Abstract
Many fish have, in addition to IgM and IgD, a third isotype called IgZ or IgT. The ζ-chain locus is embedded among the Ig heavy chain V-, D- and J-elements in a manner reminiscent of the TcR δ/α locus. Isotype selection thus occurs during VDJ recombination, a process that is facilitated by intralocus transcription. Using in silico analyses and enhancer reporter vectors we identified 3 new regions within the zebrafish IgH locus through which transcription can be activated in catfish B-cell lines. Two of these, termed Eζi (Jζ to Cζ1 intronic) and Eζ3′ regions flank the ζ-chain constant domain exons. A third region, Eδ3′, resides downstream of the δ-chain exons. All regions contain predicted binding sites for transcription factors that contribute to B-cell specific transcription in fish and mammals. Each region also has proximal matrix attachment regions, which may further contribute to transcriptional activation and chromatin remodeling. We discuss possible roles for these regions during VDJ recombination.
1. Introduction
Mammalian immunoglobulin (Ig) genes contain multiple transcriptional enhancers and cryptic promoters throughout the locus. Transcriptional regulation through Ig gene enhancers and promoters confer cell-type specific production of mRNA, and also regulate the modifications that occur to immunoglobulin genes during the course of B-cell development and activation. VDJ recombination, somatic hypermutation (SH), class switch recombination, and gene conversion are all preceded by production of sterile transcripts across the region of the gene being modified. Sterile transcripts appear only to be an unutilized by-product of a process that may make the transcribed region accessible to chromatin remodeling enzymes, RAG recombinases or the Ig mutator AID [reviewed in (Abarrategui and Krangel, 2009; Bolland et al., 2009; Chaudhuri and Alt, 2004)]. Among the transcriptional regulators driving sterile transcription perhaps the best characterized is the Ig heavy chain (IgH) JH to Cμ1 intronic enhancer, Eμ. Eμ is centrally located in the locus at the one location that is normally exempt from deletion during either VDJ or class switch recombination. The constituent components of Eμ transcriptional regulation have been extensively studied [reviewed in (Calame and Sen, 2004)], and involve a complex array of transcriptional activators, suppressors and scaffold binding proteins that jointly confer B-cell specificity to the enhancer. Among the core transcription factors involved are Oct1/2, E2A (E12 and E47), Ets-1, TFE3, YY1 and PU.1, and many of these work synergistically within the context of Eμ. In non-B-cells the transcription factors may be inactivated through dimerization with negative regulators such as Id (Calame and Sen, 2004), or the binding sites may be occupied by suppressors. Additional regulation of this enhancer may be conferred through the flanking matrix/scaffold attachment regions (MARs), which are AT-rich regions that can bind scaffold-binding proteins such as the B-cell regulator of IgH transcription – Bright. MARs can also be bound by transcriptional repressors such as the special AT-binding proteins (SATB) or the CCAAT-displacement protein (CDP). MARs have been shown to extend the accessibility of the Eμ enhancer over large distances in B-cells (Jenuwein et al., 1997), which could facilitate the production of sterile transcripts at various locations within the locus.
The recognition that Eμ had been deleted from the productive IgH locus of an antibody expressing B-cell line led to the discovery of additional regulatory regions 3′ of the locus (Pettersson et al., 1990). The four enhancers 3′ of the locus (α3′) span nearly 200 kb and appear to play a role in class switch recombination and in antibody production by plasma cells [reviewed in (Khamlichi et al., 2000)] These α3′ enhancers, as well as those associated with the two light chain loci, employ many of the same transcription factor binding sites as Eμ.
Until fairly recently it was believed that fish possessed just a simplified form of the mammalian IgH locus, with multiple V-, D- and J-elements upstream of exons encoding μ- and δ-chains, but lacking downstream isotype clusters and associated class switch capabilities. We previously identified a single enhancer (Eμ3′) between the μ- and δ-chain genes of the channel catfish IgH locus that is B-cell specific when tested in transient transfection transcription reporter assays in both fish and mammalian cell lines (Magor et al., 1994). The Eμ3′ enhancer has many of the same transcription factor binding sites as the mammalian Ig enhancers, including E2A, Oct1/2, PU.1 and TFE3 binding sites (Magor et al., 1997). While the location of the enhancer 3′ of the μ-chain would not be compatible with the evolution of class switching (Magor et al., 1999), it is reasonably well positioned to perform other functions equivalent to the mammalian Eμ enhancer. Zebrafish also have a functional Eμ3′ enhancer in their IgH locus (Ellestad and Magor, 2005) and there is some evidence that equivalent enhancers exist in the IgH locus of other bony fishes (Ellestad and Magor, 2005; Hikima et al., 2006a).
In the last decade genome sequencing has revealed that the IgH locus of fish is more complex than earlier realized. The IgH locus of cyprinids (Danilova et al., 2005), salmonids (Danilova et al., 2005; Hansen et al., 2005) and pufferfishes (Danilova et al., 2005; Savan et al., 2005) all have a third heavy chain encoding exon cluster embedded amongst the V-, D-, and J-elements. This third isotype has been dubbed IgZ, for it’s discovery in zebrafish (Danilova et al., 2005), or IgT, for being apparently unique to ‘Teleost’ fishes (Hansen et al., 2005). IgZ and IgT appear to be orthologs that are common to many, but not all teleost fishes examined thus far (Bengten et al., 2006). The organization of the Ig heavy chain in fishes having the ζ/τ-chain exons has converged on that of the TcR δ/α locus. The 5′ end of the fish IgH locus has common VH-elements followed by Dζ- and Jζ-elements and then the ζ-chain constant domain exons. Downstream of the ζ-chain is a second set of D- and J-elements followed by the μ- and δ-constant domain exons. Isotype selection, as with the TcR δ/α locus, will then be dependent on alternative VDJ recombination, rather than alternative pre-mRNA splicing (as for μ- and δ-chains) or bona fide class switch recombination as occurs in tetrapods.
It is not clear whether expression of IgZ versus IgM/D is based on specific stimuli, developmental programs, or is simply stochastic selection of recombining V-, D- and J-elements. In the original description of IgZ in zebrafish it was observed that ζ-chain expression preceded that of μ-chain during early development (Danilova et al., 2005). However the orthologs of the ζ-chain in carp, fugu and salmonids appear to begin expression at the same time as μ-chain (Hansen et al., 2005; Ryo et al., 2010; Savan et al., 2005). In the TcR δ/α locus expression of δ- versus α-chain is clearly developmentally regulated through differential activation of stage specific enhancers associated with the δ-chain and α-chain exons, that drive sterile transcription across those V-, D- and J- elements that will be recombined [reviewed in (Krangel, 2009)].
We hypothesized that if expression of IgZ versus IgM or IgD is regulated, then there should be transcriptional regulatory regions proximal to the ζ-chain exons or the upstream Dζ- and Jζ-elements. With the availability of improved search tools and genomic data we sought out additional (to the Eμ3′ enhancer) transcriptional regulatory regions that might regulate which isotype is expressed in a given B-cell. Using an established enhancer reporter vector and transient transfection system using fish lymphocyte lines, we have confirmed that several additional predicted transcriptional enhancers are active in fish B-cells.
