Abstract
In vertebrates, the conversion of fibrinogen into fibrin is an essential process that underlies the establishment of the supporting protein framework required for coagulation. In invertebrates, fibrinogen-domain-containing proteins play a role in the defense response generated against pathogens; however, they do not function in coagulation, suggesting that this role has been recently acquired. Molecules containing fibrinogen motifs have been identified in numerous invertebrate organisms, and most of these molecules known to date have been linked to defense. Moreover, recent genome projects of invertebrate animals have revealed surprisingly high numbers of fibrinogen-like loci in their genomes, suggesting important and perhaps diverse functions of fibrinogen-like proteins in invertebrates. The ancestral role of molecules containing fibrinogen-related domains (FReDs) with immunity is the focus of this review, with emphasis on specific FReDs called fibrinogen-related proteins (FREPs) identified from the schistosome-transmitting mollusc Biomphalaria glabrata. Herein, we outline the range of invertebrate organisms FREPs can be found in, and detail the roles these molecules play in defense and protection against infection.
Key Words: Biomphalaria, Coagulation, Defense, Fibrinogen, Fibrinogen-related protein, Fibrinogen-related domain, Invertebrate
Evolutionary History of Fibrinogen Domains
Fibrinogen (FBG) domains are important components of invertebrate and vertebrate molecules that have been evolutionarily conserved from at least as far back as the single-celled eukaryotes, the choanoflagellates [1]. Their association with protection against infection has been demonstrated in many vertebrate and invertebrate organisms. Mammalian FBG-containing proteins such as p35 and Hakata antigen (also called L-ficolin and H-ficolin respectively) have played a role in vertebrate innate immune responses by acting as pattern recognition receptors, as well as functioning to activate the complement system [2]. The involvement of fibrinogen in coagulation is phylogenetically recent, with the first description being in the urochordate Botryllus schlosseri [3], and the cephalochordate Branchiostoma floridae[4], which are both deuterostomes. At least in part, the lack of fibrinogen-mediated coagulation in protostome invertebrates is due to the complete absence of molecules involved in the coagulation cascade, however in some, such as molluscs, a process for blood coagulation has yet to be described [5]. Although there are numerous genes (∼400) that contain fibrinogen-related domains (FReDs) in the amphioxus genome [6], none of the other components of the coagulation cascade have been identified [7]. Recent evidence put forth after analysis of the amphioxus genome suggests that it is the most ancestral chordate [6], this hypothesis is supported by the observation that it was not until the rise of urochordates and jawless fish that the basic ingredients needed for fibrinogen-mediated coagulation appeared. Therefore, it is at this point, between cephalochordates and urochordates, that we begin to see a true coagulation response generated by the conversion of fibrinogen to fibrin [3,8].
Despite the fact that invertebrate FBG domain-containing molecules seem not to function in coagulation, many invertebrates do possess other equally complex processes that form clots. The process of hemolymph coagulation has been best characterized in the horseshoe crab and other arthropods. In these organisms, pathogen-associated molecules stimulate the rapid production of a gel formed by the cleavage of coagulogen into coagulin, which then interacts with proxins to form a matrix that immobilizes the pathogen in a network of hemocytes and coagulin polymers [9]. Although the cascade that leads to gel formation differs slightly between the horseshoe crab and other arthropods, some molecules such as transglutaminase [10], which is dependant on calcium for protein cross-linking that occurs during coagulation [11] are conserved. Other molecules, such as lipophorin and lipophorin-like lipoproteins have also been identified as clotting factors common to insect and non-insect arthropods respectively [11]. For example, lipophorin was identified as one of the molecules prominently found in hemolymph clots of the American cockroach Periplaneta americana[11] (the specific mechanisms underlying arthropod coagulation are reviewed elsewhere in this issue and in [11]). It is likely that other cascades that ultimately lead to coagulation of hemolymph factors exist in arthropods and other invertebrate phyla; however, there is limited – yet growing – data on these processes.
Based on genomic evidence, genes that encode for proteins possessing FBG domains are found in varying numbers throughout invertebrates and ancient chordates (table 1). This may be due to genome-wide duplication events and subsequent selection pressure on the duplicated genes [8,12]. The observation that the number of FBG-encoded domains tends to be higher in organisms thought to be more closely related to vertebrates and in vertebrates themselves may reflect an increasing importance of FBG in more complex organisms. It is not until early chordates, in which we can find more than 400 FBG domains encoded in the genome [6], that we begin to see a role for FBG in coagulation. It seems likely that fibrinogen-mediated coagulation is observed in these organisms because of the expansion of the number of FBG-domain-containing molecules, a phenomenon that is first seen in the cephalochordate (Branchiostoma floridae). After this expansion, some part of FBG from the pool of FBG molecules likely gained new functions, such as coagulation, and some still maintain primary immune function. Congruent with the hypothesis that genomic duplications have paved the way for the complexity of the vertebrate immune system [13,14], it is likely that some of these same duplications made it possible for FBG to acquire a new role in coagulation. Prior to these more recent functions, invertebrate FBG-domain-containing molecules played significant roles in immunological defense. That we see increased expression of a FBG-domain-encoding transcript after exposure of the sponge Suberites domuncula, a common ancestor of deuterostomes and protostomes [15], to 1–3-beta-D-glucan suggests the FBG has an ancestral role in defense processes, possibly functioning as a lectin [16].
Table 1.
Number of identified FBG-containing proteins found in invertebrates and ancient chordates
| Lancelets | Branchiostoma floridae | 428 |
| Ascidians | Ciona savignyi | 106 |
| Ciona intestinalis | 68 | |
| Echinoderms | Strongylocentrotus purpuratus | 102 |
| Annelida | Helobdella robusta | 116 |
| Capitella spp. | 139 | |
| Arthropods | Bombyx mori | 3 |
| Nasonia vitripennis | 1 | |
| Apis mellifera | 3 | |
| Drosophila grimshawi | 43 | |
| Drosophila willistoni | 34 | |
| Drosophila pseudoobscura | 29 | |
| Drosophila persimilis | 28 | |
| Drosophila yakuba | 14 | |
| Drosophila simulans | 14 | |
| Drosophila sechellia | 15 | |
| Drosophila melanogaster | 22 | |
| Drosophila erecta | 16 | |
| Drosophila ananassae | 41 | |
| Drosophila virilis | 34 | |
| Drosophila mojavensis | 22 | |
| Culex pipiens quinquefasciatus | 93 | |
| Anopheles gambiae | 59 | |
| Aedes aegypti | 35 | |
| Tribolium castaneum | 7 | |
| Pediculus humanus corporis | 2 | |
| Acyrthosiphon pisum | 3 | |
| Ixodes scapularis | 27 | |
| Daphnia pulex | 37 | |
| Nematoda | Pristionchus pacifiais | 3 |
| Meloidogyne incognita | 0 | |
| Meloidogyne hapla | 0 | |
| Brugia malayi | 1 | |
| Caenorhabditis elegans | 14 | |
| Caenorhabditis briggsae | 5 | |
| Caenorhabditis remanei | 8 | |
| Caenorhabditis brennen | 6 | |
| Caenorhabditis japónica | 6 | |
| Flatworm | Schistosoma mansoni | 0 |
| Cnidarians | Nematostella vectensis | 136 |
| Hydra magnipapillata | 21 | |
| Placozoans | Trichoplax adhaerens | 0 |
| Mollusca | Lottia gigantea | 70 |
| Choanoflagellates | Proterospongia spp. | 6 |
Data extracted from http://supfam.cs.bris.ac.uk/SUPERFAM-ILY/cgibin/taxonomic_gen_list.cgi
Molecules containing fibrinogen domains have emerged as important immune and developmental factors. Since the first identification of an invertebrate FBG domain in the sea cucumber Parastichopus parvimensis [17], a myriad of molecules have been found, and some characterized from organisms spanning almost all invertebrate phyla. From an evolutionary perspective, FBG domains appear to be highly conserved. These domains have been identified in colonial choanoflagellates [1], and sponges [16], which represent some of the most phylogenetically basal multicellular animals. These domains possess 24 canonical residues [17] that allow for them to be easily identified, and it has been demonstrated that using antibodies generated against mammalian fibrinogen one can detect FBG domains in invertebrates at the protein level [18]. However, the other structural properties of FReDs identified in different invertebrate groups are quite diverse. For example, molluscan FREPs structurally pair a FBG domain with one or two immunoglobulin (Ig) domains [19], an organizational blueprint that is not seen in FReDs from any other invertebrate phyla. Although the N-terminal region of FReDs can be varied, the C-terminal region of all FBG-containing proteins known to date is always the FBG domain.
