We took advantage of Chlmaydomonas flagellar mutant strains lacking either the 1α or 1β motor domain in I1 dynein to distinguish the functional role of each. The 1β motor domain is an effective motor required for control of microtubule sliding, whereas the 1α motor domain may restrain microtubule sliding driven by other dyneins.
Abstract
The Chlamydomonas I1 dynein is a two-headed inner dynein arm important for the regulation of flagellar bending. Here we took advantage of mutant strains lacking either the 1α or 1β motor domain to distinguish the functional role of each motor domain. Single- particle electronic microscopic analysis confirmed that both the I1α and I1β complexes are single headed with similar ringlike, motor domain structures. Despite similarity in structure, however, the I1β complex has severalfold higher ATPase activity and microtubule gliding motility compared to the I1α complex. Moreover, in vivo measurement of microtubule sliding in axonemes revealed that the loss of the 1β motor results in a more severe impairment in motility and failure in regulation of microtubule sliding by the I1 dynein phosphoregulatory mechanism. The data indicate that each I1 motor domain is distinct in function: The I1β motor domain is an effective motor required for wild-type microtubule sliding, whereas the I1α motor domain may be responsible for local restraint of microtubule sliding.
INTRODUCTION
Eukaryotic cilia and flagella are conserved organelles required for motile and sensory functions vital for development and the function of most organs (Satir and Christensen, 2007). Failure in assembly or regulation of cilia results in a wide range of human diseases called “ciliopathies” (Badano et al., 2006; Fliegauf et al., 2007; Marshall, 2008; Pazour and Witman, 2008; Gerdes et al., 2009; Leigh et al., 2009; Nigg and Raff, 2009), yet our understanding of the assembly and mechanism of cilia is incomplete. Here we focus on the motile ciliary/flagellar axoneme and the mechanism and functional interactions of the dynein motors that power motility (Kamiya, 2002; Oiwa and Sakakibara, 2005; King and Kamiya, 2008).
Motile cilia and flagella share a common “9 + 2” structure, in which nine peripheral doublet microtubules surround two central singlet microtubules. Outer and inner dynein arms projecting from each peripheral doublet microtubule are capable of extending to the neighboring doublet microtubule and, coupled with ATP hydrolysis, induce microtubule sliding (Oiwa and Sakakibara, 2005; King and Kamiya, 2008). Based on analysis of mutant phenotypes, the outer dynein arms regulate beat frequency and power motility, whereas the inner dynein arms regulate the size and shape of the bend (Brokaw and Kamiya, 1987; King and Kamiya, 2008). This view of independent functions for different axonemal dyneins may be an oversimplification, however. We are just beginning to understand the functional interactions among the different dyneins (Kamiya, 2002; Brokaw, 2008; Kikushima, 2009) and between each dynein heavy chain (HC) motor (e.g., Furuta et al., 2009).
The present study focuses on the inner arm dynein I1, also called dynein-f (Goodenough et al., 1987; Piperno et al., 1990; Kamiya et al., 1991; Kagami and Kamiya, 1992; Porter et al., 1992). I1 dynein is an exceptionally interesting dynein: It is required for normal regulation of axonemal bending, and, unlike the other inner dynein arms, is composed of two distinct motor domains (Porter and Sale, 2000; Wirschell et al., 2007). Studies of flagellar mutants from Chlamydomonas have demonstrated that cells either lacking I1 dynein or exhibiting altered I1 dynein intermediate chain (IC) phosphorylation have defects in flagellar waveform (Brokaw and Kamiya, 1987) and phototaxis (King and Dutcher, 1997; Okita et al., 2005). Thus I1 dynein plays important roles in the regulation of motility. The isolated I1 complex does not efficiently translocate microtubules in in vitro motility assays (Smith and Sale, 1991; Kagami and Kamiya, 1992; Smith and Sale, 1992b; Kotani et al., 2007). Moreover, in vitro evidence indicates that I1 dynein can function to inhibit microtubule translocation, possibly indicating a novel role for I1 dynein in the local control of microtubule sliding and regulation of axonemal bending (Kotani et al., 2007). Additional tests of this idea, however, require a detailed understanding of the molecular structure and functional capability of each motor domain.
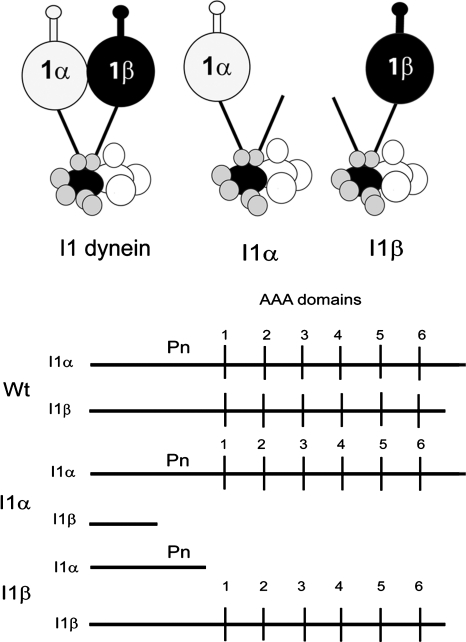
I1 dynein is located near the base of the S1 radial spoke, at the proximal end of the axonemal 96-nm repeat (Goodenough and Heuser, 1985; Piperno et al., 1990; Mastronarde et al., 1992; Porter et al., 1992; Smith and Sale, 1992a; Nicastro et al., 2006; Bui et al., 2008, 2009; Heuser et al., 2009), and is composed of two HCs (1α-HC and 1β-HC), three ICs (IC140, IC138, and IC97), FAP120, and several light chains (LCs) (Figure 1 and reviewed in Wirschell et al., 2007; King and Kamiya, 2008). Relative to HCs of other dyneins, the sequences of the 1α- and 1β-HCs of I1 dynein are highly conserved (Morris et al., 2006; Wilkes et al., 2008; Yagi, 2009). In Chlamydomonas, four independent loci, when defective, result in a failure to assemble the I1 dynein complex in the axoneme (reviewed in Myster et al., 1997; Perrone et al., 1998; Perrone et al., 2000; Wirschell et al., 2007; King and Kamiya, 2008). Two of these loci encode the I1 dynein HC subunits; IDA1(PF9) encodes the 1α-HC, and IDA2 encodes the 1β-HC (Table 1; Myster et al., 1997; Perrone et al., 2000). Importantly, mutant strains containing genes that express truncated 1α-HC or 1β-HC HCs lacking the motor domains but retaining the tail domains still assemble the remaining I1 dynein subunits (Figure 1 and Myster et al., 1999; Perrone et al., 2000). Thus these I1 dynein motor domain mutants offer an opportunity to examine the role of each motor domain in the regulation of axonemal bending.
FIGURE 1:
Predicted structure of I1 dynein and the I1α and I1β complexes. Top panel, the two-headed, heterodimeric I1 dynein (left); I1α, with an C-terminal truncation that deletes the 1β-HC motor domain (middle) (Perrone et al., 2000); I1β, with a C-terminal truncation that deletes the 1α-HC motor domain (right) (Myster et al., 1999). IC140 (light gray), the IC138 regulatory subcomplex (dark gray), and the I1 LCs (medium gray) are illustrated at the base of the I1 dynein complex (Bower et al., 2009; Ikeda et al., 2009; Wirschell et al., 2009). Bottom panel, linear diagrams of the I1 dynein 1α- and 1β-HC structure, including the positions of AAA domains 1–6 and “Pn”—an additional predicted P-loop found in the 1α-HC (Myster et al., 1999). Notably, I1α retains an intact 1α-HC but only the N terminus of the 1β-HC (Perrone et al., 2000). In contrast, I1β retains an intact 1β-HC but only the N terminus of the 1α-HC (Myster et al., 1999). Thus, in I1 dynein, the N-terminal domains of each HC are important for I1 dynein assembly.
TABLE 1:
Strains used in this study
| Strain name/genotype | Molecular phenotype | Motility | References |
|---|---|---|---|
| WT (137c) | – | WT | Harris, 1989 |
| oda2+ pf28−(CC-1877) | Lacks outer dynein arm | Slow, jerky | Kamiya 1988; Mitchell and Rosenbaum, 1985 |
| pf9–2+ (CC-3899) | Lacks I1 dynein, mutation in 1α-HC gene (Dhc1) | Slow, smooth | Porter et al., 1992 |
| pf17 (CC-1035) | Lacks radial spoke heads | Paralyzed | Lewin, 1954 |
| ida2–6 (27B3; CC-3922) | lacks I1 dynein, mutation in 1β-HC gene (Dhc10) | Slow, smooth | Perrone et al., 2000 |
| ida2–7 (J6H9; CC-3923) | Lacks I1 dynein, mutation in 1β-HC gene (Dhc10) | Slow, smooth | Perrone et al., 2000 |
| ida4 (CC-2670) | Lacks inner arm dyneins a, c, d—mutation in the inner arm p28 LC | Medium, smooth swimming | Kamiya et al., 1991; LeDizet and Piperno, 1995 |
| I1α (D11; CC-3925)/ida2–6::IDA2ΔN | Truncated I1β-HC; restores I1α | Faster than ida2–6 | Perrone et al., 2000 |
| I1α x oda (9A; CC-4079)/pf28 ida2–7 ::IDA2ΔN | Truncated I1β-HC, restores I1α, lacks the outer dynein arm | Slow, jerky | Perrone et al., 2000 |
| I1α × pf17/pf17ida2–6::IDA2ΔN | Truncated I1β-HC lacks radial spoke heads | Paralyzed | This study |
| I1β (G4–1a; CC-3920)/pf9–2::PF9ΔN | Truncated I1α-HC, retains the I1β-HC | Faster than pf9–2 | Myster et al., 1999 |
| I1β x oda (G4; CC-3917)/pf28 pf9–2 ::PF9ΔN | Truncated I1α-HC, retains the I1β-HC, lacks outer dynein arm | Slow, jerky | Myster et al., 1999 |
| I1β × pf17 / pf17 pf9–2::PF9ΔN | Tuncated I1α-HC, retains the I1β-HC, lacks the radial spoke heads | Paralyzed | This study |
| ida7–1 (5b10; CC-3921) | Lacks I1 dynein, mutation in the IC140 gene | Slow, smooth | Perrone et al., 1998 |
| bop5–1 (CC-4080) | Truncated IC138 | Medium, smooth | Hendrickson et al., 2004 |
| Dutcher et al., 1988 |
Here we take advantage of these Chlamydomonas mutant strains that assemble I1 dynein lacking either one or the other HC motor domain (see Table 1 and Figure 1). We refer to each mutant strain based on the full-length dynein HC remaining in the I1 complex (i.e., the mutant strain with a truncated 1β-HC and an intact 1α-HC is referred to as “I1α”). Double mutants also lacking the outer dynein arm, used for isolation of I1 dynein protein complexes, are referred to as I1α x oda or I1β x oda (Table 1).
