20-1 strain (RSY809). This culture, along with wild-type and vector controls, were induced to enter meiosis at the permissive temperature (23°C). After 4.5 h, and before the first meiotic division, the cells were switched to the restrictive temperature (34.5°C). Cells harboring the wild-type plasmid progressed through the meiotic program normally and produced viable spores (Figure 4A). Cells harboring either the vector or Cdc20pCBΔ/IRΔ-expressing plasmids arrested before the first meiotic division. As cdc20 mutants arrest before meiosis I with high Pds1p levels (Salah and Nasmyth, 2000), this result indicates that the CB and IR motifs are required for APC/CCdc20 activity during meiosis. To verify that the CB and IR motifs direct Cdc20p binding to the APC/C during meiosis, coimmunoprecipitation experiments were performed 9 h after the cells entered meiosis. These studies indicate that Cdc20pCBΔ/IRΔ is defective in APC/C binding (Figure 4B). Coimmunoprecipitation studies with the single CB or IR motif mutants revealed a subtle reduction in binding efficiency. These findings indicate that, as expected, the CB and IR motifs are required for Cdc20p-APC/C association during meiosis.
FIGURE 4:
Cdc20p binding to the APC/C is not required for its destruction. (A) The CB and IR regions are required for Cdc20p function. A cdc20-1 strain (RSY801) harboring plasmids expressing wild-type (WT) CDC20 or the CDC20CBΔ/IRΔ alleles were induced to enter meiosis and then shifted to the restrictive temperature to inactivate Cdc20-1p. After 24 h, samples were taken for DAPI analysis. (B) The CB and IR regions are required for Ama1p binding to the APC/C. The Cdc16-TAP–expressing strains (RSY1055) harboring either vector, WT (pMSC8), or mutant versions of Cdc20p (CBΔ/IRΔ = pKC5022, CBΔ = pKC5020, IRΔ = pKC5021) were induced to enter meiosis and cells harvested for analysis at 9 h, when both CDC16 and CDC20 are expressed (Cooper et al., 2000). Western blot analysis was conducted to detect the presence of Cdc16p-TAP in Cdc20p-18myc immunoprecipitates. The top and middle panels control for protein expression. (C) WT (RSY1055) strains harboring either Cdc20p-18myc (pMSC7) or Cdc20CBΔ/IRΔ-18myc (pKC5065) centromere plasmids were induced to enter meiosis and time points taken as indicated. Western blot analysis of protein extracts was conducted to detect Cdc20p. The blots were stripped and reprobed for Tub1p, which served as a loading control.
We next examined the expression profile of Cdc20pCBΔ/IRΔ during meiosis. No differences were observed in the accumulation or degradation of Cdc20pCBΔ/IRΔ compared with the wild type (Figure 4C). These results indicate that Cdc20p destruction occurs independently of activator-binding motifs. In addition, no defect in meiotic progression, as measured by the appearance of bi- and tetranucleated cells, was noted, indicating that Cdc20pCBΔ/IRΔ did not exhibit a dominant-negative effect (unpublished data). These observations are consistent with a model in which Ama1p recognizes Cdc20p as a substrate, recruiting it to the APC/C for degradation as cells exit from meiosis II.
Two destruction motifs are utilized for APC/CAma1-mediated destruction of Cdc20p
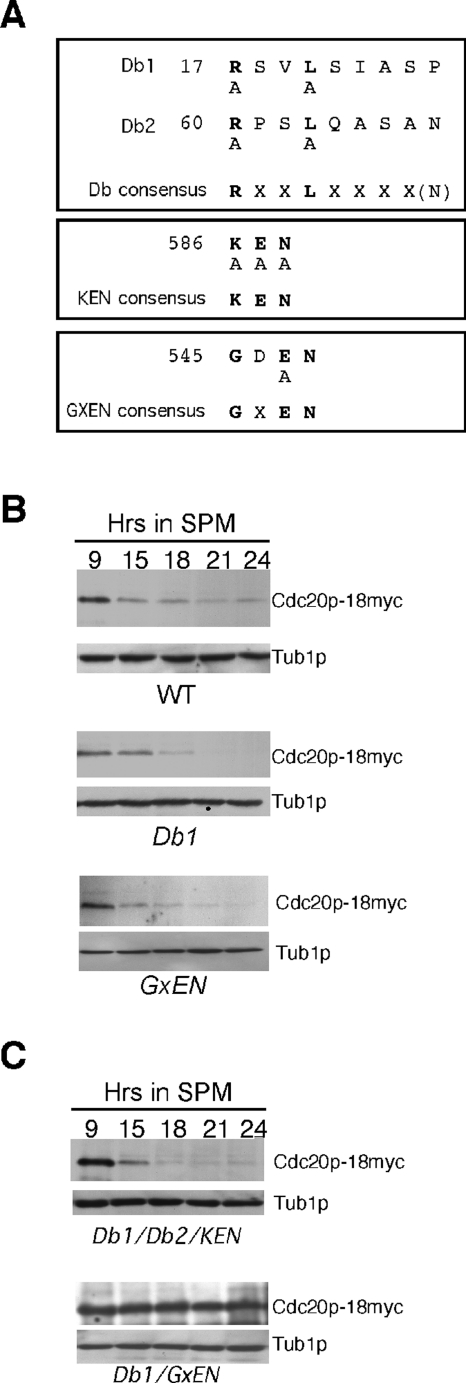
Cdc20p contains four motifs (two destruction boxes, one KEN box, and GxEN; see Figure 5A) previously implicated in APC/C-directed ubiquitylation (Glotzer et al., 1991; Pfleger and Kirschner, 2000; Littlepage and Ruderman, 2002). Destruction box 1 (Db1) mediates partial Cdc20p degradation during mitotic divisions (Prinz et al., 1998; Goh et al., 2000). To address which of these motifs are required for Cdc20p proteolysis during meiotic progression, the mutations described in Figure 5A were introduced individually into CDC20-18myc (pMsc7). Wild-type cells expressing these various mutant proteins were then induced to enter meiosis and their degradation profiles monitored by Western analysis. The results show that individually mutating these elements did not alter Cdc20p-18myc degradation profiles (Figure 5B; Supplemental Figure S2, A and B; and quantitated in Supplemental Figure S2C), indicating that none of these motifs are solely responsible for Cdc20p degradation.
FIGURE 5:
Identification of Cdc20p degrons. (A) Location of destruction boxes, KEN and GxEN motifs, in Cdc20p. Amino acid substitutions corresponding to the mutations generated are shown below the sequence. (B) Cdc20p deleted for either Db1 or GxEN is destroyed during meiosis with the same kinetics as wild type. Wild-type cells (RSY335) harboring the Cdc20p-18myc centromere plasmids indicated (see Supplemental Table S3 for details) were induced to enter meiosis and samples taken at the time points shown for Western analysis. The blots were stripped and reprobed for Tub1p as a loading control. (C) Cdc20p utilizes two degrons (Db1 and GxEN) for its destruction during meiosis. Cells were treated as described above except that Cdc20p harbored the mutations indicated.
A subset of APC/C substrates (e.g., Clb2p, Hsl1p) can require more than one degron for their efficient destruction (Burton and Solomon, 2001; Hendrickson et al., 2001). To determine whether this is also the case for meiotic Cdc20p degradation, wild-type cultures harboring plasmids bearing mutations in different combinations were examined as described above. The results show that combining the Db1, Db2, and KEN degrons does not significantly affect the rate of Cdc20p degradation compared with the wild type (Figure 5C and quantitated in Supplemental Figure S2D). However, combining the GxEN and Db1 mutations stabilized Cdc20p similarly to that observed in ama1Δ cells (compare to Figure 1A). These results indicate that either the Db1 or GxEN degron can mediate the meiotic destruction of Cdc20p. As before, meiotic progression was assessed by the appearance of bi- and tetranucleated cells. Again, no difference in the rate of meiotic progression was noted in cultures expressing the Db1/GxEN double mutant (unpublished data), suggesting that Cdc20p destruction is not essential for meiotic progression (discussed below).
