Abstract
Thioester-containing proteins (TEPs) are conserved proteins among insects that are thought to be involved in innate immunity. In Drosophila, the Tep family is composed of 6 genes named Tep1–Tep6. In this study, we investigated the phylogeny, expression pattern and roles of these genes in the host defense of Drosophila. Protostomian Tep genes are clustered in 3 distinct branches, 1 of which is specific to mosquitoes. Most D. melanogaster Tep genes are expressed in hemocytes, can be induced in the fat body, and are expressed in specific regions of the hypodermis. This expression pattern is consistent with a role in innate immunity. However, we find that TEP1, TEP2, and TEP4 are not strictly required in the body cavity to fight several bacterial and fungal infections. One possibility is that Drosophila TEPs act redundantly or that their absence can be compensated by other components of the immune response. TEPs may thus provide a subtle selective advantage during evolution. Alternatively, they may be required in host defense against specific as yet unidentified natural pathogens of Drosophila.
Key Words: Drosophila melanogaster, Gene disruption, Genome evolution, Genomic sequence analysis, In situ hybridization, Innate immunity, Insects, Invertebrates, Thioester-containing proteins
Introduction
The identification of pathogenic threats, either by pattern recognition receptors or by sensing of microbial virulence factors, is crucial to innate immune function, which only relies on germ-line-encoded receptors [1]. In vertebrates, complement factors directly bind to the surface of pathogens thereby initiating their destruction [2]. In the alternative activation pathway of vertebrate complement, surfaces are bound nonspecifically by complement proteins. The subsequent activation of complement is prevented by specific inhibitors expressed on self surfaces such as CD46 and CD55, which inhibit the formation of the C3 convertase responsible for the activation of the C3 complement factor [3]. These inhibitors are missing on nonself surfaces and complement activation ensues. Thus, the alternative complement pathway has been suggested to recognize missing self [3]. Complement proteins belong to the thioester-containing protein (TEP) family, which is present in a wide variety of species, ranging from deuterostomes to protostomes, suggesting that the complement system has evolved and adapted over a period of up to 1 billion years [4]. Phylogenetically, TEPs from insects share sequence similarities with both the vertebrate complement factors C3/C4/C5 and the α2-macroglobulin family of protease inhibitors. These TEPs contain a central hypervariable region, which corresponds to the bait domain of α2-macroglobulin and to the anaphylatoxin domain in C3. Most TEPs share the common 4-amino-acid sequence (CGEQ) defining the thioester site, which allows the formation of a covalent bond to microbial surfaces.
Drosophila melanogaster is a valuable model to decipher the mechanisms of insect innate immunity [5,6]. The TEP family was first investigated at a time when pattern recognition receptors able to trigger the systemic immune response had not yet been identified. Its relative similarity to the complement-like system described in invertebrates such as the sea urchin, tunicates and arthropods made them good candidates for missing self receptors [7]. The Tep family in Drosophila is composed of six genes (Tep1–Tep6), one of which, Tep5, does not appear to be expressed [7]. TEP6 (also known as macroglobulin-complement related, MCR) is an exception in this Drosophila family because the thioester binding site is likely to be nonfunctional as the cysteine residue of the motif has been replaced by serine.
TEPs have been mostly studied in the mosquito Anopheles gambiae where TEP1 (aTEP1) was shown to act as a bona fide opsonin to promote phagocytosis of Gram-positive and -negative bacteria [8]. aTEP1 is structurally related to the C3 complement factor, is expressed in hemocytes, and secreted in the hemolymph, where it can bind to the surface of bacteria [8,9]. Other studies have highlighted the role of aTEP1 in the defense against Plasmodium infections [9]. aTEP1 binds to the surface of ookinetes and promotes their lysis and melanization. In D. melanogaster, Tep genes are expressed at basal levels and are upregulated upon immune challenge [7,10,11]. The only functional data available so far revealed that TEP2, TEP3, and TEP6 bound to Escherichia coli, Staphylococcus aureus, and Candida albicans, respectively, thereby promoting their phagocytosis by cultured S2 cells [12]. Unexpectedly, the binding of TEP6 to C. albicans indicated that a functional thioester motif is not essential to interact with microbial surfaces [12].
Here the function of the Drosophila Tep family was investigated using several approaches. First, a phylogenetic analysis of protostomian Tep members revealed the existence of three major subgroups, one of which appears to be specific to mosquitoes. Second, we examined the expression pattern of Tep genes. We found that Tep genes are essentially expressed in hemocytes, in some barrier epithelia, and in the fat body. Finally, we tested mutants affecting one or several Tep genes in various infection models and failed to detect a specific susceptibility phenotype. Taken together, our results are compatible with a role for TEPs in innate immunity, even though functional redundancy and/or the lack of a relevant infection model may have prevented us from identifying their exact role in vivo.
Materials and Methods
Bioinformatics and Phylogenetic Analysis
Sequences of the TEPs or TEP homologues presented have been retrieved from the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/), Ensembl (www.ensembl.org), FlyBase (http://flybase.org/blast/), and JGI (http://genome.jgi-psf.org/) data bases using the sequence retrieval system or/and BLAST (basic local alignment search tool) [13]. Alignments were carried out using clustal W [14], MUSCLE 3.7 [15]; www.phylogeny.fr] or COBALT (constraint-based multiple alignment tool; www.ncbi.nlm.nih.gov/tools/cobalt). The identification numbers and the detailed species are presented in online supplementary tables S1 and S2 (www.karger.com/doi/10.1159/000321554).
Phylogenetic trees were constructed on the basis of amino acid differences using PhyML 3.0, applying the approximate likelihood ratio test and bootstrapping procedure with a minimum of 100 bootstraps [15]; http://www.phylogeny.fr/version2_cgi/phylogeny.cgi], fast minimum evolution, neighbor joining, and cobalt tree [16]; www.ncbi.nlm.nih.gov/blast/treeview]. Reliability of the trees was assessed by bootstrapping and comparison between the methods. The median bootstrap values for the phylogenetic trees were not <98%.
Fly Strains
Flies were raised on standard cornmeal agar at 25°C. w− A5001 flies were used as wild-type controls for most of the experiments [17]. Tep2 (d11521) and Tep3 (d03976) have been generated in an A5001 background [17]. The Tep4 mutant belongs to another P-insertion mutant library (EY04656) [18]. The Tep2,3 double mutant was generated by deleting the region between Tep2 and Tep3 by recombining the FRT sites contained in the P-transposable elements (Flp flippase) inserted in the respective genes. We checked the deletion by PCR using appropriate sets of primers. The Tep2,3,4 triple mutant was obtained by recombination of the Tep2,3 double mutant with the Tep4 mutant. UAS-Tep1 RNAi (ML2D) transgenic flies were generated by Dr. Marie Lagueux. Further fly strains include keyc02831 mutants (A5001 background) [Ferrandon, unpubl. data], Dif1 mutants [19], and spzrm7. The hsp-Gal4 driver was used to overexpress ubiquitously the different transgenes under heat shock conditions. Tep1-GFP are transgenic flies in which the promoter of the Tep1 gene is fused to GFP-coding sequences.
Microbial Strains
The following microbes were used: E. coli (1106), Micrococcus luteus (CIP A270), Enterobacter cloacae, S. aureus, Enterococcus faecalis, Listeria monocytogenes, Mycobacterium marinum (a kind gift from Dr. Herman Spaink), Streptococcus pyogenes, and Beauveria bassiana (80.2 strain). Infections were performed at 25°C with the exception of M. marinum and B. bassiana, which were done at 29°C.