Structural and functional analyses of the individual I1α- and I1β-dynein complexes reveal distinct roles for each HC motor domain in I1 dynein. These studies demonstrate that the 1β-HC is an effective microtubule motor required for wild-type (WT) microtubule sliding in the axoneme. Surprisingly, the 1β-HC motor also appears to functionally interact with outer arm dynein for control of microtubule sliding in the axoneme. Furthermore, assembly of the I1β complex is required for regulation of microtubule sliding by the central pair– radial spoke–phosphorylation pathway (Wirschell et al., 2007; Bower et al., 2009; Wirschell et al., 2009). In contrast, the I1α complex is not an efficient motor, and its presence is not sufficient for regulation of microtubule sliding by the axonemal phosphorylation pathway. The results are consistent with those of a model in which modification of I1 dynein on a subset of doublet microtubules locally regulates microtubule sliding, thus contributing to control of axonemal bending.
RESULTS
Purification of I1 dynein and the I1 dynein HC mutants
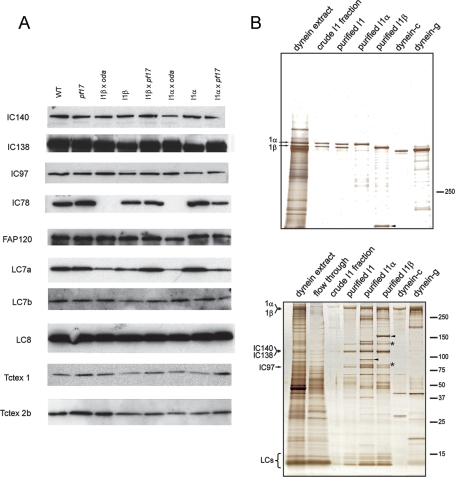
To characterize the structure, enzymatic properties, and motility of I1 dynein, we took advantage of mutant strains that lack the motor domain from either the 1α or 1β dynein HCs, yet still assemble the remaining I1 dynein subunits (Myster et al., 1997, 1999; Perrone et al., 2000). The mutant strains are listed in Table 1, and the structures of I1 dynein, the truncated motor complexes, and motor domains are illustrated in Figure 1. As described earlier in text, we refer to each mutant strain based on the intact HC remaining in the I1 complex (i.e., the protein complex and the mutant strain that contains an intact 1α-HC is referred to as “I1α”). In assessing the functional capability of each motor domain, it was important to determine whether the motor domain mutations were also accompanied by defects in the assembly of other subunits in I1 dynein. Assembly of the I1 dynein and truncated HCs in the axoneme was confirmed as shown previously for I1α (Myster et al., 1999) and I1β (Perrone et al., 2000). Additionally, Western blot analyses confirmed that the I1 dynein ICs and LCs are fully assembled in axonemes from I1α, I1β, and the double mutants I1α x oda and I1β x oda (Figure 2A). Thus the only known difference between WT and the I1α and I1β mutants is the absence of either the 1β- or 1α-HC motor domain. These results indicated that the ICs and LCs in I1 dynein are not directly associated with the motor domains (see also Myster et al., 1999; Perrone et al., 2000). This organization is in contrast to the LC1 subunit of the Chlamydomonas outer dynein arm that interacts with the γ HC motor domain (Patel-King and King, 2009).
FIGURE 2:
Composition of I1 dynein in isolated axonemes and isolated I1 dynein complexes from WT and mutant cells. (A) Immunoblot analysis of isolated axonemes from WT and the indicated mutant cells probed with the antibodies to the IC and LC subunits of I1 dynein. The antibody to IC78, an IC of the outer arm dynein, was used to assess the absence of the outer dynein arm in the mutants I1β x oda and I1α x oda. Notably, all I1 dynein subunits (IC140, IC138, IC97, FAP120, LC7a, LC7b, LC8, Tctex1, and Tctex2b) are assembled in axonemes from each cell (see Table 1 for description of cell types). (B) Silver-stained SDS–PAGE band patterns of purified I1, I1α, and I1β dyneins. Dynein subunits from the different cell strains were analyzed in either 3% polyacrylamide gels (top panel) to define the HC composition or 5–20% polyacrylamide gradient gels (bottom panel) to identify truncated HCs, ICs, and LCs. Positions of molecular weight markers are shown to the right of each gel. Top panel, lanes contain: [1] dynein extract: crude solutions of dyneins extracted from axoneme by 0.6M KCl; [2] crude I1 fraction after first ion exchange chromatography step; [3] purified I1 dynein; [4] I1α and [5] I1β after second ion exchange chromatography step. The lanes labeled as dynein-c [6] and dynein-g [7] indicate purified dynein-c and dynein-g fractions following the second ion exchange chromatography step. The I1 1α- and 1β-HCs are indicated (arrows); arrowhead indicates the C-terminal truncated 1α-HC. Bottom panel, the HCs (1α and 1β), the ICs, and the LCs of I1 dynein are identified. As predicted, the I1α fraction contains the truncated 1β-HC (arrowhead), and I1β fraction contains the truncated 1α-HC (arrowhead). The asterisk indicates unknown contaminating proteins.
I1 dynein and the truncated motor complexes were isolated from oda mutant strains (lacking the outer dynein arms) using the ion exchange procedure described previously (Kotani et al., 2007). The I1 dynein complex (dynein-f) eluted at approximately 325 mM KCl, as described before (Sakakibara et al., 1999; Kotani et al., 2007), and contains the two distinct 1α- and 1β-HCs (Figure 2B, top panel, “purified I1”). The truncated HC complexes also eluted at approximately 325 mM KCl (the dynein-f peak). SDS–PAGE confirmed that the I1α complex contains the full-length 1α-HC (Figure 2B, top panel, “purified I1α”) and that I1β contains the full-length 1β-HC (Figure 2B, top panel, “purified I1β”). The N-terminal fragment of the truncated 1α-HC was also observed in the I1β fraction (Figure 2B, arrowheads) and the N-terminal fragment of the 1β-HC was also observed in the I1α fraction (Figure 2B, bottom panel, arrowhead). The I1, I1α, and I1β dynein complexes each contain IC140, IC138, and IC97 (Figure 2B, bottom panel). Dynein-c and dynein-g fractions are included as controls (Figure 2B), and, judging from these observations, we conclude that there was no significant contamination of the I1 dyneins with these other dynein subspecies.
Structural analysis of the I1α and I1β head domains
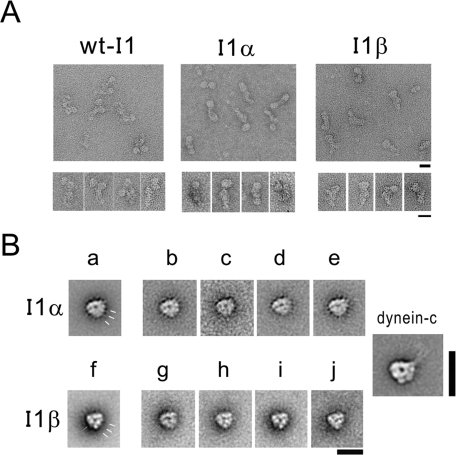
Negative stain electron microscopy was used to assess the structure of the isolated I1 dynein and truncated motor domain complexes. As described previously, electron microscopic analysis revealed that I1 dynein is a two-headed structure with a prominent tail domain (Figure 3A, left panels, and Goodenough et al., 1987; Smith and Sale, 1991; Kotani et al., 2007). Electron microscopy of the purified mutant I1 complexes revealed that they are single-headed dynein structures with a tail domain that is morphologically similar to intact I1 dynein (Figure 3B, middle and right panels). These observations are consistent with the structures predicted from HC sequence analysis and seen previously by transmission electron microscopy and image analysis of I1 mutant axonemes (Figure 1) (Myster et al., 1999; Perrone et al., 2000). The results are also consistent with the model in which the N-terminal domains of both HCs are necessary and sufficient for assembly and docking of I1 dynein in the axoneme.
FIGURE 3:
Structure of I1, I1α, and I1β dynein complexes. (A) Electron micrographs of purified I1, I1α, and I1β negatively stained with 1% uranyl acetate. A general view of WT I1, I1α, and I1β dynein is shown (top panel). Double (I1) and single (I1α and I1β) globular particles with elongated tails were selected from throughout the field (bottom panel). Calibration bar = 20 nm. (B) Single-particle image processing of the motor domain of purified I1α (top panel) and I1β (bottom panel). Red bar indicates stain-filled groove. At the right side of the head, pronounced stain-excluding globular domains exist (yellow bars). The “right” view of axonemal dynein-c is shown for comparison (right panel) (Burgess et al., 2003; Roberts et al., 2009).
