Abstract
In molecular epidemiological studies of drug resistant Mycobacterium tuberculosis (TB) in Sweden a large outbreak of an isoniazid resistant strain was identified, involving 115 patients, mainly from the Horn of Africa. During the outbreak period, the genomic pattern of the outbreak strain has stayed virtually unchanged with regard to drug resistance, IS6110 restriction fragment length polymorphism and spoligotyping patterns. Here we present the complete genome sequence analyses of the index isolate and two isolates sampled nine years after the index case as well as experimental data on the virulence of this outbreak strain. Even though the strain has been present in the community for nine years and passaged between patients at least five times in-between the isolates, we only found four single nucleotide polymorphisms in one of the later isolates and a small (4 amino acids) deletion in the other compared to the index isolate. In contrast to many other evolutionarily successful outbreak lineages (e.g. the Beijing lineage) this outbreak strain appears to be genetically very stable yet evolutionarily successful in a low endemic country such as Sweden. These findings further illustrate that the rate of genomic variation in TB can be highly strain dependent, something that can have important implications for epidemiological studies as well as development of resistance.
Introduction
Tuberculosis (TB) is a major global health concern and the increasing drug resistance makes TB-control even more demanding. Without adequate chemotherapy transmission of drug resistant TB will continue, and the suffering of the individual patients will increase. Concomitantly with the introduction of modern TB drugs over half a century ago, morbidity and mortality was dramatically reduced in countries like Sweden. Yet, TB remains a global epidemic, with one-third of the world's population infected, at least 9.9 million new active cases and 1.8 million deaths in 2008 [1]. In a recent survey, drug- and multidrug resistance (MDR, i.e. resistance to at least rifampicin (RIF) and isoniazid (INH)) was assessed, and the median global prevalence of drug resistance was estimated to be 20% [2]. The distribution of resistance varies substantially world wide with resistance to at least one drug ranging from 0% in some small Western European countries to 56.3% in Azerbaijan, and MDR TB ranging from 0% to 22.3%, again with highest frequency in Azerbaijan. An estimated 2.9% of all new TB cases worldwide have MDR-TB. Strains resistant also to the agents used in the therapy of MDR-TB such as the fluoroquinolone ofloxacin (OFL) and the injectable second-line drugs amikacin (AMI) were more recently described and named extensively drug resistant (XDR) [3]. A growing proportion of such XDR-TB cases will seriously obstruct TB control globally [4], [5].
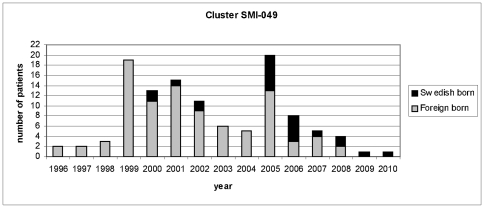
In molecular epidemiological studies of drug resistant TB in Sweden a large outbreak of INH resistant TB was identified, mainly involving patients originating from the Horn of Africa. One exceptionally large cluster, SMI-049, was identified during the study period [6]. Beginning in 1996, two patients belonging to this cluster were identified per year, until 1999 when suddenly 19 new patients were identified. The identification of isolates belonging to cluster SMI-049 during several years and the large accumulation of new cases in 1999 indicated an active spread of TB in Sweden. For this reason a more thorough investigation was initiated in 2000 by the Swedish National Board of Health and Welfare. Yet, in spite of awareness of the outbreak, and strengthened contact investigations, new cases continued to appear (Fig. 1), first at a decreasing pace, then in 2005 again rapidly increasing. Up to date (November 2010) 115 patients infected with the cluster SMI-049 strain have been identified. Twenty-two patients belonging to cluster SMI-049 were Swedish born, and appeared in the later phase of the outbreak, indicating that the epidemic had gradually taken hold in the Swedish born population. Despite the extensive spread of this particular M. tuberculosis strain in the community it has stayed genetically unchanged since its discovery with regard to drug resistance, IS6110 restriction fragment length polymorphism (RFLP)- and spoligotyping patterns. The apparent genetic stability together with the extensive spread of this strain in a low endemic country like Sweden led us to further characterize if the strain possess any particular genetic factors that could explain its evolutionary success and genetic stability.
Figure 1. Number of new cases in cluster SMI-049 identified per year.
The number of new cases belonging to the SMI-049 cluster is displayed for each year. Gray portions of bars represent foreign born patients, black portions of bars represent Swedish born patients.
Here we present a detailed analysis of the M. tuberculosis strain causing the cluster SMI-049 outbreak. The genomes of the isolate of the index case, and two SMI-049 isolates with isolation dates differing from the index strain by 9 years, were sequenced by massive parallel DNA sequencing using the 454-platform. Comparisons were made with previously sequenced M. tuberculosis strains and regions of known importance to antibiotic resistance and virulence were studied more closely. Phylogenetic studies and studies of M. tuberculosis sequence variation have previously mainly focused on unrelated isolates and not looked at whole genome sequences of clinically traceable strains over prolonged time. Interestingly, although the three isolates were obtained with 9 years in-between the only differences found on the whole genome scale were four single nucleotide polymorphisms (SNP) in one of the later isolated strains and a four amino-acids in-frame deletion in the second strain compared to the index strain. This shows that the cluster SMI-049 strain is exceptionally stable genetically and yet evolutionarily very successful and that clonally disseminated M. tuberculosis strains can stay virtually unchanged over many years and multiple transmission cycles.
Methods
Bacterial isolates
During the years 1994–2010, drug resistant (DR) Mycobacterium tuberculosis isolates were obtained from all the six Swedish TB laboratories in Gothenburg, Linköping, Malmö/Lund, Stockholm and Umeå. In Sweden, all isolates are tested for susceptibility to the first-line drugs isoniazid (INH), ethambutol (EMB) and rifampicin (RIF). During the major part of the study all isolates were also tested for susceptibility to streptomycin (SM), except for the years 2004–2010, when the Linköping and Stockholm laboratories stopped testing for SM-resistance, since SM no longer was used for treatment of TB patients in Sweden. All laboratories had taken part in the external quality assurance program for drug susceptibility testing of M. tuberculosis offered by the Swedish TB reference laboratory at the Swedish Institute for Infectious Disease Control (SMI), Solna, Sweden. The first isolate from each patient was included in the study.
RFLP
The isolates were cultured on Löwenstein-Jensen medium. DNA was extracted by resuspending two blue loops of bacteria in 450 µL TE buffer pH 8.0 (10 mM Tris-HCl, 1 mM EDTA) and then heated at 80°C for 20 min. The cells were further freeze thawed twice and 30 µL of lysozyme (20 mg/mL) was added and incubated for 2 h at 37°C. Seventy µL of 10% sodium dodecyl sulfate (SDS) and 5 µL of proteinase K (10 mg/mL) were added to the lysate, vortexed and incubated for 10 min at 65°C followed by addition of 100 µL of 5 mol/L NaCl and 100 µL of 10% N-cetyl-N,N,N-trimethyl ammonium bromide. The tubes were vortexed until the solution was white and incubated for 10 min at 65°C. DNA was extracted by two chloroform-isoamyl alcohol (24∶1, vol/vol) treatments, precipitated by addition of cold isopropanol and the pellet was redissolved in TE buffer. RFLP typing was performed using the insertion sequence IS6110 as probe and PvuII as restriction enzyme [7], [8]. Visual bands were analyzed using the BioNumerics software version 6.01 (Applied Maths, Kortrijk, Belgium). Strains with identical RFLP patterns (100% similarity) and consisting of five bands or more were judged to belong to a cluster. On the basis of the molecular sizes of the hybridizing fragments and the number of IS6110 copies of each isolate, fingerprint patterns were compared by the un-weighted pair-group method of arithmetic averaging using the Jaccard coefficient. Dendrograms were constructed to show the degree of relatedness among strains according to a previously described algorithm [9] and similarity matrixes were generated to visualize the relatedness between the banding pattern of all isolates.
Spoligotyping
All isolates were also characterized by spoligotyping [10]. The patterns obtained by spoligotyping were compared by visual examination and by sorting the results in BioNumerics. Spoligotypes in binary format were entered in the SITVIT2 database (Pasteur Institute of Guadeloupe), which is an updated version of the previously released SpolDB4 database [11], and were assigned to the major phylogenetic lineages according to signatures provided in SITVIT2, which defines 62 genetic lineages/sublineages of M. tuberculosis complex strains. At the time of the present study, SITVIT2 contained global genotyping information on about 73,000 M. tuberculosis complex clinical isolates from 160 countries of origin, with more than 3000 spoligo-international-types (SITs).
Three isolates, one from the index case, and two isolated nine years after the index case, were subjected to further analysis as follows:
Drug susceptibility testing
Drug susceptibility testing for the drugs SM, EMB, RIF, AMI and OFL was performed at SMI by the radiometric BACTEC 460 assay according to the instructions of the manufacturer (Becton Dickinson Biosciences, Sparks, MD). The results were in accordance with those previously obtained at the regional laboratory.