This review will highlight the diversity and functional properties of invertebrate FBG-domain-containing molecules that have been identified to date. Although we will touch on all identified molecules possessing FReDs, we will discuss in detail those identified in the gastropod mollusc Biomphalaria glabrata, from which numerous FReDs that are structurally unique to molluscs and known as FREPs have been identified and characterized functionally.
Functions of FBG Domains in Invertebrates
Functional properties attributed to FReDs are extremely varied. FReDs have been shown to be important for defense processes such as agglutination and bacterial defense, developmental processes, and allorecognition, all of which will be discussed in detail below.
FReDs and the Invertebrate Defense Response
Agglutination
Not long after the initial identification of an FBG domain in the sea cucumber [17], FReDs from various other invertebrates were identified. Initial characterization of these FReDs indicated that they exhibited increased transcriptional expression patterns after stimulation with pathogens or pathogen-associated compounds [20,21,22,23,24,25]. By comparing the expression patterns of FReDs to the observed responses that occurred in each organism, researchers were able to hypothesize the possible functions of FReDs and to design studies to address their roles in defense. Today, it is clear that many invertebrate FReDs possess lectin-like qualities and agglutinate bacteria [21,23,26]. The first direct evidence of FReDs being involved in agglutination came from the horseshoe crab Tachypleus tridentatus [26]. In this study, by Gokudan et al., a FBG-containing plasma lectin named tachylectin was purified and shown to recognize molecules containing acetyl groups, and to agglutinate human erythrocytes as well as both Gram-positive and Gram-negative bacteria. These molecules contained a cystidine-rich N-terminal segment connected to a C-terminal FBG domain that shared its highest homology at the time with human ficolin [26], and has high similarity to a recently identified FBG-domain-containing plasma lectin from the ticks Ornithodoros moubata and Ixodes ricinus, named Dorin M and Ixoderin, respectively [24]. As mentioned above, the horseshoe crab coagulation process is well characterized and known to be initiated by proteolytic cleavage of the molecule coagulogen [27]. The crystal structure of tachylectin 5A revealed that it is structurally related to the fibrinogen γ fragment, with high conservation of the overall 3-dimensional structure as well as the calcium binding site. The acetyl-group-recognition site of tachylectin 5A structurally corresponds to the polymerization pocket ‘a’ of the fibrinogen γ fragment, which is the primary site for the non-covalent polymerization of fibrin. Despite structural similarities to the fibrinogen γ fragment, tachylectins share functional similarities with human L-ficolin/P35, in that they both recognize pathogen associated carbohydrates via their C-terminal FBG-like domain [28]. The characterization of horseshoe crab tachylectins demonstrates that fibrinogen-containing molecules of invertebrates are not involved in coagulation, yet they possess the required structural framework to function in this capacity under the correct conditions.
More recently, a FReD identified in the bay scallop Argopecten irradians was shown to have agglutinating properties [29]. This molecule was able to agglutinate chicken and human erythrocytes as well as Gram-negative and Gram-positive bacteria. Akin to tachylectins, the FBG domain of the scallop FBG molecule shared its highest identity with ficolins and other FReD-containing molecules of invertebrates. Its expression was increased by challenge of A. irradians with Gram-negative bacteria, but remained unchanged after Gram-positive bacterial challenge [29]. Although the mechanisms that lead to agglutination of the targets of these FReDs is unknown, as FReDs from other invertebrate lineages are identified and characterized it is beginning to appear as though the FReDs act as pattern recognition receptors of pathogen-associated patterns. This hypothesis is further supported by observations of FReDs in other invertebrates demonstrating a role in the direct lysis of bacteria (see below).
Anti-Bacterial Properties
The antibacterial properties of FReDs have been best characterized using fruit fly and mosquito model organisms. From these organisms, a number of FReDs have been identified – up to 43 individual FReD genes in certain species of Drosophila[30], 14 in D. melanogaster [30], and as many as 59 FREP genes in Anopheles [31]. Although not all of these FReDs [also called fibrinogen domain immunolectins (FBNs)] have been functionally characterized, many of them are responsive to immunological stimulation using bacteria, fungi or even protozoan Plasmodium parasites [31]. RNAi-mediated knockdown of specific mosquito FBN molecules demonstrated that a number of them were important for successful defense against infection, and in the maintenance of homeostasis. For example, knockdown of FBN22 and FBN39 resulted in mosquitos losing the ability to clear bacterial infections. Moreover, FBN9 was shown to interact with the surfaces of Gram-positive and Gram-negative bacteria, as well as Plasmodium falciparum and P. berghei ookinetes. In addition, it was shown to dimerize when interacting with bacterial cell surfaces which may allow for synergism between FBN molecules [31]. An overview of the known roles that FBNs play in Anopheles summarizes the current knowledge of their functional properties [32]. Identification of another FBG-domain-containing molecule (AL-1) in the mosquito Armigeres subalbatus demonstrated that the AL-1 molecule was located primarily in the hemolymph of an adult mosquito and that it was capable of recognizing N-acetyl-D-glucosamine, and thereby bind to both Gram-positive and Gram-negative bacteria [22]. Many of the FReDs identified in Drosophila spp. share their highest predicted amino acid (aa) identity with FBNs identified in mosquitos implying that they too may have anti-bacterial properties [30].