To examine the conformations of the two heads, single-particle image analysis was performed using electron micrographs of negatively stained I1α and I1β head domains (Figure 3B). Class averages of the “right-view” images were used for comparison of the two heads because the right-view images of other dynein head domains are well characterized and reveal conserved, structural landmarks (Burgess et al., 2003, 2004; Roberts et al., 2009). Figure 3B shows class averages of the right view of the I1α head domains (Figure 3B, b–e) and I1β head domains (Figure 3B, g–j) and shows global averages of the right view of the I1α head domain (Figure 3B, a) and I1β head domain (Figure 3B, f). Alignment of the head domains of negatively stained molecules clearly shows that they display an asymmetric ringlike morphology similar to that of Chlamydomonas flagellar inner arm subspecies dynein c, cytoplasmic dyneins, and the head domains of intact I1 dynein (Figure 3B) (Burgess et al., 2003; Kotani et al., 2007; Roberts et al., 2009). The diameter of the globular heads was approximately 15 nm, and the deposit of stain in the center of the head domain is clearly observed as reported in dynein-c (compare Figure 3B, right panel, and Burgess et al., 2003). The groove on the left side, observed on cytoplasmic dynein (Roberts et al., 2009), is also observed in I1α and I1β head domains (Figure 3B, red bars). Similar to cytoplasmic dynein and dynein-c, the three globular domains are observed on the right side (Figure 3B, yellow bars; compare to Burgess et al., 2003, 2004; Roberts et al., 2009). Thus the purified, truncated I1 dyneins have retained their molecular configuration, and the globular motor domains of each HC are similar to each other and to other dyneins (see Discussion and Burgess et al., 2003, 2004; Mizuno et al., 2004; Samso and Koonce, 2004; Roberts et al., 2009). Although the right view of the I1β head has features similar to other dyneins, in several of particle class images examined (Figure 3B, f, h, i, and j), the pattern of stain density surrounding the dynein head is different from that of other dyneins: Stain density at the bottom of the I1β head is heavier than that at the top. This observation may reveal subtle differences in the structure of the I1β motor-head domain compared to the motor domain in other dyneins.
Isolated I1 dynein can induce the formation of microtubule bundles
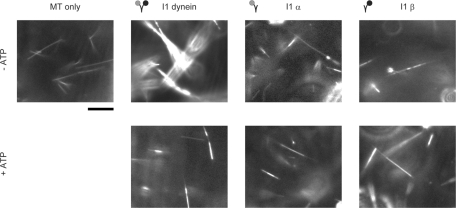
A microtubule bundling assay was used to assess the ability of the I1 dynein, I1α, and I1β complexes to interact with microtubules in an ATP-sensitive manner (Haimo et al., 1979; Smith and Sale, 1991; Moss et al., 1992b; Sakakibara and Nakayama, 1998; Toba and Toyoshima, 2004). As described previously, dark field light microscopy can be used to resolve individual microtubules in the absence of added dyneins (Figure 4, top left panel). Addition of the purified, two-headed WT I1 dynein resulted in the rapid cross-linking of microtubules into large bundles, but these bundles were dispersed into single microtubules following the addition of ATP (Figure 4, I1 dynein). These observations are consistent with previous analysis of I1 dynein-microtubule bundling by electron microscopy (Smith and Sale, 1991) and indicated that the two-headed dynein cross-links microtubules through the ATP-sensitive microtubule binding site in each HC. When microtubules were mixed with the single-headed I1 dynein complexes, however, microtubule bundles were never formed, irrespective of the presence or absence of ATP.
FIGURE 4:
Dark-field microscopy of I1 dynein induced microtubule bundles: Single-headed complexes I1α and I1β do not induce microtubule bundling. Microtubules polymerized in vitro (top left; microtubules only). The addition of intact I1 dynein formed microtubule bundles in the absence of ATP (top panel). Microtubule bundles formed by I1 dynein dissociated into single microtubules upon addition of 2 mM ATP (bottom panel; I1 dynein). In contrast, microtubules are not bundled in the presence of the single-headed I1α and I1β irrespective of ATP addition (right panels; I1α and I1β). Bar = 10 μm.
The simplest interpretation of these data is that the two-headed I1 dynein is capable of cross-bridging microtubules through the microtubule-binding domains present in both the 1α and 1β dynein HCs. Presumably, the I1α and I1β single-headed dyneins fail to cross-link microtubules because each single-headed complex has only one microtubule-binding site. These observations also suggest that the ATP-insensitive microtubule-binding site observed in the axoneme requires a docking protein or complex specialized for binding the base of the I1 dynein. This docking complex is apparently not present in microtubules assembled from purified tubulin (Smith and Sale, 1991).
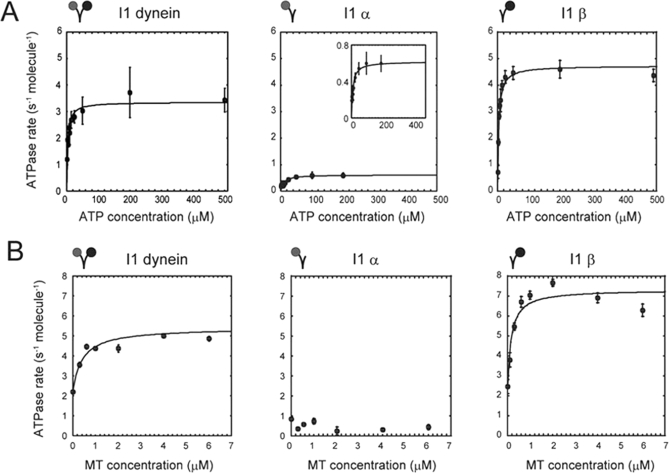
Distinct MgATPase activities of the I1α and I1β complexes
The basal ATPase activities of the isolated I1 dynein, I1α, and I1β complexes were measured at various ATP concentrations in the absence of microtubules (Figure 5A). The steady-state ATPase rates were fitted with Michaelis–Menten-type kinetics. I1β has a higher maximal velocity (kcat) compared to the two-headed, WT I1 dynein, whereas the maximal velocity of I1α is approximately one eighth the maximal velocity of I1β. The I1α complex also has a higher Km value than either the I1 or I1β dynein complex, indicating a lower affinity for ATP. Thus I1β has a much higher basal ATPase activity than I1α. Importantly, the ATPase activity of the two-headed I1 dynein is lower than the combined value of its I1α and I1β motor domains. These observations suggest that the I1α motor domain may exert an inhibitory effect on the ATPase activity of the I1β complex motor domain in the intact I1 dynein complex.
FIGURE 5:
ATPase activity of I1 mutants. (A) ATPase activity of I1, I1α, and I1β dyneins at various ATP concentrations in the absence of microtubules. The Mg-ATPase activities of I1, I1α, and I1β were fitted by the Michaelis–Menten equation: k = (kcat ⋅ [ATP])/(Km + [ATP]). I1: kcat = 3.36 ± 0.17 s−1 molecule−1, Km = 3.22 μM, 1α-HC: kcat = 0.62 ± 0.06 s−1 molecule−1, Km = 8.96 μM, 1β-HC: kcat = 4.73 ± 0.12 s−1 molecule−1, Km = 3.77 μM). The intact I1 dynein shows a characteristic increase in ATPase activity with increasing concentrations of ATP (I1 dynein, left panel). I1β shows higher ATPase activity in the absence of the I1α-HC (I1β, right panel). In contrast, I1α shows low ATPase activity (I1α, middle panel). Error bars indicate standard deviations. The inset is an expanded graph of the activity of I1α. (B) Microtubule activation of ATPase activity. The ATPase assay was performed exactly as for basal ATPase activity, and the activities were measured at various microtubule concentrations. The microtubule-activated Mg-ATPase activities of I1 and I1β were fitted by the modified Michaelis–Menten equation: k = {(kcat – kbase)⋅[MT]}/(Km,MT + [MT]) + kbase. I1: kcat = 5.41 ± 0.31 s−1 molecule−1, kbase = 2.23 ± 0.43 s−1 molecule−1, Km, MT = 0.42 μM, I1β : kcat = 7.32 ± 0.41 s−1 molecule−1, kbase = 2.31 ± 0.62 s−1 molecule−1, Km, MT = 0.16 μM). The intact I1 dynein and I1β both show microtubule-stimulated ATPase activity, whereas I1α does not. The average value of ATPase activity over the whole range of microtubule concentration is 0.52 ± 0.22 s−1 molecule−1. kbase is the basal ATPase activity. Error bars indicate standard deviations.
The ATPase activities of the two-headed I1 dynein and the single-headed I1β dynein were activated by microtubules (Figure 5B). Again, the kcat of I1β is higher than that of the intact I1 dynein. This difference is due primarily to its higher basal ATPase activity, as in both cases the addition of microtubules stimulated ATPase activity only 1.5-fold (in I1β) to 1.6-fold (in I1 dynein). Km, MT is the microtubule concentration at the half saturation of microtubule-activated ATPase activity. I1β has a lower Km, MT value than I1 has, indicating that I1β alone has a higher affinity for microtubules than does the two-headed I1 dynein. These results suggest that the I1α motor domain modulates the ATPase and microtubule affinity of the I1β motor domain in the WT I1 dynein complex. Surprisingly, I1α did not show any activation of ATPase activity by the addition of microtubules, even though I1α can support the translocation of microtubules (see next section). The failure to stimulate I1α activity further reveals novel properties of this dynein HC motor domain. Thus, although the structure of I1α-HC is strikingly similar to that of other dyneins, the structural analysis alone is not sufficient for assessing functional capability.