Quantitative RT-PCR
This analysis was done as previously described [20]. Each experiment was done at least three times. For quantitative PCR, we used the following primers shown from 5′ (left) to 3′ (right): (i) Tep1: forward: AGTCCCATAAAGGCCGACTGA, reverse: CACCTGCATCAAAGCCATATTG; (ii) Tep2: forward: TGTTCTGCACCAACAGCGATAC, reverse: CTGGCGATCCATCAACATTCTT; (iii) Tep4: forward: GCTGCAGAACCAGATCGAAATC, reverse: ATGACTTTGGCGACGTCTTGAT, and (iv) Tep6: forward: CGCCTTCCTGAACGAAACAA, reverse: GAGGCTTATCGGTCTGCACAA. The following plasmids containing full-length cDNA (generated by Dr. Marie Lagueux) served as standards: Tep1: pML115 in pUASt, Tep2: pML91 in pOT2, and Tep4: pML208 in PGEM easy. Tep6: LD23292 in pOT2 was purchased from the Berkeley Drosophila Genome Project. Quantitative PCR data obtained using these primer couples were then normalized against the values obtained by measuring the expression of the gene encoding ribosomal protein 49.
In situ Hybridization
In situ hybridization (ISH) on larval whole-mount tissues and on circulating hemocytes has been reported previously [7,10,11]. In situ RNA hybridization on sections was carried out as described [21]. The same plasmids described for quantitative RT-PCR were also used for ISH. We generated sense and antisense probes for Tep2, Tep4, and Tep6. Experiments shown are representative of at least two independent experiments.
Septic Injury and Survival Experiments
Survival experiments were performed as previously detailed [20]. Batches of 20 wild-type and mutant flies were challenged by septic injury using a needle previously dipped in a concentrated solution of an overnight bacterial culture. For some bacterial infections, we injected specific dilutions for S. aureus (OD600 = 0.1), S. pyogenes (≈OD600 = 17), and M. marinum (500 CFU). Survival experiments were performed with the fungus B. bassiana as previously described [22]. The vials containing the challenged flies were then transferred to an incubator and kept at 25 or 29°C (according to the microbial strain) and surviving flies were counted every few hours. Experiments shown are representative of at least three independent experiments (for M. marinum infections, refer to [23]). Survival data have been analyzed using the log-rank test provided in the Prism® software.
Phagocytosis Assay
Batches of 10 wild-type (A5001) or Tep2,3,4 mutant flies were injected with either PBSx1 or latex beads; 16 h later, flies were injected with pHrodo™ E. coli BioParticles® conjugate for phagocytosis (Invitrogen), prepared according to the manufacturer's instructions. Thirty minutes after injection, flies were dissected, carcasses were rinsed with PBS, mounted in Vectashield and observed with a fluorescent microscope (Axio Scope; Zeiss).
Experiments shown are representative of two independent experiments. For each experiment, several flies were scored independently by two persons in a ‘blind’ manner.
Results
Phylogenesis of Tep Genes among Protostomes
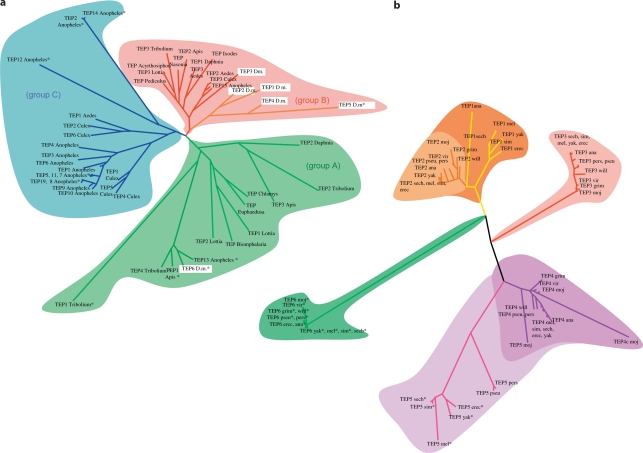
We have analyzed the phylogeny of Teps among various ecdysozoa (insects, chelicerates, and crustaceans), and lophotrochozoa (gastropods and bivalves) for which genome data are available (online suppl. table S1; online suppl. fig. S1). Three main groups emerged from the phylogenetic analysis (fig. 1a). Group A includes the D. melanogaster Tep6 and A. gambiae Tep13 (fig. 1a). It contains Tep genes from both ecdysozoa and lophotrochozoa, suggesting that this pan-protostomian Tep group has emerged first during evolution. Indeed, the only Tep genes retrieved from the gastropod sequences present in our panel, i.e. Biomphalaria glabrata, Euphaedusa tau, and Chlamys farreri, belong to this group. Note also that all Tep genes from Tribolium castaneum and Apis mellifera, except Tribolium-Tep3 and Apis-Tep2, belong to group A. In contrast, no Tep6 gene sequence was found in the chelicerate Ixodes scapularis, possibly because its recently sequenced genome might still have been incomplete. Alternatively, this branch of arthropods may have lost this gene family, as may be the case for Aedes aegypti, Culex pipiens, Nasonia vitripennis, Acyrthosiphon pisum, and Pediculus humanus corporis. The absence of a functional thioester motif in DmTEP6, Anopheles TEP13, Apis TEP1, and Tribolium TEP4 is not a defining feature of group A, as other members of the group, including Tribolium Tep2 and Apis Tep3, encode a functional motif.
Fig. 1.
Phylogenetic analysis of Tep genes. a Phylogenetic analysis of available protostomian Tep sequences. b Phylogenetic analysis of Tep sequences issued from the 12 sequenced Drosophila species. The references for all sequences used to generate the phylogenetic trees are provided in online suppl. table S1 (see also online suppl. fig. S1 for a phylogenetic tree of the species used for the interpretation of the Tep phylogenetic trees). Sequences indicated by an asterisk encode a nonfunctional thioester motif.
Group B can be divided into two branches, one related to D. melanogaster Tep3, and one restricted only to D. melanogaster Tep1, Tep2, Tep4, and Tep5 (fig. 1a). The DmTep3-related branch clusters species from both ecdysozoa and one lophotrochozoa, but in contrast to group A, these sequences have been well conserved during evolution. This suggests that the Tep genes belonging to this branch share a common ancestral function that may be under stronger selective pressure in these various organisms.
Most strikingly, group C is solely composed of Tep genes from mosquito species namely, A. gambiae, A. aegypti, and C. pipiens. Importantly, Anopheles Tep1, which plays an essential role in the mosquito host defense against parasites [9], belongs to this cluster of mosquito-related Tep genes. Of note, many TEP members from A. gambiae (TEP2, 5, 7, 8, 11, 12, 14, and 19) also contain a potentially nonfunctional thioester site, as reported for some TEPs in the two other groups.
The second branch of group B contains only D. melanogaster Tep genes (Tep1, Tep2, Tep4, and Tep5) and therefore seems restricted to Brachycera, and possibly only to the Drosophilidae (fig. 1b; online suppl. fig. S1). This suggests that these Tep genes appeared more recently during evolution and thus may reflect a specific adaptation of Brachycera/Drosophilidae. Additionally, these genes are phylogenetically clustered together, suggesting that they emerged recently, most likely from a duplication of the DmTep3 ancestor.