Sanger sequencing
Sanger DNA sequencing of the 980-bp fragment of inhA promoter region was performed using primers previously described [12], sequencing of the cluster SMI-049 specific variations in the polA, PPE55 and cyp138 genes was performed using the following primers: polA_Fwd 5′-CTGCAACTGGTCAGTGACGA, polA_Rev 5′-CGCAACACCCGAAACTCCA, PPE55_Fwd 5′-GTTGACATTGCCAGGGTTGA, PPE55_Fwd2 5′-GGATGTCGAACAGCGACATG, PPE55_Rev 5′- CAATGTCGGGTTCGGCAACT, PPE55_Rev2 5′-AACCTGGGCAACCACGTGTC, PPE55_Rev3 5′-ACCTGAATCCGCTGAACATC, CYP138_Fwd 5′-ATGGCGATCCCGACGTCTTC, CYP138_Rev 5′-CCAGCCTTTCACCCGAACTC. All Sanger sequencing was performed using The BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems).
Whole genome sequencing
Massive parallel sequencing using the 454-technology (Roche) was performed at the sequencing facility at the SMI. Chromosomal DNA of the S96-129, BTB05-552 and BTB05-559 isolates was extracted as for the RFLP analysis except for two additional extractions with chloroform-isoamylalcohol (24∶1, vol/vol). DNA was sequenced using the 454-FLX technology using the manufacturer's instructions (www.454.com) and data were processed with the accompanying software package. This generated 464949 and 502523 reads giving mean genome coverage of 23 and 24 times for S96-129 and BTB05-552, respectively. Isolate BTB05-559 was sequenced both using 454-FLX technology and also using the 454-Titanium upgrade generating a combined number of 373176 reads and a 32 times mean coverage. Gapped genome sequences of the three isolates have been deposited at GenBank as whole genome shotgun projects under accession numbers AEGB00000000 (S96-129), AEGC00000000 (BTB05-552) and AEGD00000000 (BTB05-559). The versions described in this paper are the first versions, xxxx01000000.
Bioinformatics
Assembly and analysis of 454-sequencing data were performed with the CLC Genomics Workbench v3.7 (CLC Bio, Aarhus, Denmark). Assemblies were performed for each isolate individually and also as a combined assembly including all sequence data for a deeper coverage of the common features of the cluster SMI-049 isolates. Large sequence polymorphisms (LSP) between cluster SMI-049 isolates and H37Rv (NC_000962) were identified by detecting assembly positions with partially matching reads and the source of variation elucidated by a combination of de novo assembly of non-assembled reads, reference assembly of the SMI-049 data on the other three fully sequenced M. tuberculosis genomes H37Ra (NC_009525), CDC1551 (NC_002755) and F11 (NC_009565) and manual sequence identification of partially matching reads. SNP analysis was performed for each sequenced isolate after the large sequence polymorphisms had been introduced into a modified H37Rv-SMI049 reference genome to avoid false calls related to assembly problems at locations of large rearrangements. Analysis parameters were as follows: average base quality filter cutoff 15, central base quality filter cutoff 20, minimum variation frequency cutoff 75%, maximum variation (ploidy) 2, minimum sequence coverage 3. The SNP results for the three sequenced genomes were compared and all SNP calls that were not found in all three or had a variation frequency <90% were manually verified in the assemblies. Absence of sequence coverage at the position of an SNP in one or two of the sequences is denoted in Table S3.
Growth rate and TNF induction in macrophages
Peripheral blood mononuclear cells (PBMCs) were isolated from the buffy coats of blood samples from healthy donors by density gradient centrifugation (Lymphoprep, Axis-Shield, Oslo, Norway). Monocytes were separated from PBMCs using CD14-positive magnetic beads (Miltenyi, Bergisch Gladbach, Germany) and were more than 98% pure. Monocytes were cultured in DMEM supplemented with 10% FCS, 1 mM sodium pyruvate, 50 U/ml penicillin, 50 µg/ml streptomycin (all from Gibco, Paisley, UK) and 40 ng/ml rhM-CSF (Peprotech, Rocky Hill, NJ, USA). On day 2 in culture, culture medium was exchanged, antibiotics withdrawn, and the cells were cultured for another three days to differentiate into macrophages.
Macrophages were seeded in 24-well plates (Nunc, Roskilde, Denmark) at a density of 5×105/well or in Lab-Tek chamber slides (Nunc, Roskilde, Denmark) at a density of 3×105/well. After 24 h cells were infected with mycobacteria at a multiplicity of infection of 1∶1. Three different mycobacterial strains were used: H37Rv (ATCC 27294), and the two cluster SMI-049 isolates S96-129 (isolate from index case) and BTB05-552 (late isolate). After 3 h, the cells were extensively washed to remove extracellular bacteria and were cultured for 6 days in DMEM.
On day 0, 1, 3 and 6 infected macrophages were lysed with 0.05% Triton X-100, lysates were plated on 7H11 Middlebrook agar and colony-forming units (cfu) were enumerated after 3–4 weeks. Macrophages in chamber slides were fixed with 4% paraformaldehyde and intracellular acid-fast bacteria were visualized using Kinyoun stain. Cells containing different numbers of bacteria were counted under 1000-fold magnification in consecutive microscope fields until a total of at least 103 cells was attained and expressed as proportion of total number of infected cells.
TNF was quantified in culture supernatants harvested at different time points, using an ELISA according to the manufacturer's protocol (BD OptEIA Sets, BD Biosciences, San Diego, CA, USA).
Results
Cluster SMI-049
During the years 1996-2010 115 cluster SMI-049 isolates from the same number of patients were isolated and characterized by drug susceptibility testing, IS6110 RFLP, and spoligotyping. The cluster is defined by a low-level resistance to INH, a 14-band IS6110 RFLP pattern and a spoligotyping pattern belonging to SIT52, of the T2 lineage, according to the SITVIT2 database (Fig. 2). Three isolates from three different patients were selected for further characterization. The first isolate (S96-129) from August 1996 was from the source case, a 19-year old male originating from Zaire (now Democratic Republic of the Congo). The second isolate (BTB05-552) from September 2005 was from a 28-year old Swedish woman and the third isolate (BTB05-559) from August 2005 was from a 6-year old Swedish born girl. In a timetable of cluster occurrence based on the estimated time for the development of first symptoms consistent with TB, isolate S96-129 is believed to be the source of the cluster (number 1), while BTB05-559 is number 88 and BTB05-552 is number 95 [13].
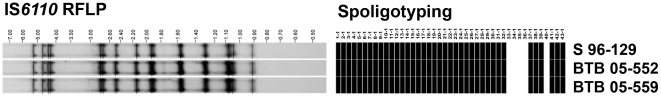
Figure 2. Stability in IS6110 RFLP and spoligotyping patterns.
IS6110 RFLP and spoligotyping patterns of the three SMI-049 isolates S96-129, BTB 05-552 and BTB 05-559. Approximate molecular weights are indicated in kilo base pairs next to the RFLP gel picture. Spoligotype products 1–43 are depicted as black (positive) and white (negative) boxes.
The cluster SMI-049 strain had been circulating in the population for about nine years between the time of diagnosis of the source case and the two other cases. In a chain of transmission the isolate BTB05-552 had been transmitted through at least three patients, more probably four patients, excluding the source case and case number 95 from which it was isolated, and the isolate BTB05-559 had been transmitted from the source case through two or three patients before infecting case number 88.
Whole genome sequencing
Massive parallel sequencing of the genomes of M. tuberculosis isolates S96-129, BTB05-552 and BTB05-559 was performed using the Roche 454-sequencing technology. The mean sequence coverage of the genomes were 23, 24 and 32 times respectively and the completeness of the genome sequences were estimated to be 98% for all isolates. Zero coverage regions that could not be ascribed to deletions compared to the H37Rv reference genome were all located at loci with higher than average GC content, mainly in PE-PGRS genes (Polymorphic GC Rich Sequences), that are known to be difficult to sequence due to their high GC content [14], [15].
Differences between isolates of cluster SMI-049
The three sequenced isolates had identical IS6110 RFLP- and spoligotyping patterns like all 115 cluster SMI-049 isolates (Fig. 2). This pattern was very stable, in only one case a strain isolated from a patient belonging to the cluster in 2000 demonstrated a RFLP pattern with one extra band indicative of a new IS6110 insertion event. The sequenced genomes each contain 18 copies of IS6110 but the theoretical RFLP pattern fits well with the obtained 14-band pattern since some fragments have very similar sizes and will migrate together thus resulting in a lower than expected number of visible bands on the gel. However, the PvuII site in gene Rv2017 (with an IS6110 inserted downstream) appears to be partially resistant to cleavage and only results in a weak band at 1735 bp. Instead, cleavage at the next following PvuII site results in a clear band at 2728 bp.
Comparison of the generated genome sequences reveals that the three sequenced isolates do not differ by any large sequence polymorphisms (LSPs). The only differences found were four SNPs in BTB05-559 and a four amino acids in-frame deletion in BTB05-552 compared to the index isolate S96-129. All differences were verified by manual Sanger sequencing. The SNPs in BTB05-559 consist of a synonymous C ->T transition at nucleotide position 579 in the polA gene (encoding DNA polymerase I) and three SNPs in the PPE55 gene that all result in amino acid changes (N1496Y, T1517A, I1520V). PPE55 belongs to a group of highly variable M. tuberculosis specific proteins with unknown function but that are highly immunogenic and secreted by the Type VII secretion pathway [16], [17]. The in-frame deletion in BTB05-552 removes amino acids 404-407 (out of 441) of the putative cytochrome P450 protein Cyp138.