Microtubule gliding produced by purified I1α or I1β complexes
To characterize the mechanical properties of the I1 dynein complexes, we used conventional in vitro motility assays in which microtubules glide over glass surfaces coated with dyneins. The velocity of I1 dynein (1.8 ± 0.6 μm/s) is similar to the value reported previously (Kotani et al., 2007). The I1α dynein translocates microtubules at a considerably reduced speed (0.7 ± 0.2 μm/s), whereas the microtubule gliding velocity of I1β (3.3 ± 0.7 μm/s) is nearly two times higher than that of the intact I1 dynein. The velocity of two-headed I1 dynein is close to the average of the individual heads. These observations suggest that the I1α motor domain may suppress the activity of I1β in the intact I1 dynein.
Curiously, I1α did not show any microtubule activation on its ATPase activity. We then tested the possibility that the microtubule gliding activity observed with the I1α was due to contamination with other dynein isoforms. On the basis of silver-stained SDS–PAGE gels, we estimate that contamination of the I1 fraction with other dynein isoforms is <1% of I1α. We then performed the microtubule gliding assay at a low protein concentration using the I1, I1β, and dynein-g diluted to ∼1% of the concentration of I1α. None of the diluted dyneins (I1, I1β, and dynein-g) supported robust microtubule gliding. Rather, with the diluted samples, microtubules exhibited back-and-forth movement, pivoting, and stop-and-go gliding, although a few microtubules stuck to the glass surface. In contrast, the purified I1α fraction supported smooth and continuous, albeit slow, microtubule gliding. We concluded that microtubule gliding by I1α is not caused by contamination with other dynein species but by I1α itself.
The 1β-HC motor domain is required for normal microtubule sliding in situ
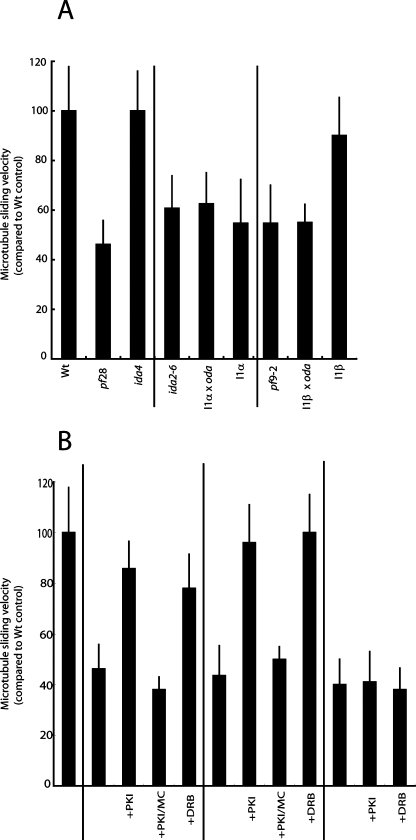
We previously showed that each I1 dynein motor domain contributes to forward swimming speed in Chlamydomonas (Myster et al., 1999; Perrone et al., 2000). In particular, the deletion of the 1β-HC motor domain reduces forward swimming velocity more significantly than does the deletion of the 1α-HC motor (see Table 2). To further assess the relative contributions of each I1 motor domain, we measured microtubule sliding in isolated axonemes, where dynein activity is uncoupled from the production of flagellar bending. In 1 mM MgATP, microtubules in WT axonemes slide at ∼18 μm/s (Figure 6A; Table 2). Similarly, microtubules slide rapidly in axonemes from the inner arm dynein mutant ida4 (Figure 6A; Table 2). As previously described (Okagaki and Kamiya, 1986; Smith and Sale, 1992a), microtubule sliding velocity is greatly reduced in axonemes lacking the outer dynein arms (pf28; Table 2; Figure 6A) or defective in radial spoke assembly (pf17; Table 2; Figure 6B). Similarly, microtubule sliding is reduced in mutants lacking I1 dynein (ida2–6, ida2–7, ida7, and pf9–2; Table 2; Figure 6A, and see Discussion), in I1 dynein mutants that lack the 1β-HC motor domain (I1α, Table 2; Figure 6A), or in double mutants that lack the outer dynein arms as well as either the 1α-HC or 1β-HC motor domain (Table 2; Figure 6A). Microtubule sliding velocity is nearly WT, however, in axonemes from the I1β strain that lacks only the 1α motor domain (Table 2; Figure 6A), suggesting that the 1α motor domain does not contribute significantly to microtubule sliding velocities. Moreover, despite the assembly of outer dynein arms in the I1α strain, its microtubule sliding velocity is slow, equivalent to complete loss of I1 dynein (compare I1α or I1β in Figure 6A). These observations suggest that the 1α-HC motor domain does not contribute significantly to microtubule sliding in the absence of the 1β-HC motor domain.
TABLE 2:
Swimming and microtubule sliding velocity.
| Strain name | Microtubule sliding velocity (μm/s) ±SD | Swimming velocity (μm/s) ±SD |
|---|---|---|
| WT | 17.3 ± 2.5 | 144.2 ± 17.1A |
| oda2 (pf28) | 7.5 ± 0.4* | 51.5 ± 6.9A |
| pf17 | 8.2 ± 0.8 | N/AB |
| ida4 | 18.5 ± 2.3 | 102 ± 11.0C |
| ida7–1 | 9.6 ± 1.7 | 81.5 ± 14.0A |
| bop5–1 | 12.3 ± 1.2 | 92D |
| ida2–6 | 9.8 ± 0.6 | 77.6 ± 15.4E |
| ida2–7 | 11.4 ± 1.9 | 53.7 ± 10.7E |
| I1α | 10.5 ± 2.0 | 107.9 ± 15.3E |
| I1α x oda | 9.8 ± 3.0* | NDB |
| pf9–2 | 8.5 ± 0.1 | 73.4 ± 12.4F |
| I1β | 16.0 ± 2.5 | 136.9 ± 16.5F |
| I1β x oda | 9.3 ± 2.0* | 41.2 ± 5.6F |
A Velocity determined in Perrone et al., 1998.
B N/A = not applicable; ND = not determined.
C Swimming velocity determined in Kamiya et al., 1991.
D Velocity determined in Hendrickson et al., 2004.
E Velocity determined in Perrone et al., 2000.
F Velocity measured in Myster et al., 1999.
*Based on the sliding assay, there is no significant difference the slow sliding in axonemes from oda2 and the double mutants I1α x oda and I1β x oda.
FIGURE 6:
The 1β-HC motor domain is required for regulated microtubule sliding in the axoneme. (A) Microtubule sliding measurements reveal that assembly of the 1β-HC motor domain is required for WT microtubule sliding velocity. In particular, microtubule sliding velocity is greatly reduced in axonemes from the I1α mutant (lacking the 1β-HC motor domain; I1α or I1α oda). In contrast, microtubule sliding velocity in the I1β axonemes (lacking the 1α-HC motor domain) is nearly WT. Note that, based on these assays, there is no significant difference in the slow sliding in axonemes from oda2 and the double mutants I1α x oda and I1β x oda. (B) The 1β-HC motor domain is required for rescue of microtubule sliding by kinase inhibitors. Compared to WT axonemes, microtubule sliding velocity in pf17 axonemes (defective in the radial spokes) or in the triple mutants (I1α × pf17 and I1β × pf17) is reduced by ∼50%. As previously shown, the kinase inhibitors PKI and DRB rescue sliding in pf17 axonemes, and the phosphatase inhibitor MC blocks rescue (Howard et al., 1994; Yang et al., 2000; Gokhale et al., 2009). In the I1β × pf17 mutant, kinase inhibitors also rescue sliding. In the I1α × pf17 mutant (lacking the 1β-HC motor domain), however, kinase inhibitors fail to rescue microtubule sliding. The average microtubule sliding velocity for each sample was calculated from three independent experiments with a total sample size of at least 80 axonemes and plotted as a percentage of the sliding velocity relative to WT axonemes. Values shown are means and standard deviations.
The 1β-HC motor is required for regulation of microtubule sliding by the axonemal radial spoke–phosphorylation pathway
Several lines of evidence indicate that the assembly of I1 dynein, in particular its regulatory ICs (IC138 and IC97), is required for regulation of microtubule sliding by a signaling pathway that involves the central pair apparatus, radial spokes, and axonemal kinases and phosphatases (Bower et al., 2009; Gokhale et al., 2009; Wirschell et al., 2009). The regulatory pathway was revealed by functional and pharmacological analysis of microtubule sliding in paralyzed axonemes from central pair or radial spoke mutants (reviewed in Porter and Sale, 2000; Smith and Yang, 2004; Wirschell et al., 2007).