A more detailed analysis of the phylogeny of the Drosophilidae branch is possible thanks to the sequencing of 12 Drosophila species (online suppl. fig. S1) [24]. Tep6 genes, followed to a lesser extent by Tep3 genes, display a high level of sequence similarity (fig. 1b). Taking into account the phylogeny described above (fig. 1a: groups A and B), this indicates that these two genes may encode the ancestral functions of Tep genes. It is likely that a gene duplication event generated both the current Drosophilidae Tep3 gene and a precursor that underwent further duplications to generate Tep1 and Tep2 on the one hand, and Tep4 and Tep5 on the other hand. The duplication event that generated Tep1 occurred just before the diversification of the melanogaster group as Tep1 is lacking in other Drosophilidae.
Tep5 (CG13079) likely emerged from a duplication of Tep4 before the separation of the obscura and melanogaster groups (fig. 1b; online suppl. fig. S1), as this gene is absent in the willistoni group of Sophophora (which includes D. melanogaster) and in the other branch of Drosophilidae, namely Drosophila. Interestingly, this gene does not encode an active thioester site in the melanogaster group, while no mutation of this site occurred in the other Drosophila species. This suggests that the loss of this thioester site occurred secondarily in the melanogaster group. Interestingly, there is no evidence that the D. melanogaster Tep5 gene is expressed as no hints of its expression could be found in FlyBase where large-scale genomic expression data, including RNAseq data, are available (http://flybase.org/). We cannot however exclude that this gene may be expressed only in a few cells or under very specific conditions, such as the encounter with a specific pathogen. Thus, it will be interesting to determine whether the expression of other Drosophila species Tep5 genes is more readily detectable. It will also be worthy to examine, when released, the genome of other Brachycera, such as Musca domestica or Glossina morsitans, to determine whether the Tep1,2,4 branch of group B is specific to Drosophilidae or Brachycera.
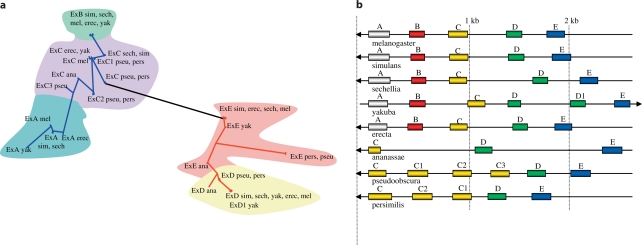
The Tep2 gene is transcribed into five isoforms (A–E) by alternative splicing of exon 5, which corresponds to the variable region of TEP2 [7], that is the region orthologous to the bait domain of the α2-macroglobulins or the anaphylatoxin of C3 complement factor (C3a). These five isoforms are found only in the Sophophora [melanogaster (D. melanogaster, D. simulans, D. sechellia, D. yakuba, D. ananassae,and D. erecta)] and obscura (D. pseudoobscura and D. persimilis) groups (fig. 2). The phylogeny of exon 5 within these species suggests that exons C and E might have arisen from a single duplication from an ancestral exon (fig. 2a). Exon E underwent an early duplication event before the separation of the melanogaster and obscura groups, thus yielding exon D. In contrast, exon C appears to have undergone independent duplication events in the obscura and melanogaster subgroups. Interestingly, exon C did not undergo such an event in D. ananassae, a member of the melanogaster group but not of the melanogaster subgroup (online suppl. fig. S1), unless duplicated genes were subsequently lost (fig. 2b). In the melanogaster subgroup, exon C yielded independently (fig. 2a) two further exons A and B. It is worth underscoring the possible adaptive value of such an evolution of exon C that generated multiple additional exons during the differentiation of the Drosophilidae.
Fig. 2.
Phylogenesis of exon 5 of Tep2. a Phylogenetic analysis of the exons 5 of Drosophila Tep2s. b Positions of the exons 5 on the genome of Drosophila species. The letters A, B and C refer to those given in FlyBase for melanogaster species; we have inverted D and E to follow the disposition on the chromosome.
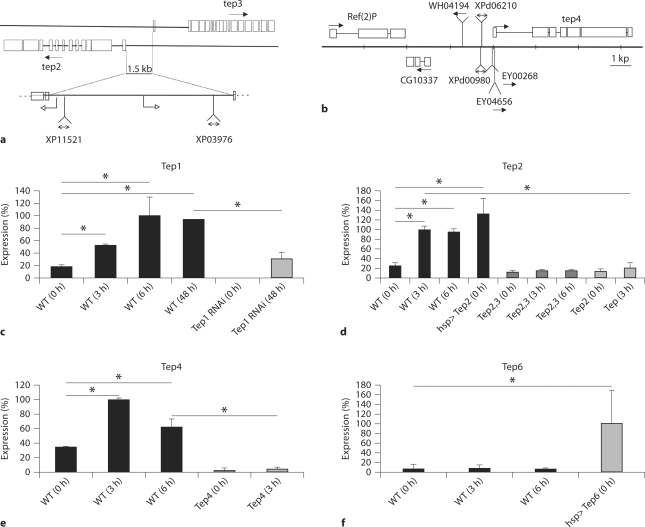
The Induction of Tep Gene Expression Is Blocked in the Corresponding Mutants
To investigate the function of TEPs in innate immunity of Drosophila, we took advantage of loss of function fly lines described below. Tep2 and Tep3 are 1.5 kb apart from each other in an opposite orientation and transposable elements are inserted next to each transcription start site (fig. 3a). Transposable elements are also inserted in the Tep4 genomic region (fig. 3b). We also used an RNAi fly line to interfere with Tep1 transcription. Previous transcriptomic studies [7,10,11] have shown that Tep2 and Tep4 genes are highly upregulated in adults upon bacterial challenge. In addition, we also report here an increased expression of the Tep1 and Tep3 genes (fig. 3c; online suppl. fig. S2). Moreover, we found that only Tep6 expression did not increase after septic injury with a mixture of E. coli and M. luteus (fig. 3f). The increased expression of Tep1, Tep2, Tep3, and Tep4 genes after an immune challenge was impaired in Tep2 and Tep4 loss-of-function fly lines (fig. 3d, e; online suppl. fig. S2), as well as in Tep1 RNAi flies where the expression of Tep1 was reduced by 70% (fig. 3c). The Tep6 loss-of-function allele is larval lethal and therefore we did not attempt any further investigation in this study.
Fig. 3.
Modulation of the expression of Tep genes in adults after septic injury. a, b Drosophila Teps are located on the left arm of the second chromosome. a Tep2 (CG7052) and Tep3 (CG7068) map to cytogenetic position 28C1. Tep2 and Tep3 are 1.5 kb apart. The modified XP transposable element XP11521 is inserted 22 bp upstream of the transcription-starting site of Tep2, whereas the XP03976 element is inserted 121 bp downstream of the transcription- starting site of Tep3. We have generated a double mutant for Tep2 and Tep3, Tep2, 3, by FLP-mediated recombination between the FRT elements carried on the XP elements. b Tep4 (CG10363) maps to cytogenetic position 37F1. Transposable elements inserted in this region are shown. We have used EY04656 as a mutant for Tep4 as this modified P element is inserted in the initiation codon of the Tep4 gene. In the following experiments, we used an RNAi transgenic line to knock down Tep1 (CG18096). c-f Steadystate transcript levels of D. melanogaster Tep genes were measured by quantitative RT-PCR before and after infection with a mix of E. coli and M. luteus. These experiments are representative of at least two independent experiments. 0 h = Non-infected flies; 3 h/6 h/48 h = flies 3/6/48 h after infection; WT = wild type. Flies were frozen for experiments 3, 6, and 48 h after infection. Gene expression was normalized against rp49 gene expression and the results are expressed as percentage of maximal expression: 6 (Tep1; c) and 3 h after infection (Tep2 and Tep4; d, e). Expression of a Tep6 transgene under the control of a heat shock promoter using the UAS-GAL4 system (hsp>Tep6) after heat shock (f). ∗ p<0.05. c The induction of Tep1 is decreased by the expression of an RNAi transgene targeting specifically this gene.
