This study reveals novel roles for the focal adhesion proteins paxillin and Hic-5 in regulating breast cancer invasion strategies and metastasis. Depletion of paxillin promotes a hypermesenchymal phenotype while dysregulating 3D adhesion dynamics. In contrast, RNAi of Hic-5 induces a hyperamoeboid phenotype with dysregulated RhoA/pMLC signaling.
Abstract
Individual metastatic tumor cells exhibit two interconvertible modes of cell motility during tissue invasion that are classified as either mesenchymal or amoeboid. The molecular mechanisms by which invasive breast cancer cells regulate this migratory plasticity have yet to be fully elucidated. Herein we show that the focal adhesion adaptor protein, paxillin, and the closely related Hic-5 have distinct and unique roles in the regulation of breast cancer cell lung metastasis by modulating cell morphology and cell invasion through three-dimensional extracellular matrices (3D ECMs). Cells depleted of paxillin by RNA interference displayed a highly elongated mesenchymal morphology, whereas Hic-5 knockdown induced an amoeboid phenotype with both cell populations exhibiting reduced plasticity, migration persistence, and velocity through 3D ECM environments. In evaluating associated signaling pathways, we determined that Rac1 activity was increased in cells devoid of paxillin whereas Hic-5 silencing resulted in elevated RhoA activity and associated Rho kinase–induced nonmuscle myosin II activity. Hic-5 was essential for adhesion formation in 3D ECMs, and analysis of adhesion dynamics and lifetime identified paxillin as a key regulator of 3D adhesion assembly, stabilization, and disassembly.
INTRODUCTION
Malignant cancer cells are able to invade through the tissue extracellular matrix (ECM) microenvironment either as collective multicellular sheets or as individual cells, thereby contributing to lymphatic and hematogenous infiltration, respectively (Friedl et al., 1995; Sahai and Marshall, 2003; Wolf et al., 2003, 2007; Carragher et al., 2006; Wyckoff et al., 2006; Gadea et al., 2008; Sanz-Moreno et al., 2008; Friedl and Wolf, 2009; Giampieri et al., 2009; Parri et al., 2009). In addition, the individual cells can use two distinct and interchangeable modes of motility referred to as mesenchymal and amoeboid migration (Wolf et al., 2003). The spontaneous interconversion between the two phenotypes, known as plasticity, is considered to be one of the primary reasons for cancer cell evasion of current invasion-targeted therapeutics, which are designed to prevent mesenchymal invasion mechanisms (Zucker et al., 2000; Coussens et al., 2002; Overall and Lopez-Otin, 2002; Friedl and Wolf, 2003).
The mesenchymal mode of cancer cell migration is characterized by an elongated cell morphology requiring extracellular proteolysis and dynamic integrin-mediated interactions with the three-dimensional (3D) ECM. In contrast, amoeboid motility is independent of protease activity and is associated with a rounded or ellipsoid morphology (Wolf et al., 2003). Cells exhibiting amoeboid migration squeeze through pre-existing gaps in the heterogeneous ECM milieu using actin-rich membrane blebs or filopodia-like projections and display highly transient, weak interactions with the surrounding matrix proteins (Sahai and Marshall, 2003; Wolf et al., 2003; Yoshida and Soldati, 2006; Sanz-Moreno et al., 2008).
The Rho GTPase family (in particular, Rac1, RhoA, and Cdc42) consists of well-established regulators of two-dimensional (2D) cell migration, coordinating adhesion dynamics and cytoskeleton remodeling (Nobes and Hall, 1999; Bristow et al., 2009). More recently, they have been identified as critical determinants of cancer cell migration through 3D ECM environments. In this context, elevated Rac1 activity promotes the elongated mesenchymal morphology and motility (Sanz-Moreno et al., 2008), whereas elevated RhoA activity drives the amoeboid mode of invasion by stimulating membrane blebbing through Rho kinase (ROCK) and nonmuscle myosin II activity and thereby actomyosin contractility (Sahai and Marshall, 2003, 2008; Wyckoff et al., 2006; Sahai et al., 2007). Cdc42 appears to regulate both amoeboid and mesenchymal invasion strategies in melanoma cells (Gadea et al., 2008), but the mechanisms by which the activity of Rho family GTPases are coordinated and counterbalanced, particularly during 3D migration, are poorly understood.
Paxillin and the closely related family member Hic-5, localize to the cytoplasmic face of integrin-mediated adhesion sites where they function as molecular scaffolds for the coordination of Rho GTPase signaling during cell migration on planar 2D surfaces (Brown and Turner, 2004; Tumbarello and Turner, 2007; Deakin and Turner, 2008). The two proteins have a similar overall domain structure, including amino-terminal LD motifs and carboxyl-terminal LIM domains and consequently associate with many of the same binding partners, such as focal adhesion kinase (FAK), GIT1/2, ILK, and parvin (reviewed in Brown and Turner, 2004), suggesting overlapping or redundant functions. Differences in binding affinities for several of these interacting proteins, as well as unique phosphorylation profiles created in response to 2D ECM adhesion and growth factor stimulation that generate specific binding sites for additional proteins (Nishiya et al., 2002; Brown and Turner, 2004; Rathore et al., 2007), however, are indicators of distinct signaling functions. Indeed, the embryonic lethality of the paxillin knockout mouse, despite the presence of Hic-5 (Hagel et al., 2002), combined with examples of differential expression (e.g., during epithelial-to-mesenchymal transition [EMT] in which Hic-5 is upregulated) (Tumbarello et al., 2005; Tumbarello and Turner, 2007), point to a more complex interrelationship between the two proteins (Nishiya et al., 2001; Brown and Turner, 2004). Furthermore, Hic-5 can interact with Smad proteins (Wang et al., 2005; Inui et al., 2010) and has a well-established role as a nuclear receptor coactivator downstream of the glucocorticoid and androgen receptors in breast and prostate cancers (reviewed in Heitzer and DeFranco, 2006), highlighting potential tissue-specific and disease-specific functions for the two proteins. Importantly, to our knowledge the respective roles of paxillin and Hic-5 in the regulation of cell phenotype and migration in 3D environments have not been described.
Herein we have identified unique functions for paxillin and Hic-5 in regulating cell morphology and 3D migration mechanisms associated with breast cancer cell invasion, plasticity, and metastasis. Using a combination of in vitro and in vivo systems, we show that paxillin and Hic-5 impact these processes through differential modulation of Rac1, RhoA, FAK, and nonmuscle myosin II activity. Moreover, we demonstrate that a balance of paxillin and Hic-5 signaling is necessary for coordinating cancer cell morphology and invasion mechanisms. We further show that paxillin and Hic-5 are both required for persistent migration in 3D ECM and for the first time have evaluated individual adhesion dynamics in 3D microenvironments, identifying a role for paxillin in regulating mesenchymal cell adhesion contact assembly, stabilization, and disassembly. Furthermore, we provide evidence that Hic-5 expression is required for integrin-mediated adhesion formation in 3D matrices. These data provide evidence for distinct and unique roles for paxillin and Hic-5 in 3D migration strategies and in coordinating breast cancer invasion and metastasis.
RESULTS
Paxillin and Hic-5 are required for MDA-MB-231 cancer cell invasion
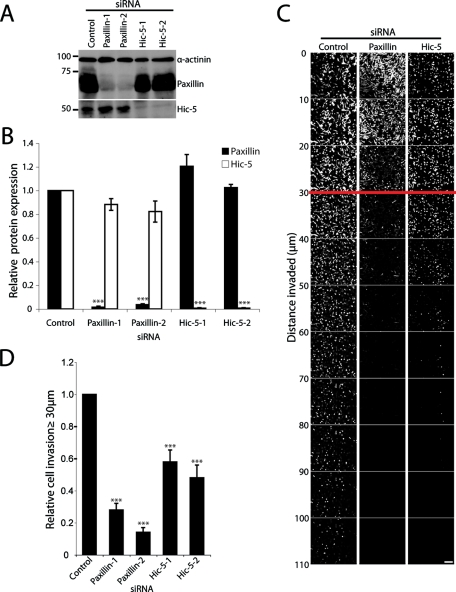
Paxillin and Hic-5 have been identified as key mediators of both normal cell and cancer cell migration when analyzed in vitro on 2D ECM-coated surfaces (Petit et al., 2000; West et al., 2001; Iwasaki et al., 2002; Brown and Turner, 2004; Azuma et al., 2005; Avraamides et al., 2007; Tumbarello and Turner, 2007; Deakin and Turner, 2008; Yu et al., 2009). However, their potential functions in coordinating tumor cell migration and invasion through more in vivo relevant 3D microenvironments have yet to be evaluated. To assess the roles of paxillin and Hic-5 in breast cancer invasion we used RNA interference (RNAi)-mediated knockdown in the highly invasive and metastatic MDA-MB-231 breast cancer cell line. Using two independent siRNAs for each protein, paxillin expression was decreased by 98.5% ± 0.97 (paxillin-1) and 96.4% ± 0.74 (paxillin-2) relative to control small interfering RNA (siRNA)-treated cells, whereas Hic-5 protein expression was decreased by 99.3% ± 0.42 (Hic-5–1) and 99.4% ± 0.38 (Hic-5–2) (Figure 1, A and B). Importantly, no significant effect on the expression of Hic-5 was observed in cells treated with paxillin siRNA or vice versa (Figure 1, A and B). Using collagen and fibronectin (FN)-rich plugs as an in vitro model for cancer cell 3D ECM invasive migration (Hennigan et al., 1994; Caswell et al., 2007) both paxillin and Hic-5 RNAi cells showed a significant reduction in the invasive capacity of the MDA-MB-231 cells (Figure 1, C and D, and Supplemental Figure 1).
FIGURE 1:
Paxillin and Hic-5 mediate breast cancer cell invasion through 3D ECM. (A) Representative Western blot of total cell lysates from MDA-MB-231 cells treated with siRNA as stated for 72 h. (B) Quantitation of paxillin and Hic-5 relative protein expression after siRNA-mediated knockdown. Data represent the relative mean ± SEM of a minimum of three individual knockdowns. Densitometric quantification of band intensity was measured using ImageJ and was normalized relative to the band intensity of the α-actinin loading control. (C) Invasive migration of MDA-MB-231 cells treated with siRNA (as indicated) migrating through 3D collagen with FN at 25 μg/ml was determined by using an inverted invasion assay. Cells were stained with Calcein acetoxymethyl ester (AM) and visualized by laser scanning confocal microscopy. Optical sections were taken at 10-μm intervals. (D) Cell invasion was quantified using ImageJ by measuring the number of cells, as indicated by Calcein AM staining, in each optical section. The total number of cells invading >30 μm into the collagen and FN were quantified relative to the total number of cells counted. Data represent mean ± SEM from a minimum of eight individual experiments. A Student’s t test was used for statistical analysis *** p < 0.0005. Scale bar = 150 μm.
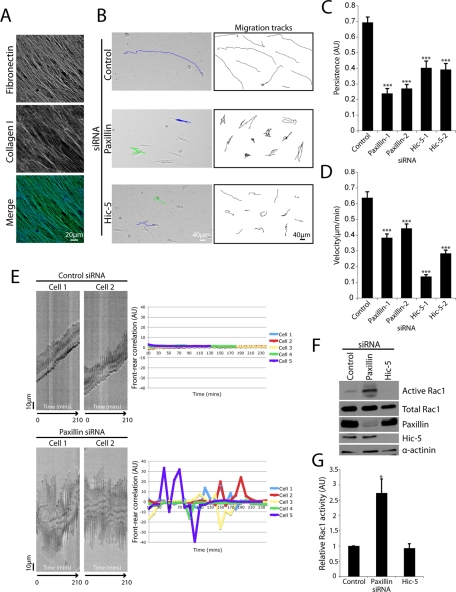
To begin to evaluate the cause of the observed defects in invasion in paxillin and Hic-5–depleted cells we performed real-time migration analysis using the 3D cell-derived matrix (CDM) model system (Cukierman et al., 2001; Cukierman, 2004; Castello-Cros and Cukierman, 2009). CDMs are composed of highly organized parallel collagen and FN fibers (Figure 2A) reminiscent of the in vivo matrix organization observed at the tumor–stroma interface (Provenzano et al., 2006, 2009). Control RNAi-treated cells exhibited strikingly linear migration tracks (Figure 2B and Supplemental Figure 2A) and thus a high degree of directional persistence (see Materials and Methods for details) (Figure 2, B and C, and Supplemental Movie 1). In contrast, paxillin RNAi cells exhibited markedly reduced persistent migration (Figure 2, B and C, and Supplemental Figure 2A) as well as decreased overall velocity (Figure 2D and Supplemental Movie 2). Hic-5 suppression also resulted in a significant decrease in persistence (Figure 2, B and C, and Supplemental Figure 2A) and migration velocity (Figure 2D and Supplemental Movie 3) relative to control siRNA-treated cells.