Apart from the identification of arthropod FReDs that play a role in bacterial clearance, a FReD with strong bacteriolytic activity against both Gram-positive and Gram-negative bacteria has been identified in the cephalochordate Branchiostoma belcheri [33]. This molecule was shown to act as a pattern recognition receptor, recognizing pathogen associated molecular patterns such as lipopolysaccharide, peptidoglycan, and lipoteichoic acid. It possesses FBG domains that are most similar to human fibrinogen β and γ chains, and is highly expressed in the hepatic caecum and hind-gut, and upregulated after stimulation with the bacterial compounds lipopolysaccharide and lipoteichoic acid [33]. Another molecule with similarities to human FBG was identified in the urochordate Halocynthia roretzi[34]. This is the most phylogenetically basal organism in which an FBG domain is found coupled to a collagen domain, creating what is structurally known as a ficolin. Additionally, it shares the ability of human ficolin to recognize N-acetyl groups. Furthermore, based on its amino acid and structural similarities with human ficolin it is believed to function in a similar fashion and in similar processes [34].
FReDs can also function as important barriers to infection, as well as play roles in regulating important enzymatic pathways involved in defense. For example, epiphragmin is a protein identified from the mucus of the snail Cernuella virgata[35]. Structurally, this protein has a C-terminal FBG domain that has 39% amino acid identity to the FBG domains found in FREPs of gastropod hemolymph [19]. Because of its acidic properties, epiphragmin is thought to function primarily as a glue for adhesion of the organism to a substrate; however, it is likely to also act in forming a barrier to protect the snail from microbial assault [35]. Additionally, FReDs have been shown to function in the melanization reaction of the crayfish Pacifastacus leniusculus[36]. Specifically, the molecule melanization inhibition protein (MIP), was shown to be structurally unique from MIPs in other invertebrate species in that it has an FBG domain similar in sequence to the FBG domain of vertebrate ficolins. How crayfish MIP functions is still unknown; however, it does not possess hemagluttinating activity, and a mutation in which 4 Asp amino acids are missing from the FBG domain results in a loss of function, thereby indicating that the FBG region is important for regulating melanization [36].
Development
In addition to being important molecules in host defense, invertebrate FReDs have also been shown to play critical roles in development. FReD-containing molecules have been shown to be involved in eye and organ development of D. melanogaster [37,38], and notochord development and patterning in the urochordate Ciona intestinalis [39]. In Drosophila, the FBG-containing molecule scabrous has been shown to complex with Notch [40], a process that facilitates proper proneuronal development in the eye [41]. Scabrous is also important for omatidial rotation during eye development [42]. It has been shown that the C-terminal FBG-domain of scabrous is critical for proper function, likely via binding to other components of the bristle determination pathway, which is responsible for proper eye development, thereby facilitating and increasing the activity of the N-terminal region of the molecule [37].
Similarly, a FBG-domain-containing protein expressed by the notochord cells in C. intestinalis, named Ci-fibrn, has been shown to interact with Notch, which is expressed in the developing central nervous system. Mutants with disrupted expression of Ci-fibrn display abnormal axon extension patterns and improper positioning of neuronal cells, indicating that Ci-fibrn and its interaction with Notch is a critical process that must occur for proper patterning of the central nervous system [39].
Allorecognition
One of the unique roles that FReD-containing molecules play is in allorecognition and gamete recognition in urochordates. In a proteomic analysis of C. intestinalis egg coat, 2 FBG-like molecules were identified, v-Themis-A and -B [43]. Analysis of the v-Themis-A and -B proteins showed that they were polymorphic proteins that possessed a C-terminal FBG domain. These proteins mapped to the same loci that had previously been identified as being responsible for self-sterility in C. intestinalis[44], a phenomenon that prevents self-fertilization by making the vitelline coat of the egg less likely to bind self sperm in the place of non-self sperm [43,44].
In another urochordate, Botryllus schlosseri, encounters between 2 histoincompatible colonies results in an inflammatory-like response [3]. At the site of the response, which in many ways resembles a coagulation response, many genes that have similarities to vertebrate coagulation molecules were identified. Immunohistochemistry studies using antibodies to FBG revealed a small population of cells that contained molecules with FBG domains. Also among the transcripts identified by EST analysis were orthologs of thrombin, thrombin inhibitors, coagulation-like serine proteases and orthologs of coagulation factors V and VIII. Thus, these points of contact between 2 incompatible colonies represent perhaps the first primitive form of coagulation using the traditional coagulation molecules. This hypothesis was further supported by demonstrating that the formation of cell clots in B. schlosseri was sensitive to addition of heparin, a compound used in vertebrates to prevent coagulation; however, the melanization reaction resulting from the encounter was not affected [3].
FREPs and the Corresponding Genes from the Gastropod Biomphalaria glabrata
The FREPs of gastropod molluscs are well-characterized invertebrate FBG-containing molecules. Their structure is unique amongst FReDs, in that they not only possess a C-terminal FBG domain, but also either one or two N-terminal immunoglobulin superfamily (IgSF) domains. Although they have been identified recently in other gastropods [45], they were initially identified and are best characterized in the planorbid snail Biomphalaria glabrata[19].
The gastropod mollusc B. glabrata is an intermediate host of the human blood fluke Schistosoma mansoni, the causative agent of human schistosomiasis. Schistosomiasis is a chronic, debilitating parasitic disease afflicting as many as 207 million people worldwide [46]. The digenean trematode S. mansoni has a complex life cycle involving snail intermediate and vertebrate definitive hosts. In order to better understand the snail-trematode interactions, 2 models, B. glabrata-S. mansoni and B. glabrata-Echinostoma paraensei have been applied for comparative studies. The immune responses of the snail intermediate hosts to the 2 trematodes are quite different; E. paraensei typically downregulates a number of immune-related transcripts, including some FREPs, as compared to S. mansoni, which initially stimulates an immune response that is later suppressed by the parasite [47,48,49].
Upon infection with E. paraensei, B. glabrata produces 3 diffusely banded sets of plasma proteins (molecular weights: 180–200, 80–120, and 50–70 kDa) [50]. Further studies have demonstrated that the corresponding proteins have the properties of calcium-dependent lectins, and are able to bind and precipitate parasitic antigens, suggestive of a role in internal defense [51]. In order to understand the molecular nature of the proteins, the 50–70 kDa bands have been sequenced and the partial peptide sequences obtained were used to identify the corresponding genes. Analysis of the nucleotide sequences revealed that the proteins encoded by the sequences possessed an N-terminal region that encodes for an IgSF domain(s), separated from a C-terminal β/γ FBG domain by an interceding region [19].