2. Methods and Materials
2.1 Programs for in Silico analyses of predicted regulatory regions
There are a number of web based prediction tools of transcription factor binding sites, and these often use different prediction algorithms and binding site matrices to determine possible sites. Initial scans of the zebrafish IgH locus were done using MatInspector (Cartharius et al., 2005). Potentially conserved regulatory regions were analyzed among the IgH locus of: zebrafish (Danio rerio – accession #’s BX649502 and BX510335), salmonids (Salmo salar – accession # GU129139 and Oncorhynchus mykiss – accession # AY872256S1), and pufferfishes (Fugu rubripes – accession # CAAB01001037 and Tetraodon nigroviridis – accession # BX629354). zPicture (Ovcharenko et al., 2004) and GLAM (gapless local alignment of multiple sequences; (Frith et al., 2004) were used to look for conserved sequence alignments among the fish IgH locus, while FrameWorker (Cartharius et al., 2005) was used to search for conserved order of predicted transcription factor binding sites.
Subsequent analysis of functional transcriptional regulatory regions employed the programs Promo (Farre et al., 2003) and TESS (Schug, 2008), allowing for inclusion of non-canonical binding sites. Predicted sites were then edited to remove non-canonical motifs that are known not to be functional, and inclusion of motifs known to be functional in fish and mammalian IgH regulatory regions (Cioffi et al., 2001; Ellestad and Magor, 2005; Ghosh, 1990; Hikima et al., 2006b).
The regions within or proximal to the functional enhancers were analyzed for the presence of scaffold/matrix attachment regions using the commercial S/MARtest (Genomatix; (Frisch et al., 2002).
2.2 Enhancer Constructs
A modified pGL3 firefly luciferase reporter (pGL3-FPr-FPL) containing a goldfish VH promoter and a polylinker from pBluescript was used for testing potential enhancer regions. The fish VH promoter (bases 23 to 199 of genbank X61312) was Pfu amplified with linkered primers (forward 5′-CCA TTA ATC CCA GTT CCA TGG TTT CC-3′; reverse 5′-CAC CGG TCA GCC AAA AAC AGT CAA AGG-3′) and blunt end cloned into klenow filled ends from Acc65 I and Hind III digested pGL3-basic (Promega). The pBluescript II KS(+) polylinker and LacZ gene were then Pfu amplified (5′-GTG AAC CAT CAC CCT AAT CAA G-3′ and 5′-CGT ATT ACC GCC TTT GAG TG-3′) and blunt end cloned into the pGL3-FPr vector that had been digested with Sal I and then blunted by klenow fill-in. In the forward polylinker (FPL) version (pGL3-FPr-FPL) the insert is in the same 5′ to 3′ orientation as the vector.
Regions of the zebrafish IgH locus containing clusters of potential transcription factor binding sites were amplified from genomic DNA, sub-cloned into pCR2.1 (InVitrogen) and then directionally cloned into the polylinker of pGL3-FPr-FPL. The regions examined for enhancer activity are tabulated below (Table 1). The primers used for sub-cloning are represented by the approximately 20 nucleotides on the flanks of the indicated sequences (Table 1). The long stretch of transcription factor binding sites downstream of the ζ-chain exons was divided into 4 discrete regions for testing. Individual testing of putative enhancer regions allowed us to roughly maintain the size, and thus transfection efficiency of the enhancer reporter constructs.
Table 1.
Relative positions of the tested enhancers within the IgH locus
| Feature | Position in BX649502a |
|---|---|
| Eζi enhancer | 238,487 to 240,951 |
| 5′ end of Cζ1 exon | 241,016 |
| 3′ end of Cζ4 exon | 242,871 |
| Eζ3′-1 enhancer | 242,976 to 246,593 |
| Eζ3′-2 enhancer | 246,754 to beyond end |
| Position in BX510335 | |
| Eζ3′-2 enhancer | 1,368 to 3,273 |
| Eζ3′-3 enhancer | 4,436 to 4,920 |
| Eζ3′-4 enhancer | 6,672 to 9,733 |
| 5′ most Dμ-element | 13,141 |
| 3′ end of Cμ4 exon | 27,814 |
| Eμ3′ enhancer | 30,015 to 31,866 |
| δ-chain transmembr exon | 43,481 |
| Eδ3′ enhancer | 44,335 to 47,766 |
Genbank accession number.
Control constructs included the catfish and zebrafish Eμ3′ enhancers (fragments 11 and 6 respectively; (Ellestad and Magor, 2005; Magor et al., 1994) cloned into pGL3-FPr-FPL. The identities and integrity of all reporter vector inserts were verified by sequencing.
2.3 Cell Lines and Transfection
Currently the only B-cell lines available from non-homeothermic vertebrates are from the channel catfish (Ictalurus punctatus), which have IgM and IgD, but apparently not an IgZ ortholog (Bengten et al., 2006). We used two B-cell lines representing a mature naïve phenotype (IgM+/IgD+; 3B11) and a post-activation phenotype (IgM+/IgD−; 1B10) as have been previously described (Miller et al., 1994b). The maintenance (Miller et al., 1994a) and electroporation parameters (Ellestad and Magor, 2005) for these cells have been described, with the exception that the cell lines were maintained using carp serum in place of catfish serum. Each transfection included 3.5 pmol of experimental reporter plasmid, 0.8 pmol of the constitutively active Renilla luciferase reporter vector phRG-TK (Promega) and carrier DNA (pBluescript plasmid) to a final mass of 20 μg total DNA. Transfections were typically done in triplicates (per experimental construct), and each construct plasmid was prepared using commercial plasmid prep kits (Qiagen or Marligen) on at least two independent occasions. Cells were harvested 36 to 48 hours after transfection and processed for the dual luciferase assay according to manufacturers protocol (Promega). Luciferase was measured on a GloMax 20/20 luminometer (Promega). To normalize for differences in transfection efficiencies each experimental firefly luciferase value was divided by the Renilla luciferase value from the background plasmid. The previously characterized catfish IgH enhancer, Eμ3′ (ELF11 version; (Magor et al., 1994) was used as a reference standard for transcriptional activation. The zebrafish Eμ3′ enhancer (Ellestad and Magor, 2005) was included in some transfection series for comparison purposes.
3. Results
3.1 Putative enhancers are only predicted through clustering of transcription factor binding sites
A variety of comparative genomics approaches were taken to identify possible additional (to the Eμ3′ enhancer) sites directing transcriptional regulation within the IgH locus of pufferfishes, salmonids and cyprinids. None of the gapless alignment of multiple sequences (GLAM), zpicture alignments, nor searches for conserved order or context of transcription factors (FrameWorker) identified probable regulatory regions that were conserved among the fishes examined. However the equivalent regions of the IgH locus of these fishes did have clusters of transcription factor binding sites (as predicted by MatInspector) of the sort found in Ig transcriptional regulatory regions (data not shown).