Dynein-driven microtubule sliding is globally inhibited in isolated, paralyzed axonemes from radial spoke mutants such as pf14 or pf17, and normal microtubule sliding velocity can be rescued by pretreating these axonemes with kinase inhibitors such as PKA inhibitor (PKI), casein kinase 1–7 (CK1–7), or 5,6-dichloro-β-d-ribofuranosylbenzimidazole (DRB) (Smith and Sale, 1992a; Howard et al., 1994; Yang and Sale, 2000; Gokhale et al., 2009) (Figure 6B, pf17 + PKI and pf17 + DRB). Rescue of microtubule sliding requires assembly of I1 dynein, indicating that I1 dynein plays an essential role in this pathway (Habermacher and Sale, 1997; Yang and Sale, 2000; Bower et al., 2009). The mechanism of inhibition and the rescue of microtubule sliding correlate with phosphorylation and dephosphorylation of IC138 (Smith and Sale, 1992a; Howard et al., 1994; Habermacher and Sale, 1996, 1997; Yang and Sale, 2000; Hendrickson et al., 2004; Bower et al., 2009; Wirschell et al., 2009). To test whether either of the I1 motor domains is required for regulation of microtubule sliding by the central pair–radial spoke phosphorylation pathway, we crossed the I1α and I1β strains to the paralyzed, radial spoke mutant, pf17, to recover triple mutants containing the original HC mutant allele and expressing the truncated motor domain mutant constructs in a radial spoke–defective background (I1α × pf17, containing the ida2–6 mutant allele, the Dhc10 transgene lacking the 1β-HC motor domain and the radial spoke heads, and I1β × pf17, containing the pf9–2 mutant allele, the Dhc1 transgene lacking the 1α-HC motor domain and the radial spoke heads). Molecular and biochemical analyses were performed to confirm the genotypes and phenotypes of the triple mutant strains (Supplemental Figure S1). We then measured microtubule sliding velocities in axonemes in the absence or presence of the kinase inhibitors PKI or DRB or the phosphatase inhibitor microcystin-LR (MC).
Microtubule sliding is greatly reduced in pf17 axonemes, and the addition of PKI or DRB restores microtubule sliding to WT levels (Figure 6B), whereas MC blocks rescue by PKI (PKI/MC, Figure 6B). As expected, microtubule sliding velocity is also greatly reduced in axonemes from the mutants I1α × pf17 and I1β × pf17 (Figure 6B). PKI or DRB treatment, however, only increases microtubule sliding velocity in axonemes from I1β × pf17 and fails to rescue sliding velocity in axonemes from I1α × pf17 (Figure 6B). These results demonstrate that the 1β-HC motor domain is required for regulation of I1 dynein–mediated microtubule sliding by the central pair–radial spoke phosphorylation pathway.
DISCUSSION
Here we took advantage of mutant cells that lack one or the other I1 dynein motor domain to address whether the 1α- and 1β-HC motor domains play distinct roles in control of flagellar movement. We determined that although the 1α- and 1β-HC motor domains are similar in structure, they display different activities. In vitro analysis revealed that the I1β complex is similar to other dynein motors with significant ATPase and microtubule translocation capability, thus possibly contributing to net microtubule sliding in the axoneme. Additionally, analysis of microtubule sliding in isolated axonemes revealed that assembly of the 1β-HC motor domain is required for regulation of microtubule sliding by the central pair–radial spoke I1 dynein phosphoregulatory pathway (reviewed in Wirschell et al., 2007). In contrast, the I1α complex displays unusually low ATPase and microtubule translocation, and assembly of the 1α motor domain is not required for regulation of microtubule sliding by phosphorylation.
The results indicate that each motor domain in I1 dynein is distinct in motor and regulatory activity. As described before (Kotani et al., 2007), I1 dynein may regulate the pattern and speed of microtubule sliding in the axoneme by locally constraining sliding driven by other axonemal dyneins. One hypothesis is that the 1α motor domain may resist the faster microtubule sliding driven by the 1β motor domain, an activity similar to that proposed for the α HC motor in the outer dynein arm from sea urchin sperm tail axonemes (Sale and Fox, 1988; Moss et al., 1992a, 1992b). This model must be tested directly, but may be consistent with observations of swimming phenotype in the I1 dynein mutants: Cells that lack the 1β-HC motor domain swim slower (107.9 ± 15.3 μm/s; Table 1, ida2–6::λC [D11]), (Perrone et al., 2000) than cells that lack the 1α-HC motor domain (136.9 ± 16.5 μm/s; Table 2 [G4 + OA]) (Myster et al., 1999).
Our results also indicate a functional interaction between I1 dynein and the outer dynein arm: Assembly of the I1 dynein and, in particular, the 1β-HC motor domain is required for full dynein activity in axonemes (Figure 6A). Thus I1 dynein may perform multiple roles, with functions segregated in each motor domain. These functions include resistance of microtubule sliding (Kotani et al., 2007), possibly a function of the 1α-HC, and regulation of the outer dynein arms through physical and/or chemical signaling, possibly a function of the 1β-HC.
Structural comparisons of each I1 dynein motor domain
Both the I1α and I1β heads retain the typical ringlike structure and the stalk structure characteristic of all dynein motors, even when the other head is missing. Negative stain microscopy coupled to single- particle analysis (Roberts and Burgess, 2009; Roberts et al., 2009) revealed that the motor domain of I1α and I1β are nearly identical to each other and to the motor domain of other inner dynein arms (Burgess et al., 2003, 2004; Nicastro et al., 2006; Bui et al., 2008, 2009), cytoplasmic dyneins (Mizuno et al., 2004; Samso and Koonce, 2004; Roberts et al., 2009), and the outer dynein arms studied by cryo-electron microscopy (cryoEM) tomography (Oda et al., 2007; Movassagh et al., 2010). The I1β head, however, tends to have different pattern of stain surrounding the head domain compared with other dyneins. Because a structure projecting from the carbon film results in accumulation of stain, we suppose that the I1β head tends to attach to the carbon film at a different angle to that of the I1α and other dyneins, possibly owing to the characteristic position and structure of the I1β tail domains. This characteristic may reflect functional specificity of I1β.
Owing to the resolution limit of negative staining electron microscopy (∼10Å), differences in ATPase, microtubule interaction, and translocation cannot be explained by structural differences. In this report, the I1α complex displays exceptionally low ATPase and motor activity. Further refinements in single-particle analysis and/or application of cryoEM tomography will resolve distinctions in motor structure of the I1α compared to other dynein motors and help define fundamentals in force production in the dyneins that are not present in I1α.
Functional domains and the role of I1 dynein in axonemal motility
Functional assays reveal large differences in activity between I1α and I1β. In particular, the I1α dynein exhibits some unusual properties. In contrast to I1β, I1α can translocate microtubules in vitro, but its ATPase activity is not activated by microtubules. Additionally, although the 1α and 1β motors have distinct properties, they do not appear to work independently in the intact I1 complex. One model is that the 1α motor domain regulates the activities of the 1β motor domain. In in vitro experiments, all the properties measured for I1β are higher than those of intact I1: i) basal ATPase activities, I1α < I1 < I1β; ii) microtubule-activated ATPase activities, 0 ≈ I1α < I1 < I1β; and iii) in vitro motility assay, I1α < I1 < I1β. Thus the activity of the 1β-HC is modulated by the presence of 1α-HC. Given the reduced ATPase activity of the 1α-HC, the primary function of the 1α-HC may be to modulate I1β activity. Refined understanding of I1 dynein structure in the axoneme and understanding of interactions between the I1 dynein motor domains and their respective stem domains is required to further test these ideas.
Diverse observations also indicate that assembly of I1 dynein and, in particular, assembly of the I1β motor domain and the IC138 regulatory complex (Bower et al., 2009) are required for full outer dynein arm activity. Thus, in addition to other unexpected functional features, I1 dynein may operate to regulate the outer dynein arms or possibly other inner dynein arms. For example, in the absence of outer dynein arms, microtubules slide slowly (compare WT and pf28, Figure 6A). This observation is consistent with other studies of microtubule sliding in axonemes indicating that the outer dynein arms are required for rapid microtubule sliding (reviewed in Kamiya, 2002; King and Kamiya, 2008). Microtubule sliding, however, is also reduced to approximately half of WT sliding velocity when I1 dynein or the 1β motor domain fails to assemble (Figure 6A). This reduced sliding velocity occurs despite full assembly of the outer dynein arms in the I1 dynein mutants (see Figure 2A, IC78). Moreover, microtubule sliding velocity is also greatly reduced in other I1 dynein mutants that fail to assemble the IC138 regulatory complex (see Figure 6A in Bower et al., 2009). In contrast, microtubule sliding velocity is nearly WT in axonemes from I1β, indicating that the outer dynein arms are fully active in I1β axonemes (Figure 6A). The mechanism for how I1 dynein could contribute to regulation of the other dynein arms is not understood. Recent reports by cryoEM tomography, however, reveal a structural linkage between the tails of the outer dynein arm and I1 dynein and also between I1 dynein and other components in the inner arm region (Nicastro et al., 2005; Bui et al., 2008; Heuser et al., 2009). These linkages through the base of the I1 dynein may be designed to relay regulatory information between dynein isoforms.
As introduced earlier in this article, assembly of I1 dynein and the IC138 complex is critical for regulation of dynein-driven microtubule sliding by the central pair–radial spoke phosphoregulatory pathway (Wirschell et al., 2007; Bower et al., 2009). Our new data also indicate that assembly of the 1β motor domain is required for regulation of the phosphoregulatory pathway, suggesting a functional interaction between the IC138 complex and the 1β-HC motor domain (Figure 6B). One model is that change in IC138 phosphorylation alters I1 dynein motor activity; however, in vitro analysis of I1 dynein motor activity, using purified I1 dyneins either in the presence or absence of phosphorylated IC138, does not support this model (unpublished data, H. Sakakibara). Thus I1 dynein phosphorylation and the 1β-HC motor domain may also regulate dynein-driven microtubule sliding through regulation of other dynein arms, including the outer dynein arm.