FIGURE 2:
Paxillin and Hic-5 regulate persistent migration in 3D microenvironments. (A) Immunofluorescence images of collagen I and FN organization in 3D CDMs. (B) Phase contrast images of siRNA-treated MDA-MB-231 cells migrating through a 3D CDM with superimposed migration tracks derived from cell centroid tracking (left panels). Ten representative migration tracks from a minimum of three individual experiments of siRNA-treated cell locomotion in 3D CDMs (right panels). Quantitation of migration (C) persistence and (D) velocity. Data represent mean ± SEM of a minimum of 30 cells from three individual experiments. Both persistence and velocity were quantified using ImageJ manual tracking software. Statistical analyses were performed using a Kruskal–Wallis test followed by Dunn’s post hoc test. *** p < 0.001. (E) Kymograph analysis of control (top panels) and paxillin (bottom panels) siRNA-treated cells migrating through 3D CDMs. Cells were visualized using a Leica deconvolution microscope and images were taken for 16 h at 10-min intervals. Kymograph indicates correlation between cell front protrusion and rear retraction during 3D migration. Line graphs (right panels) indicate front–rear correlation quantification of a minimum of six cells from three individual experiments. (F) Western blot analysis of Rac1 activity as assessed by a PAK3 CRIB pulldown assay. MDA-MB-231 cells were treated with siRNA for 56 h then allowed to migrate through 3D CDMs for 16 h in the presence of serum. (G) Quantification of active Rac1 band intensity relative to total Rac1. Data represent mean ± SEM of three individual experiments. A Student’s t test was performed to assess statistical significance. * p < 0.05.
Persistent migration has been shown previously to require the coordination of protrusive activity at the cell’s leading edge (front) combined with cell rear retraction to enable efficient, polarized locomotion (Ridley et al., 2003; Broussard et al., 2008). Kymograph analysis revealed that cells treated with control siRNA exhibited a high degree of correlation between front protrusion and rear retraction (Figure 2E; top panels), consistent with their ability to exhibit persistent migration. Conversely, paxillin RNAi cells demonstrated a distinct uncoupling of front protrusion and rear retraction as indicated by quantification of front–rear correlation (Figure 2E; bottom panels). In contrast, Hic-5–depleted cells exhibited a decrease in the kymograph slope, consistent with their reduced velocity but revealed no significant defect in front–rear correlation (Supplemental Figure 2B).
The restricted spatiotemporal regulation of the small GTPase Rac1 has been shown to be essential for persistent cell migration through 3D ECM as well as in coordinating cell protrusion (Pankov et al., 2005; Machacek et al., 2009; Petrie et al., 2009). Therefore, following knockdown of paxillin and Hic-5, the activity of Rac1 was assessed in cells migrating through the 3D CDMs. A threefold increase in Rac1 activity was observed in cells devoid of paxillin (Figure 2, F and G), whereas a small, reproducible, but not statistically significant decrease in active Rac1 was observed in cells treated with Hic-5 siRNA (Figure 2, F and G). These data suggest that both paxillin and Hic-5 are necessary for efficient MDA-MB-231 breast cancer cell invasion and that paxillin coordinates persistent 3D ECM migration, potentially through spatially restricting Rac1 activity.
Paxillin and Hic-5 exert differential effects on cancer cell morphology and plasticity during 3D ECM invasion
Many cancer cells can exhibit two different modes of invasion, referred to as mesenchymal and amoeboid migration (Friedl et al., 1995; Sahai and Marshall, 2003; Friedl and Wolf, 2009). Furthermore, individual cells can also exhibit phenotypic plasticity, frequently switching between the two morphologies (Wolf et al., 2003). Examination of the collagen/FN plugs and time-lapse movies of control siRNA versus paxillin or Hic-5 RNAi–treated MDA-MB-231 cells revealed significant differences in their cell morphology profiles.
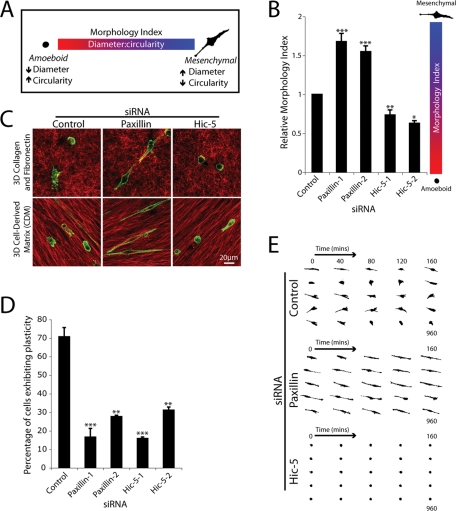
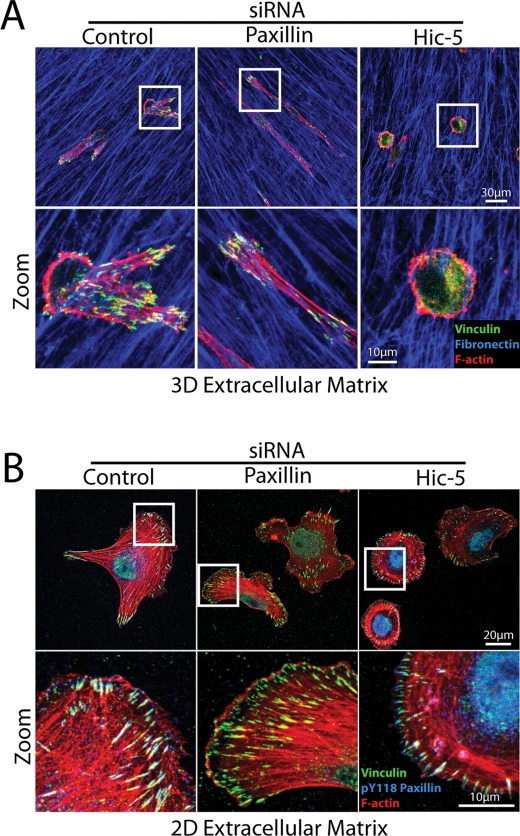
To quantify the observed morphologies, a nonsubjective Morphology Index was calculated relative to control cells, incorporating a ratiometric analysis of Feret’s dia-meter or caliper length of each individual cell versus its circularity. Amoeboid cells exhibit a small diameter and high degree of circularity (low Morphology Index), whereas mesenchymal cells display a high diameter and low circularity (high Morphology Index) (Figure 3A). Control siRNA-treated MDA-MB-231 cells displayed a mixed population of both amoeboid and mesenchymal morphologies, consistent with previous studies (Wolf et al., 2003) and so had an intermediate Morphology Index score (Figure 3B). In striking contrast, cells devoid of paxillin invading through the 3D ECM exhibited a significant increase in Morphology Index (Figure 3B), highlighting the predominantly elongated, mesenchymal morphology, similar to that observed previously in paxillin-depleted A375 melanoma cells (Pinner and Sahai, 2008). Conversely, Hic-5 siRNA-treated cells exhibited a significantly decreased Morphology Index (Figure 3B), indicative of their amoeboid phenotype. Immunofluorescence analyses using paxillin and Hic-5–specific monoclonal antibodies confirmed efficient protein knockdown in cells displaying the elongated mesenchymal and amoeboid phenotypes, respectively (Supplemental Figure 3). The distinct morphologic differences seen on siRNA knockdown of paxillin or Hic-5 appear to be independent of the microenvironment organization as similar phenotypes were observed during cell invasion through the highly disorganized 3D collagen and FN ECM of the inverted plug assay as well as the tumor-stroma interface-like 3D CDM (Figure 3C). The 3D nature of cells spread in 3D CDMs was confirmed using confocal analyses, which revealed FN-rich ECM fibers surrounding the majority (approximately 70–80%) of the cells imaged (unpublished data).
FIGURE 3:
Breast cancer cell plasticity is controlled by paxillin and Hic-5. (A) Schematic indicating the Morphology Index calculation to measure plasticity. MDA-MB-231 cell circularity and Feret’s diameter, also known as caliper length, were measured using ImageJ, and the ratio of diameter to circularity was calculated. Cells with a higher calculated Morphology Index exhibit mesenchymal characteristics, whereas lower calculated Morphology Index indicates an amoeboid morphology. (B) Quantification of Morphology Index for cells invading through collagen and FN inverted invasion assay plugs. Data represent mean ± SEM of >500 cells/siRNA from a minimum of four individual experiments. (C) Immunofluorescence images of FN (red) and F-actin (green) of cells treated with control (left panels), paxillin-1 (middle panels), and Hic-5–1 siRNA (right panels) allowed to migrate in 3D collagen and FN matrices (top panels) and CDMs (bottom panels) for 16 h. (D) Quantification of cells exhibiting plasticity during cell migration through 3D ECM. Cells were allowed to migrate in 3D CDMs for 16 h with images taken every 10 min. Data represent mean ± SEM of >50 cells from three individual experiments. (E) Representative mask images of cell morphology transitions during MDA-MB-231 cell migration through 3D CDMs. Statistical analyses of Morphology Index data was performed using a Kruskal–Wallis test followed by Dunn’s post hoc test, * p < 0.01, ** p < 0.001, and *** p < 0.0001. A Student’s t test was used for statistical analysis of plasticity data, ** p < 0.005 and *** p < 0.0005.
In addition to the dramatic effect on overall cell morphology, analysis of real-time cell invasion through 3D CDMs revealed that, whereas approximately 70% of control siRNA cells exhibited phenotypic switching during the 16-h time course of invasion, RNAi of either paxillin or Hic-5 significantly reduced the ability of the cells to exhibit migrational plasticity, with cells remaining highly mesenchymal or amoeboid, respectively (Figure 3, D and E).
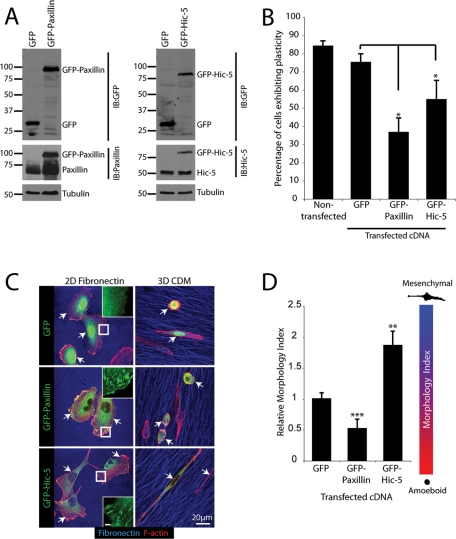
These data suggested that the balance of paxillin and Hic-5 levels may be critical for determining cell morphology in 3D microenvironments. We therefore reasoned that overexpression of paxillin or Hic-5 may produce the converse phenotypes. Accordingly, green fluorescent protein (GFP)-paxillin or GFP-Hic-5 was expressed in parental MDA-MB-231 cells (Figure 4A). As anticipated, the paxillin-expressing cells exhibited an adhesion-independent, amoeboid phenotype when spread in 3D CDMs, whereas GFP-Hic-5–expressing cells displayed a highly elongated mesenchymal morphology with GFP-Hic-5 displaying 3D adhesion contact localization (Figure 4, C and D). Furthermore, expression of either paxillin or Hic-5 significantly reduced the cells’ ability to exhibit plasticity during 16 h of 3D CDM migration relative to both untransfected and GFP-expressing cells (Figure 4B). Importantly, despite the lack of GFP-paxillin localization to adhesions in cells in 3D CDMs, both GFP-tagged proteins were able to localize to adhesion contacts when cells were spread on a 2D FN matrix (Figure 4C). Taken together, these data identify paxillin and Hic-5 as critical controllers of breast cancer cell amoeboid and mesenchymal invasion strategies and indicate the necessity for their functional balance to enable efficient plasticity-associated morphology transitions.