GxEN does not mediate mitotic Cdc20p degradation
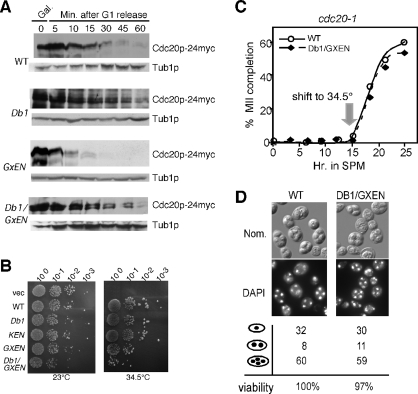
Db1 was previously shown to mediate partial destruction of Cdc20p in vegetative cells (Prinz et al., 1998). To investigate whether GxEN is also involved in the mitotic destruction of Cdc20p, we examined the stability of Cdc20pGxEN following release from α-factor–induced G1 arrest in wild-type vegetative cultures (Prinz et al., 1998). Before release, wild-type strains expressing galactose-inducible CDC20 alleles (CDC20, CDC20Db1, CDC20GxEN, CDC20Db1/GxEN) were treated with glucose and cycloheximide to stop transcription and translation of these genes, respectively (see Materials and Methods for details). Samples were taken and Western blot analysis was used to monitor Cdc20p-24myc levels. As previously reported (Prinz et al., 1998; Goh et al., 2000), partial stabilization of Cdc20p was observed in the Db1Δ strain (Figure 6A). However, mutating GxEN did not affect Cdc20p destruction compared with the wild type. Mutating both Db1 and GxEN did not appear to stabilize Cdc20p more than the single Db1Δ mutation. Interestingly, all the galactose-inducible CDC20 constructs are functional as determined by their ability to complement the temperature-sensitive growth defect associated with a cdc20-1 mutation (Shirayama et al., 1999 and Figure 6B). However, strains expressing the Db1/GxEN double mutant exhibited a reduced plating efficiency compared with strains expressing the wild type or any single mutant allele (Figure 6B), suggesting that overexpression of the double mutant is deleterious to mitotic cell division. To conclude, these experiments show that Db1 is required for normal Cdc20p destruction during mitotic cell division, but both Db1 and GxEN are active in meiotic cells.
FIGURE 6:
The GxEN degron does not function in mitotic cells. (A) Wild-type cells (RSY10) harboring either WT (pUS995), Db1 (pKC5006), GxEN (pKC5009), or Db1/GxEN (pKC5016) galactose-inducible Cdc20p-24myc plasmids were grown in raffinose to mid-log phase, arrested in G1, and then induced by the addition 2% galactose. Degradation of Cdc20p was monitored by Western analysis at the time points indicated following G1 release. The blots were stripped and reprobed for Tub1p as loading control. (B) The Cdc20p Db1, KEN, GxEN, and Db1/GxEN mutants are functional. Mid-log phase cdc20-1 cultures (RSY809) containing either vector (pRS426) or galactose-inducible Cdc20p constructs (derived from functional Cdc20p-24myc CEN plasmid [pUS995] using the oligonucleotides described in Supplemental Table S2) were grown in raffinose, serial diluted (1:10), and spotted onto 2% galactose medium selecting for plasmid maintenance at 23°C (permissive) and the restrictive temperatures (34.5°C). Images were collected following 72 h incubation. (C) Meiotic progression was monitored in cdc20-1 cultures (RSY809) expressing either WT CDC20 (pKC5071) or the DB1/GxEN mutant (pKC5072) under the control of AMA1 promotor. The cells were induced to enter meiosis at the permissive temperature (23°C) and switched to the restrictive temperature after 15 h in SPM. The cells were analyzed by DAPI staining to monitor the appearance of tetranucleated cells. (D) Nomarski imaging (top) and DAPI staining (bottom) of cells described in (C) after 24 h in SPM. The percentage of mono-, bi-, and tetranucleated cells in each culture was determined by DAPI analysis. The relative spore viability was scored with the WT value set at 100%.
Cdc20p destruction is not required for meiotic progression
Our results suggest that Cdc20p is destroyed by APC/CAma1-mediated degradation upon exit from the second meiotic division. As described in Figure 2, Ama1p is required for normal meiotic nuclear divisions and spore morphogenesis. To determine whether these phenotypes are the result of the failure to destroy Cdc20p, we placed wild-type CDC20 (pKC5069) and the Db1/GxEN double mutant (pKC5070) under the control of AMA1 promotor on single-copy plasmids. The AMA1 promotor was chosen as AMA1 and CDC20 are transcribed at similar times during meiosis. Moreover, placing CDC20 under the control of a meiotic promotor alleviates potential meiotic effects due to the vegetative growth defects associated with Cdc20pDb1ΔGxENΔ expression (see Figure 6B). This mutant CDC20 allele was able to complement the cdc20-1 meiotic phenotype, allowing the cells to form tetrads at wild-type rates at both permissive and restrictive temperatures (unpublished data). In vegetative cells, stabilizing Cdc20p impacts cell division only when combined with significant overexpression (Shirayama et al., 1999; Robbins and Cross, 2010). Thus we repeated these experiments using high-copy versions of the same plasmids (see Supplemental Table S3). In addition to examining the efficiency of spore formation, meiotic progression was also monitored by 4′,6-diamino-2-phenylindole (DAPI) analysis. There was no change in the appearance of tetranucleated cells (Figure 6C) or spore formation/viability (Figure 6D). This result was not due to plasmid loss as the vector was still present in dissected spore colonies (unpublished data). Taken together, these data indicate that Cdc20p destruction is not required for exit from meiosis II or spore formation.
The Db1 and GxEN degrons of Cdc20p are recognized by Ama1p during meiosis
If Cdc20p is a substrate of APC/CAma1, it should interact with Ama1p in vivo. As association with Ama1p could lead to Cdc20p destruction, coimmunoprecipitation studies were conduced in a strain harboring the nonfunctional allele of Ama1p (Ama1pCBΔ/IRΔ). Samples were taken following 9 h in sporulation medium (SPM), when both Ama1p and Cdc20p are expressed. The results show that Cdc20p can coimmunoprecipitate with Ama1p (Figure 7A, lanes 2 and 5), suggesting that these two proteins interact in vivo. This interaction was not simply a result of the protein extraction procedure as coimmunoprecipitation between Ama1pCBΔ/IRΔ and Cdc20p was not observed when these proteins were isolated from separate extracts and then mixed (Supplemental Figure S3). An alternative scenario is that Cdc20p and Ama1p are simply binding two independent sites on the APC/C. To rule out this explanation, the experiment was repeated with the Cdc20pCBΔ/IRΔ, which cannot associate with the APC/C (Figure 4B). These results also indicate that Ama1p can interact with Cdc20pCBΔ/IRΔ (Figure 7A, lane 3), further supporting the model that Cdc20p is a substrate of APC/CAma1.
FIGURE 7:
The Db1 and GxEN degrons direct Ama1p association. (A) An ama1Δ mutant (RSY562) harboring different Ama1p and Cdc20p expression plasmids (Ama1pCBΔ/IRΔ, pKC3048; Cdc20p, pMSC8; Ama1pCBΔ/IRΔ, pKC3048; Cdc20pCBΔ/IRΔ, pKC5022; vector, pRS424) as indicated was induced to enter meiosis and cells were harvested for analysis at 9 h, when both AMA1 and CDC20 are expressed (Cooper et al., 2000). Immunoprecipitation and Western blot analyses were conducted to detect the presence of both proteins (top and middle) or coimmunoprecipitation (bottom). (B) Coimmunoprecipitation experiments performed as described in (A) were repeated except with the inclusion of the Cdc20pDB1/GxEN-18myc (pKC5023) expression plasmid. The no-antibody mock immunoprecipitation is represented by [ ].