In addition to the previously analyzed zebrafish Jμ to Cμ1 intron and Eμ3′ enhancers, 5 other non-coding regions within the zebrafish IgH locus were found to have concentrations of potential transcription factor binding sites including: 3′ of the Dζ-elements; the Jζ to Cζ1 intron; 3′ of the ζ-chain transmembrane exons; among the 3′ Dμ-elements; 3′ of the δ-chain transmembrane exons. The putative transcriptional regulatory regions associated with the D-elements are likely cryptic promoters analogous to the DQ52 promoter in the mammalian IgH locus. We restricted our functional analyses to the three putative enhancer regions.
3.2 The zebrafish IgH locus has multiple transcriptional regulatory regions
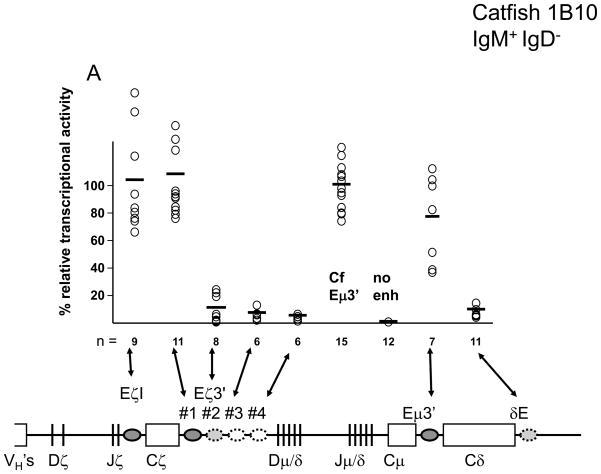
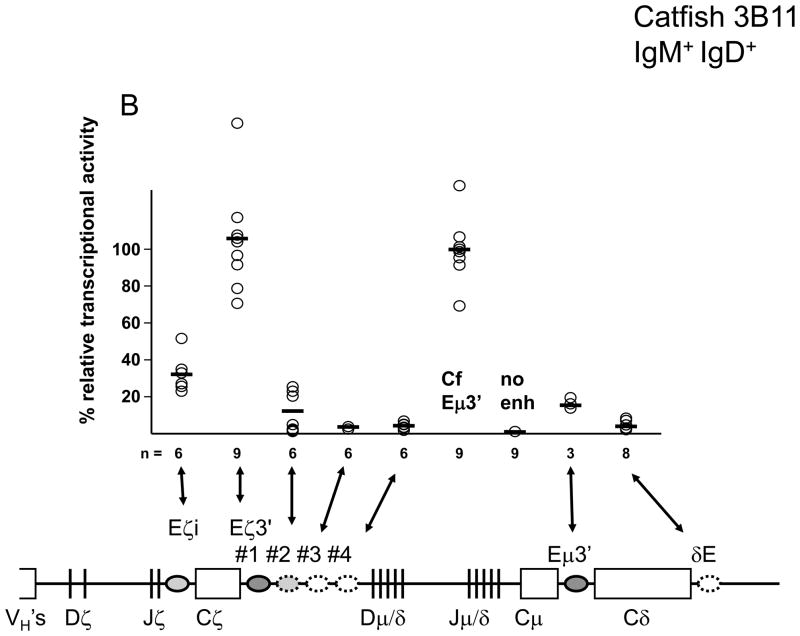
Of the six putative enhancer regions associated with the ζ- and δ-chain genes only the ζ-chain intronic enhancer (Eζi) and the first region downstream of the ζ-chain (Eζ3′-1) had strong transcriptional activity in the catfish 1B10 B-cell line (IgM+/IgD−; Fig. 1). In all cases the transcriptional activity of these ζ-chain enhancers was higher than the previously characterized zebrafish Eμ3′ enhancer which we believe controls μ-chain transcription (Ellestad and Magor, 2005). Somewhat surprisingly the zebrafish Eζi and Eμ3′ enhancers had considerably lower transcriptional activity when transfected into a cell line with a naïve B-cell phenotype (3B11; IgM+/IgD+; Fig. 1). At least some of the replicate transfections with these two reporter constructs were done using the same plasmid preparations as those used to transfect the 1B10 cells, suggesting that the low transcriptional activity was not due to a feature of the plasmids purification.
Fig. 1.
Transcriptional activities of IgH enhancers in the transiently transfected catfish B-cell lines (A) 1B10 (IgM+/IgD−), and (B) 3B11 (IgM+/IgD+). Luciferase activities from the pGL3 reporter containing the indicated enhancers were normalized against a background reporter, and then scaled relative to the mean activity generated from the catfish Eμ3′ enhancer. Individual transfections are represent by circles and mean values by bars.
In addition to the strong ζ-chain associated enhancers, two regions of the locus had modest transcriptional activity. A region downstream of Eζ3′-1 (Eζ3′-2), and a region downstream of the δ-chain, Eδ3′, both had transcriptional activities that were five to ten times lower than the Eζi or catfish Eμ3′ enhancers. The transcriptional enhancement by Eδ3′ and Eζ3′-2 were nevertheless 10 to 20 times the activity seen with the reporter containing just the fish VH-promoter.
Two other regions containing clusters of putative transcription factor binding sites (Eζ3′-3 and -4) conferred no appreciable transcriptional enhancer in either catfish B-cell line and were not analyzed further.
3.3 Composition of the zebrafish IgH enhancers
Once we established which regions were active transcriptional enhancers we revisited our attempts to find conserved features among the equivalent regions of the IgH locus of zebrafish, pufferfish and salmon. We also looked for similarities between the zebrafish Eu3′ and Eζ3′ enhancers. Even with very low stringency searches of conserved sequence regions or conservation of motifs we did not detect conserved sequence.
MARs are found proximal to several Ig gene transcriptional enhancers and VH promoters, and their presence appears to contribute to transcriptional regulation of Ig genes, as well as to somatic hypermutation and class switch recombination. The Genomatix S/MARtest algorithm detected MARs in proximity to all of the four functional transcriptional regulatory regions that we tested (Eζi, Eζ3′-1, Eζ3′-2, and Eδ3′; Table 2). In addition to the MAR upstream of the Eζi enhancer, which would be deleted following DζJζ recombination, there were 22 predicted binding sites for three MAR binding proteins, CDP, SATB1 and Bright, scattered throughout the Eζi enhancer (not shown). This would compare to the 26 predicted binding sites for CDP, SATB1 or Bright clustered within the two MARs identified within the Eζ3′-1 enhancer (data not shown). Bright would be a positive regulator of Ig transcription in B-cells, while CDP and SATB would be anticipated to repress Ig transcription in non-B-cells (Goebel et al., 2002; Liu et al., 1997; Wang et al., 1999).
Table 2.