FIGURE 4:
Overexpression of paxillin and Hic-5 promotes MDA-MB-231 morphology transitions and inhibits plasticity. (A) Representative Western blot of MDA-MB-231 cells overexpressing pEGFPC1, pEGFPC1-paxillin, and pEGFPC1-Hic-5. (B) Quantitation of cells exhibiting plasticity during cell migration through 3D CDM for 16 h. (C) Representative immunofluorescence images, F-actin (red) and FN (blue), of cells spread on 2D and 3D ECM for 16 h in the presence of serum. Arrows indicate transfected cells. Inset images of the GFP channel indicate appropriate localization of pEGFPC1-paxillin and pEGFPC1-Hic-5 to adhesion contacts upon cell spreading on 2D FN. Inset scale bar = 2 μm. (D) Quantification of Morphology Index for cells migrating through 3D CDMs. Data represent mean ± SEM of a minimum of 20 cells from a minimum of four individual experiments. Statistical analyses of Morphology Index data was performed using a Kruskal–Wallis test followed by Dunn’s post hoc test, ** p < 0.001 and *** p < 0.0001.
Hic-5 is necessary for robust adhesion formation during invasion in 3D ECM
The mesenchymal mode of cancer cell invasion is adhesion-dependent, whereas the amoeboid phenotype is characterized by a distinct lack of robust integrin-mediated adhesions, with motility being driven by the formation of cortical membrane blebs that permit the cell to squeeze between the matrix constituents (Sahai and Marshall, 2003; Pinner and Sahai, 2008). As with control cells that were exhibiting the mesenchymal morphology, the mesenchymal paxillin-depleted cells displayed robust vinculin-positive 3D adhesions (Figure 5A). In contrast, vinculin displayed a primarily cytoplasmic distribution in the amoeboid Hic-5 RNAi cells when plated in 3D ECM (Figure 5A, right panels). Importantly, both paxillin RNAi- and Hic-5 siRNA–treated cells formed robust vinculin-rich adhesion contacts upon cell spreading on 2D FN (Figure 5B) or collagen (Supplemental Figure 4). Strong phospho-paxillin staining was also observed in all Hic-5 RNAi cells plated on a 2D matrix that were negative for Hic-5 antibody staining, indicating that these cells retain the capacity for adhesion-dependent integrin signaling (Figure 5B and Supplemental Figure 4). These data confirm that the amoeboid phenotype observed in cells lacking Hic-5 is not due to an inherent defect in their ability to form adhesion contacts but rather highlight significant differences in paxillin and Hic-5 function in 2D and 3D microenvironments.
FIGURE 5:
Hic-5 silencing promotes the adhesion-independent amoeboid phenotype in 3D ECM but does not prevent adhesion contact formation on 2D ECM. Immunofluorescence images of MDA-MB-231 cells treated with siRNA as stated for 69 h and then allowed to migrate (A) in 3D CDM or (B) on 2D FN-coated coverslips for 3 h. Cells were fixed and stained as indicated. All cells analyzed in four individual experiments, which were devoid of Hic-5 as assessed using the Hic-5-specific antibody, displayed the bleb-rich, adhesion-independent amoeboid phenotype in 3D ECM and robust adhesions on 2D FN.
Hic-5 regulates morphology and invasion through RhoA signaling
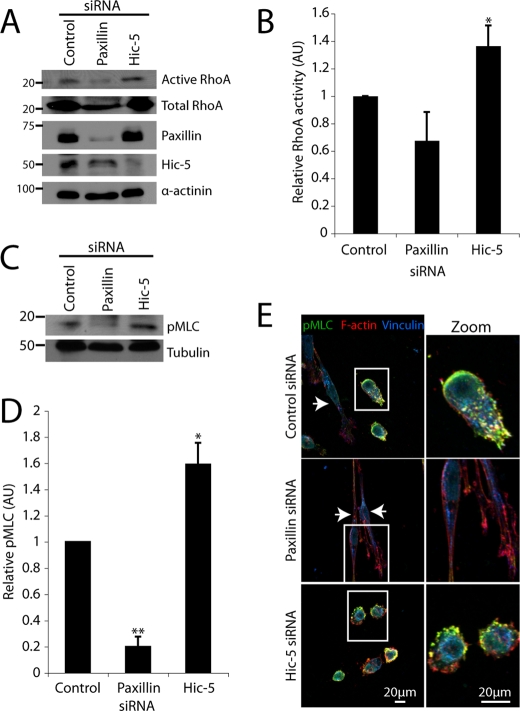
In contrast to the Rac1-mediated mesenchymal phenotype, amoeboid cell motility in 3D matrices is driven primarily by the activity of RhoA and its downstream effector ROCK (Sahai and Marshall, 2003; Yoshida and Soldati, 2006; Lammermann et al., 2008; Sanz-Moreno et al., 2008). Therefore, we hypothesized that the activity of RhoA and its effectors may be altered in the amoeboid Hic-5–deficient cells (Figure 3, B and C). Biochemical analysis of global RhoA activity in Hic-5 RNAi cells confirmed that it was significantly elevated relative to control siRNA-treated cells. A reproducible, but of low statistical significance, decrease in RhoA activity was observed in cells devoid of paxillin expression relative to controls (Figure 6, A and B).
FIGURE 6:
Silencing Hic-5 promotes an amoeboid morphology through elevation of RhoA and nonmuscle myosin II activity. (A) Representative Western blot analysis of RhoA activity in cells migrating through 3D ECM as assessed by GST-RBD (Rhotekin RhoA binding domain) pulldown assay. (B) Quantification of RhoA activity in cells treated with siRNA as indicated. Data represent mean ± SEM of three individual experiments. (C) Western blot analysis and (D) quantification of pMLC upon RNAi knockdown of paxillin or Hic-5 relative to control siRNA. (E) Immunofluorescence analysis of F-actin (red), vinculin (blue), and pMLC (green) localization in MDA-MB-231 cells treated with siRNA spread in 3D ECM for 16 h. A Student’s t test was used for statistical analyses, * p < 0.05 and ** p < 0.005.
Elevated nonmuscle myosin II light chain phosphorylation (pMLC) has also been observed in amoeboid cell populations (Wyckoff et al., 2006; Gadea et al., 2008; Sanz-Moreno et al., 2008) as a result of ROCK activation (Riento and Ridley, 2003) and is associated with increased contractility of the cortical actomyosin cytoskeleton (Chrzanowska-Wodnicka and Burridge, 1996). In accordance with the elevated RhoA activity, pMLC was also increased in cells devoid of Hic-5 (Figure 6, C and D). In contrast, pMLC was significantly decreased in cells treated with paxillin RNAi relative to control (Figure 6, C and D). Immunofluorescence localization of pMLC revealed enrichment in actin-rich blebs at the cell periphery in Hic-5 siRNA-treated cells (Figure 6E, bottom panels). Consistent with Western blot analysis (Figure 6, C and D), pMLC staining was decreased in cells devoid of paxillin expression (Figure 6E, middle panels, arrowheads).
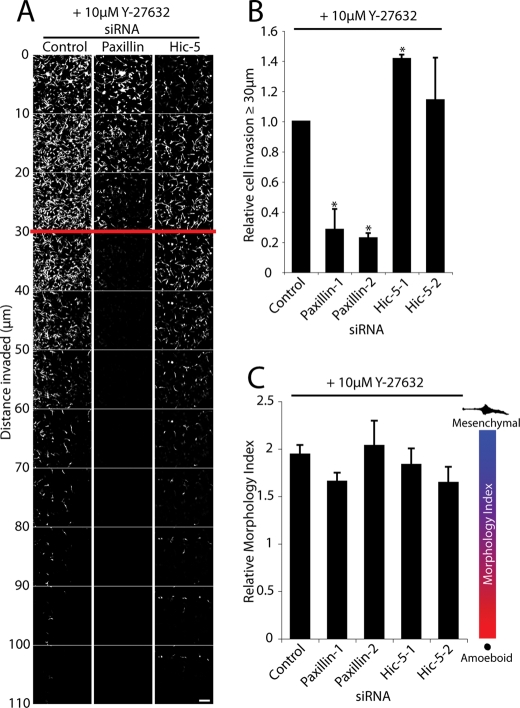
Inhibition of Rho signaling via ROCK through treatment of the Hic-5 knockdown cells with the ROCK inhibitor Y-27632 rescued their ability to invade through the ECM to levels comparable with Y-27632–treated control siRNA cells (Figure 7, A and B). Importantly, inhibition of ROCK activity also converted the Hic-5 RNAi cells to a more mesenchymal phenotype (Figure 7C), as previously observed following ROCK inhibition in other amoeboid cancer cell lines (Sahai and Marshall, 2003; Carragher et al., 2006; Wyckoff et al., 2006; Sahai et al., 2007; Gadea et al., 2008; Sanz-Moreno et al., 2008). In contrast, inhibition of ROCK activity in paxillin siRNA-treated cells did not enhance their invasive capacity (Figure 7, A and B). Treatment of control RNAi cells with Y-27632 induced a shift toward the mesenchymal phenotype with cells exhibiting a significant increase in Morphology Index, relative to control cells in the presence of vehicle. These data suggest that the hyperamoeboid phenotype and decreased motility observed in breast cancer cells lacking Hic-5 are due to the dysregulated activity of RhoA and myosin-based contractility.
FIGURE 7:
Inhibition of ROCK activity rescues invasion of Hic-5 RNAi–treated cells. (A) Invasive migration of MDA-MB-231 cells treated with siRNA in the presence of 10 μM Y-27632 (ROCK inhibitor) migrating through 3D collagen with FN at 25 μg/ml was determined by using an inverted invasion assay. Cells were stained with Calcein AM and visualized by laser scanning confocal microscopy. Optical sections were taken at 10-μm intervals. (B) Cell invasion was quantified using ImageJ by measuring the number of cells, as indicated by Calcein AM staining, in each optical section. The total number of cells invading >30 μm into the collagen and FN were quantified relative to the total number of cells counted. Data represent mean ± SEM from a minimum of three individual experiments. (C) Morphology Index calculation for RNAi-treated cells in the presence of 10 μM Y-27632. A Student’s t test was used for statistical analyses, * p < 0.05. Scale bar = 150 μm.
Paxillin coordinates 3D adhesion dynamics to regulate mesenchymal invasion
The spatiotemporal coordination of adhesion formation, maturation, and subsequent dissolution is required for efficient, persistent migration (Broussard et al., 2008). Similarly, the mesenchymal mode of migration through 3D environments requires robust, yet dynamic integrin-mediated cell–ECM interactions (Wolf et al., 2003). Importantly, the dynamics of cell–ECM adhesions during 3D mesenchymal migration and indeed the role of any adhesion regulatory protein, including paxillin, in coordinating these events has not been determined.