Comparison of cluster SMI-049 with H37Rv and other M. tuberculosis genomes
A total of 65 LSPs were identified between the SMI-049 isolates and H37Rv (33 insertions, 26 deletions, 6 combined insertion/deletions) with sizes of 9–6831 bp (Table S1). Twenty-five of these were associated with differences in the presence of the mobile insertion element IS6110. The second most common group of changes was copy number differences in regions of short intergenic repeats. The net difference in DNA content in the SMI-049 isolates is an addition of 9103 bp compared to H37Rv. In total, 33 of the polymorphisms were intergenic while 32 affected annotated genes. Twenty-four LSPs were unique to the SMI-049 cluster (i.e not found in any other M. tuberculosis genome at NCBI) and 12 of these affected annotated genes, 5 deletions and 7 IS6110 insertions (Table 1). Of the five deletions, three affected PE or PPE-genes (PE-PGRS44, PPE38, PPE39, PPE57-59). Five large regions not present in H37Rv (1674 bp, 6836 bp, 953 bp, 5000 bp and 6771 bp in length, respectively (Table S1)) results in a total of 15 extra genes that are present in the SMI-049 strains but absent from H37Rv. However, all these regions are present in the CDC1551 strain and represent regions deleted in H37Rv. None of them have been reported to be important for virulence.
Table 1. SMI-049 specific large polymorphisms.
| Reference position (H37Rv) | Allele variations | Size (bp) | Overlapping annotations | Change |
| 164574-164585 | Deletion | 12 | Gene: Rv0136 cyp138. 4 amino acids deletion specific for BTB05-552. | No frameshift |
| 888927-889020 | Deletion | 94 | Intergenic: Deletion of 94 bp compared to H37Rv | |
| 1533690 | Insertion | 1413 | Intergenic: IS6110 insertion not present in H37Rv | |
| 1541951 | Variation | 44 | Intergenic: IS6110 insertion site 44 bp downstream of corresponding IS6110 in H37Rv | |
| 1987503 | Insertion | 1362 | Gene: Rv1755 3′ part of plcD: Insertion of IS6110 199 bp upstream and in reversed orientation compared to H37Rv | Disruption |
| 1987697-1989024 | Inversion | 1299 | IS6110 in reversed orientation compared to H37Rv | |
| 2038790-2039673 | Insertion/deletion | 1355/883 | Gene: Rv1798 lppT, Putative lipoprotein: Truncated by IS6110 insertion and deletion of 883 bp | Disruption |
| 2163462 | Insertion | 1497 | Gene: Rv1917c PPE34: Insertion of IS6110 and 135 bp not present in H37Rv | Disruption |
| 2245301 | Insertion | 1358 | Gene: Rv2000, hypothetical protein: Insertion of IS6110 not present in H37Rv | Disruption |
| 2367679 | Insertion | 1359 | Intergenic: Insertion of IS6110 not present in H37Rv | |
| 2634069-2635575 | Deletion | 1507 | Genes: Rv2352c and Rv2353c, PPE38 and PPE39: Deletion of 1507 bp compared to H37Rv. Smaller than DS9 [66] | Frameshift |
| 2636931 | Insertion | 156 | Intergenic: Insertion of 156 bp compared to H37Rv | |
| 2902567-2904318 | Insertion/deletion | 122/1752 | Genes: Rv2578c and Rv2579 dhaA, hypothetical protein and haloalkane dehalogenase: Deletion of 1752 bp + insertion of 122 bp | Disruption |
| 2937529-2937530 | Deletion | 9 | Gene: Rv2591 PE-PGRS44: Deletion of 9 bp compared to H37Rv | No frameshift |
| 3125851 | Insertion | 1358 | Gene: Rv2818c, hypothetical protein: Insertion of IS6110 not present in H37Rv | Disruption |
| 3192248 | Insertion | 53 | Intergenic: Insertion of 53 bp compared to H37Rv, intergenic repeat region | |
| 3378898 | Insertion | 1359 | Gene: Rv3019c, esxR secreted ESAT-6 like protein: Insertion of IS6110 not present in H37Rv | Disruption |
| 3550843 | Insertion | 1357 | Gene: Rv3183, possible transcriptional regulatory protein: Insertion of IS6110 not present in H37Rv | Disruption |
| 3594319-3594430 | Deletion | 112 | Intergenic: Deletion of 112 bp compared to H37Rv, intergenic repeat region | |
| 3710316-3711736 | Insertion/deletion | 6771/1421 | Genes: Rv3325 and Rv3326, IS6110 transposases: IS6110 replaced by 6771 bp insertion flanked by two IS6110 in reversed direction, similar to CDC1551 | Deletion |
| 3803868-3803918 | Deletion | 51 | Intergenic: Deletion of 51 bp compared to H37Rv | |
| 3842289-3847214 | Deletion | 4926 | Genes: Deletion of Rv3425 to Rv3428c, PPE57 and PPE59 joined and IS1532 element lost through deletion of 4926 bp probably through homologous recombination via large repeat region. Similar to DS13 [66] | Deletion |
| 4172770 | Insertion | 1358 | Intergenic: IS6110 insertion not present in H37Rv | |
| 4348818 | Insertion | 59 | Intergenic: Insertion of 59 bp compared to H37Rv, intergenic repeat region |
SNP and small deletion/insertion polymorphism (DIP), 1–6 bp length, analyses were performed after the large sequence polymorphisms had been introduced into a modified H37Rv-SMI049 reference genome to avoid false calls related to assembly problems at locations of large rearrangements. The SNP and small DIP results were compared to the resequencing results of H37Rv [15], [18] and 90 SNPs and 11 DIPs resulting from identified errors in the H37Rv sequence were removed. In total, the cluster SMI-049 sequences differ from H37Rv by 777 SNPs (781 in BTB05-559, see above) and 44 small DIPs (Tables S2 and S3). Of the SNPs 73 occurred in intergenic regions and 724 in annotated genes. The majority of SNPs (56%) were non-synonymous in concordance with previous reports [18], [19], [20] illustrating the low level of purifying selection in M. tuberculosis. Premature stop-codons were found in six genes (Table 2). Of these only bglS (beta-glucosidase) and lpdA (lipoamide dehydrogenase) encode proteins with known functions and the premature stop codons occur at the very end of both genes.
Table 2. SMI-049 specific SNPs resulting in premature stop codons.
| Reference position (H37Rv) | Reference | Allele variations | Overlapping annotations | Amino acid change |
| 218204 | C | T | Gene: Rv0186 bglS, beta-glucosidase | R646 Stp (691 aa) |
| 281238 | C | T | Gene: Rv0235c, probable transmembrane protein | W459 Stp (482 aa) |
| 2129419 | G | T | Gene: Rv1879, hypothetical protein | E15 Stp (378 aa) |
| 2374245 | C | T | Gene: Rv2114, hypothetical protein | Q138 Stp (207 aa) |
| 3689523 | G | T | Gene: Rv3303 lpdA, Lipoamide dehydrogenase | C472 Stp (493aa) |
| 4351039 | G | T | Gene: Rv3872 PE35 | E99 Stp (99aa) |
Twenty-one of the identified DIPs were specific for cluster SMI-049 and 16 of these resulted in frame shifts in annotated genes, five of which have predicted functions, nanT, fadD35, ltp1, lppH and gidB (Table 3). The first four are involved in lipid biosynthesis and transport and it is not clear what effect knock out of these genes may have on cell viability. However, GidB has recently been shown to be a highly conserved 7-methylguanosine (m7G) methyltransferase responsible for methylation of position G527 of the 16S rRNA [21]. All three strains sequenced have a deletion of G102 of the gidB gene. Loss of function of GidB is associated with low-level resistance to streptomycin [21], [22]. The remaining genes affected by DIPs either encoded hypothetical proteins (N = 4) or belong to PPE or PE-PGRS gene families (N = 6).