Predicted Matrix Attachment Regions (MARs) proximal to the Ig heavy chain transcriptional enhancers tested
| Enhancer | MAR location (5′ end of MAR) | MAR size |
|---|---|---|
| Eζi | 1255 bp 5′ of Eζi | 475 bp |
| Eζ3′-1 | Bases 331 to 1375 of Eζ3′-1 | 1045 bp |
| Bases 1711 to 2680 of Eζ3′-1 | 970 bp | |
| Eζ3′-2 | 3988 bp 3′ of Eζ3′-2a | 335 bp |
| 5953 bp 3′ of Eζ3′-2a | 555 bp | |
| Eδ3′ | 2528 bp 3′ of enhancer Eδ3′ | 350 bp |
this MAR is contained within the putative enhancer Eζ3′-3
3.4 Putative enhancer motifs within the zebrafish IgH enhancers
Several algorithms and transcription factor binding matrices were used to predict possible transcription factor binding sites within the functional zebrafish enhancers. We focused on transcription factors that are known to be important for regulation of the Ig locus in mammals, as well as those specifically known to be functional in the context of the catfish and zebrafish IgH Eμ3′ enhancers. Previous studies have demonstrated that there is cross-species cell type specific transcriptional activity from the fish Eμ3′ enhancers in mammalian cells, and of the murine Eμ enhancer in fish cells (Ellestad and Magor, 2005; Magor et al., 1994; Magor et al., 1997). This is consistent with the binding sites of the respective transcription factor homologues being relatively well conserved between fish and mammals. This notion has been corroborated at least in part by the cloning of fish homologues of three Octamer binding transcription factors (Oct1, Oct2a and Oct2b) and three E-box binding proteins (CFEB1, CFEB2 and E2A1) from the channel catfish (Hikima et al., 2005; Lennard et al., 2006; Lennard et al., 2007; Ross et al., 1998). In each case these transcription factors bound nucleotide motifs similar to their mammalian counterparts, and could also reconstitute expression of reporter constructs when co-transfected in otherwise non-permissive cell lines (Hikima et al., 2005; Ross et al., 1998).
Each of the four transcriptionally active enhancers that we tested had at least two known functional (in fish Eμ3′ enhancers) transcription factor binding sites (Tables 3–5). With one exception, the transcriptional activity of the enhancers correlated with the number of E-box motifs and Oct binding sites. Eζ3′-1 was the strongest transcriptional activator and has 9 canonical E-box motifs (E2A and its products E12 and E47, as well as muEBP-C2 and the TFE3 site), including six which are known to be active. Eζ3′-1 also has six potential binding sites for Oct2, of which two are known to be active binding sites. The Eζi enhancer had only 5 canonical E-box motifs, but 9 potential Oct2 binding sites, of which one is known to be functional (Table 4). The Eδ3′, which had relatively low transcriptional activity nevertheless has 11 canonical E-box motifs, of which one is known to be utilized in fish, and three Oct2 binding sites, two of which are active canonical motifs (Table 5). Conversely Eζ3′-2 which has equivalent transcriptional activity to Eδ3′ has only one E-box and one Oct2 motif, both of which are known to be utilized in the Eμ3′ enhancer (Table 5).
Table 3.
Predicted transcription factor binding sites in the Eζ3′-1 enhancer
| Factor [Accession#]a | 5′ start in the Eζ3′-1 enhancer | Observed motifb |
|---|---|---|
| c-Ets-1 [T00112] | 72 | TTTCCCT |
| E2A [R02139] | 82 | tCAGATGc |
| Oct-2 [R02226] | 363 | ATtCAAAT |
| Oct-2 [R02226] | 803 | ATGCAAAc |
| c-Ets-1 [T00112] | 831 | ATGGAAA |
| E2A [R02139] | 898 | tCAGATG |
| Oct-2 [R02226] | 979 | ATTTGaAT |
| E2A [R02139] | 1031 | GCAGATG |
| E12 [T00204] | 1051 | CAGAGCAGGTGCA |
| c-Ets-1 [T00112] | 1072 | AAGGAAA |
| c-Ets-1 [T00112] | 1292 | GTTCCAA |
| muEBP-C2 [T00215] | 1379 | CATGTG |
| PU.1 [T02068] | 1421 | GAGGAATTTC |
| c-Ets-1 [T00111] | 1451 | TTCGGAAGCA |
| E12 [T00204] | 1498 | TACACCTGCAGAA |
| Oct-2 [R02226] | 1588 | ATaCAAAT |
| TFE3 [R02252] | 1820 | TCATtTGGCA |
| Oct-2 [R02226] | 1842 | ATGaAAAT |
| Oct-2 [R02226] | 1866 | ATGCAcAT |
| muEBP-C2 [T00215] | 1929 | CACATG |
| c-Ets-1 [T00112] | 2121 | GTGGAAC |
| c-Ets-1 [T00112] | 2308 | TTGGAAA |
| E2A [R02139] | 2558 | cCAGATG |
Accession numbers in Transfac database
small letters are non-canonical nucleotides in the enhancer sequence
bold letters are motifs whose core has been shown to be functional in fish
Table 5.
Predicted transcription factor binding sites in the Eδ3′ and Eζ3′-2 enhancers
| Factor [Accession#]a | 5′ start in the Eδ3′ Enhancer | Observed motifb |
|---|---|---|
| Oct-2 [T00646] | 94 | ATTTTCAT |
| E47 [T00207] | 133 | ACAGCTGa |
| E47 [T00207] | 267 | ACAGCTGG |
| muEBP-C2 [T00215] | 356 | CACATG |
| E47 [T00207] | 1260 | CCAGCTGGc |
| muEBP-C2 [T00215] | 1514 | CACATG |
| E12 [T00204] | 2097 | GCAGATG |
| TFE3 [T00810] | 2258 | GAAGTCATGTGGGC |
| Oct-2 [T00646] | 3020 | ATGGAAATc |
| muEBP-C2 [T00215] | 3045 | CATGTG |
| E12 [T00204] | 3217 | AGCTGCAGGTGAC |
| muEBP-C2 [T00215] | 3299 | CACATG |
| Oct-2 [T00646] | 3327 | ATGTAAAT |
| muEBP-C2 [T00215] | 3339 | CATGTG |
| Oct-2 [T00646] | 3341 | ATgTGCAT |
| 5′ start in the Eζ3′-2 Enhancer | ||
| E47 [T05421] | 26 | GCAGGTGTGC |
| PU.1 [T02068] | 126 | AgGGTTCCTC |
| Oct-2 [T00648] | 275 | ATGCAAATCCT |
| c-Ets-1 [T00112] | 359 | TTTCCTG |
| c-Ets-1 [T00111] | 942 | tTCTTCCTGG |
| c-Ets-1 [T00111] | 947 | CCTGGAAGTG |
| c-Ets-1 [T00111] | 1205 | AGCTTCCGCC |
| c-Ets-1 [T00112] | 1385 | CAGGAAT |
Accession numbers in Transfac database
small letters are non-canonical nucleotides in the enhancer sequence
bold letters are motifs whose core has been shown to be functional in fish
Table 4.