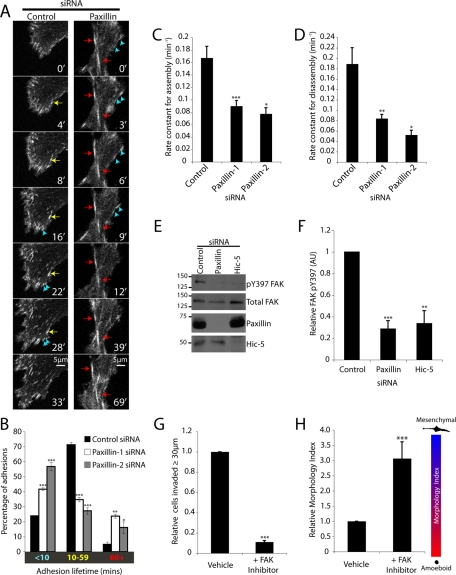
Paxillin has been reported to participate in the control of adhesion dynamics during migration on 2D ECM-coated surfaces (Webb et al., 2004). Therefore, we hypothesized that persistent 3D migration in paxillin-depleted cells may be inhibited due to altered adhesion dynamics. Paxillin RNAi-depleted MDA-MB-231 cells were subsequently transduced with a GFP-talin encoding baculovirus to enable visualization and analysis of adhesion dynamics during migration in 3D CDMs, which included evaluation of adhesion lifetime as well as determination of assembly and disassembly rate constants. Mesenchymal control RNAi cells (as defined by the presence of talin-rich adhesion contacts) demonstrated coordinated adhesion assembly and disassembly resulting in persistent cell locomotion (Figure 8A and Supplemental Movie 4). Interestingly, the adhesions in control cells were highly polarized toward the front and sides (with few adhesions toward the cell rear) and exhibited a range of lifetimes (Figure 8B). In contrast, paxillin RNAi cells produced an increase in the number of highly dynamic, peripheral, short-lived adhesions (<10 min) (Figure 8A, blue arrows, and Supplemental Movie 5) as well as a significant increase in elongated, stable adhesions located more centrally than the dynamic polar protrusions, which persisted for extended time periods (>60 min) (Figure 8A, red arrows, and Supplemental Movie 5). Quantification of adhesion lifetimes confirmed that paxillin RNAi treatment induced a significant increase in the percentage of adhesions persisting for <10 and >60 min as compared to control siRNA-treated cells (Figure 8B). In addition, paxillin knockdown resulted in highly significant decreases in both 3D adhesion assembly (Figure 8C) and disassembly (Figure 8D) rates. These data suggest that paxillin plays multiple roles in the regulation of adhesions required for persistent 3D invasion: the stabilization and/or maturation of nascent adhesions at the cell periphery and, once formed, the disassembly of more stable central adhesion contacts. Furthermore, these data also suggest potentially different functions for paxillin in 2D and 3D adhesion contact dynamics, as no decrease in assembly rate or change in adhesion populations were observed in cells devoid of paxillin migrating on 2D ECM (Webb et al., 2004). We were unable to perform a similar analysis of adhesion dynamics in Hic-5–depleted MDA-MB-231 cells due to the absence of robust integrin-mediated GFP-talin–positive adhesions in amoeboid cells (unpublished data).
FIGURE 8:
Paxillin regulates 3D adhesion dynamics and nascent adhesion stability during mesenchymal invasion. (A) Representative time-lapse images of cells exhibiting mesenchymal migration in 3D ECM expressing GFP-Talin and treated with siRNA for 72 h. Blue arrows indicate adhesions with a lifetime of <10 min, yellow arrows indicate adhesions persisting between 10 and 60 min, and red arrows indicate adhesions with lifetimes >60 min. (B) Bar graphs indicating the 3D adhesion lifetime for cells migrating through 3D ECM, cells imaged by time-lapse microscopy at 1-min intervals for 3.5 h. (C) Quantification of average adhesion assembly and (D) disassembly rates. Data represent mean ± SEM of a minimum of 56 individual adhesions from six to eight cells (from three individual experiments) for control and paxillin-1 siRNA and 12 adhesions from three cells for paxillin-2 siRNA. All adhesions that could be followed from initial assembly to dissolution were measured. (E) MDA-MB-231 cells were treated with siRNA for 56 h as indicated and were allowed to migrate in 3D ECM for 16 h. Cell lysates were then analysed by Western immunoblotting for FAK pY397 levels. (F) Bar graph indicating relative FAK phosphorylation in cells treated with RNAi as indicated. (G) Quantification of parental MDA-MB-231 cell invasion and (H) relative Morphology Index for cells treated with vehicle or 2 μM PF 573228 (FAK inhibitor). A Student’s t test was used for analysis of Western blot densitometry, adhesion lifetime, and invasion. A Mann–Whitney U test was used for analysis of Morphology Index, and a Kruskal–Wallis test followed by Dunn’s post hoc test was performed for statistical analyses of adhesion dynamics, * p < 0.01, ** p < 0.001, and *** p < 0.0001.
Previous reports have indicated that paxillin controls 2D adhesion dynamics in part through regulating the localized recruitment and activity of FAK (Webb et al., 2004; Zaidel-Bar et al., 2007). In our 3D CDM system, the activity of FAK, as assessed by Y397 phosphorylation, was decreased in cells lacking paxillin as compared to control MDA-MB-231 cells (Figure 8, E and F). Hic-5 RNAi-treated cells also exhibited a significant decrease in FAK phosphorylation (Figure 8, E and F), presumably due to the decrease in integrin signaling associated with the amoeboid phenotype. Interestingly, treatment of parental MDA-MB-231 cells with the selective FAK inhibitor PF 573228 (Slack-Davis et al., 2007; Jones et al., 2009) prevented 3D ECM invasion (Figure 8G) and converted parental cells to a highly elongated mesenchymal phenotype (Figure 8H) analogous to that observed following knockdown of paxillin (Figure 3, B and C). Although correlative, given the well-characterized physical and functional association between paxillin and FAK (Bellis et al., 1995; Tachibana et al., 1995; Bertolucci et al., 2005, 2008; Shan et al., 2009; Yu et al., 2009; Zouq et al., 2009), these data suggest that paxillin may regulate persistent 3D invasion and cell morphology in part through modulating FAK activation-dependent adhesion turnover.
Paxillin and Hic-5 mediate breast cancer cell lung metastasis
Paxillin and Hic-5 have both been linked to malignant progression in a variety of invasive and/or metastatic cancers, including breast and prostate tumors (Rodina et al., 1999; Fujimoto et al., 2001; Mestayer et al., 2003; Miyoshi et al., 2003; Nagata et al., 2003; Zhao et al., 2004; Azuma et al., 2005; Farmer et al., 2005; Radvanyi et al., 2005; Cui et al., 2006; Richardson et al., 2006; Turashvili et al., 2007; Kanteti et al., 2009; Shi et al., 2010; Yang et al., 2010). Thus, having shown distinct roles for both paxillin and Hic-5 in the regulation of invasion in 3D in vitro systems, and given the importance of ECM invasion in cancer metastasis and concomitant patient survival, we examined the metastatic potential in vivo of cells depleted of paxillin or Hic-5.
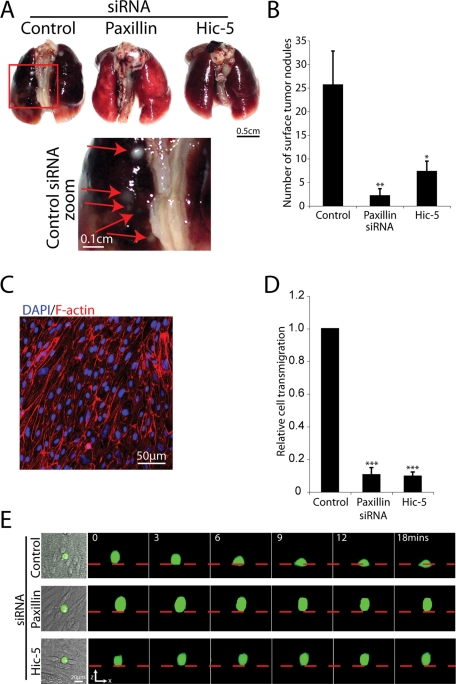
The highly invasive MDA-MB-231 cells were initially isolated from the pleural effusion of a patient with widespread metastases (Cailleau et al., 1978) and are known to metastasize to multiple sites, including the lung (Yoneda et al., 2001; Kang et al., 2003; Minn et al., 2005; Bos et al., 2009). In contrast to control RNAi cells, intravenous tail vein injection of both paxillin- and Hic-5–depleted cells resulted in a highly significant reduction in surface lung metastases (Figure 9, A and B) as assessed using the NOD/SCID experimental lung metastasis model. Histological analysis of lung tissue by hematoxylin and eosin staining revealed extensive metastases throughout the lung tissue of mice that received an injection of control siRNA-treated tumor cells, whereas the lungs isolated from mice that received MDA-MB-231 cells devoid of paxillin or Hic-5 exhibited no evidence of metastatic lesions (unpublished data).
FIGURE 9:
RNAi-mediated silencing of paxillin or Hic-5 prevents MDA-MB-231 cell transendothelial migration and metastasis to the lung. (A) Representative images of lungs removed from mice that received MDA-MB-231 cells treated with control, paxillin-1, and Hic-5–1 siRNAs as indicated. Red box indicates area of zoom in control siRNA-treated lungs. (B) Quantification of lung surface metastases (n = 9 mice/siRNA). (C) Immunofluorescence staining of F-actin (red) and DAPI (blue) of bovine aorta endothelial cell monolayer used for transendothelial migration assay. (D) Quantification of the percentage of total cells transmigrated as assessed by real-time laser scanning confocal microscopy. MDA-MB-231 cells treated with control, paxillin-1, or Hic-5–1 siRNA as indicated for 72 h were stained with the Calcein AM vital dye and were allowed to transmigrate through the confluent monolayer of endothelial cells for 4.5 h. Data represent mean ± SEM of 16 movies from four individual experiments; a Student’s t test was used for statistical analysis, *** p < 0.0005. (E) Representative 3D confocal reconstruction of MDA-MB-231 cells stained with Calcein AM (green), transmigrating through an endothelial monolayer (phase contrast). Dashed red line indicates surface of the endothelial monolayer as indicated by phase contrast imaging. Scale bar = 20 μm.
To determine whether extravasation, as well as 3D ECM invasion, was affected by paxillin or Hic-5 knockdown, RNAi-treated MDA-MB-231 cells were seeded on a confluent monolayer of bovine aorta endothelial cells, and their ability to transmigrate the endothelium to the underlying ECM was assessed (Figure 9C). Control siRNA-treated cells were able to exhibit efficient transendothelial migration (Figure 9, D and E). In contrast, cells treated with paxillin-1 or paxillin-2 displayed a 89.5% ± 4.5 (Figure 9, D and E) and 62.0% ± 4.1 (unpublished data; p < 0.0005) decrease in transendothelial migration, respectively. Treatment with Hic-5–1 or Hic-5–2 RNAi produced a similar reduction in transendothelial migration relative to control cells by 93.2% ± 2.3 (Figure 9, D and E) and 63.4% ± 2.9 (unpublished data; p < 0.0005), respectively. Therefore, as cell viability and growth analyses over the initial 144-h postinjection time frame (which encompasses both extravasation and invasion) revealed no significant differences (Supplemental Figure 5), the failure to form lung metastases in paxillin and Hic-5 RNAi cells is likely due to defects in transendothelial migration as well as 3D ECM invasion. At this time, however, we cannot discount alternative mechanisms, such as a role for paxillin or Hic-5 in regulating tumor cell adhesion to the endothelium, seeding, or growth in vivo.
DISCUSSION
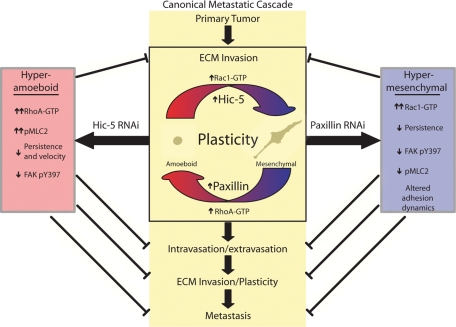
Cancer cell invasion and subsequent metastasis is considered the single worst prognostic factor and is associated with increased malignancy and decreased patient survival. Therefore, understanding the intracellular mechanisms controlling metastasis-related processes is of paramount importance. Using a combination of in vitro and in vivo model systems, we have identified novel roles for both paxillin and Hic-5 at multiple levels of the canonical metastasis cascade (summarized in Figure 10). Furthermore, using both RNAi-mediated knockdown and overexpression approaches, we have revealed that a balance of signaling through these highly related proteins is required for the coordination of breast cancer cell migration strategies through 3D ECM. Indeed, data herein reveal that although paxillin and Hic-5 serve opposing roles in determining cell morphology in 3D microenvironments, both proteins are necessary for optimal migration and/or invasion of either the amoeboid or mesenchymal cancer cell phenotype.
FIGURE 10:
Schematic identifying the respective roles of paxillin and Hic-5 in the canonical metastatic cascade. Paxillin and Hic-5 impact the canonical metastasis cascade at several levels through their ability to counterbalance Rho GTPase signaling and thus coordinate plasticity and efficient mesenchymal and amoeboid invasion, respectively.