Structure and Diversity
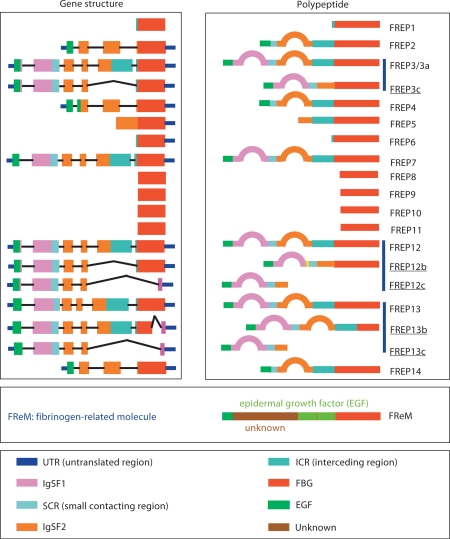
So far 4 FREP genes have been completely sequenced: 2 encoding FREPs with a single IgSF domain [52] and 2 encoding FREPs with tandem IgSF domains [53]. These studies revealed that FREPs belong to a complex gene family. At present, we know that the B. glabrata genome contains about 14 FREP subfamilies [54,55], which can be classified into 2 types; 1-IgSF and 2-IgSF FREPs (fig. 1). Recent studies uncovered a novel type of FBG-containing molecules. Different from all known FREPs, the gene encodes a protein (657aa) composed of a long N-terminal region with no sequence homology to any known protein, a middle epidermal growth factor repeat region and a C-terminal FBG domain, designated FBG-related molecule (FReM) [55]. Phylogenetic analysis indicated that the FBG-encoding region is distantly related to those of other gastropod FREP genes, suggestive of a difference in terms of origin and function [56] (fig. 1). Computational analysis indicated that all FREPs, including the alternatively spliced forms, have leader sequences, suggesting that the proteins are secreted. Sequence and Southern blot analyses have demonstrated that the FBG region, although variable, is the most conserved part of B. glabrata FREPs, whereas the IgSF and interceding regions are highly variable [52,53,54,56].
Fig. 1.
Fibrinogen-containing protein-encoded genes and their putative polypeptides identified from B. glabrata. Some members of subfamilies (FREPs 1, 5, 6, 8, 9, 10, and 11) have incomplete sequences (only FBG sequences). In the box on the right, some FREPs underlined are alternatively spliced forms.
To further explore the diversity of FREPs, the FBG region was amplified from genomic DNA using primers that targeted conserved regions of FREP FBG domains, and was used as a probe. Southern blotting analysis revealed more than 24 bands, suggesting the B. glabrata genome harbors a large number of FBG-like genes. Degenerate PCR analysis obtained 42 unique fibrinogen-encoding sequences from 180 clones derived from a single individual of the M-line strain of B. glabrata, further supporting the notion of their abundant representation in the B. glabrata genome. This suggests FREPs are indeed diverse at the genome level [56]. At the transcription level, alternatively spliced forms of FREPs 3, 12, and 13 were found, suggesting a further increase in diversity at the protein level. Western analysis revealed that most B. glabrata FREP proteins form multimers in snail hemolymph. Moreover, it has been demonstrated that FREP4 forms a multimer, whereas FREP2 forms a monomer, yet the 2 FREPs have the same structure (1 IgSF domain plus FBG domain). This observation suggests that diversity may also occur at the post-translational level [54]. Currently we do not know how the multimers form; however, when the diversity of genes, mRNA and proteins, and different molecular structure are all considered, it can be predicted that thousands of different FREP forms are present in snail hemolymph.
Diversification through Gene Conversion and Point Mutations
A particularly interesting finding is the high level of diversity that occurs in certain members of the B. glabrata FREP family. A diversified array of FREP3 IgSF1 sequences is generated from an apparently small set of source sequences. The high number of sequences obtained from a single individual snail is in sharp contrast with the low number of loci estimated by Southern analysis, suggesting the involvement of a novel mechanism such as somatic diversification [57]. This is the first evidence reported in invertebrates for the somatic diversification of innate defense molecules, implying that invertebrates are capable of producing an array of diverse molecules used in defense. By blurring the distinction between adaptive and innate immunity, this discovery offers new insights into the evolution of immune systems. It has been shown that hemocyte differentiation in Anopheles gambiae is linked to memory mediated by the innate immune response [58]. Furthermore, it is believed that the diversification and increased FREP3 expression in B. glabrata is associated with resistance, and is linked to the production of new hemocytes that exhibit proportionally higher FREP3 expression than do the resident hemocytes [59]. Evidence for diversification of defense molecules, formerly considered to be exclusively the province of the jawed vertebrates, is steadily accumulating in a wide range of organisms, from plants to jawless vertebrates, although the mechanisms and molecules involved differ from one species to another [60,61]. For those interested in the topic, please read the reviews [62,63,64,65,66].
Functional Analysis of B. glabrata FREPs
Gastropod FREPs are of particular interest because they contain 2 domains, IgSF and FBG, both of which are involved in innate defense, in both vertebrates and invertebrates. IgSF defense functions are noted with vertebrate antibodies, Drosophila hemolin [67], and Drosophila and mosquito down syndrome cell adhesion molecule (DSCAM) [68,69]. For molecules with FBG domains, a rapidly growing body of evidence indicates that they play an important role in innate immunity, as emphasized in this paper. In addition to their roles in defense, it is also likely that FREPs are involved in developmental processes. Northern blotting and qPCR analyses revealed that the expression levels of FREPs 2, 3, 4, 14 and FReM are significantly altered during the B. glabrata ontogenesis, suggesting possible roles of FREPs in development [70].
Since gastropod FREPs are a complex and large group of molecules, it will take a long time to understand the functions of all the FREPs in the family. A study using real-time quantitative PCR (qPCR) revealed a significant increase in expression of FREP2 in schistosome-resistant BS-90 snails following exposure to S. mansoni. The same parasite failed to provoke increased expression of FREP2 in susceptible M line snails [71]. This presents the interesting possibility that FREPs play a role in defense against S. mansoni infection as well as other trematodes. Remarkably, it has been suggested by recent work that FREPs may play a role in the compatibility between B. glabarata and S. mansoni. Along with the series of work on the diversity of B. glabrata FREPs, it has been found that S. mansoni is capable of producing polymorphic mucin proteins [72,73]. The protein expression patterns between S. mansoni sporocysts from strains of the parasite that are capable of infecting lab strains (C strain) of B. glabrata or not (IC strain) are different, suggesting an important role of mucins in the compatibility between the snail and parasites. Immunoprecipitation studies revealed that the mucins bind to FREPs (likely FREP2), implying that FREPs have a direct impact on the outcome of the B. glabrata-S. mansoni interaction by recognizing the parasite-associated polymorphic mucins [74]. It has long been suspected that compatibility between the snail host and parasite depends on the status of allele/genotype-match between host and parasite [75,76], in which successful detection of the parasite requires a specific combination of host and parasite alleles, also known as the Red Queen theory [77,78]. The interactions discovered between FREPs and the parasite-associated polymorphic mucins provide preliminary molecular evidence for this hypothesis. How the 2 large repertoires of molecules derived from snail host and parasite interplay will be a very interesting question, which eventually may help decipher the underlying mechanisms of host-parasite interactions in this system.