MATERIALS AND METHODS
Chlamydomonas strains
Chlamydomonas strains used for this study are listed in Table 1. Intact I1 dynein was purified from flagella of an outer-armless mutant, oda1 (Kamiya and Okamoto, 1985; Kamiya, 1988). I1 dynein with a truncated 1β-HC was purified from strain I1α x oda (9A, CC-4079) (Perrone et al., 2000), and I1 dynein with a truncated 1α-HC was purified from I1β x oda (G4, CC-3917) (Myster et al., 1999). The I1 dynein HC mutants (I1α and I1β) were crossed to pf17, and “triple” mutants (containing the original HC mutation, the truncated HC transgene mutation, and the radial spoke defect) were recovered from nonparental tetrads. These triple mutants were verified and characterized by either Western blotting or PCR as described in Supplemental Figure S1. The inner dynein arm mutants ida4 (Kamiya et al., 1991; LeDizet and Piperno, 1995), bop5–1 (Dutcher et al., 1988; Hendrickson et al., 2004), and ida7–1 (Perrone et al., 1998) were used in control experiments.
Preparation of proteins
Chlamydomonas I1 dynein was purified as described previously (Kotani et al., 2007) by using two cycles of anion exchange column chromatography in HMED buffer (30 mM HEPES–KOH, 5 mM MgSO4, 1 mM ethylene glycol tetraacetic acid [EGTA], 1 mM dithiothreitol [DTT], pH 7.4). The I1 dynein and truncated HC complexes eluted from the column at approximately 325 mM KCl. For the measurement of ATPase activity, the purified dynein fractions were pooled and assayed immediately. For other in vitro assays, 20% sucrose was added to the fractions, which were frozen in liquid nitrogen and stored at –80°C. Porcine brain microtubules were prepared by cycles of assembly and disassembly (Weingarten et al., 1975). Tubulin was separated from the microtubule-associated proteins (MAPs) by chromatography through phosphocellulose (P-11; Whatman, Maidstone, UK) chromatography (Sloboda and Rosenbaum, 1982). MAP-depleted tubulin (∼4–5 mg/ml) was assembled at 30°C for 30 min and stabilized by 20 μM Taxol (Sigma, St. Louis, MO). Protein concentrations were determined by the TONEIN-TP kit (Otsuka Pharmaceutical Co., Tokyo, Japan) based on the method of Bradford (Bradford, 1976). Protein samples were analyzed by SDS–PAGE (Laemmli, 1970).
Electron microscopy and single-particle analysis in electron micrographs
To observe the molecular configuration of I1 dynein, purified dyneins were analyzed by negative staining and electron microscopy (Burgess et al., 2003). One drop of a freshly prepared specimen containing intact or motor head-truncated dynein at ∼20 μg/ml was applied to a carbon film and stained with 1% uranyl acetate. Samples were observed with a JEM 2000EX electron microscope (JEOL, Tokyo, Japan) with a magnification of 50,000× operating at 80 kV. Electron micrographs were digitized on an EPSON GTX-700 scanner (Seiko Epson, Nagano, Japan) at 1000 dpi, corresponding to a pixel size of 0.54 nm on the grid. For investigation of precise configurations of the 1α-HC and 1β-HC head domains, the digitized images of those heads were further analyzed by using the single-particle image-processing technique (Frank, 2006). Well-isolated head images were extracted and subjected to single-particle image processing using the SPIDER software programs (Frank, 2006). The number of particles analyzed is as follows: 6877 (I1α head); 7316 (I1β head). Images were aligned by reference-free algorithms and classified into homogeneous groups as described (Burgess et al., 2004). A total of 1805 particles for I1α and 1040 particles for I1β were classified as the right view of the head domain based on positions of the tail and stalk protrusions. These images of the right view were aligned again and further classified. The averaged images in each particle class were used for comparison of configurations.
Microtubule-bundling assays
MAP-depleted and Taxol-stabilized microtubules were mixed with dynein to a final concentration of 4 μg/μl and 30 μg/ml, respectively, in HMED buffer containing 20 μM Taxol and 50 mM KCl. Microtubule bundling by dynein in the absence ATP was induced by incubating the mixture for 5 min at room temperature and then observed by dark-field microscopy. To observe the dissociation of microtubules, 2 mM ATP was added to the mixtures, and they were observed after 5 min.
Mg-ATPase activity
Mg-ATPase activities of dynein fractions were measured using the EnzChek phosphate assay kit (E-6646; Molecular Probes, Eugene, OR) in a temperature-controlled cell at 25°C. Released inorganic phosphate was measured by continuously monitoring the absorbance at 360 nm for 20 min. To measure microtubule-activated ATPase activity, we used GTP-free microtubules made by sedimenting stabilized microtubules through a 25% sucrose cushion containing HMED buffer and 10 μM Taxol. Control assays using GTP-free microtubules alone indicated that phosphate release from microtubules did not significantly contribute to the total ATPase activity of the dyneins.
In vitro microtubule gliding assays
In vitro microtubule gliding assays were performed as previously described (Kotani et al., 2007). We used untreated glass slides (#1 glass slide, 26 mm × 76 mm, S-1126; Matsunami, Osaka, Japan). 5 μl of thawed dynein was mixed with an equal volume of bovine serum albumin (BSA) at 0.5 mg/ml, then applied to a flow chamber and absorbed onto the glass for 5 min. The following were then added in sequence: i) two volumes of BSA at 0.5 mg/ml to remove unabsorbed dynein and to block the surface of the glass; ii) one volume of Taxol-stabilized microtubules (10–20 μg/ml) in HMED buffer with 1 mM ATP, 20 μM Taxol, and 0.1% methylcellulose. The gliding of the microtubules was observed by dark-field illumination with a 40× objective, captured with a digital CCD camera (excel-V; Dage-MTI, Michigan City, IN), and recorded on a personal computer. The recorded movies were analyzed by custom software (Furuta et al., 2008). All microtubules in the visual field were identified, and the displacement of microtubules was measured. The velocities of individual microtubules the length of which was > 10 μm and that traveled at least 10 μm were measured. Approximately 40 microtubules were used to measure the average and standard deviation for each dynein.
In situ axonemal microtubule sliding assay
Microtubule sliding velocities were measured using the method of Okagaki and Kamiya (Okagaki and Kamiya, 1986) and as previously described (Howard et al., 1994; Habermacher and Sale, 1996, 1997; Hendrickson et al., 2004; Bower et al., 2009; Gokhale et al., 2009; Wirschell et al., 2009). Briefly, isolated flagella were resuspended in buffer without protease inhibitors and demembranated with buffer containing 0.5% Nonidet P-40 in 10 mM HEPES, pH 7.4; 5 mM MgSO4; 1 mM DTT; 0.5 mM EDTA; 1% polyethylene glycol (20,000 MW); and 25 mM potassium acetate. The axonemes were added to a perfusion chamber, and microtubule sliding was initiated by the addition of buffer containing 1 mM ATP and subtilisin A Type VIII protease at 3 μg/ml (Sigma Aldrich, St. Louis, MO). Sliding was recorded using a Zeiss Axiovert 35 microscope equipped with dark-field optics, a 40× Plan-Apo lens (Zeiss, Thornwood, NY) and a silicon intensified camera (VE-1000; Dage-MTI). The video images were converted to a digital format using Labview 7.1 software (National Instruments, Austin, TX), and sliding velocity was determined manually by measuring microtubule displacement on tracings calibrated with a micrometer.
Supplementary Material
Acknowledgments
We are grateful to Takuo Yasunaga, Maureen Wirschell, Lea Alford, Ryosuke Yamamoto, and Rasagnya Viswanadha for helpful discussion and reading the manuscript and to Yuji Shitaka for help in defining the optimal conditions for in vitro microtubule gliding assays. We also thank Y. Sakai, M. Kawahara, and M. Nakajima for their help with Chlamydomonas cell culture. We acknowledge Catherine Perrone and Douglas Tritschler for their help with the construction of mutant strains and the verification of genotypes. This work was supported by a Grant-in-Aid for Japan Society for Promotion of Science (JSPS) Fellows to S.T. This work was also supported by a Grant-in-Aid for Scientific Research in the Priority Area “Regulation of Nano-systems in Cells” by the Ministry of Education, Science, and Culture of Japan to K.O and grants from the National Institutes of Health to M.E.P. (GM55667) and W.S.S. (GM051173).
Abbreviations used:
- HC
heavy chain
- IC
intermediate chain
- LC
light chain
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E10-10-0806) on December 9, 2010.