Efficient mesenchymal migration on 2D ECM substrata requires the scaffold function of paxillin to coordinate adhesion disassembly (Webb et al., 2004) and to spatiotemporally localize both structural and signaling components of integrin-mediated adhesion contacts (reviewed in Brown and Turner, 2004; Deakin and Turner, 2008). In contrast, the function of Hic-5 in 2D migration remains largely unexplored. Studies using Hic-5 overexpression have shown a role for the protein in preventing Rac1-driven, epidermal growth factor (EGF)-stimulated lamellipodia formation (Hetey et al., 2005) and in inhibition of cell spreading on 2D FN through preventing paxillin–FAK interaction and signaling (Nishiya et al., 2001), highlighting cross-talk between these closely related proteins. Importantly, the roles of paxillin and Hic-5 in 3D migration have yet to be elucidated. Furthermore, few studies have directly compared the relative roles of paxillin and Hic-5 in the same cell type and assays.
Consistent with 2D analyses in which cell migration was reduced (Hagel et al., 2002), MDA-MB-231 breast cancer cells devoid of paxillin exhibited a highly elongated hypermesenchymal phenotype when seeded in 3D matrices, as compared to control mesenchymal cells, resulting in inefficient 3D ECM migration, with decreases observed in both persistence and velocity (Figure 2, B–D) and culminating in decreased invasion (Figure 1, C and D). Persistent 3D migration requires moderate levels of restricted Rac1 activity to prevent peripheral protrusions frequently observed during 2D migration. Indeed, reduction of Rac1 activity during 2D migration has been shown to promote 3D-like persistence (Pankov et al., 2005). Therefore, the lack of persistence observed in paxillin RNAi-treated cells may be in part due to the significantly increased levels of active Rac1 (Figure 2, F and G), which promote increased unstable lateral protrusions (Supplemental Movie 5). Furthermore, paxillin-depleted cells exhibited a significant decrease in the activation of nonmuscle myosin II (Figure 6, C and D), pharmacologic inhibition of which has also been shown to decrease fibroblast 3D migration velocity (Even-Ram et al., 2007). Although the current study clearly demonstrates a novel role for paxillin in modulating RhoGTPase signaling during 3D cell migration, given the ability of paxillin to influence both Rac1 and RhoA activity through multiple members of its interactome in 2D systems (reviewed in Deakin and Turner, 2008), further work will be required to identify the precise mechanisms involved.
Hic-5 shares a similar interactome with paxillin due to its highly conserved domain structure (Thomas et al., 1999; Brown and Turner, 2004). Despite this fact, data presented herein indicate that, in striking contrast to paxillin knockdown, Hic-5 depletion results in an amoeboid phenotype, enhanced RhoA activity, and downstream signaling to nonmuscle myosin II and ROCK (Figures 6 and 7), indicative of nonredundant or antagonistic cellular functions during 3D ECM migration. Amoeboid motility requires actomyosin contractility-dependent cell plasma membrane bleb formation and retraction (reviewed in Fackler and Grosse, 2008). Therefore, the elevated RhoA and pMLC activity in the Hic-5–depleted cells (Figure 6) may lead to the disregulation of bleb dynamics resulting in the observed reduction in invasion (Figure 1, C and D) and 3D migration (Figure 2, C and D). Interestingly, Hic-5 has previously been implicated in suppressing actomyosin-mediated force generation through its translocation from adhesion contacts to actin stress fibers upon the stimulation of contractility in smooth muscle and osteoblast-like cells (Kim-Kaneyama et al., 2005; Guignandon et al., 2006). Conversely, increased Hic-5 expression during transforming growth factor β–induced EMT in epithelial cells cultured on 2D ECM has been shown to promote RhoA activity, increased actin stress fiber formation, and cell migration (Tumbarello and Turner, 2007), suggesting that Hic-5 function in regulating RhoA signaling may be cell type and/or context specific.
In contrast to 2D studies, in which paxillin only regulates adhesion disassembly rates (Webb et al., 2004), analysis of adhesion dynamics during 3D migration identified a role for paxillin in coordinating multiple facets of adhesion lifetime including assembly, stabilization, and disassembly (Figure 8). As with 2D adhesions, the decreased 3D adhesion disassembly rate observed in paxillin-depleted cells may be in part due to decreased FAK activity (Figure 8, E and F). The spatiotemporal activities of RhoA and MLC have also been shown to be necessary for adhesion disassembly at the cell rear (Worthylake et al., 2001). Given the highly significant decrease in pMLC and the small but consistent decrease in global RhoA activity observed in paxillin-depleted cells (Figure 6), it is conceivable that their reduced levels may influence adhesion disassembly despite the lack of a defined cell rear (Figure 2E and Supplemental Movie 2).
Non-muscle myosin II–mediated contractility, as well as the activation of FAK, is also known to regulate 2D adhesion maturation and strengthening (Bertolucci et al., 2005; Giannone et al., 2007; Choi et al., 2008; Shan et al., 2009; Dumbauld et al., 2010; Rossier et al., 2010). Therefore, it is possible that the defective adhesion stabilization and subsequent increase in short-lived adhesions observed in paxillin RNAi cells (Figure 8B) may be due to the observed decreases in both pMLC (Figure 6, C and D) and FAK activity (Figure 8, E and F). Similar to our observations of paxillin RNAi cells during 3D migration, unstable dysregulated protrusion and adhesion formation has also been reported in cells migrating on 2D ECM expressing the LD4 deletion mutant of paxillin (West et al., 2001). Furthermore, abrogated signaling through the FAK-interacting LD4 motif (Brown et al., 1996; Bertolucci et al., 2008) was also shown to promote elevated Rac1 activity (West et al., 2001). Therefore, it is possible that paxillin scaffold function through its LD4 motif is necessary for FAK and/or Rac1-driven adhesion and protrusion stabilization in 3D ECM.
Although paxillin is one of the first proteins recruited to integrin-mediated 2D adhesions (Zaidel-Bar et al., 2003; Ballestrem et al., 2006), we found that it is unable to efficiently nucleate or signal to stabilize 3D adhesions in the absence of Hic-5, resulting in an amoeboid phenotype (Figure 5A, right panels). Importantly, the MDA-MB-231 cells, when depleted of Hic-5, retain the ability to form robust integrin-mediated adhesion contacts upon spreading on 2D ECM (Figure 5B and Supplemental Figure 4), indicating ECM topography-specific functions for Hic-5. Furthermore, our analysis of adhesion dynamics during 3D migration suggests a hitherto underappreciated facet of plasticity regulation. For a cell exhibiting mesenchymal invasion to transition to the adhesion-independent amoeboid mode of motility it must efficiently disassemble its robust integrin 3D matrix adhesions. Conversely, efficient adhesion assembly and stabilization are necessary for the amoeboid-to-mesenchymal transition. To our knowledge, this is the first study to assess 3D adhesion dynamics and has identified paxillin as a key protein in controlling plasticity, possibly through coordinating efficient adhesion dynamics (Figure 8). Importantly, the fact that cells depleted of Hic-5 were unable to form robust integrin adhesions in 3D ECM (Figure 5A) and therefore could not exhibit the amoeboid-to-mesenchymal transition (Figure 3) suggests a novel role for Hic-5 in 3D adhesion formation.
Analysis of cell morphology in the highly organized CDMs revealed a clear role for paxillin and Hic-5 in controlling MDA-MB-231 cell plasticity (Figure 3). Similar phenotypes were also observed in the less organized, highly cross-linked 3D collagen and FN ECM (Figure 3C), suggesting that paxillin and Hic-5 signaling may supersede external ECM organization-driven cues to regulate cell morphology and thus plasticity. Furthermore, the significant decrease in plasticity observed upon either paxillin or Hic-5 depletion (Figure 3D) or overexpression (Figure 4B) highlights that a balance of signaling through these proteins is necessary for effective phenotypic switching (Figure 10). Given the similarity between paxillin and Hic-5 and their interactomes, their individual functions may be modulated by discrete differences in binding affinities, phosphorylation, and spatiotemporal localization. Indeed, the affinity for several shared interacting proteins, including FAK and GIT1, have been shown to differ between paxillin and Hic-5 (Fujita et al., 1998; Nishiya et al., 2002). Furthermore, Hic-5 has also been shown to influence paxillin phosphorylation and thus alter its molecular interactions (Fujita et al., 1998; Nishiya et al., 2001), suggesting that extensive cross-talk between these closely related proteins may also be required to fine-tune their cellular functions.
MATERIALS AND METHODS
Antibodies and reagents
Routine assessment of siRNA efficiency was performed using mouse anti-paxillin (clone 165) and mouse anti-Hic-5 (BD Biosciences, Franklin Lakes, NJ) for both Western blotting and immunofluorescence analyses. Other antibodies used for both Western blotting and immunofluorescence include mouse anti-Rac1 and mouse anti-FAK (BD Biosciences); rabbit anti-RhoA and rabbit anti-GFP (Santa Cruz Biotechnology, Santa Cruz, CA); mouse anti-α-actinin, mouse anti-tubulin (clone DM1A), and rabbit anti-fibronectin (Sigma Aldrich, St. Louis, MO); rabbit anti-FAK pY397 (United Biosource, Bethesda, MD); rabbit anti-pMLC (Cell Signaling Technology, Beverly, MA); and rabbit anti-collagen I (Chemicon, Temecula, CA). Rhodamine-conjugated phalloidin (Invitrogen, Carlsbad, CA) was used for fluorescent detection of F-actin. The FAK inhibitor, PF 573228, was used at a concentration of 2 μM and was purchased from Tocris Bioscience (Ellisville, MO). The ROCK inhibitor, Y-27632, was used at 10 μM and was purchased from EMD Chemicals (San Diego, CA).
siRNA and GFP-tagged protein transfection
Cells were transfected with two separate siRNAs (designated 1 or 2) to either paxillin or Hic-5 as indicated for 72 h, unless stated otherwise, using Oligofectamine (Invitrogen, Carlsbad, CA) following the manufacturer’s instructions and using a concentration of 0.1 μM for each oligonucleotide. The paxillin-1 siRNA oligo was purchased from Dharmacon (Lafayette, CO), and all other siRNA oligos used were purchased from Ambion (Austin, TX) without modifications. The siRNA sequences are as follows: paxillin-1, 5′-CCCUGACGAAAGAGAAGCCUA-3′ and 5′-UAGGCUUCUCUUUCGUCAGGG-3′; paxillin-2, 5′-GUGUGGAGCCUUCUUUGGU-3′ and 5′-ACCAAAGAAGGCUCCACAC-3′; Hic-5–1, 5′-GGAGCUGGAUAGACUGAUG-3′ and 5′-CAUCAGUCUAUCCAGCUCC-3′; Hic-5–2, 5′-GGACCAGUCUGAAGAUAAG-3′ and control, 5′-ACUCUAUCUGCACGCUGACUU-3′ and 5′-GUCAGCGUGCAGAUAGAGUUU-3′. The control nonspecific siRNA was used in all experiments as indicated.
Cells were transfected with pEGFPC1, pEGFPC1-paxillin, or pEGFPC1-Hic-5 using Lipofectamine LTX (Invitrogen, Carlsbad, CA) following the manufacturer’s instructions. Transfection efficiencies were between approximately 50% and 60% for pEGFPC1 and pEGFPC1-paxillin and between approximately 30% and 40% for pEGFPC1-Hic-5.
Inverted collagen and FN invasion assay
The inverted collagen and FN invasion assay was performed as described previously (Hennigan et al., 1994; Caswell et al., 2007). Collagen I (PureCol, Advanced Biomatrix, San Diego, CA), mixed 5:1 with 10× MEM (modified Eagles medium), supplemented with FN at 25 μg/ml, was allowed to polymerize in transwell inserts (Corning, NY) for 1 h at 37°C in the absence of CO2. Parental or 24-h siRNA-treated MDA-MB-231 cells were resuspended at 1 × 105 cells/ml in serum-free medium and were seeded on the inverted transwell, on the filter surface. Cells were allowed to adhere for 30 min at 37°C in the presence of CO2. Transwells were then washed once with PBS and placed in 1 ml of serum-free medium, with addition of inhibitors where stated. To establish a chemotactic gradient, medium supplemented with 10% fetal bovine serum, and inhibitors where stated, was placed in the upper chamber of the transwell. Five days after seeding, cells were stained with Calcein AM (Invitrogen, Carlsbad, CA) and visualized using the HC PL APO 20×/0,70 IMM CORR λ BL objective (Leica, Bannockburn, IL) and Leica SP5 laser scanning confocal microscopy. Images were acquired in 10-μm intervals up to 110 μm from the transwell surface as determined by phase contrast imaging. Total cells in all images of the confocal series were quantified, and the relative percentage of cells invading ≥30 μm into the collagen and fibronectin plug was determined.