Expression of Tep Genes in Potential Immune Tissues of D. melanogaster Adult Flies and Larvae
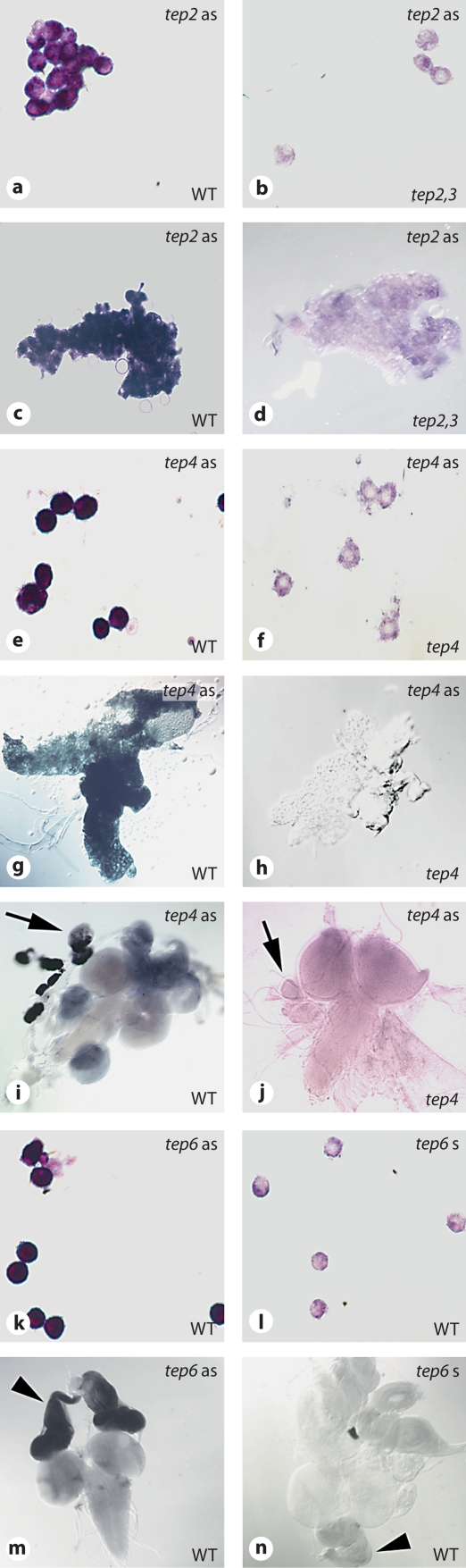
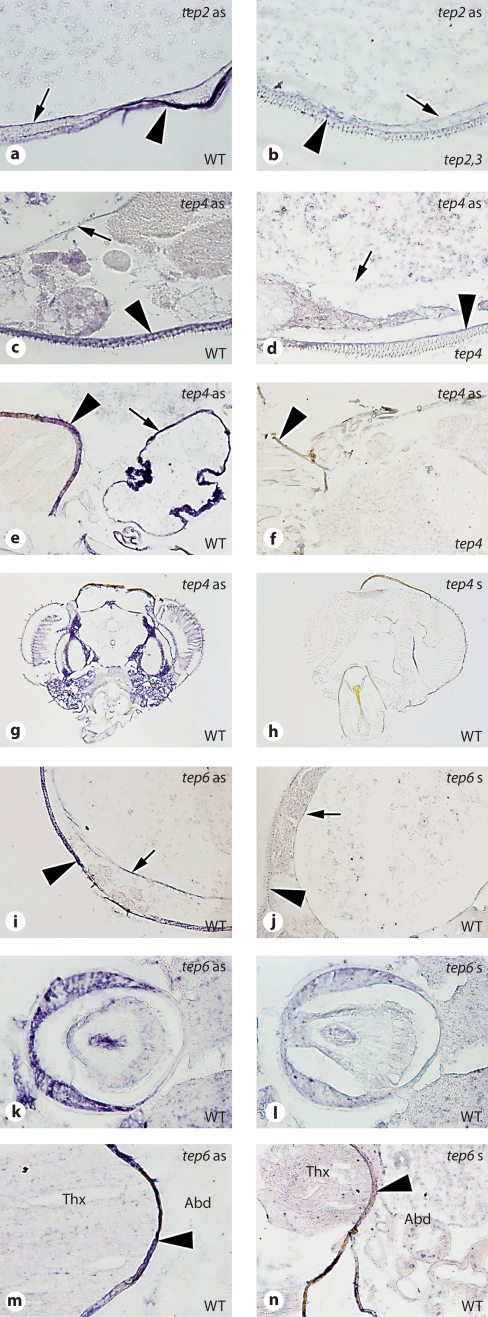
To identify the tissues in which Tep genes are expressed before or after an immune challenge, we analyzed the expression pattern of several Tep genes in adult and larval tissues. The expression pattern of Tep2, Tep3, Tep4, and Tep6 was assessed by whole-mount ISH of larvae (fig. 4; online suppl. fig. S2) and by ISH of histological sections in adult flies (fig. 5). To investigate the expression pattern of Tep1, we took advantage of a transgenic fly line expressing the GFP reporter under the control of the Tep1 promoter. We found that Tep1-GFP and Tep4 transcripts are expressed in the lymph glands (the larval hematopoietic organ; fig. 4i, j, and data not shown). Tep1-GFP was also detected in the proventriculus, the larval plasmatocytes, and the larval fat body (online suppl. fig. S3). We also found that Tep2, Tep3, Tep4, and Tep6 are expressed in larval plasmatocytes (fig. 4a, b, e, f, k, l; online suppl. fig. S2). Additionally, Tep1-GFP, Tep2, and Tep4 are expressed at basal levels in the fat body, and this expression is increased after a septic injury (E. coli + M. luteus;fig. 4c, d, g, h; online suppl. fig. S3, and data not shown). Since the fat body and plasmatocytes are responsible for the humoral and the cellular responses, respectively [6], the distributions of Tep1-GFP, Tep2, Tep3, Tep4, and Tep6 mRNA matched the main tissues involved in larval immunity. Interestingly, Tep6 is expressed in all imaginal discs (fig. 4m, n).
Fig. 4.
Expression of Tep genes in larvae. Whole-mount ISH on late third instar larvae. a-d Tep2. e-j Tep4. k-n Tep6. Left-hand panels = Tep gene expression; right-hand panels = controls. The probe used for ISH is indicated on the top right corner of each panel, whereas the genotype is indicated on the bottom right corner. The following tissues are shown: hemocytes (a, b, e, f, k, l); fat body (c, d, g, h); lymph glands (arrow), central nervous system, and imaginal discs (arrowhead; i, j, m, n). For Tep6, we used the sense probe on wild-type (WT) larvae as control, as Tep6 mutants (Mcr) are lethal.
Fig. 5.
Expression of Tep genes in adults. ISH on paraffin-embedded longitudinal and transverse sections. a, b Tep2. c-h Tep4. i-n Tep6. Left-hand panels = Tep gene expression; right-hand panels = controls. The probe used for ISH is indicated on the top right corner of each panel, whereas the genotype is indicated on the bottom right corner. The following tissues are shown: crop (arrows) and hypoderm (arrowheads; a-f, i, j); head (g, h); proventriculus (cardium; k, l); mesophragma (arrowheads; e, f, m, n). For Tep6, we used the sense probe on wild-type (WT) flies as control, as Tep6 mutants (Mcr) are lethal.