Our previous results indicate both Db1 and GxEN motifs on Cdc20p can mediate its meiotic destruction. To address whether Ama1p recognizes these degrons, the coimmunoprecipitation experiments described above were repeated with either wild-type Cdc20p or Cdc20pDb1/GxEN and Ama1CBΔ/IRΔ. The results (Figure 7B) show that the interaction between Ama1p and Cdc20pDb1/GxEN is diminished compared with wild-type Cdc20p (compare lanes 3 and 4, bottom). Given the nonspecific interaction observed between Cdc20p-18myc and protein G beads in these experiments (also see Figure 8B), it is difficult to assign any significance to the apparent association between Cdc20pDb1/GxEN and Ama1CBΔ/IRΔ. However, the genetic results presented in Figure 5 are consistent with a model that a functional interaction with Ama1p is mediated through the Db1 and GxEN degrons on Cdc20p.
FIGURE 8:
The N-terminus of Ama1p binds to Cdc20p. (A) Wild-type cultures (RSY335) containing the indicated expression plasmids (CDC20-18myc, pMsc8; GST-Ama1p1–200, pKC3070; GST, pQYAC-GST) were induced to enter meiosis. After 9 h, the cells were harvested for coimmunoprecipitation and Western blot analysis as indicated. The top and middle panels control for protein expression. The bottom panel assays coimmunoprecipitation. (B) The coimmunoprecipitation experiments described in (A) were repeated with ama1Δ (RSY562) cells harboring the indicated expression plasmids (vector, pRS426; Ama1201–596p-T7, pKC3084; Cdc20–18myc, pMsc8; Ama1pCBΔ/IRΔp-T7, pKC3048). (C) Clb1p, a previously identified substrate of Ama1p (Cooper et al., 2000), also associates with the amino-terminus of Ama1p. As (A) except that cells were harvested after 12 h and GST-Ama1p1–200 was coimmunoprecipitated with Clb1p-3HA (pKC430) after 12 h. In all panels, [°] represents the no-antibody mock immunoprecipitation.
The N-terminal domain of Ama1p directs Cdc20p association
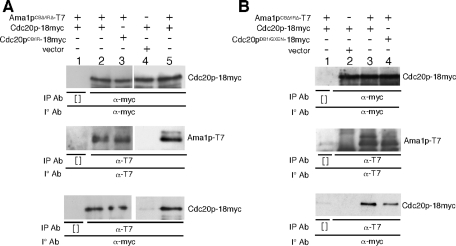
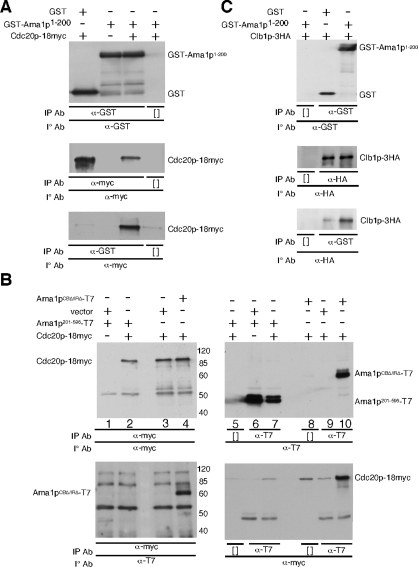
To identify the Ama1p domain(s) sufficient for Cdc20p interaction, expression constructs were generated containing either the diverged amino third of Ama1p (codons 1–200) or the more conserved carboxyl region containing WD40 repeats (codons 201–596). As the 1–200 portion of Ama1p is unstable when expressed independently (unpublished data), this region was fused to glutathione S-transferase (GST) under the control of the ADH1 promoter. These plasmids, and the appropriate controls, were transformed into a wild-type strain carrying an integrated copy of CDC20-18myc. After 9 h in SPM, samples were collected and protein extracts prepared for coimmunoprecipitation studies. These experiments revealed that GST-Ama1p1–200 associated with Cdc20p-18myc, whereas GST could not (Figure 8A). These results indicate that the amino-terminal region of Ama1p is sufficient for Cdc20p association.
Previous studies have indicated that the Cdh1p WD40 domain plays a role in degron recognition in Xenopus extracts (Kraft et al., 2005; Kimata et al., 2008). To determine whether the Ama1p WD40 region also recognizes Cdc20p, coimmunoprecipitation experiments were repeated with the Ama1p201–596 deletion mutant. Extracts were prepared as just described and immunoprecipitated with the myc monoclonal antibody (mAb). Western blot analysis of these immunoprecipitates did not detect Ama1p201–596 although Ama1pCBΔ/IRΔ was observed as before (Figure 8B, bottom, lanes 2 and 4). No difference in the expression levels of Ama1p201–596 and Ama1pCBΔ/IRΔ was detected (compare lanes 6 and 7 with lane 10), suggesting that the WD region did not associate directly with Cdc20p. To verify this conclusion, these experiments were repeated except that the Ama1p derivative immunoprecipitates were probed for the presence of Cdc20p-18myc. As expected, a strong Cdc20-18 myc signal was observed in Ama1pCBΔ/IRΔ-containing extracts. However, the Ama1p201–596 signal was not above the no-antibody control (compare lanes 7 and 8). These negative results do not exclude the possibility that the WD40 region interacts with Cdc20p. However, they do show that if an interaction exists, it is below the limits of detection using coimmunoprecipitation analysis of protein extracts.
Previously we have shown that APC/CAma1 mediates the destruction of the B-type cyclin Clb1p as cells complete the meiotic nuclear divisions (Cooper et al., 2000). To determine whether the amino-terminal domain of Ama1p also binds Clb1p, the coimmunoprecipitation experiments described in Figure 8A were repeated using Clb1p-3HA as the substrate. The results (Figure 8C) show that Clb1p does coimmunoprecipitate with GST-Ama1p1–200 above the background level with GST alone. Taken together, these data indicate that the amino terminus of Ama1p is sufficient to bind Cdc20p and Clb1p. However, GST-Ama11–200 is unable to complement the sporulation defect of an ama1Δ strain (unpublished data), indicating that the WD40 region is also necessary for Ama1p function (discussed below).