Generation of CDMs
Three-dimensional CDMs were derived as previously described (Bass et al., 2007). Briefly, glass-bottom dishes (MatTek, Ashland, MA), 10-cm dishes, and glass coverslips were coated with 0.2% sterile gelatin for 30 min at 37°C. Glass-bottom dishes and glass coverslips were subsequently treated with 1% glutaraldehyde to cross-link the gelatin and then quenched with 1 M glycine. After incubation with complete growth medium, 1 × 105 cells/ml primary human foreskin fibroblasts (HFF) were seeded onto each dish and cultured for 10 days. Cell growth medium was changed every 2 days and supplemented with ascorbic acid at 50 μg/ml to stabilize the collagen matrix. The matrices were denuded of cells by treating the HFF monolayers with 20 mM NH4OH, 0.5% Triton X-100 (vol/vol), and PBS for 2 min at 37°C. After repeated washings with PBS, CDMs were treated with DNase I at 10 μg/ml (Roche Diagnostics, Indianapolis, IL) in PBS containing magnesium and calcium.
CDM migration assays and analyses
Parental or siRNA-treated MDA-MB-231 as indicated were allowed to spread on CDMs for 4 h in the presence of serum (± vehicle or inhibitors as indicated). Time lapse imaging was performed on a Leica AF6000 Deconvolution with images acquired every 10 min for 16 h using a HCX PL FLUOTAR 10×/0.30 objective (Leica, Bannockburn, IL). ImageJ software was used to track cell centroid movement and to calculate migration velocity and persistence of all cells present in the field throughout the time course. The persistence was calculated by dividing the linear displacement of the cell centroid over the 16-h time period by the total net distance migrated (Pankov et al., 2005). Cell front–rear correlation analysis was also performed using the ImageJ software and was calculated in each frame of the time series by measuring the relative distance moved by the cell front and rear from the origin.
RhoGTPase activity assays
MDA-MB-231 cells were treated with siRNA as indicated for 56 h and then were seeded at 1 × 105 cells/ml in the presence of serum on 35-mm dishes coated in CDM for 16 h. Rac activity assays were performed as previously described using the Cdc42 and Rac1-interacting binding (CRIB) domain of PAK3 (Deakin et al., 2009). For RhoA analysis, cells were treated with siRNA as described, resuspended at 2 × 105 cells/ml, and seeded on 10-cm dishes coated in CDM in the presence of serum for 16 h. RhoA activity assays were performed as previously described (Tumbarello and Turner, 2007).
Immunofluorescence
Glass coverslips were coated with FN at 10 μg/ml for 4 h at 4°C in PBS containing magnesium and calcium or collagen I at 10 μg/ml for 2D analyses, or CDMs were grown on 12-mm glass coverslips as described earlier in text for 3D analyses. Coverslips (both 2D and 3D) were blocked with heat-denatured bovine serum albumin (BSA) at 10 mg/ml. MDA-MB-231 cells in complete growth medium at 5 × 104 cells/ml were incubated on 2D or 3D coverslips for 16 h at 37°C, unless otherwise stated. Cells were washed once in PBS, fixed, and permeabilized simultaneously using 4% paraformaldehyde with 1% Triton X-100 in PBS, quenched with 0.1 M glycine in PBS, and blocked overnight at 4°C with 3% (wt/vol) BSA. Fixed cells were stained with primary antibodies as indicated. PBS with 0.05% Tween-20 was used for subsequent washes. Cells were imaged using the Leica SP5 scanning confocal with a HCX PL APO 63×/1.40–0.60 OIL λ BL objective (Leica, Bannockburn, IL)
3D adhesion dynamics quantification
Adhesion assembly and disassembly rates were quantified as previously described (Webb et al., 2004). MDA-MB-231 cells treated with stated siRNA for 48 h were transduced with the Cellular Lights GFP-Talin (Invitrogen) baculovirus following the manufacturer’s instructions. Cells at 5 × 104 cells/ml were spread on 35-mm glass-bottom dishes coated with 3D CDMs for 16 h at 37°C, and then 3D adhesions were imaged for 3.5 h with images acquired every minute using the Leica SP5 laser scanning confocal microscope equipped with a HCX PL APO 63×/1.40–0.60 OIL λ BL objective (Leica, Bannockburn, IL) and climate-controlled chamber. Rate constants of assembly and disassembly were calculated from the increases or decreases in mean fluorescence intensity (MFI) of individual adhesions on a semilogarithmic scale versus time. Adhesion lifetime was measured from the time at which the individual adhesion was visible to its dissolution to background levels. Only adhesions that could be tracked from initial formation to complete disassembly were used for dynamics calculations. Dynamics were measured for >56 individual adhesions for control and paxillin-1 siRNA treatment from a minimum of six to eight cells and three individual experiments and 12 adhesions from three cells for paxillin-2 RNAi.
Experimental lung metastasis assay
Analysis of lung metastasis in mice was performed in accordance with institutionally approved protocols. Invasive breast cancer cells were treated with siRNA for 72 h and resuspended at 5 × 106 cells/ml in serum-free medium. Cells (5 × 105) were injected intravenously into the tail vein of 3- to 6-wk-old NOD/SCID mice. Metastases were allowed to form for 12 wk, then lungs were removed and fixed and the number of visible surface metastatic nodules were counted. Lung tissue was then analyzed by histological staining for evidence of tissue micrometastases and lymphovascular invasion.
Transendothelial migration assay
Transendothelial migration assays were performed as described previously (Estecha et al., 2009). Briefly, bovine aorta endothelial cells were seeded on 35-mm glass-bottom dishes coated with >100-μm-thick 3D collagen gels for at least 48 h. MDA-MB-231 cells were treated with siRNA oligos as stated for 72 h, and 2 × 105 cells were seeded on the surface of the confluent endothelium monolayer. Transendothelial migration was visualized in real time by confocal microscopy for 3.5 h using the Leica SP5 laser scanning confocal microscope system using the HC PL APO 20×/0,70 IMM CORR λBL objective (Leica, Bannockburn, IL). Image processing and quantification were performed using ImageJ and Leica LAS AF software, respectively. Cells transmigrating to or below the plane of the endothelial monolayer, as assessed by relative z position, during the time-lapse imaging were quantified as exhibiting transendothelial migration.
Cell viability and growth analysis
To assess cell viability and growth characteristics, 0.5 × 104 RNAi-treated cells were cultured on 12-mm glass coverslips and fixed at 24-h time points up to 144 h. Cells were fixed and stained as described earlier in the text (immunofluorescence) using 4’,6-diamidino-2-phenylindole (DAPI) to assess cell viability and cell number and either mouse anti-paxillin specific mAb or mouse anti-Hic-5 specific mAb to indicate the knockdown longevity. Viable cells were counted blind from 30 random fields, and cell number versus time after knockdown was analyzed and displayed relative to the initial time point.
Statistical analysis
Independent data sets exhibiting a normal distribution, as determined using the Shapiro–Wick and Kolmogorov–Smirnov tests of normality, were subjected to an unpaired Student’s t test. Where stated, nonparametric unpaired data sets displaying non-Gaussian distributions were subjected to a Mann–Whitney U test. Primary statistical analysis of non-Gaussian data sets used a nonparametric Kruskal–Wallis test to determine whether there was a significant treatment effect, followed by a Dunn’s post-hoc test to compare each of the treatment groups to the control group (with the significance threshold set at p < 0.01 to control the family-wise error rate). To complement the nonparametric analysis, we also performed a one-way analysis of variance followed by Dunnett’s post-hoc test, with the same significance threshold (p < 0.01). Both of these analyses produced equivalent findings in all data sets tested, demonstrating highly significant differences. All statistical analyses were performed using GraphPad Prism software.
Acknowledgments
We thank D. Pruyne for critical appraisal of the manuscript and members of the Turner lab for insightful discussion. We also thank A. Tatum for his assistance and guidance with tissue histology; F. Middleton and D. Robertson for their expert advice regarding statistical analyses; G. Feuer, P. Banerjee, and L. Crawford for assistance with the animal studies; and K. Maier and V. Gahtan for providing bovine arterial endothelial cells. This work was supported by National Institutes of Health Grant RO1 GM47607 (C.E.T.) and by a Postdoctoral Fellowship from Susan G. Komen for the Cure, KG091360 (N.O.D).
Abbreviations used:
- CDM
cell-derived matrix
- ECM
extracellular matrix
- EGF
epidermal growth factor
- EMT
epithelial-to-mesenchymal transition
- FAK
focal adhesion kinase
- FN
fibronectin
- GFP
green fluorescent protein
- pMLC
nonmuscle myosin II light chain phosphorylation
- RNAi
RNA interference
- ROCK
Rho kinase
- siRNA
small interfering RNA
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E10-09-0790) on December 9, 2010.