In adults, we found that the digestive tract was a major site of expression of Tep genes. The epithelium of the crop, a diverticulum of the digestive tract that is used to store food, expressed Tep2, Tep3, Tep4, and Tep6 (fig. 5a–f, i, j; online suppl. fig. S2). The proventriculus, the valve-like structure that guards the midgut entrance, was found to be positive for Tep1-GFP and Tep6 expression (fig. 5k, l; online suppl. fig. S3). However, we did not detect the expression of these Tep genes in the midgut itself. In addition to the epithelia of the digestive tract, we found that the epidermis underlying the cuticle in the abdominal part was a major site for Tep gene expression. Interestingly, Tep2, Tep3, Tep4, and Tep6 expression was restricted to the ventral side of the abdominal epidermis (fig. 5a–d, i, j; online suppl. fig. S2). In addition, Tep4 and Tep6 expression was detected in the mesophragma, the structure found at the border between thorax and abdomen and to which flight muscles are attached posteriorly (fig. 5e, f, m, n). Surprisingly, in contrast to larvae, no expression could be detected in the abdominal fat body (before or after immune challenge) of adult flies, except in the fat body of the head region where Tep4 (fig. 5g, h) was strongly induced. Interestingly, upon a septic injury, the expression pattern of Tep2, Tep4, and Tep6 remained unchanged in adults (data not shown).
TEPs Appear Not to Be Required for D. melanogaster Survival upon Microbial Challenge
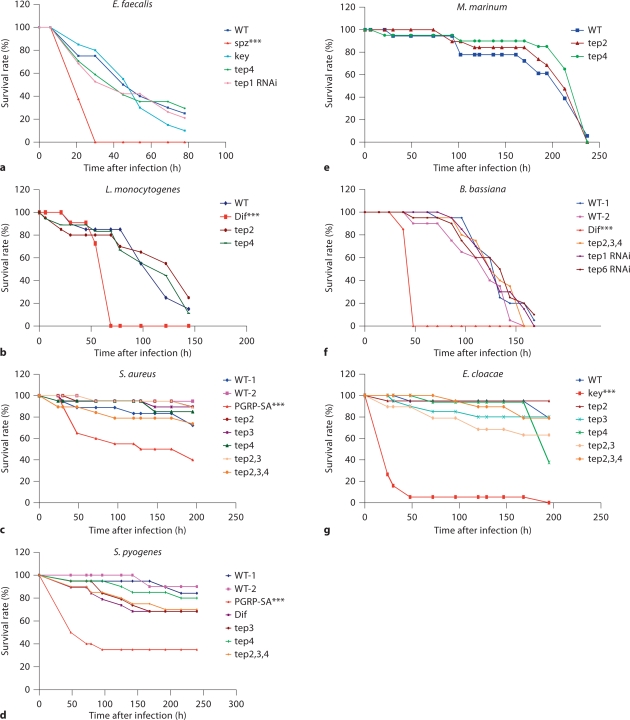
Since most Tep genes are expressed in hemocytes (fig. 4a, b, e, f, k, l) and are predicted to act as opsonins [12], we decided to investigate their function in the defense against pathogens. We thus challenged Tep mutants with different microbes introduced in the hemocoel by a septic injury (fig. 6). Survival experiments were performed in adult flies with either Gram-positive (S. pyogenes, S. aureus, E. faecalis, L. monocytogenes, and M. marinum) or Gram-negative bacteria (E. coli and E. cloacae). S. pyogenes was chosen because patients deficient in the complement system are commonly suffering from recurrent pyogenic infections. M. marinum was used as Tep2 and Tep4 are the most strongly induced genes in this infection model [11]. As phagocytosis plays a critical role in the clearance of E. faecalis and S. aureus [25], [26], Nehme et al., submitted], infections with these bacteria are more likely to reveal an involvement of TEPs in phagocytosis.
Fig. 6.
Tep mutants survive as wild-type (WT) flies to different types of infection. Survival experiments after distinct challenges in the septic injury model are presented and are representative of at least two independent experiments. The appropriate controls for the different microbes have been used: Gram-positive bacteria, fungi: mutants of the Toll pathway [Dif, spätzle (spz), PGRP-SA] and IMD pathway: kenny (key). None of the Tep mutants shows a reproducible susceptibility or resistance to infection phenotype, either in homozygous or hemizygous conditions. We used the logrank test to determine the significance between wild-type and mutant survival curves. ∗∗∗ p < 0.001 was the only constantly measured p value in several experiments.
Our results showed that Tep1-, Tep2-, Tep3-, and Tep4-deficient flies were not more susceptible to bacteria than wild-type flies (fig. 6a–e, g, and data not shown). Additionally, we studied the survival rate of Tep mutants after a fungal infection using spores from the entomopathogenic fungus B. bassiana[22] and we found that Tep mutant flies for either Tep1 (RNAi), Tep2, Tep4, or Tep6 (RNAi) survive as well as wild-type flies to B. bassiana natural infection (fig. 6f). We also observed a normal resistance to microbial infections in flies hemizygous for Tep2, Tep3, and Tep4. Collectively, these results showed that none of the tested Tep genes is required on its own for the defense against the bacterial and fungal infections we have tested.
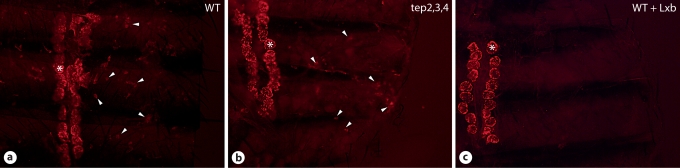
Given (i) the expression pattern of multiple Tep genes in the hemocytes and the epidermis underlying the cuticle; (ii) that TEPs are secreted into the hemolymph, and (iii) that Tep1, Tep2, and Tep4 belong to the same phylogenetic group, we hypothesized that the function(s) of TEPs in the hemolymph might be redundant, at least to some extent. To test this hypothesis, we generated flies doubly mutant for Tep2 and Tep3 or triply mutant for Tep2, Tep3, and Tep4, and then assessed their susceptibility to septic injuries with different microbes (E. cloacae, S. aureus, E. faecalis, L. monocytogenes, S. pyogenes, and B. bassiana). These compound mutants resisted normally to these infections (fig. 6c, d, g, and data not shown). Accordingly, a phagocytosis assay performed on compound Tep2, Tep3, and Tep4 mutant flies failed to reveal any qualitative defects in the activity of sessile hemocytes (fig. 7).
Fig. 7.
Tep mutants display normal phagocytosis activity. Adult Drosophila carcasses from WT (a, c) and Tep 2,3,4 mutant(b). pHrodo™ E. coli BioParticles® produce a red fluorescence when exposed to an acidic environment such as that present in the phagosome. It labeled sessile hemocytes (arrowheads; a, b) and pericardial cells (asterisk; a-c). Injection of latex beads, prior to pHrodo E. coli BioParticles injection (WT + Lxb), blocked phagocytosis. Note that the pericardial cells were unspecifically labeled.
Discussion
Do TEPs Function in D. Melanogaster Innate Immunity?
The finding that Tep2, Tep3, or Tep4 mutant flies resist the microbial infections we have tested in wild-type flies for example is somewhat unexpected because: (i) it was shown in Drosophila S2 cells that TEP2 and TEP3 are required for the phagocytosis of E. coli and S. aureus, respectively [12], and (ii) phagocytosis in flies is essential to defend against S. aureus and E. faecalis [25], [26], Nehme et al., submitted]. It has been shown that TEP6binds to C. albicans and is required for its phagocytosis by cultured S2 cells [12]. We did not assess the survival of Tep6 mutant flies to this infection as null mutants are larval lethal.