Ama1-Cdh1 hybrid protein promotes APC/CAma1-specific activity
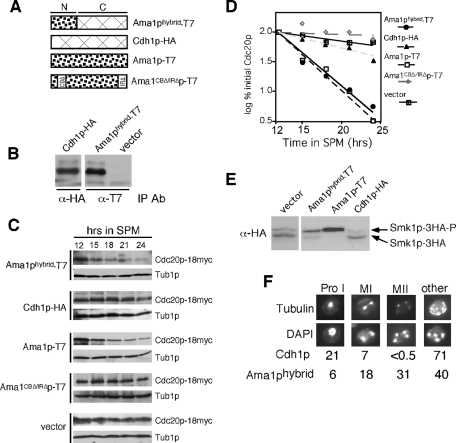
To further investigate the role of the conserved WD40 domain in Ama1p activity, a domain swap experiment was performed. A fusion protein was made that contained the amino-terminal domain of Ama1p fused to the WD40-containing region of Cdh1p (Ama1phybrid; see Figure 9A). As a control, hemagglutinin (HA) epitope–tagged CDH1 was placed under the control of the Ama1p promoter (Cdh1p-HA). Both of these constructs produced full-length protein at the same time during meiosis (Figure 9B). These constructs, plus plasmids expressing wild-type Ama1p, Ama1pCBΔ/IRΔ, or vector, were introduced into an ama1Δ mutant host, and three APC/CAma1-specific activities were assayed. The results show that the hybrid protein was able to direct Cdc20p-18myc destruction with kinetics similar to the wild type (Figure 9C and quantitated in Figure 9D). Next, the ability of the hybrid protein to activate Smk1p was determined. Ama1phybrid expression was able to stimulate Smk1p activation as determined by the increased ratio of phosphorylated to unphosphorylated species of the MAP kinase above vector control levels (Figure 9E) but not to the extent observed in the wild type. We also examined the ability of the hybrid protein to promote normal meiotic nuclear divisions and spore wall assembly. Expression of Ama1phybrid was able to stimulate meiosis I and meiosis II well above levels observed in the vector control (31% vs. <0.5%, respectively). However, this level was still below the wild-type value (75%; see Figure 3A), suggesting again that the hybrid protein was able to provide partial Ama1p function. However, the hybrid protein did not allow spore wall assembly (Figure 9F), as scored by the appearance of natural fluorescence observed when yeast spore walls are exposed to UV light (unpublished data). These data suggest that either the hybrid protein cannot sufficiently activate Smk1p to promote spore wall assembly or that another, unknown function of Ama1p is required that is not provided by the hybrid protein. Finally, to assess the contribution of the Cdh1p WD40 domain for Ama1p-specific activities, we tested the functionality of Cdh1p in these assays. When expressed from the Ama1p promoter, Cdh1p was unable to induce Cdc20p destruction (Figure 9C), activate Smk1p (Figure 9E), or promote meiotic nuclear divisions (Figure 9F). Likewise, Cdh1p under the control of its own promotor also could not complement the ama1Δ null defect (unpublished data). These findings suggest that the WD40 domain is interchangeable with respect to Cdc20p destruction, Smk1p activation, and nuclear divisions although the efficiency of function is reduced (see Discussion). However, the hybrid protein was unable to promote detectable spore wall formation. These results suggest that specific information included in the Ama1p WD40 region is required for function.
FIGURE 9:
The Ama1-Cdh1p fusion protein partially complements ama1Δ phenotypes. (A) Diagram depicting the proteins tested. N represents the amino-terminal divergent region, C the carboxyl terminus–conserved WD40 repeat domain. All constructs are under the control of the AMA1 promotor and terminator. (B) The expression of the AMA1-hybrid and AMA1pro-CDH1 constructs were verified in meiotic cultures. Extracts prepared from samples harvested 9 h following transfer to SPM were immunoprecipitated, and then Western blots were performed with the indicated antibodies. (C) Cdc20p levels were followed in meiotic RSY853 (ama1Δ, Cdc20-18myc) harboring either the Ama1-hybrid (pKC3077), Cdh1p-HA (pKC3078), Ama1p-T7 (pKC3056), or Ama1CboxΔ/IRΔp-T7 (pKC3057) expression plasmids or vector control. (D) The Cdc20p signals obtained in (C) were quantitated and graphed. All Cdc20p values were compared with Tub1p levels to control for protein quantitation. (E) Smk1p activation assays were performed on CMY15 containing either an empty vector, AMA1-T7, AMA1-hybrid, or AMA1pro-CDH1–expressing plasmids as described in Figure 3B. (F) An ama1Δ culture (RSY562) harboring plasmids expressing either Cdh1p-HA or Ama1p-hybrid was induced to enter meiosis and harvested for analysis after 12 h in SPM. Nuclei and spindle microtubules were visualized by DAPI (bottom) and indirect immunofluorescence (top), respectively. The number of cells seen at each stage of meiosis was scored as a percentage of the total cells counted. 1000× final magnification.
DISCUSSION
In this study, we demonstrate that the meiosis-specific APC/CAma1 activity destroys Cdc20p at the end of meiosis II. Moreover, unlike mitotic cell division, APC/C-dependent ubiquitylation is the primary pathway leading to Cdc20p degradation. We show that Ama1p activity is mediated through its conserved APC/C-binding CB and IR motifs, although individually neither domain is essential for Ama1p function. Using a combination of genetic and biochemical assays, we demonstrate that Ama1p recognizes two degrons, Db1 and GxEN, to dock with Cdc20p. The N-terminal domain of Ama1p mediates this activator/substrate interaction as well as interaction with another APC/CAma1 substrate, Clb1p. Finally, domain swap experiments reveal that specific information is contained within the WD region of Ama1p that is required for spore wall formation.
Model for substrate recognition by APC/C activators
It has been proposed that APC/C activators may have two functionally distinct roles: APC/C activation and substrate recognition (Kimata et al., 2008). This protein family is structurally similar in that they contain a divergent amino third of the protein, with the remaining two-thirds containing the conserved WD repeats. Currently there is no consensus as to which region orchestrates substrate binding (Pfleger et al., 2001a; Sorensen et al., 2001; Burton et al., 2005). The results from several studies support a model that the N-terminal region dictates target specificity while the C-terminal domain is more involved in interacting with APC/C components (Zhang and Lees, 2001; Burton et al., 2005). Consistent with this possibility, our data demonstrate that the amino-terminal region of Ama1p binds Cdc20p through the Db1 and GxEN degrons. However, in vitro cross-linking studies provided evidence that the carboxyl-terminal WD40 region directly interacts with degrons (reviewed in Thornton et al., 2006; Benanti and Toczyski, 2008). However, we could not detect an interaction between Cdc20p and the C-terminal region of Ama1p (Figure 8C). One explanation is that the interaction strength between the WD40 repeats and the degrons is below the limits of detection using this assay. Alternatively, the different methods used to detect protein–protein interactions may be the source of this discrepancy. Interestingly, domain swap experiments in which we combined the amino-terminal domain of Ama1p with the Cdh1p WD40 region provided most, but not all, of APC/CAma1 function in three different assays. These findings would suggest that the WD40 region plays a more general role in providing APC/C activator function. However, expression of the hybrid protein was not able to complement the spore wall assembly defect found in ama1Δ mutants. This finding strongly argues against the notion that the WD40 domains are completely interchangeable. Rather, it supports the notion that the WD40 domain of Ama1p also plays a specific role in substrate recognition. Therefore the precise role the C-terminal WD40 region plays with respect to Ama1p activity remains undefined. One possibility is that this region could be required for binding to the chaperonin-containing TCP1 (Valpuesta et al., 2002; Camasses et al., 2003), an event that is essential in activating Cdc20p and Cdh1p during mitotic cell divisions (Camasses et al., 2003).
Relationship between Cdh1p and Ama1p during meiosis
Our results show that APC/CAma1, and not APC/CCdh1, down-regulates Cdc20p as cells complete meiosis II. However, Cdh1p is expressed during meiosis, with increased amounts of the activator being present as cells execute the meiotic divisions (our unpublished data and Chu et al., 1998). How APC/CCdh1 activity is regulated during meiosis remains unknown. One possibility is that Acm1p, an inhibitor of Cdh1p (Martinez et al., 2006; Choi et al., 2008; Enquist-Newman et al., 2008; Ostapenko et al., 2008) whose mRNA is also expressed during meiosis, plays a role (Chu et al., 1998). In support of this idea is the observation that ACM1 has two Ndt80p consensus sites, suggesting that it is up-regulated after meiotic recombination is executed (Chu et al., 1998). Another intriguing observation resulting from our studies is that progression through the meiotic program is similar in wild-type and cdh1 strains (as assessed by DAPI and mRNA profiles). Despite this, Cdc20p levels are significantly down-regulated earlier in meiotic cdh1Δ cells. If APC/CAma1 triggers this event, this would suggest that, in cdh1Δ cells, APC/CAma1 is precociously activated similar to what we observe in an mnd2Δ mutant. A possible mechanism for this could be that APC/CCdh1 regulates a protein whose activity regulates Mnd2p. In this scenario, Mnd2p is precociously inactivated in cdh1Δ cells, thus allowing APC/CAma1 to destroy Cdc20p before the meiotic divisions are completed. Alternatively, APC/CCdh1 could regulate a protein that directly activates Ama1p. Thus, in the absence of APC/CCdh1 activity, Ama1p is precociously activated to destroy Cdc20p. However, it has been proposed that both the removal of Mnd2p and the inactivation of Cdk1 are required for APC/CAma1 activation as cells exit meiosis (Diamond et al., 2009). So do both these events have to occur for APC/CAma1 to be precociously active? The answers to this question may be addressed by studying the role Cdh1p plays during meiotic divisions in more detail.