Moreover, injection of bacterial lipopolysaccharide leads to increased expression of FREP3, and injection with Gram-positive bacteria results in the upregulated expression of FREP4 and FREP7, suggesting that FREPs may take part in complex responses to a wide range of pathogenic agents [48]. Western blot analyses revealed that expression of multiple FREPs, including FREP4 in plasma from M line and BS-90 snails is upregulated significantly after infection with the trematode E. paraensei. Studies have demonstrated that FREPs are able to bind E. paraensei sporocysts and their secretory/excretory products, and a variety of microbes (Gram-positive and Gram-negative bacteria and yeast). This binding capability shows evidence of specificity with respect to pathogen type; for example, 65–75-kDa FREPs (mainly FREP4) bind to E. paraensei sporocysts and their secretory/excretory products whereas 95-kDa and 125-kDa FREPs bind the microbes assayed. These results suggest that FREPs can recognize a wide range of pathogens, from prokaryotes to eukaryotes, and different categories of FREPs seem to exhibit functional specialization with respect to the pathogen encountered [70].
Finally, recent studies have demonstrated that RNAi-mediated knockdown of FREP3 resulted in disruption of snail size-related resistance to infection by E. paraensei. Normally, adult snails (>12 mm diameter) are resistant to E. paraensei; however, about 30% FREP3 RNAi-treated adult snails became infected. No snails were infected in the green fluorescent protein (GFP) RNAi controls. This study provides direct evidence indicating an anti-trematode role of FREPs in B. glabrata[79].
Conclusions
Based on evidence from invertebrate and vertebrate models, it is clear that FBG and molecules containing FBG domains play important roles in defense. That FBG domains are found in such a wide range of organisms, and in such high numbers attests to the versatility of FBG and its importance. Evidence from invertebrates implicates FBG-domain-containing molecules in important defense processes such as development, agglutination, pathogen recognition, bacterial lysis, histocompatability, and parasite defense. In addition, the FBG domain can be coupled to a variety of N-terminal domains such as IgSF or collagen domains, providing the possibility for diversification of the molecule (IgSF) or adding new functional possibilities. With such a diversity of functions it is no surprise that this molecule also plays an essential role in coagulation, a process first linked to FBG in cephalochordates, and then further developed in jawless fish. Most likely, gene duplication events allowed for the expansion of molecules essential for FBG-mediated coagulation in chordates, thereby facilitating the development of this role. The existence in vertebrates of FBG-domain-containing molecules, such as ficolins, that do not participate in coagulation further reinforces its importance in defense against pathogens, a capacity in which FBG-containing molecules played a role even in early protostome invertebrates.
Acknowledgements
The authors would like to acknowledge Coen M. Adema for editorial assistance and critique of the manuscript. Support for this project came from NIHAI067686, NIHAI24340, and NCRR P20RR18754.
Abbreviations used in this paper
- FBG
fibrinogen
- FREP
fibrinogen-related protein: molluscan molecules containing a fibrinogen domain coupled to a immu-noglobulin superfamily domain
- FReD
fibrinogen-related domain: refers to all other inver-tebrate fibrinogen domain-containing molecules described in this manuscript
- FBN
fibrinogen domain immunolectins: specific fibrino-gen molecules of mosquitos
- FReM
fibrinogen-related molecule: snail molecules com-posed of a long N-terminal region with no sequence homology to any known protein, a middle epidermal factor repeat region and a C-terminal FBG domain
References
- 1.King N, Westbrook MJ, Young SL, Kuo A, Abedin M, Chapman J, Fairclough S, Hellsten U, Isogai Y, Letunic I, Marr M, Pincus D, Putnam N, Rokas A, Wright KJ, Zuzow R, Dirks W, Good M, Goodstein D, Lemons D, Li W, Lyons JB, Morris A, Nichols S, Richter DJ, Salamov A, Sequencing JG, Bork P, Lim WA, Manning G, Miller WT, McGinnis W, Shapiro H, Tjian R, Grigoriev IV, Rokhsar D. The genome of the choanoflagellate Monosiga brevicollis and the origin of metazoans. Nature. 2008;451:783–788. doi: 10.1038/nature06617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Matsushita M, Fujita T. The role of ficolins in innate immunity. Immunobiology. 2002;205:490–497. doi: 10.1078/0171-2985-00149. [DOI] [PubMed] [Google Scholar]
- 3.Oren M, Escande ML, Paz G, Fishelson Z, Rinkevich B. Urochordate histoincompatible interactions activate vertebrate-like coagulation system components. PLoS ONE. 2008;3:e3123. doi: 10.1371/journal.pone.0003123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Putnam NH, Butts T, Ferrier DE, Furlong RF, Hellsten U, Kawashima T, Robinson-Rechavi M, Shoguchi E, Terry A, Yu JK, Benito-Gutierrez EL, Dubchak I, Garcia-Fernandez J, Gibson-Brown JJ, Grigoriev IV, Horton AC, de Jong PJ, Jurka J, Kapitonov VV, Kohara Y, Kuroki Y, Lindquist E, Lucas S, Osoegawa K, Pennacchio LA, Salamov AA, Satou Y, Sauka-Spengler T, Schmutz J, Shin IT, Toyoda A, Bronner-Fraser M, Fujiyama A, Holland LZ, Holland PW, Satoh N, Rokhsar DS. The amphioxus genome and the evolution of the chordate karyotype. Nature. 2008;453:1064–1071. doi: 10.1038/nature06967. [DOI] [PubMed] [Google Scholar]
- 5.Millar DA, Rateliffe NA. Invertebrate. In: Turner RJ, editor. Immunology, a Comparative Approach. New York: Wiley; 1994. pp. 29–60. [Google Scholar]
- 6.Huang S, Yuan S, Guo L, Yu Y, Li J, Wu T, Liu T, Yang M, Wu K, Liu H, Ge J, Huang H, Dong M, Yu C, Chen S, Xu A. Genomic analysis of the immune gene repertoire of amphioxus reveals extraordinary innate complexity and diversity. Genome Res. 2008;18:1112–1126. doi: 10.1101/gr.069674.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Doolittle RF. Step-by-step evolution of vertebrate blood coagulation. Cold Spring Harb Symp Quant Biol. 2009;74:35–40. doi: 10.1101/sqb.2009.74.001. [DOI] [PubMed] [Google Scholar]
- 8.Doolittle RF, Jiang Y, Nand J. Genomic evidence for a simpler clotting scheme in jawless vertebrates. J Mol Evol. 2008;66:185–196. doi: 10.1007/s00239-008-9074-8. [DOI] [PubMed] [Google Scholar]