Table 3. SMI-049 specific small deletion and insertion polymorphisms (DIP).
| Reference position (H37Rv) | Reference | Allele variations | Overlapping annotations | Change |
| 33203 | G | - | Intergenic | |
| 336684 | A | - | Gene: Rv0279c PE_PGRS4 | Frameshift |
| 336694 | - | C | Gene: Rv0279c PE_PGRS4 | Frameshift |
| 671259 | -- | TG | Gene: Rv0577 TB27.3, hypothetical protein | Frameshift |
| 789689 | G | - | Intergenic | |
| 839545 | - | G | Gene: Rv0747 PE_PGRS10 | Frameshift |
| 1178876 | AT | -- | Intergenic | |
| 1242287 | G | - | Gene: Rv1118c, hypothetical protein | Frameshift |
| 1280683 | ------ | CGAAGT | Gene: Rv1153c omt, Putative O-methyltransferase | No frameshift |
| 1537711 | AC | -- | Intergenic | |
| 1982962 | C | - | Gene: Rv1753c PPE24 | Frameshift |
| 2046760 | C | - | Gene: Rv1803c PE_PGRS32 | Frameshift |
| 2149855 | - | A | Gene: Rv1902c nanT, sialic acid-transport integral membrane protein | Frameshift |
| 2821079 | GG | -- | Gene: Rv2505c fadD35, Putative acyl-CoA synthetase | Frameshift |
| 3006362 | G | - | Gene: Rv2689c, hypothetical protein | Frameshift |
| 3100153 | - | A | Gene: Rv2790c ltp1, Putative lipid -transfer protein | Frameshift |
| 3742352 | C | - | Gene: Rv3345c PE_PGRS50 | Frameshift |
| 4018414 | - | A | Gene: Rv3576 lppH, Putative lipoprotein | Frameshift |
| 4117362 | C | - | Gene: Rv3677c, Putative hydrolase | Frameshift |
| 4408101 | C | - | Gene: Rv3919c gidB, 7-methylguanosine (m7G) methyltransferase | Frameshift |
Positional effects of IS6110 insertions
M. tuberculosis contains several mobile insertion sequences (IS) [23]. IS6110 is a member of the IS3-family and is found in almost all members of the M. tuberculosis complex in varying number of copies. Integration of IS6110 can, depending on the site of insertion, result in phenotypic changes in the host bacterium, both detrimental as well as occasionally beneficial (Reviewed in [24]). To determine if any obvious effects of IS6110 insertions could be predicted, we analyzed the insertions of IS6110 in the SMI-049 cluster genomes in more detail. Eight IS6110 insertions occur in annotated genes in SMI-049 and the affected open reading frames are most likely completely disrupted (Table 1). As expected, none of the affected genes are reported to be essential for M. tuberculosis. Notably, the PPE34 and plcD genes are the genes with most detected individual IS6110 insertions in clinical isolates of M. tuberculosis [25] and disruption of these genes have been speculated to result in changes in the immunological interaction with the host [26], [27], [28], [29]. Of the remaining genes with insertions of IS6110, two have predicted functions, one in lipid biosynthesis (lppT) and one as a secreted ESAT-6-like gene (esxR) and four are hypothetical genes. Over-representation of IS6110 insertion in hypothetical genes has been noted earlier [25].
Apart from disrupting genes, IS6110 insertions have also been found to activate gene expression of nearby genes by an outward directed promoter (OP6110) at the 3′ end of the insertion element [30], [31], [32]. To assess if any IS6110 insertions in the SMI-049 genomes could account for differences in virulence by the OP6110 promoter we mapped what genes were present nearby inserted IS6110 (Table 4). Apart from the truncated genes that harbored an IS6110, and most likely are non-functional, five genes were found at a reasonable distance from an IS6110 and oriented in the forward direction compared to OP6110, an IS1547 transposase, PPE36, PE27A, moaC (molybdenum cofactor biosynthesis protein C) and an oxidoreductase with unknown specificity. Of these genes, changed expression of PPE36 and PE27A might result in changes in the immunogenic response.
Table 4. Putative genes where IS6110 insertions may alter promoter activity.
| Position of OP6110 in SMI-049 | Downstream gene | Function | Distance | Orientation of downstream gene | Comment |
| 890769-890838 | Rv0797 | IS1547 Transposase | 94 bp | Forward | |
| 1537121-1537190 | Rv1362c | Hyp. Prot., unknown function | 398 bp | Reversed | |
| 1545719-1545788c | Rv1368 | lprF, probable lipoprotein | 272 bp | Reversed | |
| 1992219-1992288 | Rv1755c | plcD 5′, Phospholipase C - truncated | 86 bp | Reversed | IS6110 inserted into plcD |
| 2000585-2000654 | Rv1758 | cut1 3′, Serine esterase, hydrolysis of cutin - truncated | 66 bp | Forward | IS6110 inserted in cut1, Non-functional [79] |
| 2051273-2051342 | Rv1799 | lppt 3′, probable lipoprotein - truncated | 82 bp | Forward | IS6110 inserted into lppT |
| 2175810-2175879c | Rv1917c | PPE34 3′, unknown function - truncated | 81 bp | Forward | IS6110 inserted into PPE34 |
| 2260061-2260130 | Rv2000 | Hyp. Prot. 3′, unknown function - truncated | 85 bp | Forward | IS6110 inserted into Rv2000 |
| 2280108-2280177 | Rv2117 | Transcriptional regulator protein | 155 bp | Reversed | |
| 2387585-2387654 | Rv2108 | PPE36, unknown function | 119 bp | Forward | |
| 2652563-2652632 | Rv2356c | PPE40, unknown function | 994 bp | Reversed | |
| 3132243-3132312 | Rv2813 | Hyp. Prot., unknown function | 1495 bp | Reversed | |
| 3137724-3137793 | Rv2818c | Hyp. Prot. 3′, unknown function - truncated | 82 bp | Forward | IS6110 inserted into Rv2818c |
| 3392133-3392202c | Rv3019c | esxR 5′, secreted ESAT-6 like protein - truncated | 82 bp | Reversed | IS6110 inserted into esxR |
| Rv3018A | PE27A, unknown function | 564 bp | Forward | ||
| 3565496-3565565 | Rv3182 | Hyp. Prot., unknown function | 210 bp | Reversed | |
| 3723555-3723624c | Rv3324A | Pseudogen | 84 bp | Forward | IS6110 inserted into Rv3324A |
| Rv3324c | moaC, molybdenum cofactor biosynthesis protein C | 194 bp | Forward | ||
| 3728662-3728732c | MT3429 | Hyp. Prot., unknown function | 79 bp | Reversed | Identical to MT3429 of CDC1551, not present in H37Rv |
| 4184858-4184927 | Rv3727 | Oxidoreductase | 269 bp | Forward |
Growth rate and TNF induction in human macrophages
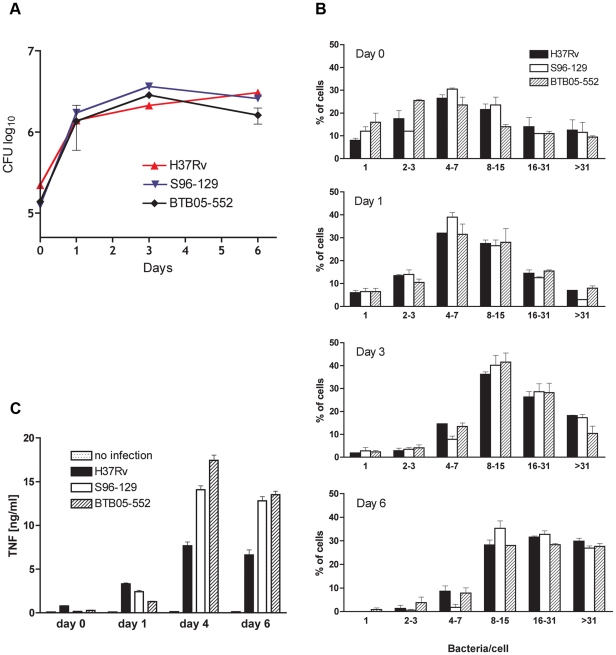
Cluster SMI-049 isolates S96-129 and BTB05-552 grew in macrophages as indicated by a successive increase in cfu in macrophage lysates (Fig. 3A) and an increase of proportion of cells containing higher numbers of bacteria over time (Fig. 3B). The growth rate of both SMI-049 cluster isolates was similar and comparable to that of the H37Rv laboratory strain until day 3. Of note, at day 6 less bacteria were recovered from cluster SMI-049-infected macrophages as compared with day 3, while macrophages infected with H37Rv showed further increment in cfu at that time point (Fig. 3A). Microscopic analysis of chamber slide cultures revealed that there was augmented death of macrophages infected with the SMI-049 isolates on day 6 as indicated by increased number of cells with fragmented cytoplasm and pycnotic nuclei; no increased cell death was observed in H37Rv-infected macrophages (not shown). Interestingly, from the day 4 post-infection SMI-049-infected macrophages produced much greater amounts of TNF than macrophages infected with H37Rv (Fig. 3C).
Figure 3. Virulence of SMI-049 isolates in human macrophages.
Monocyte-derived human macrophages were infected with strain H37Rv or SMI-049 isolates S96-129 or BTB05-552, at multiplicity of infection 1∶1. Intracellular bacterial growth was determined by enumeration of cfu in plated macrophage lysates (A) or by estimation of proportion of cells containing varying amounts of bacteria (B) over time. TNF induction in infected macrophages was quantified by ELISA using culture supernatants harvested at different time points post-infection (C).
Drug resistance genes
When tested by the BACTEC 460 reference method all three isolates were resistant to isoniazide (INH) at 0.2 mg/L. S96-129 was resistant to streptomycin (SM) at 4 mg/L whereas BTB05-552 and BTB05-559 were SM-susceptible to this concentration. All three isolates were susceptible to the first line drugs rifampicin (RIF), ethambutol (EMB) and pyrazinamide (PZA). These isolates were also susceptible to the second line drugs ofloxacin (OFL) (at 2 mg/L) and amikacin (AMI) (1 mg/L). Genes previously reported to be involved in resistance to antibiotics used to treat M. tuberculosis infections (rpoB – RIF; katG, inhA, oxyR-ahpC, iniBAC and furA – INH; rrs, rpsL and gidB – SM; pncA – PZA; embB – EMB; gyrA and gyrB –OFL) were specifically monitored for non-synonymous mutations that may change the tolerance to these antibiotics. Low-level INH resistance of cluster SMI-049 strains is explained by a -15 C to T transition in the promoter region of the inhA gene, which encodes an enoyl acyl carrier protein reductase involved in fatty acid synthesis [33]. This mutation has been reported as the second most common resistance mutation to INH and is most often associated with low (<10 ug/ml) level resistance [34], [35], [36], [37], [38]. Twenty-five randomly chosen isolates belonging to the SMI-049 cluster were also found to contain this mutation (data not shown). Only one other change was found in a gene previously reported to be involved in INH resistance, iniA Q394E, a substitution that has never been reported to alter drug susceptibility.