REFERENCES
- Badano JL, Mitsuma N, Beales PL, Katsanis N. The ciliopathies: an emerging class of human genetic disorders. Annu Rev Genomics Hum Genet. 2006;7:125–148. doi: 10.1146/annurev.genom.7.080505.115610. [DOI] [PubMed] [Google Scholar]
- Bower R, VanderWaal K, O’Toole E, Fox L, Perrone C, Mueller J, Wirschell M, Kamiya R, Sale WS, Porter ME. IC138 defines a subdomain at the base of the I1 dynein that regulates microtubule sliding and flagellar motility. Mol Biol Cell. 2009;20:3055–3063. doi: 10.1091/mbc.E09-04-0277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Brokaw CJ. Thinking about flagellar oscillation. Cell Motil Cytoskeleton. 2008;66(8):425–436. doi: 10.1002/cm.20313. [DOI] [PubMed] [Google Scholar]
- Brokaw CJ, Kamiya R. Bending patterns of Chlamydomonas flagella: IV. Mutants with defects in inner and outer dynein arms indicate differences in dynein arm function. Cell Motil Cytoskeleton. 1987;8:68–75. doi: 10.1002/cm.970080110. [DOI] [PubMed] [Google Scholar]
- Bui KH, Sakakibara H, Movassagh T, Oiwa K, Ishikawa T. Molecular architecture of inner dynein arms in situ in Chlamydomonas reinhardtii flagella. J Cell Biol. 2008;183:923–932. doi: 10.1083/jcb.200808050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bui KH, Sakakibara H, Movassagh T, Oiwa K, Ishikawa T. Asymmetry of inner dynein arms and inter-doublet links in Chlamydomonas flagella. J Cell Biol. 2009;186:437–446. doi: 10.1083/jcb.200903082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess SA, Walker ML, Sakakibara H, Knight PJ, Oiwa K. Dynein structure and power stroke. Nature. 2003;421:715–718. doi: 10.1038/nature01377. [DOI] [PubMed] [Google Scholar]
- Burgess SA, Walker ML, Thirumurugan K, Trinick J, Knight PJ. Use of negative stain and single-particle image processing to explore dynamic properties of flexible macromolecules. J Struct Biol. 2004;147:247–258. doi: 10.1016/j.jsb.2004.04.004. [DOI] [PubMed] [Google Scholar]
- Dutcher SK, Gibbons W, Inwood WB. A genetic analysis of suppressors of the PF10 mutation in Chlamydomonas reinhardtii. Genetics. 1988;120:965–976. doi: 10.1093/genetics/120.4.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fliegauf M, Benzing T, Omran H. When cilia go bad: cilia defects and ciliopathies. Nat Rev Mol Cell Biol. 2007;8:880–893. doi: 10.1038/nrm2278. [DOI] [PubMed] [Google Scholar]
- Frank J. Three-dimensional Electron Microscopy of Macromolecular Assemblies. New York: Oxford University Press; 2006. [Google Scholar]
- Furuta A, Yagi T, Yanagisawa HA, Higuchi H, Kamiya R. Systematic comparison of in vitro motile properties between Chlamydomonas wild-type and mutant outer arm dyneins each lacking one of the three heavy chains. J Biol Chem. 2009;284:5927–5935. doi: 10.1074/jbc.M807830200. [DOI] [PubMed] [Google Scholar]
- Furuta K, Edamatsu M, Maeda Y, Toyoshima YY. Diffusion and directed movement: in vitro motile properties of fission yeast kinesin-14 Pkl1. J Biol Chem. 2008;283:36465–36473. doi: 10.1074/jbc.M803730200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes JM, Davis EE, Katsanis N. The vertebrate primary cilium in development, homeostasis, and disease. Cell. 2009;137:32–45. doi: 10.1016/j.cell.2009.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gokhale A, Wirschell M, Sale WS. Regulation of dynein-driven microtubule sliding by the axonemal protein kinase CK1 in Chlamydomonas flagella. J Cell Biol. 2009;186:817–824. doi: 10.1083/jcb.200906168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodenough UW, Gebhart B, Mermall V, Mitchell DR, Heuser JE. High-pressure liquid chromatography fractionation of Chlamydomonas dynein extracts and characterization of inner-arm dynein subunits. J Mol Biol. 1987;194:481–494. doi: 10.1016/0022-2836(87)90676-0. [DOI] [PubMed] [Google Scholar]
- Goodenough UW, Heuser JE. Substructure of inner dynein arms, radial spokes, and the central pair/projection complex of cilia and flagella. J Cell Biol. 1985;100:2008–2018. doi: 10.1083/jcb.100.6.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habermacher G, Sale WS. Regulation of flagellar dynein by an axonemal type-1 phosphatase in Chlamydomonas. J Cell Sci. 1996;109(7):1899–1907. doi: 10.1242/jcs.109.7.1899. [DOI] [PubMed] [Google Scholar]
- Habermacher G, Sale WS. Regulation of flagellar dynein by phosphorylation of a 138-kD inner arm dynein intermediate chain. J Cell Biol. 1997;136:167–176. doi: 10.1083/jcb.136.1.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haimo LT, Telzer BR, Rosenbaum JL. Dynein binds to and crossbridges cytoplasmic microtubules. Proc Natl Acad Sci USA. 1979;76:5759–5763. doi: 10.1073/pnas.76.11.5759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris EH. San Diego, CA: Academic Press; 1989. The Chlamydomonas Sourcebook. [Google Scholar]
- Hendrickson TW, Perrone CA, Griffin P, Wuichet K, Mueller J, Yang P, Porter ME, Sale WS. IC138 is a WD-repeat dynein intermediate chain required for light chain assembly and regulation of flagellar bending. Mol Biol Cell. 2004;15:5431–5442. doi: 10.1091/mbc.E04-08-0694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuser T, Raytchev M, Krell J, Porter ME, Nicastro D. The dynein regulatory complex is the nexin link and a major regulatory node in cilia and flagella. J Cell Biol. 2009;187:921–933. doi: 10.1083/jcb.200908067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard DR, Habermacher G, Glass DB, Smith EF, Sale WS. Regulation of Chlamydomonas flagellar dynein by an axonemal protein kinase. J Cell Biol. 1994;127:1683–1692. doi: 10.1083/jcb.127.6.1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda K, Yamamoto R, Wirschell M, Yagi T, Bower R, Porter ME, Sale WS, Kamiya R. A novel ankyrin-repeat protein interacts with the regulatory proteins of inner arm dynein f (I1) of Chlamydomonas reinhardtii. Cell Motil Cytoskeleton. 2009;66:448–456. doi: 10.1002/cm.20324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagami O, Kamiya R. Translocation and rotation of microtubule caused by multiple species of Chlamydomonas inner-arm dynein. J Cell Sci. 1992;103:653–664. [Google Scholar]
- Kamiya R. Mutations at twelve independent loci result in absence of outer dynein arms in Chylamydomonas reinhardtii. J Cell Biol. 1988;107:2253–2258. doi: 10.1083/jcb.107.6.2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiya R. Functional diversity of axonemal dyneins as studied in Chlamydomonas mutants. Int Rev Cytol. 2002;219:115–155. doi: 10.1016/s0074-7696(02)19012-7. [DOI] [PubMed] [Google Scholar]
- Kamiya R, Kurimoto E, Muto E. Two types of Chlamydomonas flagellar mutants missing different components of inner-arm dynein. J Cell Biol. 1991;112:441–447. doi: 10.1083/jcb.112.3.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiya R, Okamoto M. A mutant of Chlamydomonas reinhardtii that lacks the flagellar outer dynein arm but can swim. J Cell Sci. 1985;74:181–191. doi: 10.1242/jcs.74.1.181. [DOI] [PubMed] [Google Scholar]
- Kikushima K. Central pair apparatus enhances outer-arm dynein activities through regulation of inner-arm dyneins. Cell Motil Cytoskeleton. 2009;66:272–280. doi: 10.1002/cm.20355. [DOI] [PubMed] [Google Scholar]
- King SJ, Dutcher SK. Phosphoregulation of an inner dynein arm complex in Chlamydomonas reinhardtii is altered in phototactic mutant strains. J Cell Biol. 1997;136:177–191. doi: 10.1083/jcb.136.1.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King SM, Kamiya R. Axonemal dyneins: Assembly, structure, and force generation. In: Stern D, Harris EH, Witman GB, editors. In: The Chlamudomonas Sourcebook, vol. III, ed. Amsterdam: Elsevier; 2008. pp. 131–208. [Google Scholar]
- Kotani N, Sakakibara H, Burgess SA, Kojima H, Oiwa K. Mechanical properties of inner-arm dynein-f (dynein I1) studied with in vitro motility assays. Biophys J. 2007;93:886–894. doi: 10.1529/biophysj.106.101964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- LeDizet M, Piperno G. The light chain p28 associates with a subset of inner dynein arm heavy chains in Chlamydomonas axonemes. Mol Biol Cell. 1995;6:697–711. doi: 10.1091/mbc.6.6.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leigh MW, Pittman JE, Carson JL, Ferkol TW, Dell SD, Davis SD, Knowles MR, Zariwala MA. Clinical and genetic aspects of primary ciliary dyskinesia/Kartagener syndrome. Genet Med. 2009;11:473–487. doi: 10.1097/GIM.0b013e3181a53562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall WF. The cell biological basis of ciliary disease. J Cell Biol. 2008;180:17–21. doi: 10.1083/jcb.200710085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastronarde DN, O’Toole ET, McDonald KL, McIntosh JR, Porter ME. Arrangement of inner dynein arms in wild-type and mutant flagella of Chlamydomonas. J Cell Biol. 1992;118:1145–1162. doi: 10.1083/jcb.118.5.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell DR, Rosenbaum JL. A motile Chlamydomonas flagellar mutant that lacks outer dynein arms. J Cell Biol. 1985;100:1228–1234. doi: 10.1083/jcb.100.4.1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno N, Toba S, Edamatsu M, Watai-Nishii J, Hirokawa N, Toyoshima YY, Kikkawa M. Dynein and kinesin share an overlapping microtubule-binding site. EMBO J. 2004;23(13):2359–2467. doi: 10.1038/sj.emboj.7600240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris RL et al. Analysis of cytoskeletal and motility proteins in the sea urchin genome assembly. Dev Biol. 2006;300:219–237. doi: 10.1016/j.ydbio.2006.08.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss AG, Gatti JL, Witman GB. The motile beta/IC1 subunit of sea urchin sperm outer arm dynein does not form a rigor bond. J Cell Biol. 1992a;118:1177–1188. doi: 10.1083/jcb.118.5.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss AG, Sale WS, Fox LA, Witman GB. The alpha subunit of sea urchin sperm outer arm dynein mediates structural and rigor binding to microtubules. J Cell Biol. 1992b;118:1189–1200. doi: 10.1083/jcb.118.5.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Movassagh T, Bui KH, Sakakibara H, Oiwa K, Ishikawa T. Nucleotide-induced global conformational changes of flagellar dynein arms revealed by in situ analysis. Nat Struct Mol Biol. 2010;17:761–767. doi: 10.1038/nsmb.1832. [DOI] [PubMed] [Google Scholar]