- 9.Osaki T, Kawabata S. Structure and function of coagulogen, a clottable protein in horseshoe crabs. Cell Mol Life Sci. 2004;61:1257–1265. doi: 10.1007/s00018-004-3396-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cerenius L, Kawabata SI, Lee BL, Nonaka M, Soderhall K. Proteolytic cascades and their involvement in invertebrate immunity. Trends Biochem Sci. 2010;35:575–583. doi: 10.1016/j.tibs.2010.04.006. [DOI] [PubMed] [Google Scholar]
- 11.Dushay MS. Insect hemolymph clotting. Cell Mol Life Sci. 2009;66:2643–2650. doi: 10.1007/s00018-009-0036-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dehal P, Satou Y, Campbell RK, Chapman J, Degnan B, De Tomaso A, Davidson B, Di Gregorio A, Gelpke M, Goodstein DM, Harafuji N, Hastings KE, Ho I, Hotta K, Huang W, Kawashima T, Lemaire P, Martinez D, Meinertzhagen IA, Necula S, Nonaka M, Putnam N, Rash S, Saiga H, Satake M, Terry A, Yamada L, Wang HG, Awazu S, Azumi K, Boore J, Branno M, Chin-Bow S, DeSantis R, Doyle S, Francino P, Keys DN, Haga S, Hayashi H, Hino K, Imai KS, Inaba K, Kano S, Kobayashi K, Kobayashi M, Lee BI, Makabe KW, Manohar C, Matassi G, Medina M, Mochizuki Y, Mount S, Morishita T, Miura S, Nakayama A, Nishizaka S, Nomoto H, Ohta F, Oishi K, Rigoutsos I, Sano M, Sasaki A, Sasakura Y, Shoguchi E, Shin-i T, Spagnuolo A, Stainier D, Suzuki MM, Tassy O, Takatori N, Tokuoka M, Yagi K, Yoshizaki F, Wada S, Zhang C, Hyatt PD, Larimer F, Detter C, Doggett N, Glavina T, Hawkins T, Richardson P, Lucas S, Kohara Y, Levine M, Satoh N, Rokhsar DS. The draft genome of Ciona intestinalis: insights into chordate and vertebrate origins. Science. 2002;298:2157–2167. doi: 10.1126/science.1080049. [DOI] [PubMed] [Google Scholar]
- 13.Okada K, Asai K. Expansion of signaling genes for adaptive immune system evolution in early vertebrates. BMC Genomics. 2008;9:218. doi: 10.1186/1471-2164-9-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kasahara M. The 2R hypothesis: an update. Curr Opin Immunol. 2007;19:547–552. doi: 10.1016/j.coi.2007.07.009. [DOI] [PubMed] [Google Scholar]
- 15.Telford MJ. Resolving animal phylogeny: a sledgehammer for a tough nut? Dev Cell. 2008;14:457–459. doi: 10.1016/j.devcel.2008.03.016. [DOI] [PubMed] [Google Scholar]
- 16.Perovic-Ottstadt S, Adell T, Proksch P, Wiens M, Korzhev M, Gamulin V, Muller IM, Muller WE. A (1→3)-beta-D-glucan recognition protein from the sponge Suberites domuncula: mediated activation of fibrinogen-like protein and epidermal growth factor gene expression. Eur J Biochem. 2004;271:1924–1937. doi: 10.1111/j.1432-1033.2004.04102.x. [DOI] [PubMed] [Google Scholar]
- 17.Xu X, Doolittle RF. Presence of a vertebrate fibrinogen-like sequence in an echinoderm. Proc Natl Acad Sci USA. 1990;87:2097–2101. doi: 10.1073/pnas.87.6.2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Adema CM, Hertel LA, Loker ES. Evidence from two planorbid snails of a complex and dedicated response to digenean (echinostome) infection. Parasitology. 1999;119:395–404. doi: 10.1017/s0031182099004850. [DOI] [PubMed] [Google Scholar]
- 19.Adema CM, Hertel LA, Miller RD, Loker ES. A family of fibrinogen-related proteins that precipitates parasite-derived molecules is produced by an invertebrate after infection. Proc Natl Acad Sci USA. 1997;94:8691–8696. doi: 10.1073/pnas.94.16.8691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ramirez-Gomez F, Ortiz-Pineda PA, Rivera-Cardona G, Garcia-Arraras JE. LPS-induced genes in intestinal tissue of the sea cucumber Holothuria glaberrima. PLoS ONE. 2009;4:e6178. doi: 10.1371/journal.pone.0006178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gorbushin AM, Iakovleva NV. A new gene family of single fibrinogen domain lectin in Mytilus. Fish Shellfish Immunol. 2010 doi: 10.1016/j.fsi.2010.10.002. DOI: 10.1016/j.fsi.2010.10.002. [DOI] [PubMed] [Google Scholar]
- 22.Wang X, Rocheleau TA, Fuchs JF, Hillyer JF, Chen CC, Christensen BM. A novel lectin with a fibrinogen-like domain and its potential involvement in the innate immune response of Armigeres subalbatus against bacteria. Insect Mol Biol. 2004;13:273–282. doi: 10.1111/j.0962-1075.2004.00484.x. [DOI] [PubMed] [Google Scholar]
- 23.Kurachi S, Song Z, Takagaki M, Yang Q, Winter HC, Kurachi K, Goldstein IJ. Sialic-acid-binding lectin from the slug Limax flavus: cloning, expression of the polypeptide, and tissue localization. Eur J Biochem. 1998;254:217–222. doi: 10.1046/j.1432-1327.1998.2540217.x. [DOI] [PubMed] [Google Scholar]
- 24.Rego RO, Kovar V, Kopacek P, Weise C, Man P, Sauman I, Grubhoffer L. The tick plasma lectin, Dorin M, is a fibrinogen-related molecule. Insect Biochem Mol Biol. 2006;36:291–299. doi: 10.1016/j.ibmb.2006.01.008. [DOI] [PubMed] [Google Scholar]
- 25.Rego RO, Hajdusek O, Kovar V, Kopacek P, Grubhoffer L, Hypsa V. Molecular cloning and comparative analysis of fibrinogen-related proteins from the soft tick Ornithodoros moubata and the hard tick Ixodes ricinus. Insect Biochem Mol Biol. 2005;35:991–1004. doi: 10.1016/j.ibmb.2005.04.001. [DOI] [PubMed] [Google Scholar]
- 26.Gokudan S, Muta T, Tsuda R, Koori K, Kawahara T, Seki N, Mizunoe Y, Wai SN, Iwanaga S, Kawabata S. Horseshoe crab acetyl group-recognizing lectins involved in innate immunity are structurally related to fibrinogen. Proc Natl Acad Sci USA. 1999;96:10086–10091. doi: 10.1073/pnas.96.18.10086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kawasaki H, Nose T, Muta T, Iwanaga S, Shimohigashi Y, Kawabata S. Head-to-tail polymerization of coagulin, a clottable protein of the horseshoe crab. J Biol Chem. 2000;275:35297–35301. doi: 10.1074/jbc.M006856200. [DOI] [PubMed] [Google Scholar]
- 28.Kairies N, Beisel HG, Fuentes-Prior P, Tsuda R, Muta T, Iwanaga S, Bode W, Huber R, Kawabata S. The 2.0-A crystal structure of tachylectin 5A provides evidence for the common origin of the innate immunity and the blood coagulation systems. Proc Natl Acad Sci USA. 2001;98:13519–13524. doi: 10.1073/pnas.201523798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang H, Wang LL, Song LS, Song XY, Wang B, Mu CK, Zhang Y. A fibrinogen-related protein from bay scallop Argopecten irradians involved in innate immunity as pattern recognition receptor. Fish Shellfish Immunol. 2009;26:56–64. doi: 10.1016/j.fsi.2008.07.019. [DOI] [PubMed] [Google Scholar]