According to the phylogenetic analysis described above, we speculate that Tep1 and Tep2 are likely to act redundantly. Thus, the deletion of both genes will be required to test this hypothesis. The combinatorial role of TEPs remains therefore to be wholly investigated.
Most Tep genes are constitutively expressed in barrier epithelia and especially in weak points of the cuticle (fig. 5). Thus, these proteins may serve to anticipate infections by pathogens attacking through the cuticle, i.e. some entomophtorales fungi, or microbes transmitted by blood-sucking mites or wasps. Thus, we may not have tested the relevant pathogens.
Because many Tep genes (fig. 3, and data not shown) are expressed in portions of the digestive tract of adult flies and larvae, it will be of interest in the future to combine the inactivation of these Tep genes with an oral infection model to unravel more subtle or physiological functions of TEPs.
Tep Genes during Evolution
The failure to observe an innate immunity phenotype for D. melanogaster Tep mutants contrasts the results obtained with mosquito A. gambiae TEP1 [8,9]. Nevertheless, analysis of the phylogeny of Tep genes reveals that Tep1 from Anopheles belongs to the group of Tep genes (group C) that is restricted to mosquito species. The founder gene of group C may have arisen by duplication from Anopheles Tep13 (group A) or Tep15 (group B) and selected for its adaptation to the mosquito life cycle and ecology (fig. 1). Mosquitoes are blood-sucking insects that have an aquatic life during their larval stage and an aerial life during their imaginal stage. We did not find any Tep genes belonging to group C in other blood-sucking pestspresent in our phylogenetic analysis panel, such as I. scapularis and P. humanus. It is thus unlikely that the blood meal provides on its own a significant selective pressure for the appearance or maintenance of group C Tep genes. However, the Anophelinae A. gambiae and the Culicinae A. aegypti and C. pipiens may have evolved more recently in blood-sucking insects than the chelicerate I. scapularis and the Phthirapter P. humanus. Therefore, group C-related Tep genes might reflect a more recent adaptation to blood feeding. It will be of interest to analyze the genome of the tsetse fly G. morsitans when released. Another possibility is that the appearance of this Tep group mirrors an adaptation to the dual aquatic and aerial biotopes that mosquitoes encounter during their life cycle. Again, the genomes of insects displaying a similar life cycle such as the Odonata Libellula may provide the answer to this question.
Additionally, our phylogenetic analysis shows that Tep6 is highly conserved within Drosophilidae, which suggests that the function of this gene is under a strong selective pressure. Also, we found that Tep6 is expressed in tissues required for metamorphosis (e.g. imaginal discs). This may reflect a role for Tep6 either during metamorphosis (or development), which may account for the lethality of Tep6 mutants. To a lower extent, Tep3 genes are also significantly conserved within the Drosophilidae group. This suggests that the function of Tep3 is highly selected for. It is of interest to note that the Tep3-related genes from group B are also highly conserved in ecdysozoa and lophotrochozoa (fig. 1a). This further suggests that the function of D. melanogaster Tep3 is likely to be (i) reminiscent of the Tep ancestral function, and (ii) conserved among protostomes.
In conclusion, our study defines three clusters of Tep genes among protostomes. Each of these clusters most probably reflects a particular biological function of TEPs. We failed to identify a role for Tep1, Tep2, Tep3, and Tep4 in D. melanogaster innate immunity using a septic injury model or a ‘natural’ fungal infection. We cannot exclude that the immune function of TEPs is masked by other defense mechanisms. Interestingly, we found that the expression pattern of Drosophila Tep genes is compatible with a defense function against pathogens in barrier epithelia [27,28,29]. Additionally, a population genetic study predicted that the rapid evolution of D. melanogaster TEP1 and TEP2 is likely to be the signature of host-pathogen co-evolution, indicating a specific role of TEPs against a restricted subset of pathogens [30]. Thus, it is likely that the investigation of infection models with natural pathogens such as Serratia marcescens, Pseudomonas entomophila, Providencia sneebia, Providencia burhodogranariea, Tubulinosema ratisbonensis (microsporidia), or Heterorhabditis bacteriophora (nematode symbiotically associated with the entomopathogenic bacterium Photorhabdus luminescens) is likely to yield novel insights into the potential immune functions of Tep family genes [31,32,33,34,35,36].
Disclosure Statement
We report no conflict of interest.
Acknowledgements
We thank Dr. M. Lagueux and Dr. M. Meister for scientific discussions. We are indebted to François Bonnay for his help with injection of latex beads and pHrodo E. coli BioParticles. We are grateful to Dr. H. Spaink and Dr. M. Lagueux for the gift of strains and reagents. We thank the Berkeley Drosophila Genome Project for the Tep6 cDNA clone. This work and R.B.A. were supported by funding from the US National Institutes of Health, grant PO1 AI44220, and CNRS (French National Center for Scientific Research). D.F.'s team is an Equipe Fondation pour la Recherche Médicale.
References
- 1.Sansonetti PJ. The innate signaling of dangers and the dangers of innate signaling. Nat Immunol. 2006;7:1237–1242. doi: 10.1038/ni1420. [DOI] [PubMed] [Google Scholar]
- 2.Sottrup-Jensen L, Hansen HF, Mortensen SB, Petersen TE, Magnusson S. Sequence location of the reactive thiol ester in human alpha 2-macroglobulin. FEBS Lett. 1981;123:145–148. doi: 10.1016/0014-5793(81)80039-7. [DOI] [PubMed] [Google Scholar]
- 3.Medzhitov R, Janeway CA., Jr Decoding the patterns of self and nonself by the innate immune system. Science. 2002;296:298–300. doi: 10.1126/science.1068883. [DOI] [PubMed] [Google Scholar]
- 4.Nonaka M, Kimura A. Genomic view of the evolution of the complement system. Immunogenetics. 2006;58:701–713. doi: 10.1007/s00251-006-0142-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lemaitre B, Hoffmann J. The host defense of Drosophila melanogaster. Annu Rev Immunol. 2007;25:697–743. doi: 10.1146/annurev.immunol.25.022106.141615. [DOI] [PubMed] [Google Scholar]
