Abstract
The cleavage of the A2-domain of Von Willebrand Factor (VWF) by the metalloprotease ADAMTS13 regulates VWF size and platelet thrombosis rates. Reduction or inhibition of this enzyme activity leads to thrombotic thrombocytopenic purpura (TTP). We generated a set of novel molecules called VWF-A2 FRET proteins’, where variants of YFP (Venus) and CFP (Cerulean) flank either the entire VWF-A2 domain (175 amino acids) or truncated fragments (141, 113, 77 amino acids) of this domain. These proteins were expressed in E. coli in soluble form, and they exhibited Fluorescence/Förster Resonance Energy Transfer (FRET) properties. Results show that introduction of Venus/Cerulean itself did not alter the ability of VWF-A2 to undergo ADAMTS13 mediated cleavage. The smallest FRET protein, XS-VWF, detected plasma ADAMTS13 activity down to 10% of normal levels. Tests of acquired and inherited TTP could be completed within 30 min. VWF-A2 conformation changed progressively, and not abruptly, upon increasing urea concentration. While proteins with 77 and 113 VWF-A2 residues were cleaved in the absence of denaturant, 4M urea was required for the efficient cleavage of larger constructs. Overall, VWF-A2 FRET proteins can be applied both for the rapid diagnosis of plasma ADAMTS13 activity, and as a tool to study VWF-A2 conformation dynamics.
Keywords: ADAMTS13, coagulation, fluorescence, platelet, thrombotic thrombocytopenic purpura, VWF
INTRODUCTION
Von Willebrand Factor (VWF) is a large, multimeric glycoprotein found in human circulation. The hemostatic function of this protein increases with protein molecular mass. The size of VWF is regulated by the constitutively active blood metalloprotease ADAMTS13 [1; 2; 3] (a disintegrin and metalloprotease with thrombospondin type 1 motif 13). This enzyme cleaves the Tyr1605-Met1606 bond, which is constitutively buried within the A2-domain of VWF. The rate of VWF cleavage is enhanced by the application of hydrodynamic/mechanical forces that expose the cleavage site [4]. The binding of VWF to platelet GPIbα [5], endothelial αvβ3 [6] and thrombus formation [7; 8] further augment this rate of mechanoenzymatic cleavage by enhancing the ability of shear forces to expose the Tyr-Met scissile bond. In this regard, VWF binding to cells/surfaces may be important for proteolysis since, at a given shear rate, the force applied on an immobilized protein is 10–100 fold more than that in circulation [9; 10].
Due to its potential use as a diagnostic tool, there is currently interest in developing simple and efficient methods that can quantify VWF proteolysis rates and ADAMTS13 activity in circulation [11]. Inherited/familial or acquired defects in ADAMTS13 function result in the inefficient cleavage of VWF and the presence of high molecular mass VWF in circulation. While inherited defects are associated with severe deficiency of ADAMTS13, acquired disease is typically characterized by the presence of inhibitory auto-antibodies. The presence of high molecular mass VWF, under these conditions, contributes to the spontaneous binding of platelet receptor GpIbα to VWF via the VWF-A1 domain, platelet activation, aggregation and microthrombi formation. Such microthrombi can cause vessel occlusion, ischemia and organ failure leading to a condition called thrombotic thrombocytopenic purpura (TTP). Accurate and rapid strategies to quantify ADAMTS13 activity can thus serve as a prognostic marker of TTP.
In the current manuscript, we incorporated in the A2 domain of VWF, variants of the cyan (CFP) and yellow fluorescent protein (YFP) that are called Cerulean [12] and Venus [13] respectively. Although 77–175 amino acids of the VWF A2 domain separate Cerulean and Venus in the primary sequence of these molecules, the insertion sites of these green fluorescent protein (GFP) variants lie spatially within 2.5nm in VWF-A2 crystal structure [14]. Due to this design, the novel family of proteins exhibit Fluorescence/Förster Resonance Energy Transfer (FRET) properties. Together, these molecules are called “VWF-A2 FRET” proteins. The smallest FRET molecule we generated (‘XS-VWF’) detects ADAMTS13 in blood within 5–10 min in the absence of denaturant (KM =4μM), and it can readily detect plasma protease levels down to <10% of normal levels. This sensitivity rivals or exceeds the resolution of FRETS-VWF73, another fluorescence based biosensor of ADAMTS13 activity [15; 16; 17]. Other advantages of the VWF-A2 FRET proteins compared to FRETS-VWF73 include (Table 1): i) They are produced in large amounts by E. coli in soluble form, unlike FRETS-VWF73 that requires chemical synthesis. The latter synthesis is complicated since the synthetic peptide is long with 73 amino acids; ii) VWF-A2 FRET proteins contain two fluorophores (Venus and Cerulean) unlike FRETS-VWF73 which has one fluorophore and a quencher. Thus, data generated by our molecule can be presented in the form of “FRET ratio” (defined in Methods). While the absolute value of this parameter can vary between instruments since detector sensitivity settings at individual wavelengths can be tuned by users, the FRET ratio of the ADAMTS13 cleaved substrate to that of the original intact protein will be approximately 2.75 ± 0.15, regardless of the measuring device and instrument settings. Since FRETS-VWF73 is based on the principles of quenching, however, cleavage measurements vary with instrument settings and assay conditions. Importantly, it is not straightforward to translate the measured fluorescence change to estimate % substrate cleavage. iii) Due to the large separation between fluorophores in VWF-A2 FRET, unlike FRETS-VWF73 where the fluorophore and quencher are separated by ten amino acids, our proteins can be applied to study changes in VWF-A2 structure under a variety of conditions in addition to proteolysis. iv) The excitation and emission wavelengths used for XS-VWF are higher compared to the FRETS-VWF73 substrate, and this results in lower auto-fluorescence due to plasma proteins in the former case.