- 30.Middha S, Wang X. Evolution and potential function of fibrinogen-like domains across twelve Drosophila species. BMC Genomics. 2008;9:260. doi: 10.1186/1471-2164-9-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dong Y, Dimopoulos G. Anopheles fibrinogen-related proteins provide expanded pattern recognition capacity against bacteria and malaria parasites. J Biol Chem. 2009;284:9835–9844. doi: 10.1074/jbc.M807084200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cirimotich CM, Dong Y, Garver LS, Sim S, Dimopoulos G. Mosquito immune defenses against Plasmodium infection. Dev Comp Immunol. 2010;34:387–395. doi: 10.1016/j.dci.2009.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fan C, Zhang S, Li L, Chao Y. Fibrinogen-related protein from amphioxus Branchiostoma belcheri is a multivalent pattern recognition receptor with a bacteriolytic activity. Mol Immunol. 2008;45:3338–3346. doi: 10.1016/j.molimm.2008.04.019. [DOI] [PubMed] [Google Scholar]
- 34.Kenjo A, Takahashi M, Matsushita M, Endo Y, Nakata M, Mizuochi T, Fujita T. Cloning and characterization of novel ficolins from the solitary ascidian, Halocynthia roretzi. J Biol Chem. 2001;276:19959–19965. doi: 10.1074/jbc.M011723200. [DOI] [PubMed] [Google Scholar]
- 35.Li D, Graham LD. Epiphragmin, the major protein of epiphragm mucus from the vineyard snail, Cernuella virgata. Comp Biochem Physiol B Biochem Mol Biol. 2007;148:192–200. doi: 10.1016/j.cbpb.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 36.Soderhall I, Wu C, Novotny M, Lee BL, Soderhall K. A novel protein acts as a negative regulator of prophenoloxidase activation and melanization in the freshwater crayfish Pacifastacus leniusculus. J Biol Chem. 2009;284:6301–6310. doi: 10.1074/jbc.M806764200. [DOI] [PubMed] [Google Scholar]
- 37.Lee EC, Yu SY, Hu X, Mlodzik M, Baker NE. Functional analysis of the fibrinogen-related scabrous gene from Drosophila melanogaster identifies potential effector and stimulatory protein domains. Genetics. 1998;150:663–673. doi: 10.1093/genetics/150.2.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mlodzik M, Baker NE, Rubin GM. Isolation and expression of scabrous, a gene regulating neurogenesis in Drosophila. Genes Dev. 1990;4:1848–1861. doi: 10.1101/gad.4.11.1848. [DOI] [PubMed] [Google Scholar]
- 39.Yamada S, Hotta K, Yamamoto TS, Ueno N, Satoh N, Takahashi H. Interaction of notochord-derived fibrinogen-like protein with Notch regulates the patterning of the central nervous system of Ciona intestinalis embryos. Dev Biol. 2009;328:1–12. doi: 10.1016/j.ydbio.2008.12.037. [DOI] [PubMed] [Google Scholar]
- 40.Powell PA, Wesley C, Spencer S, Cagan RL. Scabrous complexes with Notch to mediate boundary formation. Nature. 2001;409:626–630. doi: 10.1038/35054566. [DOI] [PubMed] [Google Scholar]
- 41.Lee EC, Hu X, Yu SY, Baker NE. The scabrous gene encodes a secreted glycoprotein dimer and regulates proneural development in Drosophila eyes. Mol Cell Biol. 1996;16:1179–1188. doi: 10.1128/mcb.16.3.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chou YH, Chien CT. Scabrous controls ommatidial rotation in the Drosophila compound eye. Dev Cell. 2002;3:839–850. doi: 10.1016/s1534-5807(02)00362-3. [DOI] [PubMed] [Google Scholar]
- 43.Yamada L, Saito T, Taniguchi H, Sawada H, Harada Y. Comprehensive egg coat proteome of the ascidian Ciona intestinalis reveals gamete recognition molecules involved in self-sterility. J Biol Chem. 2009;284:9402–9410. doi: 10.1074/jbc.M809672200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Harada Y, Takagaki Y, Sunagawa M, Saito T, Yamada L, Taniguchi H, Shoguchi E, Sawada H. Mechanism of self-sterility in a hermaphroditic chordate. Science. 2008;320:548–550. doi: 10.1126/science.1152488. [DOI] [PubMed] [Google Scholar]
- 45.Gorbushin AM, Panchin YV, Iakovleva NV. In search of the origin of FREPs: characterization of Aplysia californica fibrinogen-related proteins. Dev Comp Immunol. 2010;34:465–473. doi: 10.1016/j.dci.2009.12.007. [DOI] [PubMed] [Google Scholar]
- 46.King CH. Toward the elimination of schistosomiasis. N Engl J Med. 2009;360:106–109. doi: 10.1056/NEJMp0808041. [DOI] [PubMed] [Google Scholar]
- 47.Loker ES, Adema CM. Schistosomes, Echinostomes and snails: comparative immunobiology. Parasitology Today. 1995;11:120–124. [Google Scholar]
- 48.Adema CM, Hanington PC, Lun CM, Rosenberg GH, Aragon AD, Stout BA, Lennard Richard ML, Gross PS, Loker ES. Differential transcriptomic responses of Biomphalaria glabrata (Gastropoda, Mollusca) to bacteria and metazoan parasites, Schistosoma mansoni and Echinostoma paraensei (Digenea, Platyhelminthes) Mol Immunol. 2010;47:849–860. doi: 10.1016/j.molimm.2009.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hanington PC, Lun CM, Adema CM, Loker ES. Time series analysis of the transcriptional responses of Biomphalaria glabrata throughout the course of intramolluscan development of Schistosoma mansoni and Echinostoma paraensei. Int J Parasitol. 2010;40:819–831. doi: 10.1016/j.ijpara.2009.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Monroy FP, Loker ES. Production of heterogeneous carbohydrate-binding proteins by the host snail Biomphalaria glabrata following exposure to Echinostoma paraensei and Schistosoma mansoni. J Parasitol. 1993;79:416–423. [PubMed] [Google Scholar]
- 51.Hertel LA, Stricker SA, Monroy FP, Wilson WD, Loker ES. Biomphalaria glabrata hemolymph lectins: binding to bacteria, mammalian erythrocytes, and to sporocysts and rediae of Echinostoma paraensei. J Invertebr Pathol. 1994;64:52–61. doi: 10.1006/jipa.1994.1068. [DOI] [PubMed] [Google Scholar]
- 52.Leonard PM, Adema CM, Zhang SM, Loker ES. Structure of two FREP genes that combine IgSF and fibrinogen domains, with comments on diversity of the FREP gene family in the snail Biomphalaria glabrata. Gene. 2001;269:155–165. doi: 10.1016/s0378-1119(01)00444-9. [DOI] [PubMed] [Google Scholar]
- 53.Zhang SM, Leonard PM, Adema CM, Loker ES. Parasite-responsive IgSF members in the snail Biomphalaria glabrata: characterization of novel genes with tandemly arranged IgSF domains and a fibrinogen domain. Immunogenetics. 2001;53:684–694. doi: 10.1007/s00251-001-0386-8. [DOI] [PubMed] [Google Scholar]