The APC/C and exit from meiosis
APC/C regulation is critical for proper execution of the meiotic divisions as unscheduled APC/C activity can lead to missegregated chromosomes and aneuploid gametes. Recently it was suggested that APC/C inactivation at the end meiosis is critical for embryonic development in Drosophila (Pesin and Orr-Weaver, 2007). Here the meiosis-specific APC/C activator CORT (also known as CORTEX; Chu et al., 2001) is destroyed by APC/CFZY (Cdc20p homologue) at the completion of meiosis in the early embryo (Pesin and Orr-Weaver, 2007). Here we provide a mechanism to inactivate APC/CCdc20 as cells exit from meiosis II in S. cerevisiae. In this model system, Cdc20p is destroyed by APC/CAma1-mediated degradation, utilizing either the destruction box and/or GxEN motifs as degrons. However, the destruction of this activator is not essential, as introducing stabilized alleles of CDC20 did not affect spore viability. Intriguingly, FZY levels are not reduced in the early embryo (Pesin and Orr-Weaver, 2007). If inhibiting APC/C is important for normal gametogenesis, then these results suggest that APC/CCdc20 can be inactivated by alternative mechanisms. For example, dephosphorylation of core APC/C subunits, possibly by PP1 or PPa2, decreases APC/C activity, although the mechanism of this inhibition is not known (reviewed in Harper et al., 2002). Alternatively, or in addition, the association of APC/C inhibitors may play a role in down-regulating this ubiquitin ligase. Taken together, these results suggest a model in which inhibiting APC/CCdc20 activity at the end of meiosis is important for normal development and that this task is accomplished using multiple mechanisms including Ama1p-dependent destruction in yeast. Interestingly, Ama1p protein levels also decrease as cells exit from meiosis II (Cooper et al., 2000). It will be of interest to address whether this down-regulation is also APC/C mediated.
MATERIALS AND METHODS
Yeast strains and culture conditions
The strains used in this study (Supplemental Table S1) are isogenic to RSY335 (Cooper et al., 1999) except RSY10, a W303 derivative (see Figure 6, A and B) (Strich et al., 1989; Cooper et al., 1997), and CMY15, an SK1 strain used for Smk1 assays (E. Winter) (McDonald et al., 2005). Our wild-type parent strain (RSY335) is derived from the SK1 and W303 backgrounds. Strains harboring epitope-tagged CDC20-18myc were constructed using the integrating plasmid from W. Zachariae or derivatives thereof. RSY1210 (cdh1::LEU2) was made using pWS176 (W. Seufert). All other deletion strains were made by the method outlined by Longtine et al. (1998). Details are available upon request. Meiotic time course and mitotic culture conditions experiments were conducted as previously described (Cooper et al., 1997). To permit cdc20-1 cultures to exit mitosis and enter meiosis, the cells were maintained at the permissive temperature (23°C) and then shifted to the restrictive temperature (34.5°C) for the times indicated in the text. Cdc20p stability experiments in G1-arrested cultures were performed as described (Prinz et al., 1998).
Plasmids
Supplemental Tables S2 and S3 list the oligonucleotides and plasmids described in this study, respectively. Details of plasmid constructions are available on request. In brief, the Cdc20-myc18 plasmids pMSC7 (CEN) and pMSC8 (2μ) were derived from pCDC20-myc18 (W. Zachariae). From these plasmids, mutations were introduced using the QuikChange Site-directed Mutagenesis (SDM) Kit (Stratagene) according to the manufacturer’s protocol. All introduced mutations were verified by DNA sequencing (MWG/Operon). For the galactose induction experiments (Figure 6), mutations were introduced into pUS995 (galactose-inducible Cdc20p-24myc) (U. Surana). The constitutive GST-Ama11–200 fusion construct (ADH1 promotor; Figure 8A) was made by inserting the PCR-amplified AMA11–600 open reading frame into pQYAC (Quattromed) (oligonucleotides KCO171 and KCO173) to make pKC3070. The ADH1 GST vector (pQYAC-GST) was made by inserting the BamHI fragment from pKC3070, containing just the GST open reading frame, into pQYAC. Galactose-inducible GST-Ama1 fusion constructs were made by introducing AMA1 into pEG[KT], which contains GST under the control of the galactose promotor (a gift from M. Solomon).
The epitope-tagged derivative of AMA1 (pKC3036) was constructed by inserting a single 10–amino acid T7 epitope tag at the amino terminus by SDM. The T7-tagged AMA1-CDH1 hybrid gene was constructed by introducing a PstI site into pKC3036 at amino acid 200 using oligonucleotides KCO204 and KCO205. Next, the PstI restriction site in the promoter of AMA1 was deleted using oligonucleotides KCO217 and KCO218 to form pKC3061. The carboxyl terminus of AMA1 was then removed by excising the internal PstI fragment to form pKC3062. The carboxyl terminus of CDH1 was inserted into pKC3062 by introducing a PstI restriction site into pVZCDH1 (W. Seufert) at amino acid 234 using oligonucleotides KCO230 and KCO231 to make pKC6000. The 900–base pairs fragment encoding Cdh1p234–566 was then inserted into the PstI site of pKC3062 to make pKC3064 (called Ama1phybrid-T7). To place CDH1 under the control of AMA1 promoter, the SpeI/SacI fragment from pWS216 (W. Seufert) containing functional CDH1-3HA was placed downstream of the AMA1 promoter in pRS424 (Christianson et al., 1992) to form pKC3067 (called Cdh1p-HA).
Protein extract preparation, coimmunoprecipitation, and Western analysis
Protein extracts for coimmunoprecipitation and Western blot analyses were prepared as described (Cooper and Strich, 1999), except recombinant Protein G agarose beads (Invitrogen) were used instead of Protein A resin. Western blot analysis and coimmunoprecipitation experiments were conducted with 100 μg and 1 mg soluble protein, respectively. Epitope-tagged derivatives were visualized as follows: Myc and HA by using mouse mAb (Roche) at a final concentration (FC) of 2 μg/ml; T7 with mouse mAb (Novagen) at an FC of 0.1 ng/ml; TAP with rabbit polyclonal antibody (Open Biosystems) at an FC of 0.4 μg/ml; and Tub1p (tubulin) using rabbit polyclonal antibody (V. Guacci) at an FC of 0.1 μl/ml. Signals were quantitated using a Kodak Image Station 4000R.
Other techniques
Quantitation of meiosis I and II execution was achieved by analyzing DAPI-stained cells as described (Akamatsu et al., 1998; Cooper and Strich, 2002). At least 200 cells were counted per time point. For determining percent sporulation of ama1Δ cells harboring various mutant plasmids, at least three independent isolates were sporulated as described above and the results presented as the mean with standard deviation (SD). Spindle morphology was determined using indirect immunofluorescence as described previously (Cooper et al., 2000) on meiotic cells fixed in 4% paraformaldehyde and 3.4% sucrose for 15 min. Cells were scored as follows: prophase I, spindle smaller than nucleus; metaphase I, spindle the same width as nucleus; pseudo-anaphase, spindle masses separate with elongated, but not separated, nuclei; and fragmented nuclei, more than four DAPI-staining bodies, irregular-sized DAPI staining bodies, and/or misoriented (more than two poles) spindle formation. The results presented are the mean of the three strains with SD. Zymolyase was purchased from US Biological (Z1004), and the rat α-tubulin mAb and Alexa Fluor 594 anti-rat immunoglobulin G secondary antibody were obtained from Harlan Sera-Lab (MAS 078) and Molecular Probes (A11007), respectively. An Olympus PROVIS AX70 fluorescence microscope was used for all experiments at a final magnification of 1000×. Northern blots were performed as described previously (Cooper et al., 2009).