Predicted transcription factor binding sites in the Eζi enhancer
| Factor [Accession#]a | 5′ start in the Eζi enhancer | Observed motifb |
|---|---|---|
| c-Ets-1 [T00112] | 163 | TAGGAAG |
| Oct 2 [T00648] | 230 | ATGCAAAcAAT |
| Oct 2 [T00648] | 523 | ATGCAAAAAAG |
| Oct 2 [R02226] | 558 | ATTTtCAT |
| PU.1 [R02238] | 575 | TTCCTC |
| c-Ets-1 [T00112] | 573 | TTTCCTC |
| Oct 2 [R02226] | 682 | ATGCcAAT |
| E47 [T05421] | 741 | ACAGGTGCACc |
| Oct 2 [R02226] | 754 | ATaTGCAT |
| Oct 2 [R02226] | 941 | tTGCAAAT |
| c-Ets-1 [T00112] | 1220 | TAGGAAA |
| c-Ets-1 [T00112] | 1449 | GTGGAAA |
| c-Ets-1 [T00112] | 1459 | TTTCCAA |
| Oct 2 [R02226] | 1537 | ATGaAAAT |
| c-Ets-1 [T00112] | 1581 | CTTCCAG |
| muEBP-C2 [T00215] | 1750 | CACATG |
| TFE3 [R02139] | 1817 | CACTTG |
| E47 [T00207] | 1815 | cCATCTG |
| c-Ets-1 [T00112] | 1879 | TTGGAAA |
| Oct 2 [R02226] | 1924 | AcGCAAAT |
| TFE3 [R02139] | 1929 | GCAACTGTG |
| muEBP-C2 [T00215] | 2114 | CACATG |
| Oct 2 [R02226] | 2118 | ATGCAAAa |
| YY1 [T00278] | 2359 | CCATCCT |
| c-Ets-1 [T00112] | 2441 | TTTCCAA |
Accession numbers in Transfac database
small letters are non-canonical nucleotides in the enhancer sequence
bold letters are motifs whose core has been shown to be functional in fish
In the mouse Eμ enhancer the coordinated bind of 4 sites within the enhancer core is sufficient to confer cell type specific transcriptional activity (Nikolajczyk et al., 1997). These sites, μA (bound by Ets-1), μB (bound by PU.1) and μE3 (bound by TFE3) have specific relative spatial requirements, in order for there to be active transcriptional activity, though these spatial requirements have only been assessed within the context of the Eμ enhancer, and some functional redundancy has been noted among contributing transcription factors (Dang et al., 1998). Canonical binding sites for all PU.1, Ets-1 and TFE3 are found in each of the functional zebrafish enhancers with the exception of Eζ3′- 2 which appears to be lacking a TFE3 binding site and Eδ3′ which lacks sites for Ets-1 and PU.1. In addition to the listed putative binding sites there are also numerous canonical binding sites for C/EBP binding proteins and for YY1 in each of the enhancers (data not shown).
4. Discussion
We report here that in addition to the Eμ3′ enhancer that is attributed with controlling expression of the μ- and δ-chains, there are at least 3 other regions of the IgH locus containing potential transcriptional regulatory regions. Flanking the 3′ end of the locus is a relatively weak enhancer that we’ve called Eδ3′. Given the number of canonical or known active transcription factor binding sites that Eδ3′ contains at its 3′ end, we might have anticipated greater transcriptional activity. However this enhancer did lack binding sites for Ets-1 and PU.1, which can act synergistically with TFE3 to confer B-cell specific activity (Nikolajczyk et al., 1997). Thus Eδ3′ may need to act synergistically with other enhancers within the locus, perhaps aided by the matrix attachment region located 2.5 kb downstream of Eδ3′. While some insights might be gained by extending the enhancer regions, elucidating the true contributions of Eδ3′ (and other enhancers) will have to await development of technologies that allow long nucleotide deletions from fish genomes. The other two regions of the zebrafish IgH locus that were demonstrated to harbor putative transcriptional regulatory regions flank the ζ-chain constant domain (and transmembrane) exons. In order to keep the reporter plasmids at an optimal size the region downstream of the ζ-chain was broken into four regions containing clusters of possible transcription factor binding sites. Of these regions the most ζ-chain proximal segment, Eζ3′-1, had by far the strongest transcriptional activity. Eζ3′-2 also had measurable transcriptional activity, while the more distal sites, Eζ3′-3 and -4, had nominal if any transcription activating activity by themselves. Presumably the entire region would act as a single regulatory unit perhaps in conjunction with the other upstream ζ-chain enhancer. Eζi was also a strong transcriptional activator when tested in catfish 1B10 B-cells, which possess what appears to be a post-activation phenotype (secretory and membrane IgM+; IgD−), but Eζi was less able to drive transcription in the catfish 3B11 B-cells, which have a mature naïve phenotype ((IgM+/IgD+). Diminished transcriptional activity within the 3B11 cells was also noted for the zebrafish Eμ3′ enhancer, but not for Eζ3′-1 or the catfish Eμ3′ enhancers. The only readily recognizable trait shared by the zebrafish Eμ3′ and Eζi, that is not contained within either of the constructs for catfish Eμ3′ and zebrafish Eζ3′-1 enhancers, is binding sites for the Mit family member TFE3, which in the original characterization of mammalian Ig transcriptional regulators was referred to as C2 or μE3 (Tsao et al., 1988). However the μE3 site has only ever been recognized as a transcriptional activator, and although an alternative splice form of TFE3 can abrogate activation through this site (Roman et al., 1991), it has not been shown to suppress activation through other sites within mammalian Ig enhancers or promoters, thus leaving us without a ready explanation for the observed differences.
What is apparent from the observed differences between the transcriptional activities of the zebrafish Eμ3′, Eζi and Eζ3′-1 is that the enhancers tested have at least one difference in which transcriptional activating or suppressing protein is necessary for strong activation. If this holds true in vivo, then these enhancers may be performing distinct functions in the cell during development of the fish or the B-cells, including dictating which isotype is expressed in a cell. And while we cannot knockout the individual enhancers, efforts are underway to develop zebrafish containing reporter transgenes under control of the respective enhancers. To that end preliminary observations on zebrafish carrying a GFP reporter under control of the zebrafish Eμ3′ enhancer indicate that fluorescent cells are IgM positive and IgZ negative (Dawne Page, personal communication). If this observation continues to hold true, then one interpretation would be that Eμ3′ does not contribute to the transcriptional regulation of ζ-chain expression. There is more emerging evidence fore there being differential regulation of IgZ, IgM, and IgD expression. Both catfish and mammals have a population of IgD+/IgM− cells that produce both secretory and membrane IgD (Chen et al., 2009). In both groups of vertebrates the secreted IgD seems to act via Ig receptors on the surface of granulocytes. Two recent publications provide evidence that IgZ and IgM expressing cells are differentially distributed through the tissues of carp (Ryo et al., 2010) and trout (Zhang et al., 2010). While Sunyer and colleagues provide compelling evidence that IgZ/T is important in mucosal immunity, they have not yet examined whether cytokine cues from mucosal infections can influence lineage development of IgZ versus IgM B-cells in kidney. The question of regulation of B-cell lineages was also examined in part by Boehm and colleagues (Schorpp et al., 2006). In zebrafish homozygous for an ikaros allele predicted to be missing the C-terminus zinc fingers there is no apparent productive or non-productive VDζJζ recombination events. Conversely there was expression of IgM, though this was developmentally delayed, and there was a significant reduction in the diversity of the VDμJμ repertoire (Schorpp et al., 2006). As Boehm and colleagues point out (Schorpp et al., 2006), if isotype choice was a result of stochastic processes then at the least some of the surviving B-cells should have had VDζJζ rearrangements, even if they were unable to encode productive transcripts. The apparent inactivity of the ζ-chain enhancers within cells destined to express μ-chain may result from the state of chromatin accessibility at the enhancers, anδ/or the availability of enhancer specific regulatory transcription factors. Though the ζ-chain enhancers were active in IgM expressing catfish B-cells, catfish do not appear to have a ζ-chain ortholog (Bengten et al., 2006) and so may not have developed or retained a means of differentially utilizing ζ-chain enhancers.