- 6.Ferrandon D, Imler JL, Hetru C, Hoffmann JA. The Drosophila systemic immune response: sensing and signalling during bacterial and fungal infections. Nat Rev Immunol. 2007;7:862–874. doi: 10.1038/nri2194. [DOI] [PubMed] [Google Scholar]
- 7.Lagueux M, Perrodou E, Levashina EA, Capovilla M, Hoffmann JA. Constitutive expression of a complement-like protein in Toll and Jak gain-of-function mutants of Drosophila. Proc Natl Acad Sci USA. 2000;97:11427–11432. doi: 10.1073/pnas.97.21.11427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Levashina EA, Moita LF, Blandin S, Vriend G, Lagueux M, Kafatos FC. Conserved role of a complement-like protein in phagocytosis revealed by dsRNA knockout in cultured cells of the mosquito, Anopheles gambiae. Cell. 2001;104:709–718. doi: 10.1016/s0092-8674(01)00267-7. [DOI] [PubMed] [Google Scholar]
- 9.Blandin S, Shiao SH, Moita LF, Janse CJ, Waters AP, Kafatos FC, Levashina EA. Complement-like protein TEP1 is a determinant of vectorial capacity in the malaria vector Anopheles gambiae. Cell. 2004;116:661–670. doi: 10.1016/s0092-8674(04)00173-4. [DOI] [PubMed] [Google Scholar]
- 10.Irving P, Ubeda JM, Doucet D, Troxler L, Lagueux M, Zachary D, Hoffmann JA, Hetru C, Meister M. New insights into Drosophila larval haemocyte functions through genome-wide analysis. Cell Microbiol. 2005;7:335–350. doi: 10.1111/j.1462-5822.2004.00462.x. [DOI] [PubMed] [Google Scholar]
- 11.Dionne MS, Pham LN, Shirasu-Hiza M, Schneider DS. Akt and foxo dysregulation contribute to infection-induced wasting in Drosophila. Curr Biol. 2006;16:1977–1985. doi: 10.1016/j.cub.2006.08.052. [DOI] [PubMed] [Google Scholar]
- 12.Stroschein-Stevenson SL, Foley E, O'Farrell PH, Johnson AD. Identification of Drosophila gene products required for phagocytosis of Candida albicans. PLoS Biol. 2006;4:e4. doi: 10.1371/journal.pbio.0040004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 14.Thompson JD, Higgins DG, Gibson TJ. Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard JF, Guindon S, Lefort V, Lescot M, Claverie JM, Gascuel O. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008;36:W465–W469. doi: 10.1093/nar/gkn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Papadopoulos JS, Agarwala R. COBALT: Constraint-based alignment tool for multiple protein sequences. Bioinformatics. 2007;23:1073–1079. doi: 10.1093/bioinformatics/btm076. [DOI] [PubMed] [Google Scholar]
- 17.Thibault ST, Singer MA, Miyazaki WY, Milash B, Dompe NA, Singh CM, Buchholz R, Demsky M, Fawcett R, Francis-Lang HL, Ryner L, Cheung LM, Chong A, Erickson C, Fisher WW, Greer K, Hartouni SR, Howie E, Jakkula L, Joo D, Killpack K, Laufer A, Mazzotta J, Smith RD, Stevens LM, Stuber C, Tan LR, Ventura R, Woo A, Zakrajsek I, Zhao L, Chen F, Swimmer C, Kopczynski C, Duyk G, Winberg ML, Margolis J. A complementary transposon tool kit for Drosophila melanogaster using P and piggyBac. Nat Genet. 2004;36:283–287. doi: 10.1038/ng1314. [DOI] [PubMed] [Google Scholar]
- 18.Bellen HJ, Levis RW, Liao G, He Y, Carlson JW, Tsang G, Evans-Holm M, Hiesinger PR, Schulze KL, Rubin GM, Hoskins RA, Spradling AC. The BDGP gene disruption project: single transposon insertions associated with 40% of Drosophila genes. Genetics. 2004;167:761–781. doi: 10.1534/genetics.104.026427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rutschmann S, Jung AC, Hetru C, Reichhart JM, Hoffmann JA, Ferrandon D. The Rel protein DIF mediates the antifungal, but not the antibacterial, response in Drosophila. Immunity. 2000;12:569–580. doi: 10.1016/s1074-7613(00)80208-3. [DOI] [PubMed] [Google Scholar]
- 20.Gobert V, Gottar M, Matskevich A, Rutschmann S, Royet J, Belvin M, Hoffmann JA, Ferrandon D. Dual activation of the Drosophila Toll pathway by two pattern recognition receptors. Science. 2003;302:2126–2130. doi: 10.1126/science.1085432. [DOI] [PubMed] [Google Scholar]
- 21.Dupe V, Matt N, Garnier JM, Chambon P, Mark M, Ghyselinck NB. A newborn lethal defect due to inactivation of retinaldehyde dehydrogenase type 3 is prevented by maternal retinoic acid treatment. Proc Natl Acad Sci USA. 2003;100:14036–14041. doi: 10.1073/pnas.2336223100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lemaitre B, Reichhart JM, Hoffmann JA. Drosophila host defense: differential display of antimicrobial peptide genes after infection by various classes of microorganisms. Proc Natl Acad Sci USA. 1997;94:14614–14619. doi: 10.1073/pnas.94.26.14614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dionne MS, Ghori N, Schneider DS. Drosophila melanogaster is a genetically tractable model host for Mycobacterium marinum. Infect Immun. 2003;71:3540–3550. doi: 10.1128/IAI.71.6.3540-3550.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Clark AG, Eisen MB, Smith DR, Bergman CM, Oliver B, Markow TA, Kaufman TC, Kellis M, Gelbart W, Iyer VN, Pollard DA, Sackton TB, Larracuente AM, Singh ND, Abad JP, Abt DN, Adryan B, Aguade M, Akashi H, Anderson WW, Aquadro CF, Ardell DH, Arguello R, Artieri CG, Barbash DA, Barker D, Barsanti P, Batterham P, Batzoglou S, Begun D, Bhutkar A, Blanco E, Bosak SA, Bradley RK, Brand AD, Brent MR, Brooks AN, Brown RH, Butlin RK, Caggese C, Calvi BR, Bernardo de Carvalho A, Caspi A, Castrezana S, Celniker SE, Chang JL, Chapple C, Chatterji S, Chinwalla A, Civetta A, Clifton SW, Comeron JM, Costello JC, Coyne JA, Daub J, David RG, Delcher AL, Delehaunty K, Do CB, Ebling H, Edwards K, Eickbush T, Evans JD, Filipski A, Findeiss S, Freyhult E, Fulton L, Fulton R, Garcia AC, Gardiner A, Garfield DA, Garvin BE, Gibson G, Gilbert D, Gnerre S, Godfrey J, Good R, Gotea V, Gravely B, Greenberg AJ, Griffiths-Jones S, Gross S, Guigo R, Gustafson EA, Haerty W, Hahn MW, Halligan DL, Halpern AL, Halter GM, Han MV, Heger A, Hillier L, Hinrichs AS, Holmes I, Hoskins RA, Hubisz MJ, Hultmark D, Huntley MA, Jaffe DB, Jagadeeshan S, Jeck WR, Johnson J, Jones CD, Jordan WC, Karpen GH, Kataoka E, Keightley PD, Kheradpour P, Kirkness EF, Koerich LB, Kristiansen K, Kudrna D, Kulathinal RJ, Kumar S, Kwok R, Lander E, Langley CH, Lapoint R, Lazzaro BP, Lee SJ, Levesque L, Li R, Lin CF, Lin MF, Lindblad-Toh K, Llopart A, Long M, Low L, Lozovsky E, Lu J, Luo M, Machado CA, Makalowski W, Marzo M, Matsuda M, Matzkin L, McAllister B, McBride CS, McKernan B, McKernan K, Mendez-Lago M, Minx P, Mollenhauer MU, Montooth K, Mount