As noted above, all three isolates contain a deletion of G102 in the gidB gene that previously has been reported to result in low-level SM resistance [21], [22]. The low-level of resistance reported earlier for gidB mutants is just at the clinical break point and this may explain why S96-129 was found to be resistant while BTB05-552 and BTB05-559 were susceptible. No changes were found in rrs or rpsL genes that could explain the differences in SM susceptibility between the isolates.
The gyrA gene of all three isolates contain five non-synonymous mutations (E21Q, T80A, A90G, S95T and G668D) compared to H37Rv. Interestingly, change of alanine at position 90 to valine is commonly reported in quinolone resistant isolates [39], [40], [41] but change to a glycine in combination with the T80A mutation has instead been reported to result in hypersensitivity to quinolones [42]. This finding led us to determine the MIC for OFL more precisely by testing all three isolates at 2, 1, 0.5 and 0.25 mg/L. No growth was detected at any of these concentrations (only without antibiotics) and the MIC for OFL is therefore < = 0.125, well below the average wild-type distribution MIC of 0.5 [43]. This is interesting since the T80A mutation is a marker for the Uganda genotype [44]. The S95T mutation is known as a naturally occurring polymorphism among quinolone susceptible strains [45], [46] and E21Q and G668D are attributed to H37Rv specific differences and present in many other susceptible strains and should therefore not contribute any resistance.
Phylogenetic background of SMI-049
Phylogenetically the SMI-049 isolates were of the modern principal genetic group 2 (PGG2) defined by katG463 and gyrA95 SNPs (katG463 CGG (Arg) and gyrA95 ACC (Thr)) [45]. By extended SNP analysis (Filliol 2006) they were found to belong to SNP cluster group 5 (SCG-5), which is predominant in Uganda [47]. The RFLP pattern of the cluster SMI-049 strain matches (92%) with a strain (BEA000007341) from Rwanda recorded in the international database of IS6110 RFLP patterns maintained at RIVM, Bilthoven, the Netherlands, also indicating that the strain may have its origin in sub-Saharan Africa. According to the phylogeny in [20] and [44] SNPs of the three sequenced genomes placed the SMI-049 strain in the Lineage 4 (red) Europe and Americas lineages, with 2/3 matches to the “Uganda” cluster within the PGG2 group.
By spoligotyping all cluster SMI-049 isolates were of the T2 lineage, which is geographically linked to East Africa [11]. All isolates had the spoligotype international type SIT52. The sequenced isolates also lacked the region of difference 724 (RD724), a polymorphism that defines one major sub-lineage of M. tuberculosis commonly seen in the central African human host population, and which is common in isolates from Uganda [48], [49]. Together with a T2 specific spoligotype pattern [18] RD724 defines the “Ugandan genotype” [48], again supporting the concept that the cluster SMI-049 strain has its origin in Central/Eastern Africa.
Discussion
The SMI-049 outbreak is one of the largest outbreaks of TB ever reported from low endemic countries. A few other outbreaks of this size have been reported from Denmark, England, The Canary Islands and The Netherlands [50], [51], [52], [53], the largest being two TB clusters comprising 184 and 272 cases respectively found over a ten years period in Denmark [54].
In spite of the long transmission time and many carriers of the cluster SMI-049 strain in the community, the IS6110 RFLP pattern has stayed virtually unchanged and we found extremely few genomic changes (4 SNPs and one small deletion) between the three sequenced isolates that were isolated 9 years apart. This indicates that the SMI-049 lineage is genetically very stable both with respect to point mutations and to transpositional activity of insertion sequences, in contrast with reports of other M. tuberculosis lineages e.g. the Beijing lineage. Differences in the level of genetic variability have to be considered in view of the already very low genetic variability among members of the M. tuberculosis complex. Our genome sequences are lacking coverage of several PE-PGRS genes most likely as a result of sequencing bias due to high GC-content in those genes. It is therefore possible that there exists more variation between SMI-049 strains than we have detected in this study.
Niemann et al [18] found that two isolates of the Beijing lineage from an outbreak in Uzbekistan with identical IS6110 RFLP fingerprinting pattern exhibited considerable genomic diversity (130 SNPs and one large deletion). The fact that strains with identical genotyping patterns can accumulate significant amounts of genetic diversity indicates that epidemiological links between strains with identical genotyping data can be more remote and are likely to represent older transmission events rather than cases of recent transmission among patients in one RFLP cluster. The two isolates studied by Niemann et al actually exhibited differences in MIRU/VNTR as well as in drug resistance patterns, indicating that they were genetically more remote than indicated by their identical RFLP pattern. In Estonia genetically closely related isolates of the Beijing lineage showed a range from full susceptibility to four-drug resistance, indicating that drug resistance had developed recently and independently in different clones of Beijing strains [55].
Stability in RFLP
Yeh et al. found that about 30% of serial isolates of patients whose cultures spanned at least 90 days had changed IS6110 RFLP patterns [56]. In contrast, here we found that the cluster SMI-049 strain has maintained the same stable IS6110 RFLP profile over 9 years with only one isolate as an exception. This may indicate that the SMI-049 strain has a very low transposition activity of IS6110 despite the presence of 18 copies in the genome. Low transposition activity is generally thought to occur in genomes with low number of IS6110 copies (1–5 copies) [57]. However, transpositional activity has been linked to the transcriptional activity of the genomic location of IS6110 elements with increased overall transpositional activity occurring when an IS6110 element is inserted at a genomic position with strong transcriptional activity [58]. There also seems to be an upper limit of about 25 IS6110 elements per genome, possibly regulated by a trans acting repressor expressed from each element that limits the transpositional activity more the more elements are present [24]. If any of these factors keep the IS6110 pattern of the SMI-049 cluster so stable is hard to speculate upon.
Stability in drug resistance
Although members of the M. tuberculosis complex have accumulated a very low general genetic variability since their last common ancestor, the modern use of anti-TB drugs presents a new selective pressure that can increase the genetic variability in genes that render the bacteria drug resistant by selecting those over their susceptible counterparts. In clinical isolates exposed to therapy, increased genetic variation should therefore be expected in drug resistance genes. However, some resistance mutations have been associated with fitness costs that might result in a reduced virulence/transmissibility (reviewed in [59]). All initial isolates of the 115 patients belonging to cluster SMI-049 were resistant to INH, but susceptible to EB and RIF. In only one instance a strain developed further resistance: in 2003 a patient was found to be infected with a strain belonging to the SMI-049 clone and resistant to INH, in 2004 another isolate with the same fingerprint from the same patient was in addition resistant to RIF, making the strain MDR [6]. This mutant variant with increased resistance did not spread further. It is intriguing that in spite of the long passage of this strain in the community there has been no further development of drug resistance. Resistance to INH has been coupled with various chromosomal mutations but those in katG and in the promoter regions of inhA have been most frequently associated with clinical isolates [60], [61]. The INH resistance of the cluster SMI-049 isolates was low level, with a MIC of 0.4 mg/L, which agrees with the fact that the isolates had the inhA promoter mutation. Strains with the inhA promoter mutation or the KatG S315T mutation were more likely to spread than strains with other mutations, suggesting that these mutations have a low fitness cost [62] which is in agreement with the stability in INH resistance of cluster SMI-049.
All three isolates contain the G102 deletion in gidB reported earlier to give low-level resistance to SM [21], [22]. This level of resistance is just at the clinical resistance break-point and this is probably why some isolates belonging to the cluster SMI-049 have been reported as resistant while most isolates are considered susceptible. We could not find any genomic difference between isolate S96-129 (SMR) and BTB05-552 (SMS) and BTB05-559 (SMS) that can explain this difference in resistance. Use of SM to treat infections of SMI-049 should therefore be avoided, especially since the gidB mutation has been reported to dramatically elevate the frequency of high-level mutations to SM in M. tuberculosis [22]. On the other hand, SMI-049 isolates are highly sensitive to quinolones and this group of antibiotics might therefore be useful for treatment.
Effects of polymorphisms on evolutionary success and virulence
Large sequence polymorphisms (LSPs) such as deletions and insertions of IS-elements (mainly IS6110) represent a substantial source of genetic variation among M. tuberculosis lineages [63], [64]. How LSPs in the M. tuberculosis complex may affect the evolutionary success of certain lineages has been discussed frequently [64], [65], [66], [67]. There is increasing evidence that certain M. tuberculosis strains are particularly prone to disseminate and/or to cause disease [60], [68], [69], [70], [71]. There are also several examples of hypervirulence resulting from gene deletion in M. tuberculosis [72]. However, the potential pathogenic relevance of most specific regions of difference is unknown. Interestingly, the Beijing lineage that is causing concern due to its global distribution and its involvement in severe outbreaks does not seem to spread in Sweden [73]. Since the SMI-049 cluster strain has been extensively spread in Sweden, a country with low incidence of TB, questions have been raised whether it contains genotypic determinants that make it particularly prone to disseminate. The fact that transmission was very high during certain periods [13] around particular individuals might on the other hand indicate that part of the extensive transmission may be explained by socio-demographic factors.