- 54.Zhang SM, Loker ES. The FREP gene family in the snail Biomphalaria glabrata: additional members, and evidence consistent with alternative splicing and FREP retrosequences. Fibrinogen-related proteins. Dev Comp Immunol. 2003;27:175–187. doi: 10.1016/s0145-305x(02)00091-5. [DOI] [PubMed] [Google Scholar]
- 55.Zhang SM, Nian H, Zeng Y, Dejong RJ. Fibrinogen-bearing protein genes in the snail Biomphalaria glabrata: characterization of two novel genes and expression studies during ontogenesis and trematode infection. Dev Comp Immunol. 2008;32:1119–1130. doi: 10.1016/j.dci.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang SM, Loker ES. Representation of an immune responsive gene family encoding fibrinogen-related proteins in the freshwater mollusc Biomphalaria glabrata, an intermediate host for Schistosoma mansoni. Gene. 2004;341:255–266. doi: 10.1016/j.gene.2004.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang SM, Adema CM, Kepler TB, Loker ES. Diversification of Ig superfamily genes in an invertebrate. Science. 2004;305:251–254. doi: 10.1126/science.1088069. [DOI] [PubMed] [Google Scholar]
- 58.Rodrigues J, Brayner FA, Alves LC, Dixit R, Barillas-Mury C. Hemocyte differentiation mediates innate immune memory in Anopheles gambiae mosquitoes. Science. 2010;329:1353–1355. doi: 10.1126/science.1190689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stout BA, Adema CM, Zhang S-M, Loker ES. Biology of FREPs: diversified lectins with fibrinogen-related domains from the freshwater snail Biomphalaria glabrata. In: Vasta GR, Ahmed H, editors. Animal Lectins: A Functional View. Boca Raton: CRC Press; 2009. pp. 475–491. [Google Scholar]
- 60.Cooper MD, Alder MN. The evolution of adaptive immune systems. Cell. 2006;124:815–822. doi: 10.1016/j.cell.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 61.Litman GW, Cannon JP, Rast JP. New insights into alternative mechanisms of immune receptor diversification. Adv Immunol. 2005;87:209–236. doi: 10.1016/S0065-2776(05)87006-3. [DOI] [PubMed] [Google Scholar]
- 62.Litman GW, Dishaw LJ, Cannon JP, Haire RN, Rast JP. Alternative mechanisms of immune receptor diversity. Curr Opin Immunol. 2007;19:526–534. doi: 10.1016/j.coi.2007.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Herrin BR, Cooper MD. Alternative adaptive immunity in jawless vertebrates. J Immunol. 2010;185:1367–1374. doi: 10.4049/jimmunol.0903128. [DOI] [PubMed] [Google Scholar]
- 64.Jones JD, Dangl JL. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 65.Kurtz J, Armitage SA. Alternative adaptive immunity in invertebrates. Trends Immunol. 2006;27:493–496. doi: 10.1016/j.it.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 66.Loker ES, Adema CM, Zhang SM, Kepler TB. Invertebrate immune systems: not homogeneous, not simple, not well understood. Immunol Rev. 2004;198:10–24. doi: 10.1111/j.0105-2896.2004.0117.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sun SC, Lindstrom I, Boman HG, Faye I, Schmidt O. Hemolin: an insect-immune protein belonging to the immunoglobulin superfamily. Science. 1990;250:1729–1732. doi: 10.1126/science.2270488. [DOI] [PubMed] [Google Scholar]
- 68.Watson FL, Puttmann-Holgado R, Thomas F, Lamar DL, Hughes M, Kondo M, Rebel VI, Schmucker D. Extensive diversity of Ig-superfamily proteins in the immune system of insects. Science. 2005;309:1874–1878. doi: 10.1126/science.1116887. [DOI] [PubMed] [Google Scholar]
- 69.Dong Y, Taylor HE, Dimopoulos G. AgDscam, a hypervariable immunoglobulin domain-containing receptor of the Anopheles gambiae innate immune system. PLoS Biol. 2006;4:e229. doi: 10.1371/journal.pbio.0040229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang SM, Zeng Y, Loker ES. Expression profiling and binding properties of fibrinogen-related proteins (FREPs), plasma proteins from the schistosome snail host Biomphalaria glabrata. Innate Immun. 2008;14:175–189. doi: 10.1177/1753425908093800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hertel LA, Adema CM, Loker ES. Differential expression of FREP genes in two strains of Biomphalaria glabrata following exposure to the digenetic trematodes Schistosoma mansoni and Echinostoma paraensei. Dev Comp Immunol. 2005;29:295–303. doi: 10.1016/j.dci.2004.08.003. [DOI] [PubMed] [Google Scholar]
- 72.Roger E, Gourbal B, Grunau C, Pierce RJ, Galinier R, Mitta G. Expression analysis of highly polymorphic mucin proteins (Sm PoMuc) from the parasite Schistosoma mansoni. Mol Biochem Parasitol. 2008;157:217–227. doi: 10.1016/j.molbiopara.2007.11.015. [DOI] [PubMed] [Google Scholar]
- 73.Roger E, Grunau C, Pierce RJ, Hirai H, Gourbal B, Galinier R, Emans R, Cesari IM, Cosseau C, Mitta G. Controlled chaos of polymorphic mucins in a metazoan parasite (Schistosoma mansoni) interacting with its invertebrate host (Biomphalaria glabrata) PLoS Negl Trop Dis. 2008;2:e330. doi: 10.1371/journal.pntd.0000330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mone Y, Gourbal B, Duval D, Du Pasquier L, Keiffer-Jaquinod S, Mitta G. A large repertoire of parasite epitopes matched by a large repertoire of host immune receptors in an invertebrate host/parasite model. PLoS Negl Trop Dis. 2010;4:e813. doi: 10.1371/journal.pntd.0000813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Richards CS. Genetic factors in susceptibility of Biomphalaria glabrata for different strains of Schistosoma mansoni. Parasitology. 1975;70:231–241. doi: 10.1017/s0031182000049696. [DOI] [PubMed] [Google Scholar]
- 76.Theron A, Coustau C. Are Biomphalaria snails resistant to Schistosoma mansoni? J Helminthol. 2005;79:187–191. doi: 10.1079/joh2005299. [DOI] [PubMed] [Google Scholar]
- 77.Agrawal AF, Lively CM. Modelling infection as a two-step process combining gene-for-gene and matching-allele genetics. Proc Royal Soc Lond B Biol Sci. 2003;270:323–334. doi: 10.1098/rspb.2002.2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Agrawal A, Lively CM. Infection genetics: gene-for-gene versus matching-alleles models and all points in between. Evol Ecol Res. 2002;4:79–90. [Google Scholar]
- 79.Hanington PC, Forys MA, Dragoo JW, Zhang SM, Adema CM, Loker ES: A role for a somatically diversified lectin in resistance of an invertebrate to parasite infection. Proc Natl Acad Sci USA 2010, in press. [DOI] [PMC free article] [PubMed]