Supplementary Material
Acknowledgments
We thank David Bardford, Wolfgang Zachariae, Uttam Surana, Wolfgang Seufert, Mark Solomon, Edward Winter, and Andrew Murray for the plasmids and Vincent Guacci for Tub1p antibodies. We thank Michael Mallory for technical assistance. We thank Randy Strich and Michael Law for helpful discussions and comments on the manuscript and Edward Winter for his help with the Smk1p assays. This work was supported by American Cancer Society grant CCG106162 to K.F.C.
Abbreviations used:
- APC/C
anaphase-promoting complex/cyclosome
- CB
C-box
- DAPI
4′,6-diamino-2-phenylindole
- Db1
destruction box degron 1
- FC
final concentration
- GST
glutathione S-transferase
- GxEN
destruction degron
- HA
hemagglutinin
- mAb
monoclonal antibody
- MAPK
mitogen-activated protein kinase
- MI
meiosis I
- MII
meiosis II
- SPM
sporulation medium
- WT
wild type
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E10-04-0360) on November 30, 2010.
REFERENCES
- Akamatsu E, Tanaka T, Kato JY. Transcription factor E2F and cyclin E-Cdk2 complex cooperate to induce chromosomal DNA replication in Xenopus oocytes. J Biol Chem. 1998;273:16494–16500. doi: 10.1074/jbc.273.26.16494. [DOI] [PubMed] [Google Scholar]
- Anghileri P, Branduardi P, Sternieri F, Monti P, Visintin R, Bevilacqua A, Alberghina L, Martegani E, Baroni MD. Chromosome separation and exit from mitosis in budding yeast: dependence on growth revealed by cAMP-mediated inhibition. Exp Cell Res. 1999;250:510–523. doi: 10.1006/excr.1999.4531. [DOI] [PubMed] [Google Scholar]
- Benanti JA, Toczyski DP. Cdc20, an activator at last. Mol Cell. 2008;32:460–461. doi: 10.1016/j.molcel.2008.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharadwaj R, Yu H. The spindle checkpoint, aneuploidy, and cancer. Oncogene. 2004;23:2016–2027. doi: 10.1038/sj.onc.1207374. [DOI] [PubMed] [Google Scholar]
- Burton JL, Solomon MJ. D box and KEN box motifs in budding yeast Hsl1p are required for APC-mediated degradation and direct binding to Cdc20p and Cdh1p. Genes Dev. 2001;15:2381–2395. doi: 10.1101/gad.917901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton JL, Tsakraklides V, Solomon MJ. Assembly of an APC-Cdh1-substrate complex is stimulated by engagement of a destruction box. Mol Cell. 2005;18:533–542. doi: 10.1016/j.molcel.2005.04.022. [DOI] [PubMed] [Google Scholar]
- Camasses A, Bogdanova A, Shevchenko A, Zachariae W. The CCT chaperonin promotes activation of the anaphase-promoting complex through the generation of functional Cdc20. Mol Cell. 2003;12:87–100. doi: 10.1016/s1097-2765(03)00244-2. [DOI] [PubMed] [Google Scholar]
- Carlile TM, Amon A. Meiosis I is established through division-specific translational control of a cyclin. Cell. 2008;133:280–291. doi: 10.1016/j.cell.2008.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll CW, Enquist-Newman M, Morgan DO. The APC subunit Doc1 promotes recognition of the substrate destruction box. Curr Biol. 2005;15:11–18. doi: 10.1016/j.cub.2004.12.066. [DOI] [PubMed] [Google Scholar]
- Carroll CW, Morgan DO. The Doc1 subunit is a processivity factor for the anaphase-promoting complex. Nat Cell Biol. 2002;4:880–887. doi: 10.1038/ncb871. [DOI] [PubMed] [Google Scholar]
- Castro A, Vigneron S, Bernis C, Labbe JC, Lorca T. Xkid is degraded in a D-box, KEN-box, and A-box-independent pathway. Mol Cell Biol. 2003;23:4126–4138. doi: 10.1128/MCB.23.12.4126-4138.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi E, Dial JM, Jeong DE, Hall MC. Unique D box and KEN box sequences limit ubiquitination of Acm1 and promote pseudosubstrate inhibition of the anaphase-promoting complex. J Biol Chem. 2008;283:23701–23710. doi: 10.1074/jbc.M803695200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson TW, Sikorski RS, Dante M, Shero JH, Hieter P. Multifunctional yeast high-copy-number shuttle vectors. Gene. 1992;110:119–122. doi: 10.1016/0378-1119(92)90454-w. [DOI] [PubMed] [Google Scholar]
- Chu S, DeRisi J, Eisen M, Mulholland J, Botstein D, Brown PO, Herskowitz I. The transcriptional program of sporulation in budding yeast. Science. 1998;282:699–705. doi: 10.1126/science.282.5389.699. [DOI] [PubMed] [Google Scholar]
- Chu T, Henrion G, Haegeli V, Strickland S. Cortex, a Drosophila gene required to complete oocyte meiosis, is a member of the Cdc20/fizzy protein family. Genesis. 2001;29:141–152. doi: 10.1002/gene.1017. [DOI] [PubMed] [Google Scholar]
- Coluccio A, Bogengruber E, Conrad MN, Dresser ME, Briza P, Neiman AM. Morphogenetic pathway of spore wall assembly in Saccharomyces cerevisiae. Eukaryot Cell. 2004;3:1464–1475. doi: 10.1128/EC.3.6.1464-1475.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper KF, Egeland DE, Mallory MJ, Jarnik M, Strich R. Ama1p is a meiosis-specific regulator of the anaphase promoting complex/cyclosome in yeast. Proc Natl Acad Sci USA. 2000;97:14548–14553. doi: 10.1073/pnas.250351297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper KF, Mallory MJ, Guacci V, Lowe K, Strich R. Pds1p is required for meiotic recombination and prophase I progression in Saccharomyces cerevisiae. Genetics. 2009;181:65–79. doi: 10.1534/genetics.108.095513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper KF, Mallory MJ, Smith JS, Strich R. Stress and developmental regulation of the yeast C-type cyclin UME3 (SRB11/SSN8). EMBO J. 1997;16:4665–4675. doi: 10.1093/emboj/16.15.4665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper KF, Mallory MJ, Strich R. Oxidative stress-induced destruction of the yeast C-type cyclin Ume3p requires the phosphatidylinositol-specific phospholipase C and the 26S proteasome. Mol Cell Biol. 1999;19:3338–3348. doi: 10.1128/mcb.19.5.3338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper KF, Strich R. Functional analysis of the yeast C-type cyclin Ume3p/Srb11p- RNA polymerase II holoenzyme interaction. Gene Exp. 1999;8:43–57. [PMC free article] [PubMed] [Google Scholar]
- Cooper KF, Strich R. Saccharomyces cerevisiae C-type cyclin Ume3p/Srb11p is required for efficient induction and execution of meiotic development. Eukaryot Cell. 2002;1:66–74. doi: 10.1128/EC.01.1.66-74.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crasta K, Lim HH, Giddings TH Jr, Winey M, Surana U. Inactivation of Cdh1 by synergistic action of Cdk1 and polo kinase is necessary for proper assembly of the mitotic spindle. Nat Cell Biol. 2008;10:665–675. doi: 10.1038/ncb1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson IA, Roth S, Artavanis-Tsakonas S. The Drosophila cell cycle gene fizzy is required for normal degradation of cyclins A and B during mitosis and has homology to the CDC20 gene of Saccharomyces cerevisiae. J Cell Biol. 1995;129:725–737. doi: 10.1083/jcb.129.3.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshaies RJ. SCF and Cullin/RING H2-based ubiquitin ligases. Annu Rev Cell Dev Biol. 1999;15:435–467. doi: 10.1146/annurev.cellbio.15.1.435. [DOI] [PubMed] [Google Scholar]