SMI-049 does not contain any unique sequences not present in other M. tuberculosis strains. On the other hand there are several specific deletions and IS6110 insertions not found elsewhere. The successful spread of SMI-049 and the fact that it is not attenuated in growth in human macrophages also shows that deletion of the genes absent from its genome compared to other sequenced strains is not likely to be detrimental to virulence. In fact our findings of the high TNF production in human macrophages infected by SMI-049 isolates in vitro paralleled by the augmented macrophage death support the concept of the sustained virulence of SMI-049; similar induction of increased TNF expression and cellular necrosis by virulent clinical strains, as compared to H37Rv, have recently been found in murine macrophages in vitro [74] and in a mouse model of TB infection [75].
In some instances deletion of genes has been associated with altered development of M. tuberculosis disease, e.g. Kong et al found a significant linkage between the deletions of RD105, RD181 and RD142 (which are markers for Beijing strains) and the occurrence of extrathoracic tuberculosis [28]. Many of the inactivating polymorphisms (deletions and IS6110 insertions in genes) specific for the SMI-049 cluster have occurred in genes belonging to the PE and PPE-gene families (PE-PGRS44, PPE34, PPE38-39, PPE57-59). Since these gene classes are thought to play important roles for the bacterium by generating antigenic variation and immune evasion, variable occurrence of PE and PPE genes may change the interaction of the bacterium with its host and thus alter the progress of the disease.
SMI-049 isolates also have a unique IS6110 inserted at a slightly different position in the plcD gene (Rv1755) compared to H37Rv. There are four phospholipase C genes (PLC) in M. tuberculosis, plcABC as an operon and plcD alone at a separate position in the genome. All four contribute to PLC activity and are involved in virulence although their exact role is not fully understood [76]. Expression of the plc-genes is upregulated after infection of macrophages and the quadruple knock out mutant is attenuated in growth in lungs and spleen of mice. Interruptions of plc-genes are frequent among clinical M. tuberculosis isolates and most frequent in plcD [27], [28], [77]. It is hypothesized that reduced PLC activity causes persistence of the bacterium inside macrophages and aid travel to distant organs. Alternatively, reduced activity limits release of arachidonic acid from the macrophage membrane and reduce immune response in the initial lung infection. It is therefore possible that the clinical presentation of the disease may be influenced by the genetic variability of the plcD region. It is not immediately obvious if any of the other genes with annotated functions affected by inactivating changes would mediate phenotypic changes in the cluster SMI-049 strains although dhaA (encoding a haloalkane dehalogenase) has been shown to be upregulated upon in vitro infection of a macrophage cell line [78].
In summary, we have found that the cluster SMI-049 strain that has caused a major outbreak of INH resistant TB in Sweden has been genetically extremely stable over extended time in the community and through many transmissions and has not developed resistance to the antibiotics used to treat patients infected with the strain. Even though we have found specific genetic features in this strain not found in other TB strains, the question why this particular strain of TB has been so successful in spreading both in the immigrant population where it originated and also in the Swedish born population remains to be answered.
Supporting Information
Complete list of LSPs compared to H37Rv.
(XLS)
Complete list of DIPs compared to H37Rv.
(XLS)
Complete list of SNPs compared to H37Rv.
(XLS)
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was financed by Swedish Heart-Lung Foundation (www.hjart-lungfonden.se/); Swedish Research Council (www.vr.se/); Swedish International Development Cooperation Agency (www.sida.se/) (to GK). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.World Health Organization. Global tuberculosis control: a short update to the 2009 report. Geneva: World Health Organization. 2009:vi, 39. [Google Scholar]
- 2.WHO/IUATLD Global Project on Anti-Tuberculosis Drug Resistance Surveillance. Geneva: World Health Organization.; 2008. Anti-tuberculosis drug resistance in the world: fourth global report.142 [Google Scholar]
- 3.Emergence of Mycobacterium tuberculosis with extensive resistance to second-line drugs–worldwide, 2000-2004. MMWR Morb Mortal Wkly Rep. 2006;55:301–305. [PubMed] [Google Scholar]
- 4.Beijing/W genotype Mycobacterium tuberculosis and drug resistance. Emerg Infect Dis. 2006;12:736–743. doi: 10.3201/eid1205.050400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gandhi NR, Moll A, Sturm AW, Pawinski R, Govender T, et al. Extensively drug-resistant tuberculosis as a cause of death in patients co-infected with tuberculosis and HIV in a rural area of South Africa. Lancet. 2006;368:1575–1580. doi: 10.1016/S0140-6736(06)69573-1. [DOI] [PubMed] [Google Scholar]
- 6.Ghebremichael S, Petersson R, Koivula T, Pennhag A, Romanus V, et al. Molecular epidemiology of drug-resistant tuberculosis in Sweden. Microbes Infect. 2008;10:699–705. doi: 10.1016/j.micinf.2008.03.006. [DOI] [PubMed] [Google Scholar]
- 7.van Embden JD, Cave MD, Crawford JT, Dale JW, Eisenach KD, et al. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J Clin Microbiol. 1993;31:406–409. doi: 10.1128/jcm.31.2.406-409.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van Soolingen D, Qian L, de Haas PE, Douglas JT, Traore H, et al. Predominance of a single genotype of Mycobacterium tuberculosis in countries of east Asia. J Clin Microbiol. 1995;33:3234–3238. doi: 10.1128/jcm.33.12.3234-3238.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van Soolingen D, Hermans PW, de Haas PE, Soll DR, van Embden JD. Occurrence and stability of insertion sequences in Mycobacterium tuberculosis complex strains: evaluation of an insertion sequence-dependent DNA polymorphism as a tool in the epidemiology of tuberculosis. J Clin Microbiol. 1991;29:2578–2586. doi: 10.1128/jcm.29.11.2578-2586.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kamerbeek J, Schouls L, Kolk A, van Agterveld M, van Soolingen D, et al. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J Clin Microbiol. 1997;35:907–914. doi: 10.1128/jcm.35.4.907-914.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brudey K, Driscoll JR, Rigouts L, Prodinger WM, Gori A, et al. Mycobacterium tuberculosis complex genetic diversity: mining the fourth international spoligotyping database (SpolDB4) for classification, population genetics and epidemiology. BMC Microbiol. 2006;6:23. doi: 10.1186/1471-2180-6-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guo H, Seet Q, Denkin S, Parsons L, Zhang Y. Molecular characterization of isoniazid-resistant clinical isolates of Mycobacterium tuberculosis from the USA. J Med Microbiol. 2006;55:1527–1531. doi: 10.1099/jmm.0.46718-0. [DOI] [PubMed] [Google Scholar]
- 13.Kan B, Berggren I, Ghebremichael S, Bennet R, Bruchfeld J, et al. Extensive transmission of an isoniazid-resistant strain of Mycobacterium tuberculosis in Sweden. Int J Tuberc Lung Dis. 2008;12:199–204. [PubMed] [Google Scholar]
- 14.Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 15.Zheng H, Lu L, Wang B, Pu S, Zhang X, et al. Genetic basis of virulence attenuation revealed by comparative genomic analysis of Mycobacterium tuberculosis strain H37Ra versus H37Rv. PLoS One. 2008;3:e2375. doi: 10.1371/journal.pone.0002375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abdallah AM, Gey van Pittius NC, Champion PA, Cox J, Luirink J, et al. Type VII secretion–mycobacteria show the way. Nat Rev Microbiol. 2007;5:883–891. doi: 10.1038/nrmicro1773. [DOI] [PubMed] [Google Scholar]
- 17.Simeone R, Bottai D, Brosch R. ESX/type VII secretion systems and their role in host-pathogen interaction. Curr Opin Microbiol. 2009;12:4–10. doi: 10.1016/j.mib.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 18.Niemann S, Koser CU, Gagneux S, Plinke C, Homolka S, et al. Genomic diversity among drug sensitive and multidrug resistant isolates of Mycobacterium tuberculosis with identical DNA fingerprints. PLoS One. 2009;4:e7407. doi: 10.1371/journal.pone.0007407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fleischmann RD, Alland D, Eisen JA, Carpenter L, White O, et al. Whole-genome comparison of Mycobacterium tuberculosis clinical and laboratory strains. J Bacteriol. 2002;184:5479–5490. doi: 10.1128/JB.184.19.5479-5490.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hershberg R, Lipatov M, Small PM, Sheffer H, Niemann S, et al. High functional diversity in Mycobacterium tuberculosis driven by genetic drift and human demography. PLoS Biol. 2008;6:e311. doi: 10.1371/journal.pbio.0060311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Okamoto S, Tamaru A, Nakajima C, Nishimura K, Tanaka Y, et al. Loss of a conserved 7-methylguanosine modification in 16S rRNA confers low-level streptomycin resistance in bacteria. Mol Microbiol. 2007;63:1096–1106. doi: 10.1111/j.1365-2958.2006.05585.x. [DOI] [PubMed] [Google Scholar]