Table 1.
Differences between XS-VWF and FRETS-VWF73
| XS-VWF | FRETS-VWF73 |
|---|---|
| E.coli expression | Chemical synthesis |
| FRET based assay | Fluorescence quenching assay |
| Linear relationship with ADAMTS13 activity | Non-linear response |
| Large/77-amino acid separation between fluorophores | 10 amino acid separation |
| Excitation=435nm (lower plasma auto- fluorescence) | Excitation=340–350nm (high auto- fluorescence) |
| Emissions at 485 and 530nm | Emission=440–450 nm |
METHODS
VWF-A2 FRET constructs
Fusion proteins containing the A2 domain of VWF were expressed in E. coli. Many of the proteins were fused to variants of either YFP (Venus and Citrine) or CFP (Cerulean). These fluorophores are monomeric since they incorporate the A206K mutation, and they exhibit higher brightness, lower photobleaching, higher extinction coefficient and quantum yield when compared to the original CFP and YFP [12; 13]. PCR primers used for molecular biology steps are listed in Supplemental Table I.
The plasmids pCS-CG (plasmid number 12154), pVenus-GalT (11931), pCerulean-VSVG (11913) and pRSET FLII12Pglu-600u (13563) were purchased from Addgene (Cambridge, MA). Initially, pCS-CG was modified by replacing the green fluorescence protein (GFP) sequence that it originally contained with the Kozak sequence followed by the VWF signal peptide, AgeI and HpaI restriction enzyme sites, the FLAG epitope, TEV cleavage site, poly 6X-histidine tag and a stop codon. A BstBI enzyme site is present between FLAG and TEV. This vector where the CMV promoter and VWF signal peptide drive protein expression is designated pCSCG-KZK-SS-FLAG-His. Using full length VWF cDNA in pcDNA3.1 (Invitrogen) as a template [18], the A2-domain of VWF (amino acids 1481–1668) was cloned into pCSCG-KZK-SS-FLAG-His using the AgeI and HpaI sites for insertion. The resulting plasmid is called pCSCG-A2-FH. Next, the KpnI-HindIII fragment containing Citrine from pRSET FLII12Pglu-600u was cloned into pUC19. Citrine was amplified in this vector with primers flanked by AgeI, and it was inserted in the correct orientation at the N-terminus of VWF-A2 in pCSCG-A2-FH. The resulting vector is called pCSCG-Citrine-A2-FH, and it was used for mammalian protein expression (results not discussed).