The mammalian TcRδ/α locus has an organization that is analogous to that of the zebrafish IgH locus, insofar as V(D)J recombination determines whether TcRδ or TcRα is expressed. In the TcRδ/α locus the Eδ enhancer situated between the 3′ most Jδ and the Cδ-exon while a second enhancer (Eα) is situated immediately downstream of the Cα exons. During thymocyte development the Eδ enhancer becomes active and drives VDδJδ recombination during double-negative III, while Eα does not become active until the double-positive phase at which time it directs sterile transcription from the T-early alpha (TEA) promoter which is situated downstream of Cδ adjacent to the Jα-elements (Hernandez-Munain et al., 1999). Transcription from the TEA promoter initiates V- to Jα recombination, which in turn excises the intervening sequence including Eδ. In terms of positioning the zebrafish IgH Eζi enhancer is like that of the TcR Cδ enhancer, and is well positioned to direct VDζJζ recombination in the heavy chain. Furthermore some of the clusters of transcription factor binding sites situated downstream of the ζ-chain exons could be envisioned to be part of a cryptic promoter equivalent to the TEA promoter in the TcRδ/α chain (Hernandez-Munain et al., 1999) or the DQ52 promoter (Alessandrini and Desiderio, 1991) upstream of the D-elements with the mouse IgH locus. Whether the Eζi and Eμ3′ enhancers are necessarily activated sequentially as are the Eδ and Eα enhancer, or if they respond to different developmental niches or milieus remains to be determined.
As we’ve argued before (Magor et al., 1999) the presence of an intronic enhancer such as Eζi has implications both for the evolution of the IgH locus as well as for affinity maturation of antibodies expressed by a locus containing an intronic enhancer. Among the components necessary for the development of class switch recombination in the early tetrapod locus would be an enhancer in the JH to Cμ1 intron that would not be excised during class switching [reviewed in (Magor et al., 1999)]. While the Eζi enhancer seemingly does not fulfill that role it does offer the possibility that there already existed a selective advantage to developing transcriptional regulators in that region of the locus. One possible selective advantage may be improved targeting of the VDJ exon by the somatic hypermutation apparatus. Many of the original studies to suggest that transcription was as a requirement for SH and CSR to occur also indicated that these processes occurred with a higher frequency in the presence of both an intronic and a 3′ enhancers (Bachl et al., 1998; Betz et al., 1994; Papavasiliou and Schatz, 2002). Using a stably transfected reporter construct in what is now known to be a cell line constitutively expressing AID, Wabl and colleagues (Bachl et al., 1998) observed that the rate of point mutations within a specific site in the VDJ exon was 40 times greater when an intronic enhancer was added to a construct containing an enhancer 3′ of the μ-chain exons. Conversely retention of the intronic enhancer but deletion of the 3′ enhancer diminished the mutation frequency by more than 5 fold. Subsequent research questioned whether it was the enhancer proper, or flanking elements that were necessary for recruiting the mutator (Klix et al., 1998; Ronai et al., 2005). Storb and colleagues subsequent dissection of potential AID recruiting elements suggest that the binding site E47 homodimers, CAGGTG, acts in some manner to recruit the mutational apparatus to nearby transcribed targets (Michael et al., 2003). And while this motif is typically thought of as a core component of the Eμ enhancer (of mammalian IgH), additional motifs outside of the enhancer can facilitate SH (Michael et al., 2003). Consistent with Storbs’ findings, Honjo and colleagues (Kotani et al., 2005), in seeking to understand why some genes are ectopically targeted by AID in centroblasts and AID induced T-cell lymphomas, noted that most of the targeted genes contained an E47 motif within their enhancers or promoters.
All of the functional IgH enhancers of zebrafish reported here and in prior publications (Ellestad and Magor, 2005) contain at least one binding site for the fish homologue of E2A (which generates E47), CFEB1 (Hikima et al., 2004). Furthermore CFEB1 binds equally well to another E-box motif, CAGATG (Hikima et al., 2006a), that is a core component of the zebrafish Eμ3′ enhancer (Ellestad and Magor, 2005). This second CFEB1 binding site is also found in Eζi, the J to Cμ1 intron (Ellestad and Magor, 2005), and in multiple copies within the Eζ3′-1 enhancer. If AID targeting operates in fish as it does in mammals, then these CFEB1 binding sites may contribute both to transcriptional activation, as well as to targeting of AID to their proximal VDJ exons. And in the latter context it may be particularly advantageous to have the CFEB1 targeting motif immediately downstream of the VDJ exon in the J to C1 intron, as is the case for Eζi.
In summary, we have identified three additional regions with IgH locus of zebrafish that likely contribute to the transcriptional regulation of IgZ, IgM and IgD. At the least the existence of multiple enhancers suggests considerable redundancy in ensuring transcription of the IgH locus. Though the complexity of transcriptional regulation leads us to speculate that there may be distinct functions for at least some of these enhancers. While we cannot yet say whether these regions regulate alternative VDJ recombination associated with isotype selection, or somatic hypermutation associated with affinity maturation, these enhancers used to drive reporter transgenes, may prove useful in elucidating these processes.
Acknowledgments
The authors wish to thank Lisa Steiner, MIT, for the use of equipment and facilities. B.G.M. is supported by the National Science and Engineering Research Council of Canada. N.D. was supported by NIH grant R01 AI08054.