- 22.Spies FS, da Silva PE, Ribeiro MO, Rossetti ML, Zaha A. Identification of mutations related to streptomycin resistance in clinical isolates of Mycobacterium tuberculosis and possible involvement of efflux mechanism. Antimicrob Agents Chemother. 2008;52:2947–2949. doi: 10.1128/AAC.01570-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gordon SV, Heym B, Parkhill J, Barrell B, Cole ST. New insertion sequences and a novel repeated sequence in the genome of Mycobacterium tuberculosis H37Rv. Microbiology. 1999;145(Pt 4):881–892. doi: 10.1099/13500872-145-4-881. [DOI] [PubMed] [Google Scholar]
- 24.McEvoy CR, Falmer AA, Gey van Pittius NC, Victor TC, van Helden PD, et al. The role of IS6110 in the evolution of Mycobacterium tuberculosis. Tuberculosis (Edinb) 2007;87:393–404. doi: 10.1016/j.tube.2007.05.010. [DOI] [PubMed] [Google Scholar]
- 25.Yesilkaya H, Dale JW, Strachan NJ, Forbes KJ. Natural transposon mutagenesis of clinical isolates of Mycobacterium tuberculosis: how many genes does a pathogen need? J Bacteriol. 2005;187:6726–6732. doi: 10.1128/JB.187.19.6726-6732.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sampson SL, Lukey P, Warren RM, van Helden PD, Richardson M, et al. Expression, characterization and subcellular localization of the Mycobacterium tuberculosis PPE gene Rv1917c. Tuberculosis (Edinb) 2001;81:305–317. doi: 10.1054/tube.2001.0304. [DOI] [PubMed] [Google Scholar]
- 27.Kong Y, Cave MD, Yang D, Zhang L, Marrs CF, et al. Distribution of insertion- and deletion-associated genetic polymorphisms among four Mycobacterium tuberculosis phospholipase C genes and associations with extrathoracic tuberculosis: a population-based study. J Clin Microbiol. 2005;43:6048–6053. doi: 10.1128/JCM.43.12.6048-6053.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kong Y, Cave MD, Zhang L, Foxman B, Marrs CF, et al. Population-based study of deletions in five different genomic regions of Mycobacterium tuberculosis and possible clinical relevance of the deletions. J Clin Microbiol. 2006;44:3940–3946. doi: 10.1128/JCM.01146-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang Z, Yang D, Kong Y, Zhang L, Marrs CF, et al. Clinical relevance of Mycobacterium tuberculosis plcD gene mutations. Am J Respir Crit Care Med. 2005;171:1436–1442. doi: 10.1164/rccm.200408-1147OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beggs ML, Eisenach KD, Cave MD. Mapping of IS6110 insertion sites in two epidemic strains of Mycobacterium tuberculosis. J Clin Microbiol. 2000;38:2923–2928. doi: 10.1128/jcm.38.8.2923-2928.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Safi H, Barnes PF, Lakey DL, Shams H, Samten B, et al. IS6110 functions as a mobile, monocyte-activated promoter in Mycobacterium tuberculosis. Mol Microbiol. 2004;52:999–1012. doi: 10.1111/j.1365-2958.2004.04037.x. [DOI] [PubMed] [Google Scholar]
- 32.Soto CY, Menendez MC, Perez E, Samper S, Gomez AB, et al. IS6110 mediates increased transcription of the phoP virulence gene in a multidrug-resistant clinical isolate responsible for tuberculosis outbreaks. J Clin Microbiol. 2004;42:212–219. doi: 10.1128/JCM.42.1.212-219.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Banerjee A, Dubnau E, Quemard A, Balasubramanian V, Um KS, et al. inhA, a gene encoding a target for isoniazid and ethionamide in Mycobacterium tuberculosis. Science. 1994;263:227–230. doi: 10.1126/science.8284673. [DOI] [PubMed] [Google Scholar]
- 34.Brossier F, Veziris N, Truffot-Pernot C, Jarlier V, Sougakoff W. Performance of the genotype MTBDR line probe assay for detection of resistance to rifampin and isoniazid in strains of Mycobacterium tuberculosis with low- and high-level resistance. J Clin Microbiol. 2006;44:3659–3664. doi: 10.1128/JCM.01054-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Herrera-Leon L, Molina T, Saiz P, Saez-Nieto JA, Jimenez MS. New multiplex PCR for rapid detection of isoniazid-resistant Mycobacterium tuberculosis clinical isolates. Antimicrob Agents Chemother. 2005;49:144–147. doi: 10.1128/AAC.49.1.144-147.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lavender C, Globan M, Sievers A, Billman-Jacobe H, Fyfe J. Molecular characterization of isoniazid-resistant Mycobacterium tuberculosis isolates collected in Australia. Antimicrob Agents Chemother. 2005;49:4068–4074. doi: 10.1128/AAC.49.10.4068-4074.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Park H, Song EJ, Song ES, Lee EY, Kim CM, et al. Comparison of a conventional antimicrobial susceptibility assay to an oligonucleotide chip system for detection of drug resistance in Mycobacterium tuberculosis isolates. J Clin Microbiol. 2006;44:1619–1624. doi: 10.1128/JCM.44.5.1619-1624.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Piatek AS, Telenti A, Murray MR, El-Hajj H, Jacobs WR, Jr, et al. Genotypic analysis of Mycobacterium tuberculosis in two distinct populations using molecular beacons: implications for rapid susceptibility testing. Antimicrob Agents Chemother. 2000;44:103–110. doi: 10.1128/aac.44.1.103-110.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ginsburg AS, Grosset JH, Bishai WR. Fluoroquinolones, tuberculosis, and resistance. Lancet Infect Dis. 2003;3:432–442. doi: 10.1016/s1473-3099(03)00671-6. [DOI] [PubMed] [Google Scholar]
- 40.Kocagoz T, Hackbarth CJ, Unsal I, Rosenberg EY, Nikaido H, et al. Gyrase mutations in laboratory-selected, fluoroquinolone-resistant mutants of Mycobacterium tuberculosis H37Ra. Antimicrob Agents Chemother. 1996;40:1768–1774. doi: 10.1128/aac.40.8.1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xu C, Kreiswirth BN, Sreevatsan S, Musser JM, Drlica K. Fluoroquinolone resistance associated with specific gyrase mutations in clinical isolates of multidrug-resistant Mycobacterium tuberculosis. J Infect Dis. 1996;174:1127–1130. doi: 10.1093/infdis/174.5.1127. [DOI] [PubMed] [Google Scholar]
- 42.Aubry A, Veziris N, Cambau E, Truffot-Pernot C, Jarlier V, et al. Novel gyrase mutations in quinolone-resistant and -hypersusceptible clinical isolates of Mycobacterium tuberculosis: functional analysis of mutant enzymes. Antimicrob Agents Chemother. 2006;50:104–112. doi: 10.1128/AAC.50.1.104-112.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Angeby KA, Jureen P, Giske CG, Chryssanthou E, Sturegard E, et al. Wild-type MIC distributions of four fluoroquinolones active against Mycobacterium tuberculosis in relation to current critical concentrations and available pharmacokinetic and pharmacodynamic data. J Antimicrob Chemother. 2010;65:946–952. doi: 10.1093/jac/dkq091. [DOI] [PubMed] [Google Scholar]
- 44.Comas I, Homolka S, Niemann S, Gagneux S. Genotyping of genetically monomorphic bacteria: DNA sequencing in mycobacterium tuberculosis highlights the limitations of current methodologies. PLoS One. 2009;4:e7815. doi: 10.1371/journal.pone.0007815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sreevatsan S, Pan X, Stockbauer KE, Connell ND, Kreiswirth BN, et al. Restricted structural gene polymorphism in the Mycobacterium tuberculosis complex indicates evolutionarily recent global dissemination. Proc Natl Acad Sci U S A. 1997;94:9869–9874. doi: 10.1073/pnas.94.18.9869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Takiff HE, Salazar L, Guerrero C, Philipp W, Huang WM, et al. Cloning and nucleotide sequence of Mycobacterium tuberculosis gyrA and gyrB genes and detection of quinolone resistance mutations. Antimicrob Agents Chemother. 1994;38:773–780. doi: 10.1128/aac.38.4.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Filliol I, Motiwala AS, Cavatore M, Qi W, Hazbon MH, et al. Global phylogeny of Mycobacterium tuberculosis based on single nucleotide polymorphism (SNP) analysis: insights into tuberculosis evolution, phylogenetic accuracy of other DNA fingerprinting systems, and recommendations for a minimal standard SNP set. J Bacteriol. 2006;188:759–772. doi: 10.1128/JB.188.2.759-772.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Asiimwe BB, Koivula T, Kallenius G, Huard RC, Ghebremichael S, et al. Mycobacterium tuberculosis Uganda genotype is the predominant cause of TB in Kampala, Uganda. Int J Tuberc Lung Dis. 2008;12:386–391. [PubMed] [Google Scholar]