For bacterial expression, the region containing Citrine, VWF-A2, FLAG, TEV site, and poly-histidine from pCSCG-Citrine-A2-FH was amplified by PCR using primers with NdeI and HindIII overhangs, and this was cloned into pRSET-B (Invitrogen). During PCR, sequence encoding for an extra histidine was introduced and thus all bacterial proteins have a his(7) tag. Venus (Ven) from pVenus-GalT with primers containing NdeI and AgeI overhangs also replaced Citrine in pRSET-Citrine-A2-FH to produce pRSET-Ven-A2-FH. Next, Cerulean (Cer) was amplified from pCerulean-VSVG using primers with phosphorylated HpaI compatible overhangs and BstBI, and this was fused at the C-terminus of the A2 domain in pRSET-Ven-A2-FH. Insertion of Cerulean resulted in loss of the FLAG epitope. This last vector is designated pRSET-Ven-A2-Cer-H. Based on the VWF-A2 crystal structure (3GXB), regions pertaining to 1496–1670 (L-VWF), 1530–1670 (M-VWF), 1558–1670 (S-VWF) and 1594–1670 (XS-VWF) were amplified by PCR with forward primers containing AgeI and phosphorylated reverse primers containing HpaI compatible restriction site overhangs. These PCR products that encode for truncated fragments of the full A2-domain were individually cloned into pRSET-Ven-A2-Cer-H. This resulted in a series of products where truncated forms of A2 were flanked by Venus and Cerulean. A2-domain containing Cerulean alone was also created by PCR amplifying A2 (1481–1668) with primers containing NdeI and HpaI compatible restriction site overhangs and replacing Venus and L-VWF in the vector pRSET-Ven-(L-VWF)-Cer-H. This product is called pRSET-A2-Cer-H. To generate XS-VWF(AA), we mutated the 1605Tyr (Y)-1606Met (M) sequence in XS-VWF (1594–1670) to 1605Ala (A)-1606Ala (A). To this end, a mega primer was first generated using a reverse primer containing the YM to AA mutation and a forward primer encoding for the start of the XS-VWF sequence. The PCR product was purified using Qiaquick PCR kit (Qiagen, Valencia, CA), and this was used along with a reverse primer encoding for the 3′ section of XS-VWF during the second PCR step. The final PCR product was cloned into pRSET-Ven-(L-VWF)-Cer-H to replace L-VWF. DNA sequencing was performed to verify all plasmid constructs described above.
Expression and purification of VWF-A2 FRET proteins from E. coli
pRSET vectors encoding for VWF-A2 constructs were transformed into E. coli BL21 StarTM or other strains. Single colonies were scaled up to 1L in LB broth supplemented with 50μg/mL ampicillin. Cells were grown to OD600=0.6 at 37°C, and then induced with 1mM IPTG for 12h at 30°C. Following this, all protein purification steps described below were performed at 4°C. First, the cells from 1L culture were centrifuged at 2000g for 20 min, washed with phosphate buffered saline, and then resuspended in 10mL of 20mM HEPES buffer (pH-7.4) containing 300mM NaCl, 10mM imidazole and EDTA-free protease inhibitor cocktail (Roche Diagnostics, Indianapolis, IN). Cells were then lysed by 3 cycles of freeze-thaw. Lysozyme (10μg/mL) was added for additional 30 min to further facilitate lysis. Following this, the genomic DNA was sheared using a Branson sonifier (Danbury, CT). Lysate was separated from the cell debris by centrifuging at 20,000g for 30 min. The pellet was discarded and the supernatant was filtered through a 0.22μM syringe filter. The lysate was then passed through HisTrap HP column (GE Healthcare, Piscataway, NJ) that was equilibrated with HEPES buffer used for the cell lysis. The column was washed with 5–10 volumes of 20mM HEPES buffer (pH-7.4) containing 300mM NaCl and 68mM imidazole. Protein was eluted with 20mM HEPES (pH-7.4) containing 300mM NaCl and 300mM imidazole.
Plasma VWF and ADAMTS13
Multimeric human VWF was purified from plasma cryoprecipitate as described previously [19].
Recombinant human ADAMTS13 and diluted human plasma were used as a source of ADAMTS13 activity. To produce the recombinant protein, HEK 293T cells were transiently transfected with pCAGG-hADAMTS13 vector (kind gift from Dr. Kenji Soejima, The Chemo-Sero-Therapeutic Research Institute, Kaketsuken, Japan) using calcium phosphate method [20]. After 8–12h, culture medium was changed to chemically-defined, serum free medium Pro293a (Lonza, Basel, Switzerland) supplemented with GlutaMAX. Media collected 48h thereafter was concentrated 20-fold using Amicon 50 KDa cut off centrifugal filter devices (Millipore, Billerica, MA) and dialyzed against 50mM Tris (pH-8). In some cases, recombinant ADAMTS13 was also purchased from R&D Systems (Minneapolis, MN). Human plasma was obtained from blood drawn from healthy non-smoking volunteers into sodium citrate. All human subject protocols were approved by the University at Buffalo Institutional Review Board. Platelet poor plasma (PPP) was obtained as described earlier [19]. These samples were stored at −80°C and thawed at 37°C prior to use.
Proteolysis assays
Typically, FRET assays were performed in 40μL volume containing 1μM VWF-A2 FRET protein and 8μL concentrated recombinant ADAMTS13 diluted in cleavage buffer (50mM Tris pH 8.0, 12.5mM CaCl2). Urea was added to cleavage buffer in some cases. In other instances, protein was denatured with 4M urea for 1h at 37ºC prior to addition of ADAMTS13. Comparison of FRETS-VWF73 (Peptides International, Louisville, KY) and XS-VWF was performed in a 100μL cleavage reaction mixture containing 1.5μM of substrate and 6μL of plasma.