REFERENCES
- Avraamides C, Bromberg ME, Gaughan JP, Thomas SM, Tsygankov AY, Panetti TS. Hic-5 promotes endothelial cell migration to lysophosphatidic acid. Am J Physiol Heart Circ Physiol. 2007;293:H193–H203. doi: 10.1152/ajpheart.00728.2006. [DOI] [PubMed] [Google Scholar]
- Azuma K, Tanaka M, Uekita T, Inoue S, Yokota J, Ouchi Y, Sakai R. Tyrosine phosphorylation of paxillin affects the metastatic potential of human osteosarcoma. Oncogene. 2005;24:4754–4764. doi: 10.1038/sj.onc.1208654. [DOI] [PubMed] [Google Scholar]
- Ballestrem C, Erez N, Kirchner J, Kam Z, Bershadsky A, Geiger B. Molecular mapping of tyrosine-phosphorylated proteins in focal adhesions using fluorescence resonance energy transfer. J Cell Sci. 2006;119:866–875. doi: 10.1242/jcs.02794. [DOI] [PubMed] [Google Scholar]
- Bass MD, Roach KA, Morgan MR, Mostafavi-Pour Z, Schoen T, Muramatsu T, Mayer U, Ballestrem C, Spatz JP, Humphries MJ. Syndecan-4-dependent Rac1 regulation determines directional migration in response to the extracellular matrix. J Cell Biol. 2007;177:527–538. doi: 10.1083/jcb.200610076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellis SL, Miller JT, Turner CE. Characterization of tyrosine phosphorylation of paxillin in vitro by focal adhesion kinase. J Biol Chem. 1995;270:17437–17441. doi: 10.1074/jbc.270.29.17437. [DOI] [PubMed] [Google Scholar]
- Bertolucci CM, Guibao CD, Zheng J. Structural features of the focal adhesion kinase-paxillin complex give insight into the dynamics of focal adhesion assembly. Protein Sci. 2005;14:644–652. doi: 10.1110/ps.041107205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertolucci CM, Guibao CD, Zheng JJ. Phosphorylation of paxillin LD4 destabilizes helix formation and inhibits binding to focal adhesion kinase. Biochemistry. 2008;47:548–554. doi: 10.1021/bi702103n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bos PD, et al. Genes that mediate breast cancer metastasis to the brain. Nature. 2009;459:1005–1009. doi: 10.1038/nature08021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bristow JM, Sellers MH, Majumdar D, Anderson B, Hu L, Webb DJ. The Rho-family GEF Asef2 activates Rac to modulate adhesion and actin dynamics and thereby regulate cell migration. J Cell Sci. 2009;122:4535–4546. doi: 10.1242/jcs.053728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broussard JA, Webb DJ, Kaverina I. Asymmetric focal adhesion disassembly in motile cells. Curr Opin Cell Biol. 2008;20:85–90. doi: 10.1016/j.ceb.2007.10.009. [DOI] [PubMed] [Google Scholar]
- Brown MC, Perrotta JA, Turner CE. Identification of LIM3 as the principal determinant of paxillin focal adhesion localization and characterization of a novel motif on paxillin directing vinculin and focal adhesion kinase binding. J Cell Biol. 1996;135:1109–1123. doi: 10.1083/jcb.135.4.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown MC, Turner CE. Paxillin: adapting to change. Physiol Rev. 2004;84:1315–1339. doi: 10.1152/physrev.00002.2004. [DOI] [PubMed] [Google Scholar]
- Cailleau R, Olive M, Cruciger QV. Long-term human breast carcinoma cell lines of metastatic origin: preliminary characterization. In Vitro. 1978;14:911–915. doi: 10.1007/BF02616120. [DOI] [PubMed] [Google Scholar]
- Carragher NO, Walker SM, Scott Carragher LA, Harris F, Sawyer TK, Brunton VG, Ozanne BW, Frame MC. Calpain 2 and Src dependence distinguishes mesenchymal and amoeboid modes of tumour cell invasion: a link to integrin function. Oncogene. 2006;25:5726–5740. doi: 10.1038/sj.onc.1209582. [DOI] [PubMed] [Google Scholar]
- Castello-Cros R, Cukierman E. Stromagenesis during tumorigenesis: characterization of tumor-associated fibroblasts and stroma-derived 3D matrices. Methods Mol Biol. 2009;522:275–305. doi: 10.1007/978-1-59745-413-1_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caswell PT, et al. Rab25 associates with alpha5beta1 integrin to promote invasive migration in 3D microenvironments. Dev Cell. 2007;13:496–510. doi: 10.1016/j.devcel.2007.08.012. [DOI] [PubMed] [Google Scholar]
- Choi CK, Vicente-Manzanares M, Zareno J, Whitmore LA, Mogilner A, Horwitz AR. Actin and alpha-actinin orchestrate the assembly and maturation of nascent adhesions in a myosin II motor-independent manner. Nat Cell Biol. 2008;10:1039–1050. doi: 10.1038/ncb1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chrzanowska-Wodnicka M, Burridge K. Rho-stimulated contractility drives the formation of stress fibers and focal adhesions. J Cell Biol. 1996;133:1403–1415. doi: 10.1083/jcb.133.6.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coussens LM, Fingleton B, Matrisian LM. Matrix metalloproteinase inhibitors and cancer: trials and tribulations. Science. 2002;295:2387–2392. doi: 10.1126/science.1067100. [DOI] [PubMed] [Google Scholar]
- Cui W, Wang X, Liu YC, Wan YL, Guo HJ, Zhu J. Expression of HIC-5/ARA55 in colonrectal cancer and its mechanisms of action. Beijing Da Xue Xue Bao. 2006;38:280–283. [PubMed] [Google Scholar]
- Cukierman E. A visual-quantitative analysis of fibroblastic stromagenesis in breast cancer progression. J Mammary Gland Biol Neoplasia. 2004;9:311–324. doi: 10.1007/s10911-004-1403-y. [DOI] [PubMed] [Google Scholar]
- Cukierman E, Pankov R, Stevens DR, Yamada KM. Taking cell-matrix adhesions to the third dimension. Science. 2001;294:1708–1712. doi: 10.1126/science.1064829. [DOI] [PubMed] [Google Scholar]
- Deakin NO, Bass MD, Warwood S, Schoelermann J, Mostafavi-Pour Z, Knight D, Ballestrem C, Humphries MJ. An integrin-alpha4–14-3–3zeta-paxillin ternary complex mediates localised Cdc42 activity and accelerates cell migration. J Cell Sci. 2009;122:1654–1664. doi: 10.1242/jcs.049130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deakin NO, Turner CE. Paxillin comes of age. J Cell Sci. 2008;121:2435–2444. doi: 10.1242/jcs.018044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumbauld DW, Shin H, Gallant ND, Michael KE, Radhakrishna H, Garcia AJ. Contractility modulates cell adhesion strengthening through focal adhesion kinase and assembly of vinculin-containing focal adhesions. J Cell Physiol. 2010;223:746–756. doi: 10.1002/jcp.22084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estecha A, Sanchez-Martin L, Puig-Kroger A, Bartolome RA, Teixido J, Samaniego R, Sanchez-Mateos P. Moesin orchestrates cortical polarity of melanoma tumour cells to initiate 3D invasion. J Cell Sci. 2009;122:3492–3501. doi: 10.1242/jcs.053157. [DOI] [PubMed] [Google Scholar]
- Even-Ram S, Doyle AD, Conti MA, Matsumoto K, Adelstein RS, Yamada KM. Myosin IIA regulates cell motility and actomyosin-microtubule crosstalk. Nat Cell Biol. 2007;9:299–309. doi: 10.1038/ncb1540. [DOI] [PubMed] [Google Scholar]
- Fackler OT, Grosse R. Cell motility through plasma membrane blebbing. J Cell Biol. 2008;181:879–884. doi: 10.1083/jcb.200802081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer P, et al. Identification of molecular apocrine breast tumours by microarray analysis. Oncogene. 2005;24:4660–4671. doi: 10.1038/sj.onc.1208561. [DOI] [PubMed] [Google Scholar]
- Friedl P, Noble PB, Walton PA, Laird DW, Chauvin PJ, Tabah RJ, Black M, Zanker KS. Migration of coordinated cell clusters in mesenchymal and epithelial cancer explants in vitro. Cancer Res. 1995;55:4557–4560. [PubMed] [Google Scholar]
- Friedl P, Wolf K. Tumour-cell invasion and migration: diversity and escape mechanisms. Nat Rev Cancer. 2003;3:362–374. doi: 10.1038/nrc1075. [DOI] [PubMed] [Google Scholar]
- Friedl P, Wolf K. Plasticity of cell migration: a multiscale tuning model. J Cell Biol. 2009;188:11–19. doi: 10.1083/jcb.200909003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto N, Mizokami A, Harada S, Matsumoto T. Different expression of androgen receptor coactivators in human prostate. Urology. 2001;58:289–294. doi: 10.1016/s0090-4295(01)01117-7. [DOI] [PubMed] [Google Scholar]
- Fujita H, Kamiguchi K, Cho D, Shibanuma M, Morimoto C, Tachibana K. Interaction of Hic-5, A senescence-related protein, with focal adhesion kinase. J Biol Chem. 1998;273:26516–26521. doi: 10.1074/jbc.273.41.26516. [DOI] [PubMed] [Google Scholar]
- Gadea G, Sanz-Moreno V, Self A, Godi A, Marshall CJ. DOCK10-mediated Cdc42 activation is necessary for amoeboid invasion of melanoma cells. Curr Biol. 2008;18:1456–1465. doi: 10.1016/j.cub.2008.08.053. [DOI] [PubMed] [Google Scholar]
- Giampieri S, Manning C, Hooper S, Jones L, Hill CS, Sahai E. Localized and reversible TGFbeta signalling switches breast cancer cells from cohesive to single cell motility. Nat Cell Biol. 2009;11:1287–1296. doi: 10.1038/ncb1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannone G, et al. Lamellipodial actin mechanically links myosin activity with adhesion-site formation. Cell. 2007;128:561–575. doi: 10.1016/j.cell.2006.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guignandon A, Boutahar N, Rattner A, Vico L, Lafage-Proust MH. Cyclic strain promotes shuttling of PYK2/Hic-5 complex from focal contacts in osteoblast-like cells. Biochem Biophys Res Commun. 2006;343:407–414. doi: 10.1016/j.bbrc.2006.02.162. [DOI] [PubMed] [Google Scholar]
- Hagel M, George EL, Kim A, Tamimi R, Opitz SL, Turner CE, Imamoto A, Thomas SM. The adaptor protein paxillin is essential for normal development in the mouse and is a critical transducer of fibronectin signaling. Mol Cell Biol. 2002;22:901–915. doi: 10.1128/MCB.22.3.901-915.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitzer MD, DeFranco DB. Hic-5, an adaptor-like nuclear receptor coactivator. Nucl Recept Signal. 2006;4:e019. doi: 10.1621/nrs.04019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennigan RF, Hawker KL, Ozanne BW. Fos-transformation activates genes associated with invasion. Oncogene. 1994;9:3591–3600. [PubMed] [Google Scholar]
- Hetey SE, Lalonde DP, Turner CE. Tyrosine-phosphorylated Hic-5 inhibits epidermal growth factor-induced lamellipodia formation. Exp Cell Res. 2005;311:147–156. doi: 10.1016/j.yexcr.2005.08.011. [DOI] [PubMed] [Google Scholar]
- Inui S., Shono F, Noguchi F, Nakajima T, Hosokawa K, Itami S. In vitro and in vivo evidence of pathogenic roles of Hic-5/ARA55 in keloids through Smad pathway and profibrotic transcription. J Dermatol Sci. 2010;58(2):152–154. doi: 10.1016/j.jdermsci.2010.03.008. [DOI] [PubMed] [Google Scholar]
- Iwasaki T, et al. Involvement of phosphorylation of Tyr-31 and Tyr-118 of paxillin in MM1 cancer cell migration. Int J Cancer. 2002;97:330–335. doi: 10.1002/ijc.1609. [DOI] [PubMed] [Google Scholar]
- Jones ML, Shawe-Taylor AJ, Williams CM, Poole AW. Characterization of a novel focal adhesion kinase inhibitor in human platelets. Biochem Biophys Res Commun. 2009;389:198–203. doi: 10.1016/j.bbrc.2009.08.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y, Siegel PM, Shu W, Drobnjak M, Kakonen SM, Cordon-Cardo C, Guise TA, Massague J. A multigenic program mediating breast cancer metastasis to bone. Cancer Cell. 2003;3:537–549. doi: 10.1016/s1535-6108(03)00132-6. [DOI] [PubMed] [Google Scholar]
- Kanteti R, Yala S, Ferguson MK, Salgia R. MET, HGF, EGFR, and PXN gene copy number in lung cancer using DNA extracts from FFPE archival samples and prognostic significance. J Environ Pathol Toxicol Oncol. 2009;28:89–98. doi: 10.1615/jenvironpatholtoxicoloncol.v28.i2.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim-Kaneyama JR, Suzuki W, Ichikawa K, Ohki T, Kohno Y, Sata M, Nose K, Shibanuma M. Uni-axial stretching regulates intracellular localization of Hic-5 expressed in smooth-muscle cells in vivo. J Cell Sci. 2005;118:937–949. doi: 10.1242/jcs.01683. [DOI] [PubMed] [Google Scholar]
- Lammermann T, et al. Rapid leukocyte migration by integrin-independent flowing and squeezing. Nature. 2008;453:51–55. doi: 10.1038/nature06887. [DOI] [PubMed] [Google Scholar]
- Machacek M, Hodgson L, Welch C, Elliott H, Pertz O, Nalbant P, Abell A, Johnson GL, Hahn KM, Danuser G. Coordination of Rho GTPase activities during cell protrusion. Nature. 2009;461:99–103. doi: 10.1038/nature08242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mestayer C, Blanchere M, Jaubert F, Dufour B, Mowszowicz I. Expression of androgen receptor coactivators in normal and cancer prostate tissues and cultured cell lines. Prostate. 2003;56:192–200. doi: 10.1002/pros.10229. [DOI] [PubMed] [Google Scholar]