- Myster SH, Knott JA, O’Toole E, Porter ME. The Chlamydomonas Dhc1 gene encodes a dynein heavy chain subunit required for assembly of the I1 inner arm complex. Mol Biol Cell. 1997;8:607–620. doi: 10.1091/mbc.8.4.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myster SH, Knott JA, Wysocki KM, O’Toole E, Porter ME. Domains in the 1alpha dynein heavy chain required for inner arm assembly and flagellar motility in Chlamydomonas. J Cell Biol. 1999;146:801–818. doi: 10.1083/jcb.146.4.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicastro D, McIntosh JR, Baumeister W. 3D structure of eukaryotic flagella in a quiescent state revealed by cryo-electron tomography. Proc Natl Acad Sci USA. 2005;102:15889–15894. doi: 10.1073/pnas.0508274102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicastro D, Schwartz C, Pierson J, Gaudette R, Porter ME, McIntosh JR. The molecular architecture of axonemes revealed by cryoelectron tomography. Science. 2006;313:944–948. doi: 10.1126/science.1128618. [DOI] [PubMed] [Google Scholar]
- Nigg EA, Raff JW. Centrioles, centrosomes, and cilia in health and disease. Cell. 2009;139:663–678. doi: 10.1016/j.cell.2009.10.036. [DOI] [PubMed] [Google Scholar]
- Oda T, Hirokawa N, Kikkawa M. Three-dimensional structures of the flagellar dynein-microtubule complex by cryoelectron microscopy. J Cell Biol. 2007;177:243–252. doi: 10.1083/jcb.200609038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oiwa K, Sakakibara H. Recent progress in dynein structure and mechanism. Curr Opin Cell Biol. 2005;17:98–103. doi: 10.1016/j.ceb.2004.12.006. [DOI] [PubMed] [Google Scholar]
- Okagaki T, Kamiya R. Microtubule sliding in mutant Chlamydomonas axonemes devoid of outer or inner dynein arms. J Cell Biol. 1986;103:1895–1902. doi: 10.1083/jcb.103.5.1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okita N, Isogai N, Hirono M, Kamiya R, Yoshimura K. Phototactic activity in Chlamydomonas ‘non-phototactic’ mutants deficient in Ca2+-dependent control of flagellar dominance or in inner-arm dynein. J Cell Sci. 2005;118:529–537. doi: 10.1242/jcs.01633. [DOI] [PubMed] [Google Scholar]
- Patel-King RS, King SM. An outer arm dynein light chain acts in a conformational switch for flagellar motility. J Cell Biol. 2009;186:283–295. doi: 10.1083/jcb.200905083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour GJ, Witman GB. The Chlamydomonas flagellum as a model for human ciliary disease. In: Stern D, Harris EH, Witman GB, editors. In: The Chlamydomonas Sourcebook, vol III, ed. Amsterdam:: Elsevier; 2008. pp. 445–478. [Google Scholar]
- Perrone CA, Myster SH, Bower R, O’Toole ET, Porter ME. Insights into the structural organization of the I1 inner arm dynein from a domain analysis of the 1b dynein heavy chain. Mol Biol Cell. 2000;11:2297–2313. doi: 10.1091/mbc.11.7.2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrone CA, Yang P, O’Toole E, Sale WS, Porter ME. The Chlamydomonas IDA7 locus encodes a 140-kDa dynein intermediate chain required to assemble the I1 inner arm complex. Mol Biol Cell. 1998;9:3351–3365. doi: 10.1091/mbc.9.12.3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piperno G, Ramanis Z, Smith EF, Sale WS. Three distinct inner dynein arms in Chlamydomonas flagella: molecular composition and location in the axoneme. J Cell Biol. 1990;110:379–389. doi: 10.1083/jcb.110.2.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter ME, Power J, Dutcher SK. Extragenic suppressors of paralyzed flagellar mutations in Chlamydomonas reinhardtii identify loci that alter the inner dynein arms. J Cell Biol. 1992;118:1163–1176. doi: 10.1083/jcb.118.5.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter ME, Sale WS. The 9 + 2 axoneme anchors multiple inner arm dyneins and a network of kinases and phosphatases that control motility. J Cell Biol. 2000;151:F37–42. doi: 10.1083/jcb.151.5.f37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts AJ, Burgess SA. Electron microscopic imaging and analysis of isolated dynein particles. Methods Cell Biol. 2009;91:41–61. doi: 10.1016/S0091-679X(08)91002-5. [DOI] [PubMed] [Google Scholar]
- Roberts AJ et al. AAA +Ring and linker swing mechanism in the dynein motor. Cell. 2009;136:485–495. doi: 10.1016/j.cell.2008.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakakibara H, Kojima H, Sakai Y, Katayama E, Oiwa K. Inner-arm dynein c of Chlamydomonas flagella is a single-headed processive motor. Nature. 1999;400:586–590. doi: 10.1038/23066. [DOI] [PubMed] [Google Scholar]
- Sakakibara H, Nakayama H. Translocation of microtubules caused by the alphabeta, beta and gamma outer arm dynein subparticles of Chlamydomonas. J Cell Sci. 1998;111(Pt 9):1155–1164. doi: 10.1242/jcs.111.9.1155. [DOI] [PubMed] [Google Scholar]
- Sale WS, Fox LA. Isolated beta-heavy chain subunit of dynein translocates microtubules in vitro. J Cell Biol. 1988;107:1793–1797. doi: 10.1083/jcb.107.5.1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samso M, Koonce MP. 25 Angstrom resolution structure of a cytoplasmic dynein motor reveals a seven-member planar ring. J Mol Biol. 2004;340:1059–1072. doi: 10.1016/j.jmb.2004.05.063. [DOI] [PubMed] [Google Scholar]
- Satir P, Christensen ST. Overview of structure and function of mammalian cilia. Annu Rev Physiol. 2007;69:377–400. doi: 10.1146/annurev.physiol.69.040705.141236. [DOI] [PubMed] [Google Scholar]
- Sloboda RD, Rosenbaum JL. Purification and assay of microtubule-associated proteins (MAPs) Methods Enzymol. 1982;85(B):409–416. doi: 10.1016/0076-6879(82)85041-6. [DOI] [PubMed] [Google Scholar]
- Smith EF, Sale WS. Microtubule binding and translocation by inner dynein arm subtype I1. Cell Motil Cytoskeleton. 1991;18:258–268. doi: 10.1002/cm.970180403. [DOI] [PubMed] [Google Scholar]
- Smith EF, Sale WS. Regulation of dynein-driven microtubule sliding by the radial spokes in flagella. Science. 1992a;257:1557–1559. doi: 10.1126/science.1387971. [DOI] [PubMed] [Google Scholar]
- Smith EF, Sale WS. Structural and functional reconstitution of inner dynein arms in Chlamydomonas flagellar axonemes. J Cell Biol. 1992b;117:573–581. doi: 10.1083/jcb.117.3.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith EF, Yang P. The radial spokes and central apparatus: mechano-chemical transducers that regulate flagellar motility. Cell Motil Cytoskeleton. 2004;57:8–17. doi: 10.1002/cm.10155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toba S, Toyoshima YY. Dissociation of double-headed cytoplasmic dynein into single-headed species and its motile properties. Cell Motil Cytoskeleton. 2004;58:281–289. doi: 10.1002/cm.20018. [DOI] [PubMed] [Google Scholar]
- Weingarten MD, Lockwood AH, Hwo SY, Kirschner MW. A protein factor essential for microtubule assembly. Proc Natl Acad Sci USA. 1975;72:1858–1862. doi: 10.1073/pnas.72.5.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkes DE, Watson HE, Mitchell DR, Asai DJ. Twenty-five dyneins in Tetrahymena: A re-examination of the multidynein hypothesis. Cell Motil Cytoskeleton. 2008;65:342–351. doi: 10.1002/cm.20264. [DOI] [PubMed] [Google Scholar]
- Wirschell M, Hendrickson T, Sale WS. Keeping an eye on I1: I1 dynein as a model for flagellar dynein assembly and regulation. Cell Motil Cytoskeleton. 2007;64:569–579. doi: 10.1002/cm.20211. [DOI] [PubMed] [Google Scholar]
- Wirschell M, Yang C, Yang P, Fox L, Yanagisawa HA, Kamiya R, Witman GB, Porter ME, Sale WS. IC97 is a novel intermediate chain of I1 dynein that interacts with tubulin and regulates interdoublet sliding. Mol Biol Cell. 2009;20:3044–3054. doi: 10.1091/mbc.E09-04-0276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi T. Bioinformatic approaches to dynein heavy chain classification. Methods Cell Biol. 2009;92:1–9. doi: 10.1016/S0091-679X(08)92001-X. [DOI] [PubMed] [Google Scholar]
- Yang P, Fox L, Colbran RJ, Sale WS. Protein phosphatases PP1 and PP2A are located in distinct positions in the Chlamydomonas flagellar axoneme. J Cell Sci. 2000;113(1):91–102. doi: 10.1242/jcs.113.1.91. [DOI] [PubMed] [Google Scholar]
- Yang P, Sale WS. The Mr 140,000 intermediate chain of Chlamydomonas flagellar inner arm dynein is a WD-repeat protein implicated in dynein arm anchoring. Mol Biol Cell. 1998;9:3335–3349. doi: 10.1091/mbc.9.12.3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P, Sale WS. Casein kinase I is anchored on axonemal doublet microtubules and regulates flagellar dynein phosphorylation and activity. J Biol Chem. 2000;275:18905–18912. doi: 10.1074/jbc.M002134200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.