Abbreviations
- IgH
immunoglobulin heavy chain
- SH
somatic hypermutation
- MAR
matrix attachment region
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
H. L. Saunders, Email: hollys@ualberta.ca.
B. G. Magor, Email: bmagor@ualberta.ca.
References
- Abarrategui I, Krangel MS. Germline transcription: a key regulator of accessibility and recombination. Advances in experimental medicine and biology. 2009;650:93–102. doi: 10.1007/978-1-4419-0296-2_8. [DOI] [PubMed] [Google Scholar]
- Alessandrini A, Desiderio SV. Coordination of immunoglobulin DJH transcription and D-to-JH rearrangement by promoter-enhancer approximation. Molecular and cellular biology. 1991;11:2096–2107. doi: 10.1128/mcb.11.4.2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachl J, Olsson C, Chitkara N, Wabl M. The Ig mutator is dependent on the presence, position, and orientation of the large intron enhancer. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:2396–2399. doi: 10.1073/pnas.95.5.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bengten E, Quiniou S, Hikima J, Waldbieser G, Warr GW, Miller NW, et al. Structure of the catfish IGH locus: analysis of the region including the single functional IGHM gene. Immunogenetics. 2006;58:831–844. doi: 10.1007/s00251-006-0139-9. [DOI] [PubMed] [Google Scholar]
- Betz AG, Milstein C, Gonzalez-Fernandez A, Pannell R, Larson T, Neuberger MS. Elements regulating somatic hypermutation of an immunoglobulin kappa gene: critical role for the intron enhancer/matrix attachment region. Cell. 1994;77:239–248. doi: 10.1016/0092-8674(94)90316-6. [DOI] [PubMed] [Google Scholar]
- Bolland DJ, Wood AL, Corcoran AE. Large-scale chromatin remodeling at the immunoglobulin heavy chain locus: a paradigm for multigene regulation. Advances in experimental medicine and biology. 2009;650:59–72. doi: 10.1007/978-1-4419-0296-2_5. [DOI] [PubMed] [Google Scholar]
- Calame K, Sen R. Transcription of Immunoglobulin Genes. In: Honjo T, Alt FW, Neuberger MS, editors. Molecular Biology of B-cells. Elsevier; San Diego: 2004. pp. 83–100. [Google Scholar]
- Cartharius K, Frech K, Grote K, Klocke B, Haltmeier M, Klingenhoff A, et al. MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics (Oxford, England) 2005;21:2933–2942. doi: 10.1093/bioinformatics/bti473. [DOI] [PubMed] [Google Scholar]
- Chaudhuri J, Alt FW. Class-switch recombination: interplay of transcription, DNA deamination and DNA repair. Nature reviews. 2004;4:541–552. doi: 10.1038/nri1395. [DOI] [PubMed] [Google Scholar]
- Chen K, Xu W, Wilson M, He B, Miller NW, Bengten E, et al. Immunoglobulin D enhances immune surveillance by activating antimicrobial, proinflammatory and B cell-stimulating programs in basophils. Nature immunology. 2009;10:889–898. doi: 10.1038/ni.1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cioffi CC, Middleton DL, Wilson MR, Miller NW, Clem LW, Warr GW. An IgH enhancer that drives transcription through basic helix-loop-helix and Oct transcription factor binding motifs. Functional analysis of the E(mu)3′ enhancer of the catfish. The Journal of biological chemistry. 2001;276:27825–27830. doi: 10.1074/jbc.M100110200. [DOI] [PubMed] [Google Scholar]
- Dang W, Nikolajczyk BS, Sen R. Exploring functional redundancy in the immunoglobulin mu heavy-chain gene enhancer. Molecular and cellular biology. 1998;18:6870–6878. doi: 10.1128/mcb.18.11.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danilova N, Bussmann J, Jekosch K, Steiner LA. The immunoglobulin heavy-chain locus in zebrafish: identification and expression of a previously unknown isotype, immunoglobulin Z. Nature immunology. 2005;6:295–302. doi: 10.1038/ni1166. [DOI] [PubMed] [Google Scholar]
- Ellestad KK, Magor BG. Evolution of transcriptional enhancers in the immunoglobulin heavy-chain gene: functional characteristics of the zebrafish Emu3′ enhancer. Immunogenetics. 2005;57:129–139. doi: 10.1007/s00251-005-0785-3. [DOI] [PubMed] [Google Scholar]
- Farre D, Roset R, Huerta M, Adsuara JE, Rosello L, Alba MM, et al. Identification of patterns in biological sequences at the ALGGEN server: PROMO and MALGEN. Nucleic acids research. 2003;31:3651–3653. doi: 10.1093/nar/gkg605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisch M, Frech K, Klingenhoff A, Cartharius K, Liebich I, Werner T. In silico prediction of scaffold/matrix attachment regions in large genomic sequences. Genome research. 2002;12:349–354. doi: 10.1101/gr.206602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frith MC, Hansen U, Spouge JL, Weng Z. Finding functional sequence elements by multiple local alignment. Nucleic acids research. 2004;32:189–200. doi: 10.1093/nar/gkh169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh D. A relational database of transcription factors. Nucleic acids research. 1990;18:1749–1756. doi: 10.1093/nar/18.7.1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goebel P, Montalbano A, Ayers N, Kompfner E, Dickinson L, Webb CF, et al. High frequency of matrix attachment regions and cut-like protein x/CCAAT-displacement protein and B cell regulator of IgH transcription binding sites flanking Ig V region genes. J Immunol. 2002;169:2477–2487. doi: 10.4049/jimmunol.169.5.2477. [DOI] [PubMed] [Google Scholar]
- Hansen JD, Landis ED, Phillips RB. Discovery of a unique Ig heavy-chain isotype (IgT) in rainbow trout: Implications for a distinctive B cell developmental pathway in teleost fish. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:6919–6924. doi: 10.1073/pnas.0500027102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Munain C, Sleckman BP, Krangel MS. A developmental switch from TCR delta enhancer to TCR alpha enhancer function during thymocyte maturation. Immunity. 1999;10:723–733. doi: 10.1016/s1074-7613(00)80071-0. [DOI] [PubMed] [Google Scholar]
- Hikima J, Cioffi CC, Middleton DL, Wilson MR, Miller NW, Clem LW, et al. Evolution of transcriptional control of the IgH locus: characterization, expression, and function of TF12/HEB homologs of the catfish. J Immunol. 2004;173:5476–5484. doi: 10.4049/jimmunol.173.9.5476. [DOI] [PubMed] [Google Scholar]
- Hikima J, Middleton DL, Wilson MR, Miller NW, Clem LW, Warr GW. Regulation of immunoglobulin gene transcription in a teleost fish: identification, expression and functional properties of E2A in the channel catfish. Immunogenetics. 2005;57:273–282. doi: 10.1007/s00251-005-0793-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hikima J, Lennard ML, Wilson MR, Miller NW, Clem LW, Warr GW. Conservation and divergence of the Emicro3′ enhancer in the IGH locus of teleosts. Immunogenetics. 2006a;58:226–234. doi: 10.1007/s00251-006-0090-9. [DOI] [PubMed] [Google Scholar]
- Hikima J, Lennard ML, Wilson MR, Miller NW, Warr GW. Regulation of the immunoglobulin heavy chain locus expression at the phylogenetic level of a bony fish: transcription factor interaction with two variant octamer motifs. Gene. 2006b;377:119–129. doi: 10.1016/j.gene.2006.03.014. [DOI] [PubMed] [Google Scholar]
- Jenuwein T, Forrester WC, Fernandez-Herrero LA, Laible G, Dull M, Grosschedl R. Extension of chromatin accessibility by nuclear matrix attachment regions. Nature. 1997;385:269–272. doi: 10.1038/385269a0. [DOI] [PubMed] [Google Scholar]