- Minn AJ, Gupta GP, Siegel PM, Bos PD, Shu W, Giri DD, Viale A, Olshen AB, Gerald WL, Massague J. Genes that mediate breast cancer metastasis to lung. Nature. 2005;436:518–524. doi: 10.1038/nature03799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi Y, Ishiguro H, Uemura H, Fujinami K, Miyamoto H, Miyoshi Y, Kitamura H, Kubota Y. Expression of AR associated protein 55 (ARA55) and androgen receptor in prostate cancer. Prostate. 2003;56:280–286. doi: 10.1002/pros.10262. [DOI] [PubMed] [Google Scholar]
- Nagata M, et al. Identification of potential biomarkers of lymph node metastasis in oral squamous cell carcinoma by cDNA microarray analysis. Int J Cancer. 2003;106:683–689. doi: 10.1002/ijc.11283. [DOI] [PubMed] [Google Scholar]
- Nishiya N, Shirai T, Suzuki W, Nose K. Hic-5 interacts with GIT1 with a different binding mode from paxillin. J Biochem. 2002;132:279–289. doi: 10.1093/oxfordjournals.jbchem.a003222. [DOI] [PubMed] [Google Scholar]
- Nishiya N, Tachibana K, Shibanuma M, Mashimo JI, Nose K. Hic-5-reduced cell spreading on fibronectin: competitive effects between paxillin and Hic-5 through interaction with focal adhesion kinase. Mol Cell Biol. 2001;21:5332–5345. doi: 10.1128/MCB.21.16.5332-5345.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobes CD, Hall A. Rho GTPases control polarity, protrusion, and adhesion during cell movement. J Cell Biol. 1999;144:1235–1244. doi: 10.1083/jcb.144.6.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overall CM, Lopez-Otin C. Strategies for MMP inhibition in cancer: innovations for the post-trial era. Nat Rev Cancer. 2002;2:657–672. doi: 10.1038/nrc884. [DOI] [PubMed] [Google Scholar]
- Pankov R, Endo Y, Even-Ram S, Araki M, Clark K, Cukierman E, Matsumoto K, Yamada KM. A Rac switch regulates random versus directionally persistent cell migration. J Cell Biol. 2005;170:793–802. doi: 10.1083/jcb.200503152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parri M, Taddei ML, Bianchini F, Calorini L, Chiarugi P. EphA2 reexpression prompts invasion of melanoma cells shifting from mesenchymal to amoeboid-like motility style. Cancer Res. 2009;69:2072–2081. doi: 10.1158/0008-5472.CAN-08-1845. [DOI] [PubMed] [Google Scholar]
- Petit V, Boyer B, Lentz D, Turner CE, Thiery JP, Valles AM. Phosphorylation of tyrosine residues 31 and 118 on paxillin regulates cell migration through an association with CRK in NBT-II cells. J Cell Biol. 2000;148:957–970. doi: 10.1083/jcb.148.5.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrie RJ, Doyle AD, Yamada KM. Random versus directionally persistent cell migration. Nat Rev Mol Cell Biol. 2009;10:538–549. doi: 10.1038/nrm2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinner S, Sahai E. PDK1 regulates cancer cell motility by antagonising inhibition of ROCK1 by RhoE. Nat Cell Biol. 2008;10:127–137. doi: 10.1038/ncb1675. [DOI] [PubMed] [Google Scholar]
- Provenzano PP, Eliceiri KW, Campbell JM, Inman DR, White JG, Keely PJ. Collagen reorganization at the tumor-stromal interface facilitates local invasion. BMC Med. 2006;4:38. doi: 10.1186/1741-7015-4-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provenzano PP, Eliceiri KW, Keely PJ. Multiphoton microscopy and fluorescence lifetime imaging microscopy (FLIM) to monitor metastasis and the tumor microenvironment. Clin Exp Metastasis. 2009;26:357–370. doi: 10.1007/s10585-008-9204-0. [DOI] [PubMed] [Google Scholar]
- Radvanyi L, et al. The gene associated with trichorhinophalangeal syndrome in humans is overexpressed in breast cancer. Proc Natl Acad Sci USA. 2005;102:11005–11010. doi: 10.1073/pnas.0500904102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathore VB, Okada M, Newman PJ, Newman DK. Paxillin family members function as Csk-binding proteins that regulate Lyn activity in human and murine platelets. Biochem J. 2007;403:275–281. doi: 10.1042/BJ20061618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson AL, Wang ZC, De Nicolo A, Lu X, Brown M, Miron A, Liao X, Iglehart JD, Livingston DM, Ganesan S. X chromosomal abnormalities in basal-like human breast cancer. Cancer Cell. 2006;9:121–132. doi: 10.1016/j.ccr.2006.01.013. [DOI] [PubMed] [Google Scholar]
- Ridley AJ, Schwartz MA, Burridge K, Firtel RA, Ginsberg MH, Borisy G, Parsons JT, Horwitz AR. Cell migration: integrating signals from front to back. Science. 2003;302:1704–1709. doi: 10.1126/science.1092053. [DOI] [PubMed] [Google Scholar]
- Riento K, Ridley AJ. Rocks: multifunctional kinases in cell behaviour. Nat Rev Mol Cell Biol. 2003;4:446–456. doi: 10.1038/nrm1128. [DOI] [PubMed] [Google Scholar]
- Rodina A, Schramm K, Musatkina E, Kreuser ED, Tavitian A, Tatosyan A. Phosphorylation of p125FAK and paxillin focal adhesion proteins in src-transformed cells with different metastatic capacity. FEBS Lett. 1999;455:145–148. doi: 10.1016/s0014-5793(99)00794-2. [DOI] [PubMed] [Google Scholar]
- Rossier OM, et al. Force generated by actomyosin contraction builds bridges between adhesive contacts. EMBO J. 2010;29:1055–1068. doi: 10.1038/emboj.2010.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahai E, Garcia-Medina R, Pouyssegur J, Vial E. Smurf1 regulates tumor cell plasticity and motility through degradation of RhoA leading to localized inhibition of contractility. J Cell Biol. 2007;176:35–42. doi: 10.1083/jcb.200605135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahai E, Marshall CJ. Differing modes of tumour cell invasion have distinct requirements for Rho/ROCK signalling and extracellular proteolysis. Nat Cell Biol. 2003;5:711–719. doi: 10.1038/ncb1019. [DOI] [PubMed] [Google Scholar]
- Sanz-Moreno V, Gadea G, Ahn J, Paterson H, Marra P, Pinner S, Sahai E, Marshall CJ. Rac activation and inactivation control plasticity of tumor cell movement. Cell. 2008;135:510–523. doi: 10.1016/j.cell.2008.09.043. [DOI] [PubMed] [Google Scholar]
- Shan Y, Yu L, Li Y, Pan Y, Zhang Q, Wang F, Chen J, Zhu X. Nudel and FAK as antagonizing strength modulators of nascent adhesions through paxillin. PLoS Biol. 2009;7:e1000116. doi: 10.1371/journal.pbio.1000116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Wang S, Zhao E, Shi L, Xu X, Fang M. Paxillin expression levels are correlated with clinical stage and metastasis in salivary adenoid cystic carcinoma. J Oral Pathol Med. 2010;39(7):548–551. doi: 10.1111/j.1600-0714.2009.00859.x. [DOI] [PubMed] [Google Scholar]
- Slack-Davis JK, et al. Cellular characterization of a novel focal adhesion kinase inhibitor. J Biol Chem. 2007;282:14845–14852. doi: 10.1074/jbc.M606695200. [DOI] [PubMed] [Google Scholar]
- Tachibana K, Sato T, D’Avirro N, Morimoto C. Direct association of pp125FAK with paxillin, the focal adhesion-targeting mechanism of pp125FAK. J Exp Med. 1995;182:1089–1099. doi: 10.1084/jem.182.4.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas SM, Hagel M, Turner CE. Characterization of a focal adhesion protein, Hic-5, that shares extensive homology with paxillin. J Cell Sci. 1999;112(Pt 2):181–190. doi: 10.1242/jcs.112.2.181. [DOI] [PubMed] [Google Scholar]
- Tumbarello DA, Brown MC, Hetey SE, Turner CE. Regulation of paxillin family members during epithelial-mesenchymal transformation: a putative role for paxillin delta. J Cell Sci. 2005;118:4849–4863. doi: 10.1242/jcs.02615. [DOI] [PubMed] [Google Scholar]
- Tumbarello DA, Turner CE. Hic-5 contributes to epithelial-mesenchymal transformation through a RhoA/ROCK-dependent pathway. J Cell Physiol. 2007;211:736–747. doi: 10.1002/jcp.20991. [DOI] [PubMed] [Google Scholar]
- Turashvili G, et al. Novel markers for differentiation of lobular and ductal invasive breast carcinomas by laser microdissection and microarray analysis. BMC Cancer. 2007;7:55. doi: 10.1186/1471-2407-7-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Song K, Sponseller TL, Danielpour D. Novel function of androgen receptor-associated protein 55/Hic-5 as a negative regulator of Smad3 signaling. J Biol Chem. 2005;280(7):5154–5162. doi: 10.1074/jbc.M411575200. [DOI] [PubMed] [Google Scholar]
- Webb DJ, Donais K, Whitmore LA, Thomas SM, Turner CE, Parsons JT, Horwitz AF. FAK-Src signalling through paxillin, ERK and MLCK regulates adhesion disassembly. Nat Cell Biol. 2004;6:154–161. doi: 10.1038/ncb1094. [DOI] [PubMed] [Google Scholar]
- West KA, Zhang H, Brown MC, Nikolopoulos SN, Riedy MC, Horwitz AF, Turner CE. The LD4 motif of paxillin regulates cell spreading and motility through an interaction with paxillin kinase linker (PKL). J Cell Biol. 2001;154:161–176. doi: 10.1083/jcb.200101039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf K, Mazo I, Leung H, Engelke K, von Andrian UH, Deryugina EI, Strongin AY, Brocker EB, Friedl P. Compensation mechanism in tumor cell migration: mesenchymal-amoeboid transition after blocking of pericellular proteolysis. J Cell Biol. 2003;160:267–277. doi: 10.1083/jcb.200209006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf K, Wu YI, Liu Y, Geiger J, Tam E, Overall C, Stack MS, Friedl P. Multi-step pericellular proteolysis controls the transition from individual to collective cancer cell invasion. Nat Cell Biol. 2007;9:893–904. doi: 10.1038/ncb1616. [DOI] [PubMed] [Google Scholar]
- Worthylake RA, Lemoine S, Watson JM, Burridge K. RhoA is required for monocyte tail retraction during transendothelial migration. J Cell Biol. 2001;154:147–160. doi: 10.1083/jcb.200103048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyckoff JB, Pinner SE, Gschmeissner S, Condeelis JS, Sahai E. ROCK- and myosin-dependent matrix deformation enables protease-independent tumor-cell invasion in vivo. Curr Biol. 2006;16:1515–1523. doi: 10.1016/j.cub.2006.05.065. [DOI] [PubMed] [Google Scholar]
- Yang HJ, Chen JZ, Zhang WL, Ding YQ. Focal adhesion plaque associated cytoskeletons are involved in the invasion and metastasis of human colorectal carcinoma. Cancer Invest. 2010;28:127–134. doi: 10.3109/07357900903147184. [DOI] [PubMed] [Google Scholar]
- Yoneda T, Williams PJ, Hiraga T, Niewolna M, Nishimura R. A bone-seeking clone exhibits different biological properties from the MDA-MB-231 parental human breast cancer cells and a brain-seeking clone in vivo and in vitro. J Bone Miner Res. 2001;16:1486–1495. doi: 10.1359/jbmr.2001.16.8.1486. [DOI] [PubMed] [Google Scholar]
- Yoshida K, Soldati T. Dissection of amoeboid movement into two mechanically distinct modes. J Cell Sci. 2006;119:3833–3844. doi: 10.1242/jcs.03152. [DOI] [PubMed] [Google Scholar]
- Yu JA, Deakin NO, Turner CE. Paxillin-kinase-linker tyrosine phosphorylation regulates directional cell migration. Mol Biol Cell. 2009;20:4706–4719. doi: 10.1091/mbc.E09-07-0548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidel-Bar R, Ballestrem C, Kam Z, Geiger B. Early molecular events in the assembly of matrix adhesions at the leading edge of migrating cells. J Cell Sci. 2003;116:4605–4613. doi: 10.1242/jcs.00792. [DOI] [PubMed] [Google Scholar]
- Zaidel-Bar R, Milo R, Kam Z, Geiger B. A paxillin tyrosine phosphorylation switch regulates the assembly and form of cell-matrix adhesions. J Cell Sci. 2007;120:137–148. doi: 10.1242/jcs.03314. [DOI] [PubMed] [Google Scholar]
- Zhao H, et al. Different gene expression patterns in invasive lobular and ductal carcinomas of the breast. Mol Biol Cell. 2004;15:2523–2536. doi: 10.1091/mbc.E03-11-0786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zouq NK, Keeble JA, Lindsay J, Valentijn AJ, Zhang L, Mills D, Turner CE, Streuli CH, Gilmore AP. FAK engages multiple pathways to maintain survival of fibroblasts and epithelia: differential roles for paxillin and p130Cas. J Cell Sci. 2009;122:357–367. doi: 10.1242/jcs.030478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zucker S, Cao J, Chen WT. Critical appraisal of the use of matrix metalloproteinase inhibitors in cancer treatment. Oncogene. 2000;19:6642–6650. doi: 10.1038/sj.onc.1204097. [DOI] [PubMed] [Google Scholar]