- Dial JM, Petrotchenko EV, Borchers CH. Inhibition of APCCdh1 activity by Cdh1/Acm1/Bmh1 ternary complex formation. J Biol Chem. 2007;282:5237–5248. doi: 10.1074/jbc.M606589200. [DOI] [PubMed] [Google Scholar]
- Diamond AE, Park JS, Inoue I, Tachikawa H, Neiman AM. The anaphase promoting complex targeting subunit Ama1 links meiotic exit to cytokinesis during sporulation in Saccharomyces cerevisiae. Mol Biol Cell. 2009;20:134–145. doi: 10.1091/mbc.E08-06-0615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dube P, Herzog F, Gieffers C, Sander B, Riedel D, Muller SA, Engel A, Peters JM, Stark H. Localization of the coactivator Cdh1 and the cullin subunit Apc2 in a cryo-electron microscopy model of vertebrate APC/C. Mol Cell. 2005;20:867–879. doi: 10.1016/j.molcel.2005.11.008. [DOI] [PubMed] [Google Scholar]
- Enquist-Newman M, Sullivan M, Morgan DO. Modulation of the mitotic regulatory network by APC-dependent destruction of the Cdh1 inhibitor Acm1. Mol Cell. 2008;30:437–446. doi: 10.1016/j.molcel.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eytan E, Moshe Y, Braunstein I, Hershko A. Roles of the anaphase-promoting complex/cyclosome and of its activator Cdc20 in functional substrate binding. Proc Natl Acad Sci USA. 2006;103:2081–2086. doi: 10.1073/pnas.0510695103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang G, Yu H, Kirschner MW. Direct binding of CDC20 protein family members activates the anaphase-promoting complex in mitosis and G1. Molecular Cell. 1998a;2:163–171. doi: 10.1016/s1097-2765(00)80126-4. [DOI] [PubMed] [Google Scholar]
- Glotzer M, Murray AW, Kirschner MW. Cyclin is degraded by the ubiquitin pathway. Nature. 1991;349:132–138. doi: 10.1038/349132a0. [DOI] [PubMed] [Google Scholar]
- Goh P, Lim HL, Surana U. Cdc20 protein contains a destruction-box but, unlike Clb2, its proteolysis is not acutely dependent on the activity of the anaphase-promoting complex. Eur J Biochem. 2000;267:434–449. doi: 10.1046/j.1432-1327.2000.01014.x. [DOI] [PubMed] [Google Scholar]
- Hall MC, Jeong DE, Henderson JT, Choi E, Bremmer SC, Iliuk AB, Charbonneau H. Cdc28 and Cdc14 control stability of the anaphase-promoting complex inhibitor Acm1. J Biol Chem. 2008;283:10396–10407. doi: 10.1074/jbc.M710011200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper JW, Burton JL, Solomon MJ. The anaphase-promoting complex: it’s not just for mitosis any more. Genes Dev. 2002;16:2179–2206. doi: 10.1101/gad.1013102. [DOI] [PubMed] [Google Scholar]
- Hendrickson C, Meyn III M, Morabito L, Holloway S. The KEN box regulates Clb2 proteolysis in G1 and at the metaphase-anaphase transition. Curr Biol. 2001;11:1781–1787. doi: 10.1016/s0960-9822(01)00564-4. [DOI] [PubMed] [Google Scholar]
- Hilioti Z, Chung YS, Mochizuki Y, Hardy CF, Cohen-Fix O. The anaphase inhibitor Pds1 binds to the APC/C-associated protein Cdc20 in a destruction box-dependent manner [correction published in Curr Biol (2001). 11, 1643]. Curr Biol. 2001;11:1347–1352. doi: 10.1016/s0960-9822(01)00399-2. [DOI] [PubMed] [Google Scholar]
- Huang JN, Park I, Ellingson E, Littlepage LE, Pellman D. Activity of the APCCdh1 form of the anaphase promoting complex persists until S phase and prevents premature expression of Cdc20p. J Biol Chem. 2001;154:85–94. doi: 10.1083/jcb.200102007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaquenoud M, van Drogen F, Peter M. Cell cycle-dependent nuclear export of Cdh1p may contribute to the inactivation of APC/C(Cdh1). EMBO J. 2002;21:6515–6526. doi: 10.1093/emboj/cdf634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaspersen SL, Charles JF, Morgan DO. Inhibitory phosphorylation of the APC regulator Hct1 is controlled by the kinase Cdc28 and the phosphatase Cdc14. Curr Biol. 1999;9:227–236. doi: 10.1016/s0960-9822(99)80111-0. [DOI] [PubMed] [Google Scholar]
- Kimata Y, Baxter JE, Fry AM, Yamano H. A role for the Fizzy/Cdc20 family of proteins in activation of the APC/C distinct from substrate recruitment. Mol Cell. 2008;32:576–583. doi: 10.1016/j.molcel.2008.09.023. [DOI] [PubMed] [Google Scholar]
- Kraft C, Herzog F, Gieffers C, Mechtler K, Hagting A, Pines J, Peters JM. Mitotic regulation of the human anaphase-promoting complex by phosphorylation. EMBO J. 2003;22:6598–6609. doi: 10.1093/emboj/cdg627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraft C, Vodermaier HC, Maurer-Stroh S, Eisenhaber F, Peters JM. The WD40 propeller domain of Cdh1 functions as a destruction box receptor for APC/C substrates. Mol Cell. 2005;18:543–553. doi: 10.1016/j.molcel.2005.04.023. [DOI] [PubMed] [Google Scholar]
- Littlepage LE, Ruderman JV. Identification of a new APC/C recognition domain, the A box, which is required for the Cdh1-dependent destruction of the kinase Aurora-A during mitotic exit. Genes Dev. 2002;16:2274–2285. doi: 10.1101/gad.1007302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, McKenzie Ar, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Mallory MJ, Cooper KF, Strich R. Meiosis-specific destruction of the Ume6p repressor by the Cdc20-directed APC/C. Mol Cell. 2007;27:951–961. doi: 10.1016/j.molcel.2007.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez JS, Jeong DE, Choi E, Billings BM, Hall MC. Acm1 is a negative regulator of the CDH1-dependent anaphase-promoting complex/cyclosome in budding yeast. Mol Cell Biol. 2006;26:9162–9176. doi: 10.1128/MCB.00603-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matyskiela ME, Morgan DO. Analysis of activator-binding sites on the APC/C supports a cooperative substrate-binding mechanism. Mol Cell. 2009;34:68–80. doi: 10.1016/j.molcel.2009.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald CM, Cooper KF, Winter E. The Ama1-directed anaphase-promoting complex regulates the Smk1 mitogen-activated protein kinase during meiosis in yeast. Genetics. 2005;171:901–911. doi: 10.1534/genetics.105.045567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyn MA III, Melloy PG, Li J, Holloway SL. The destruction box of the cyclin Clb2 binds the anaphase-promoting complex/cyclosome subunit Cdc23. Arch Biochem Biophys. 2002;407:189–195. doi: 10.1016/s0003-9861(02)00467-8. [DOI] [PubMed] [Google Scholar]
- Morris MC, Kaiser P, Rudyak S, Baskerville C, Watson MH, Reed SI. Cks1-dependent proteasome recruitment and activation of CDC20 transcription in budding yeast. Nature. 2003;423:1009–1013. doi: 10.1038/nature01720. [DOI] [PubMed] [Google Scholar]
- Oelschlaegel T, Schwickart M, Matos J, Bogdanova A, Camasses A, Havlis J, Shevchenko A, Zachariae W. The yeast APC/C subunit Mnd2 prevents premature sister chromatid separation triggered by the meiosis-specific APC/C-Ama1. Cell. 2005;120:773–788. doi: 10.1016/j.cell.2005.01.032. [DOI] [PubMed] [Google Scholar]