- Khamlichi AA, Pinaud E, Decourt C, Chauveau C, Cogne M. The 3′ IgH regulatory region: a complex structure in a search for a function. Advances in immunology. 2000;75:317–345. doi: 10.1016/s0065-2776(00)75008-5. [DOI] [PubMed] [Google Scholar]
- Klix N, Jolly CJ, Davies SL, Bruggemann M, Williams GT, Neuberger MS. Multiple sequences from downstream of the J kappa cluster can combine to recruit somatic hypermutation to a heterologous, upstream mutation domain. European journal of immunology. 1998;28:317–326. doi: 10.1002/(SICI)1521-4141(199801)28:01<317::AID-IMMU317>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- Kotani A, Okazaki IM, Muramatsu M, Kinoshita K, Begum NA, Nakajima T, et al. A target selection of somatic hypermutations is regulated similarly between T and B cells upon activation-induced cytidine deaminase expression. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:4506–4511. doi: 10.1073/pnas.0500830102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krangel MS. Mechanics of T cell receptor gene rearrangement. Current opinion in immunology. 2009;21:133–139. doi: 10.1016/j.coi.2009.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lennard ML, Wilson MR, Miller NW, Clem LW, Warr GW, Hikima J. Oct2 transcription factors in fish--a comparative genomic analysis. Fish & shellfish immunology. 2006;20:227–238. doi: 10.1016/j.fsi.2005.01.011. [DOI] [PubMed] [Google Scholar]
- Lennard ML, Hikima J, Ross DA, Kruiswijk CP, Wilson MR, Miller NW, et al. Characterization of an Oct1 orthologue in the channel catfish, Ictalurus punctatus: a negative regulator of immunoglobulin gene transcription? BMC molecular biology. 2007;8:8. doi: 10.1186/1471-2199-8-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Bramblett D, Zhu Q, Lozano M, Kobayashi R, Ross SR, et al. The matrix attachment region-binding protein SATB1 participates in negative regulation of tissue-specific gene expression. Molecular and cellular biology. 1997;17:5275–5287. doi: 10.1128/mcb.17.9.5275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magor BG, Wilson MR, Miller NW, Clem LW, Middleton DL, Warr GW. An Ig heavy chain enhancer of the channel catfish Ictalurus punctatus: evolutionary conservation of function but not structure. J Immunol. 1994;153:5556–5563. [PubMed] [Google Scholar]
- Magor BG, Ross DA, Middleton DL, Warr GW. Functional motifs in the IgH enhancer of the channel catfish. Immunogenetics. 1997;46:192–198. doi: 10.1007/s002510050261. [DOI] [PubMed] [Google Scholar]
- Magor BG, Ross DA, Pilstrom L, Warr GW. Transcriptional enhancers and the evolution of the IgH locus. Immunology today. 1999;20:13–17. doi: 10.1016/s0167-5699(98)01380-2. [DOI] [PubMed] [Google Scholar]
- Michael N, Shen HM, Longerich S, Kim N, Longacre A, Storb U. The E box motif CAGGTG enhances somatic hypermutation without enhancing transcription. Immunity. 2003;19:235–242. doi: 10.1016/s1074-7613(03)00204-8. [DOI] [PubMed] [Google Scholar]
- Miller NW, Chinchar VG, Clem LW. Development of leukocyte cell lines from the channel catfish (Ictalurus punctatus) Journal of Tissue Culture Methods. 1994a;16:117–123. [Google Scholar]
- Miller NW, Rycyzyn MA, Wilson MR, Warr GW, Naftel JP, Clem LW. Development and characterization of channel catfish long term B cell lines. J Immunol. 1994b;152:2180–2189. [PubMed] [Google Scholar]
- Nikolajczyk BS, Cortes M, Feinman R, Sen R. Combinatorial determinants of tissue-specific transcription in B cells and macrophages. Molecular and cellular biology. 1997;17:3527–3535. doi: 10.1128/mcb.17.7.3527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovcharenko I, Loots GG, Hardison RC, Miller W, Stubbs L. zPicture: dynamic alignment and visualization tool for analyzing conservation profiles. Genome research. 2004;14:472–477. doi: 10.1101/gr.2129504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papavasiliou FN, Schatz DG. Somatic hypermutation of immunoglobulin genes: merging mechanisms for genetic diversity. Cell. 2002;109(Suppl):S35–44. doi: 10.1016/s0092-8674(02)00706-7. [DOI] [PubMed] [Google Scholar]
- Pettersson S, Cook GP, Bruggemann M, Williams GT, Neuberger MS. A second B cell-specific enhancer 3′ of the immunoglobulin heavy-chain locus. Nature. 1990;344:165–168. doi: 10.1038/344165a0. [DOI] [PubMed] [Google Scholar]
- Roman C, Cohn L, Calame K. A dominant negative form of transcription activator mTFE3 created by differential splicing. Science (New York, NY) 1991;254:94–97. doi: 10.1126/science.1840705. [DOI] [PubMed] [Google Scholar]
- Ronai D, Iglesias-Ussel MD, Fan M, Shulman MJ, Scharff MD. Complex regulation of somatic hypermutation by cis-acting sequences in the endogenous IgH gene in hybridoma cells. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:11829–11834. doi: 10.1073/pnas.0505449102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross DA, Magor BG, Middleton DL, Wilson MR, Miller NW, Clem LW, et al. Characterization of Oct2 from the channel catfish: functional preference for a variant octamer motif. J Immunol. 1998;160:3874–3882. [PubMed] [Google Scholar]
- Ryo S, Wijdeven RH, Tyagi A, Hermsen T, Kono T, Karunasagar I, et al. Common carp have two subclasses of bonyfish specific antibody IgZ showing differential expression in response to infection. Developmental and comparative immunology. 2010 doi: 10.1016/j.dci.2010.06.012. In Press. [DOI] [PubMed] [Google Scholar]
- Savan R, Aman A, Sato K, Yamaguchi R, Sakai M. Discovery of a new class of immunoglobulin heavy chain from fugu. European journal of immunology. 2005;35:3320–3331. doi: 10.1002/eji.200535248. [DOI] [PubMed] [Google Scholar]
- Schorpp M, Bialecki M, Diekhoff D, Walderich B, Odenthal J, Maischein HM, et al. Conserved functions of Ikaros in vertebrate lymphocyte development: genetic evidence for distinct larval and adult phases of T cell development and two lineages of B cells in zebrafish. J Immunol. 2006;177:2463–2476. doi: 10.4049/jimmunol.177.4.2463. [DOI] [PubMed] [Google Scholar]
- Schug J. Using TESS to predict transcription factor binding sites in DNA sequence. In: Baxevanis Andreas D, et al., editors. Current protocols in bioinformatics. Unit 26. Chapter 2. 2008. [DOI] [PubMed] [Google Scholar]
- Tsao BP, Wang XF, Peterson CL, Calame K. In vivo functional analysis of in vitro protein binding sites in the immunoglobulin heavy chain enhancer. Nucleic acids research. 1988;16:3239–3253. doi: 10.1093/nar/16.8.3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Goldstein A, Zong RT, Lin D, Neufeld EJ, Scheuermann RH, et al. Cux/CDP homeoprotein is a component of NF-muNR and represses the immunoglobulin heavy chain intronic enhancer by antagonizing the bright transcription activator. Molecular and cellular biology. 1999;19:284–295. doi: 10.1128/mcb.19.1.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YA, Salinas I, Li J, Parra D, Bjork S, Xu Z, et al. IgT, a primitive immunoglobulin class specialized in mucosal immunity. Nature immunology. 2010 doi: 10.1038/ni.1913. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]