SM, Mu X, Myers E, Negre B, Newfeld S, Nielsen R, Noor MA, O'Grady P, Pachter L, Papaceit M, Parisi MJ, Parisi M, Parts L, Pedersen JS, Pesole G, Phillippy AM, Ponting CP, Pop M, Porcelli D, Powell JR, Prohaska S, Pruitt K, Puig M, Quesneville H, Ram KR, Rand D, Rasmussen MD, Reed LK, Reenan R, Reily A, Remington KA, Rieger TT, Ritchie MG, Robin C, Rogers YH, Rohde C, Rozas J, Rubenfield MJ, Ruiz A, Russo S, Salzberg SL, Sanchez-Gracia A, Saranga DJ, Sato H, Schaeffer SW, Schatz MC, Schlenke T, Schwartz R, Segarra C, Singh RS, Sirot L, Sirota M, Sisneros NB, Smith CD, Smith TF, Spieth J, Stage DE, Stark A, Stephan W, Strausberg RL, Strempel S, Sturgill D, Sutton G, Sutton GG, Tao W, Teichmann S, Tobari YN, Tomimura Y, Tsolas JM, Valente VL, Venter E, Venter JC, Vicario S, Vieira FG, Vilella AJ, Villasante A, Walenz B, Wang J, Wasserman M, Watts T, Wilson D, Wilson RK, Wing RA, Wolfner MF, Wong A, Wong GK, Wu CI, Wu G, Yamamoto D, Yang HP, Yang SP, Yorke JA, Yoshida K, Zdobnov E, Zhang P, Zhang Y, Zimin AV, Baldwin J, Abdouelleil A, Abdulkadir J, Abebe A, Abera B, Abreu J, Acer SC, Aftuck L, Alexander A, An P, Anderson E, Anderson S, Arachi H, Azer M, Bachantsang P, Barry A, Bayul T, Berlin A, Bessette D, Bloom T, Blye J, Boguslavskiy L, Bonnet C, Boukhgalter B, Bourzgui I, Brown A, Cahill P, Channer S, Cheshatsang Y, Chuda L, Citroen M, Collymore A, Cooke P, Costello M, D'Aco K, Daza R, De Haan G, DeGray S, DeMaso C, Dhargay N, Dooley K, Dooley E, Doricent M, Dorje P, Dorjee K, Dupes A, Elong R, Falk J, Farina A, Faro S, Ferguson D, Fisher S, Foley CD, Franke A, Friedrich D, Gadbois L, Gearin G, Gearin CR, Giannoukos G, Goode T, Graham J, Grandbois E, Grewal S, Gyaltsen K, Hafez N, Hagos B, Hall J, Henson C, Hollinger A, Honan T, Huard MD, Hughes L, Hurhula B, Husby ME, Kamat A, Kanga B, Kashin S, Khazanovich D, Kisner P, Lance K, Lara M, Lee W, Lennon N, Letendre F, LeVine R, Lipovsky A, Liu X, Liu J, Liu S, Lokyitsang T, Lokyitsang Y, Lubonja R, Lui A, MacDonald P, Magnisalis V, Maru K, Matthews C, McCusker W, McDonough S, Mehta T, Meldrim J, Meneus L, Mihai O, Mihalev A, Mihova T, Mittelman R, Mlenga V, Montmayeur A, Mulrain L, Navidi A, Naylor J, Negash T, Nguyen T, Nguyen N, Nicol R, Norbu C, Norbu N, Novod N, O'Neill B, Osman S, Markiewicz E, Oyono OL, Patti C, Phunkhang P, Pierre F, Priest M, Raghuraman S, Rege F, Reyes R, Rise C, Rogov P, Ross K, Ryan E, Settipalli S, Shea T, Sherpa N, Shi L, Shih D, Sparrow T, Spaulding J, Stalker J, Stange-Thomann N, Stavropoulos S, Stone C, Strader C, Tesfaye S, Thomson T, Thoulutsang Y, Thoulutsang D, Topham K, Topping I, Tsamla T, Vassiliev H, Vo A, Wangchuk T, Wangdi T, Weiand M, Wilkinson J, Wilson A, Yadav S, Young G, Yu Q, Zembek L, Zhong D, Zimmer A, Zwirko Z, Alvarez P, Brockman W, Butler J, Chin C, Grabherr M, Kleber M, Mauceli E, MacCallum I. Evolution of genes and genomes on the Drosophila phylogeny. Nature. 2007;450:203–218. doi: 10.1038/nature06341. [DOI] [PubMed] [Google Scholar]
- 25.Defaye A, Evans I, Crozatier M, Wood W, Lemaitre B, Leulier F. Genetic ablation of Drosophila phagocytes reveals their contribution to both development and resistance to bacterial infection. J Innate Immun. 2009;1:322–334. doi: 10.1159/000210264. [DOI] [PubMed] [Google Scholar]
- 26.Charroux B, Royet J. Elimination of plasmatocytes by targeted apoptosis reveals their role in multiple aspects of the Drosophila immune response. Proc Natl Acad Sci USA. 2009;106:9797–9802. doi: 10.1073/pnas.0903971106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tzou P, Ohresser S, Ferrandon D, Capovilla M, Reichhart JM, Lemaitre B, Hoffmann JA, Imler JL. Tissue-specific inducible expression of antimicrobial peptide genes in Drosophila surface epithelia. Immunity. 2000;13:737–748. doi: 10.1016/s1074-7613(00)00072-8. [DOI] [PubMed] [Google Scholar]
- 28.Ferrandon D, Jung AC, Criqui MC, Lemaitre B, Uttenweiler-Joseph S, Michaut L, Reichhart JM, Hoffmann JA. A Drosomycin-GFP reporter transgene reveals a local immune response in Drosophila that is not dependent on the Toll pathway. EMBO J. 1998;17:1217–1227. doi: 10.1093/emboj/17.5.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Onfelt Tingvall T, Roos E, Engstrom Y. The imd gene is required for local Cecropin expression in Drosophila barrier epithelia. EMBO Rep. 2001;2:239–243. doi: 10.1093/embo-reports/kve048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jiggins FM, Kim KW. Contrasting evolutionary patterns in Drosophila immune receptors. J Mol Evol. 2006;63:769–780. doi: 10.1007/s00239-006-0005-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Franzen C, Fischer S, Schroeder J, Scholmerich J, Schneuwly S. Morphological and molecular investigations of Tubulinosema ratisbonensis gen. nov., sp. nov. (Microsporidia: Tubulinosematidae fam. nov.), a parasite infecting a laboratory colony of Drosophila melanogaster (Diptera: Drosophilidae) J Eukaryot Microbiol. 2005;52:141–152. doi: 10.1111/j.1550-7408.2005.04-3324.x. [DOI] [PubMed] [Google Scholar]
- 32.Vodovar N, Vinals M, Liehl P, Basset A, Degrouard J, Spellman P, Boccard F, Lemaitre B. Drosophila host defense after oral infection by an entomopathogenic Pseudomonas species. Proc Natl Acad Sci USA. 2005;102:11414–11419. doi: 10.1073/pnas.0502240102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nehme NT, Liegeois S, Kele B, Giammarinaro P, Pradel E, Hoffmann JA, Ewbank JJ, Ferrandon D. A model of bacterial intestinal infections in Drosophila melanogaster. PLoS Pathog. 2007;3:e173. doi: 10.1371/journal.ppat.0030173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Juneja P, Lazzaro BP. Providencia sneebia sp. nov. and Providencia burhodogranariea sp. nov., isolated from wild Drosophila melanogaster. Int J Syst Evol Microbiol. 2009;59:1108–1111. doi: 10.1099/ijs.0.000117-0. [DOI] [PubMed] [Google Scholar]
- 35.Wang Z, Wilhelmsson C, Hyrsl P, Loof TG, Dobes P, Klupp M, Loseva O, Morgelin M, Ikle J, Cripps RM, Herwald H, Theopold U. Pathogen entrapment by transglutaminase – a conserved early innate immune mechanism. PLoS Pathog. 2010;6:e1000763. doi: 10.1371/journal.ppat.1000763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hallem EA, Rengarajan M, Ciche TA, Sternberg PW. Nematodes, bacteria, and flies: a tripartite model for nematode parasitism. Curr Biol. 2007;17:898–904. doi: 10.1016/j.cub.2007.04.027. [DOI] [PubMed] [Google Scholar]