- Ostapenko D, Burton JL, Wang R, Solomon MJ. Pseudosubstrate inhibition of the anaphase-promoting complex by Acm1: regulation by proteolysis and Cdc28 phosphorylation. Mol Cell Biol. 2008;28:4653–4664. doi: 10.1128/MCB.00055-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passmore LA, Barford D. Coactivator functions in a stoichiometric complex with anaphase-promoting complex/cyclosome to mediate substrate recognition. EMBO Rep. 2005;6:873–878. doi: 10.1038/sj.embor.7400482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passmore LA, McCormack EA, Au SW, Paul A, Willison KR, Harper JW, Barford D. Doc1 mediates the activity of the anaphase-promoting complex by contributing to substrate recognition. EMBO J. 2003;22:786–796. doi: 10.1093/emboj/cdg084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penkner AM, Prinz S, Ferscha S, Klein F. Mnd2, an essential antagonist of the anaphase-promoting complex during meiotic prophase. Cell. 2005;120:789–801. doi: 10.1016/j.cell.2005.01.017. [DOI] [PubMed] [Google Scholar]
- Pesin JA, Orr-Weaver TL. Developmental role and regulation of cortex, a meiosis-specific anaphase-promoting complex/cyclosome activator. PLoS Genet. 2007;3:e202. doi: 10.1371/journal.pgen.0030202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfleger CM, Kirschner MW. The KEN box: an APC recognition signal distinct from the D box targeted by Cdh1. Genes Dev. 2000;14:655–665. [PMC free article] [PubMed] [Google Scholar]
- Pfleger CM, Lee E, Kirschner MW. Substrate recognition by the Cdc20 and Cdh1 components of the anaphase-promoting complex. Genes Dev. 2001a;15:2396–2407. doi: 10.1101/gad.918201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prinz S, Hwang ES, Visintin R, Amon A. The regulation of Cdc20 proteolysis reveals a role for APC components Cdc23 and Cdc27 during S phase and early mitosis. Curr Biol. 1998;8:750–760. doi: 10.1016/s0960-9822(98)70298-2. [DOI] [PubMed] [Google Scholar]
- Rabitsch KP, et al. A screen for genes required for meiosis and spore formation based on whole-genome expression. Curr Biol. 2001;11:1001–1009. doi: 10.1016/s0960-9822(01)00274-3. [DOI] [PubMed] [Google Scholar]
- Robbins JA, Cross FR. Regulated degradation of the APC coactivator Cdc20. Cell Div. 2010;5:23. doi: 10.1186/1747-1028-5-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salah SM, Nasmyth K. Destruction of the securin Pds1p occurs at the onset of anaphase during both meiotic divisions in yeast. Chromosoma. 2000;109:27–34. doi: 10.1007/s004120050409. [DOI] [PubMed] [Google Scholar]
- Schwab M, Neutzner M, Mocker D, Seufert W. Yeast Hct1 recognizes the mitotic cyclin Clb2 and other substrates of the ubiquitin ligase APC. EMBO J. 2001;20:5165–5175. doi: 10.1093/emboj/20.18.5165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab M, Schulze Lutum A, Seufert W. Yeast Hct1 is a regulator of Clb2 cyclin protolysis. Cell. 1997;90:683–693. doi: 10.1016/s0092-8674(00)80529-2. [DOI] [PubMed] [Google Scholar]
- Searle JS, Schollaert KL, Wilkins BJ, Sanchez Y. The DNA damage checkpoint and PKA pathways converge on APC substrates and Cdc20 to regulate mitotic progression. Nat Cell Biol. 2004;6:138–145. doi: 10.1038/ncb1092. [DOI] [PubMed] [Google Scholar]
- Shirayama M, Toth A, Galova M, Nasmyth K. APC(Cdc20) promotes exit from mitosis by destroying the anaphase inhibitor Pds1 and cyclin Clb5. Nature. 1999;402:203–207. doi: 10.1038/46080. [DOI] [PubMed] [Google Scholar]
- Shirayama M, Zachariae W, Ciosk R, Nasmyth K. The Polo-like kinase Cdc5p and the WD-repeat protein Cdc20p/fizzy are regulators and substrates of the anaphase promoting complex in Saccharomyces cerevisiae. EMBO J. 1998;17:1336–1349. doi: 10.1093/emboj/17.5.1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigrist SJ, Lehner CF. Drosophila fizzy-related down-regulates mitotic cyclins and is required for cell proliferation arrest and entry into endocycles. Cell. 1997;90:671–681. doi: 10.1016/s0092-8674(00)80528-0. [DOI] [PubMed] [Google Scholar]
- Sorensen CS, Lukas C, Kramer ER, Peters JM, Bartek J, Lukas J. A conserved cyclin-binding domain determines functional interplay between anaphase-promoting complex-Cdh1 and cyclin A-Cdk2 during cell cycle progression. Mol Cell Biol. 2001;21:3692–3703. doi: 10.1128/MCB.21.11.3692-3703.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spingola M, Ares M Jr. A yeast intronic splicing enhancer and Nam8p are required for Mer1p-activated splicing. Mol Cell. 2000;6:329–338. doi: 10.1016/s1097-2765(00)00033-2. [DOI] [PubMed] [Google Scholar]
- Strich R, Slater MR, Esposito RE. Identification of negative regulatory genes that govern the expression of early meiotic genes in yeast. Proc Natl Acad Sci USA. 1989;86:10018–10022. doi: 10.1073/pnas.86.24.10018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton BR, Ng TM, Matyskiela ME, Carroll CW, Morgan DO, Toczyski DP. An architectural map of the anaphase-promoting complex. Genes Dev. 2006;20:449–460. doi: 10.1101/gad.1396906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valpuesta JM, Martin-Benito J, Gomez-Puertas P, Carrascosa JL, Willison KR. Structure and function of a protein folding machine: the eukaryotic cytosolic chaperonin CCT. FEBS Lett. 2002;529:11–16. doi: 10.1016/s0014-5793(02)03180-0. [DOI] [PubMed] [Google Scholar]
- Visintin R, Prinz S, Amon A. CDC20 and CDH1: a family of substrate-specific activators of APC-dependent proteolysis. Science. 1997;278:460–463. doi: 10.1126/science.278.5337.460. [DOI] [PubMed] [Google Scholar]
- Vodermaier HC, Gieffers C, Maurer-Stroh S, Eisenhaber F, Peters JM. TPR subunits of the anaphase-promoting complex mediate binding to the activator protein CDH1. Curr Biol. 2003;13:1459–1468. doi: 10.1016/s0960-9822(03)00581-5. [DOI] [PubMed] [Google Scholar]
- Yamano H, Gannon J, Mahbubani H, Hunt T. Cell cycle-regulated recognition of the destruction box of cyclin B by the APC/C in Xenopus egg extracts. Mol Cell. 2004;13:137–147. doi: 10.1016/s1097-2765(03)00480-5. [DOI] [PubMed] [Google Scholar]
- Yu H. Cdc20: a WD40 activator for a cell cycle degradation machine. Mol Cell. 2007;27:3–16. doi: 10.1016/j.molcel.2007.06.009. [DOI] [PubMed] [Google Scholar]
- Zachariae W, Schwab M, Nasmyth K, Seufert W. Control of cyclin ubiquitination by CDK-regulated binding of Hct1 to the anaphase promoting complex. Science. 1998;282:1721–1724. doi: 10.1126/science.282.5394.1721. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Lees E. Identification of an overlapping binding domain on Cdc20 for Mad2 and anaphase-promoting complex: model for spindle checkpoint regulation. Mol Cell Biol. 2001;21:5190–5199. doi: 10.1128/MCB.21.15.5190-5199.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.