- 49.Kim EY, Nahid P, Hopewell PC, Kato-Maeda M. Novel hot spot of IS6110 insertion in Mycobacterium tuberculosis. J Clin Microbiol. 2010;48:1422–1424. doi: 10.1128/JCM.02210-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Caminero JA, Pena MJ, Campos-Herrero MI, Rodriguez JC, Garcia I, et al. Epidemiological evidence of the spread of a Mycobacterium tuberculosis strain of the Beijing genotype on Gran Canaria Island. Am J Respir Crit Care Med. 2001;164:1165–1170. doi: 10.1164/ajrccm.164.7.2101031. [DOI] [PubMed] [Google Scholar]
- 51.Kiers A, Drost AP, van Soolingen D, Veen J. Use of DNA fingerprinting in international source case finding during a large outbreak of tuberculosis in The Netherlands. Int J Tuberc Lung Dis. 1997;1:239–245. [PubMed] [Google Scholar]
- 52.Ruddy MC, Davies AP, Yates MD, Yates S, Balasegaram S, et al. Outbreak of isoniazid resistant tuberculosis in north London. Thorax. 2004;59:279–285. doi: 10.1136/thx.2003.010405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jenkins C. Rifampicin resistance in tuberculosis outbreak, London, England. Emerg Infect Dis. 2005;11:931–934. doi: 10.3201/eid1106.041262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lillebaek T, Dirksen A, Kok-Jensen A, Andersen AB. A dominant Mycobacterium tuberculosis strain emerging in Denmark. Int J Tuberc Lung Dis. 2004;8:1001–1006. [PubMed] [Google Scholar]
- 55.Kruuner A, Hoffner SE, Sillastu H, Danilovits M, Levina K, et al. Spread of drug-resistant pulmonary tuberculosis in Estonia. J Clin Microbiol. 2001;39:3339–3345. doi: 10.1128/JCM.39.9.3339-3345.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yeh RW, Ponce de Leon A, Agasino CB, Hahn JA, Daley CL, et al. Stability of Mycobacterium tuberculosis DNA genotypes. J Infect Dis. 1998;177:1107–1111. doi: 10.1086/517406. [DOI] [PubMed] [Google Scholar]
- 57.Warren RM, van der Spuy GD, Richardson M, Beyers N, Booysen C, et al. Evolution of the IS6110-based restriction fragment length polymorphism pattern during the transmission of Mycobacterium tuberculosis. J Clin Microbiol. 2002;40:1277–1282. doi: 10.1128/JCM.40.4.1277-1282.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wall S, Ghanekar K, McFadden J, Dale JW. Context-sensitive transposition of IS6110 in mycobacteria. Microbiology. 1999;145(Pt 11):3169–3176. doi: 10.1099/00221287-145-11-3169. [DOI] [PubMed] [Google Scholar]
- 59.Borrell S, Gagneux S. Infectiousness, reproductive fitness and evolution of drug-resistant Mycobacterium tuberculosis. Int J Tuberc Lung Dis. 2009;13:1456–1466. [PubMed] [Google Scholar]
- 60.Gagneux S, DeRiemer K, Van T, Kato-Maeda M, de Jong BC, et al. Variable host-pathogen compatibility in Mycobacterium tuberculosis. Proc Natl Acad Sci U S A. 2006;103:2869–2873. doi: 10.1073/pnas.0511240103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nikolayevskyy VV, Brown TJ, Bazhora YI, Asmolov AA, Balabanova YM, et al. Molecular epidemiology and prevalence of mutations conferring rifampicin and isoniazid resistance in Mycobacterium tuberculosis strains from the southern Ukraine. Clin Microbiol Infect. 2007;13:129–138. doi: 10.1111/j.1469-0691.2006.01583.x. [DOI] [PubMed] [Google Scholar]
- 62.Gagneux S, Burgos MV, DeRiemer K, Encisco A, Munoz S, et al. Impact of bacterial genetics on the transmission of isoniazid-resistant Mycobacterium tuberculosis. PLoS Pathog. 2006;2:e61. doi: 10.1371/journal.ppat.0020061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Alland D, Lacher DW, Hazbon MH, Motiwala AS, Qi W, et al. Role of large sequence polymorphisms (LSPs) in generating genomic diversity among clinical isolates of Mycobacterium tuberculosis and the utility of LSPs in phylogenetic analysis. J Clin Microbiol. 2007;45:39–46. doi: 10.1128/JCM.02483-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tsolaki AG, Hirsh AE, DeRiemer K, Enciso JA, Wong MZ, et al. Functional and evolutionary genomics of Mycobacterium tuberculosis: insights from genomic deletions in 100 strains. Proc Natl Acad Sci U S A. 2004;101:4865–4870. doi: 10.1073/pnas.0305634101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bifani PJ, Mathema B, Liu Z, Moghazeh SL, Shopsin B, et al. Identification of a W variant outbreak of Mycobacterium tuberculosis via population-based molecular epidemiology. JAMA. 1999;282:2321–2327. doi: 10.1001/jama.282.24.2321. [DOI] [PubMed] [Google Scholar]
- 66.Kato-Maeda M, Rhee JT, Gingeras TR, Salamon H, Drenkow J, et al. Comparing genomes within the species Mycobacterium tuberculosis. Genome Res. 2001;11:547–554. doi: 10.1101/gr166401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rhee JT, Piatek AS, Small PM, Harris LM, Chaparro SV, et al. Molecular epidemiologic evaluation of transmissibility and virulence of Mycobacterium tuberculosis. J Clin Microbiol. 1999;37:1764–1770. doi: 10.1128/jcm.37.6.1764-1770.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Valway SE, Sanchez MP, Shinnick TF, Orme I, Agerton T, et al. An outbreak involving extensive transmission of a virulent strain of Mycobacterium tuberculosis. N Engl J Med. 1998;338:633–639. doi: 10.1056/NEJM199803053381001. [DOI] [PubMed] [Google Scholar]
- 69.Caws M, Thwaites G, Stepniewska K, Nguyen TN, Nguyen TH, et al. Beijing genotype of Mycobacterium tuberculosis is significantly associated with human immunodeficiency virus infection and multidrug resistance in cases of tuberculous meningitis. J Clin Microbiol. 2006;44:3934–3939. doi: 10.1128/JCM.01181-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hanekom M, van der Spuy GD, Streicher E, Ndabambi SL, McEvoy CR, et al. A recently evolved sublineage of the Mycobacterium tuberculosis Beijing strain family is associated with an increased ability to spread and cause disease. J Clin Microbiol. 2007;45:1483–1490. doi: 10.1128/JCM.02191-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Reed MB, Domenech P, Manca C, Su H, Barczak AK, et al. A glycolipid of hypervirulent tuberculosis strains that inhibits the innate immune response. Nature. 2004;431:84–87. doi: 10.1038/nature02837. [DOI] [PubMed] [Google Scholar]
- 72.ten Bokum AM, Movahedzadeh F, Frita R, Bancroft GJ, Stoker NG. The case for hypervirulence through gene deletion in Mycobacterium tuberculosis. Trends Microbiol. 2008;16:436–441. doi: 10.1016/j.tim.2008.06.003. [DOI] [PubMed] [Google Scholar]
- 73.Ghebremichael S, Groenheit R, Pennhag A, Koivula T, Andersson E, et al. Drug resistant Mycobacterium tuberculosis of the Beijing genotype does not spread in Sweden. PLoS One. 2010;5:e10893. doi: 10.1371/journal.pone.0010893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Park JS, Tamayo MH, Gonzalez-Juarrero M, Orme IM, Ordway DJ. Virulent clinical isolates of Mycobacterium tuberculosis grow rapidly and induce cellular necrosis but minimal apoptosis in murine macrophages. J Leukoc Biol. 2006;79:80–86. doi: 10.1189/jlb.0505250. [DOI] [PubMed] [Google Scholar]
- 75.Marquina-Castillo B, Garcia-Garcia L, Ponce-de-Leon A, Jimenez-Corona ME, Bobadilla-Del Valle M, et al. Virulence, immunopathology and transmissibility of selected strains of Mycobacterium tuberculosis in a murine model. Immunology. 2009;128:123–133. doi: 10.1111/j.1365-2567.2008.03004.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Raynaud C, Guilhot C, Rauzier J, Bordat Y, Pelicic V, et al. Phospholipases C are involved in the virulence of Mycobacterium tuberculosis. Mol Microbiol. 2002;45:203–217. doi: 10.1046/j.1365-2958.2002.03009.x. [DOI] [PubMed] [Google Scholar]
- 77.Viana-Niero C, de Haas PE, van Soolingen D, Leao SC. Analysis of genetic polymorphisms affecting the four phospholipase C (plc) genes in Mycobacterium tuberculosis complex clinical isolates. Microbiology. 2004;150:967–978. doi: 10.1099/mic.0.26778-0. [DOI] [PubMed] [Google Scholar]
- 78.Ryoo SW, Park YK, Park SN, Shim YS, Liew H, et al. Comparative proteomic analysis of virulent Korean Mycobacterium tuberculosis K-strain with other mycobacteria strain following infection of U-937 macrophage. J Microbiol. 2007;45:268–271. [PubMed] [Google Scholar]
- 79.West NP, Chow FM, Randall EJ, Wu J, Chen J, et al. Cutinase-like proteins of Mycobacterium tuberculosis: characterization of their variable enzymatic functions and active site identification. FASEB J. 2009;23:1694–1704. doi: 10.1096/fj.08-114421. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Complete list of LSPs compared to H37Rv.
(XLS)
Complete list of DIPs compared to H37Rv.
(XLS)
Complete list of SNPs compared to H37Rv.
(XLS)