At fixed times, in some cases, VWF-A2 FRET fluorescence spectra was obtained using a spectrophotometer (model F-2500, Hitachi, Tokyo, Japan) with excitation wavelength/slit width of 435/20nm or 485/20nm, and emission slit width of 5nm. In other cases, proteolysis was quantified in 96/384-well black plates using either a Biotek synergy4 or FLx800 reader (Winooski, VT). These instruments were equipped with filters for Cerulean (Ex420/50nm and Em485/20nm) and Venus (Ex485/20nm and Em540/25nm). FRETS-VWF73 was read on the FLx800 reader equipped with Ex360/40 and Em 460/40 filters. A parameter termed “FRET ratio” was used to quantify the extent of VWF-A2 FRET proteolysis. This is defined as the ratio of emitted light intensity at 485nm (primarily by Cerulean) versus 540nm (primarily by Venus), when the protein was excited at 420nm. This parameter is inversely related to FRET efficiency and it increases upon VWF-A2 proteolysis.
Western blot analysis
SDS-PAGE under standard reducing conditions was performed using either 4–20% gradient gels (Thermo-Fisher, Rockford, IL) for VWF-A2 FRET or 6% resolving gels for plasma VWF. Proteins were then transferred onto nitrocellulose membranes. These were detected using either goat anti-his polyclonal Ab (Bethyl laboratories, Montgomery, TX) for VWF-A2 FRET, or rabbit anti-human VWF polyclonal Ab (Dako, Carpinteria, CA) for plasma VWF. Silver staining was performed using a kit from Thermo-Fisher.
Statistics
Unless stated otherwise, data are presented as mean + standard error of mean for ≥3 replicate experiments. ANOVA was applied for comparison between multiple treatments. * p<0.05 was considered significant.
RESULTS
Truncated VWF-A2 constructs exhibit FRET properties
The entire VWF-A2 domain or fragments of this domain were engineered to express Venus at the N-terminus, and Cerulean at the C-terminus (Fig. 1A). While proteins with Citrine were also created, these were unsuitable for the current study, since our emission spectra did not match earlier reports in literature (Supplemental Fig S1). All proteins with Venus and Cerulean were named based on the size of the inserted VWF-A2 fragment. Venus insertion sites were located in loop regions between β-sheets/α-helices based on X-ray crystal data [14] (Fig. 1B). Although the GFP variant insertion sites were located 77–175 amino acids from each other, in all proteins, these sites are located within 2.5nm in the crystal. Thus, the proteins were engineered to exhibit FRET properties. In addition, in the control protein, Y1605 and M1606 in the cleavage sequence in the smallest FRET construct (XS-VWF) were both mutated to alanine. All proteins were enriched using Ni-NTA columns to >95% purity as assessed using silver staining of SDS-PAGE gels (Fig. 1C).
Fig. 1. VWF-A2 FRET proteins.
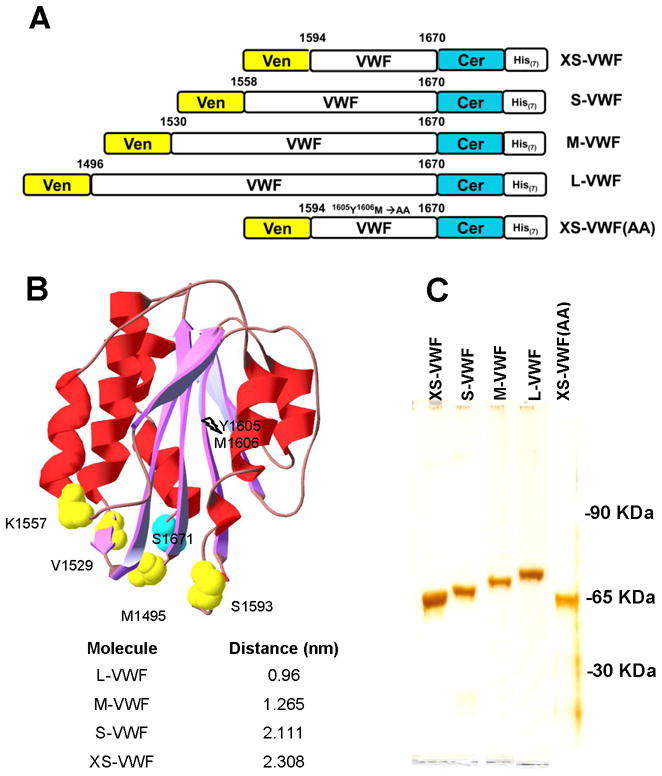
A. Five FRET proteins were expressed and purified from E. coli. In these, Venus (Ven) and Cerulean (Cer) flank truncated fragments of the VWF-A2 domain. His-tag is available for protein purification. The size of the A2-insertion increases from 77 amino acids for XS-VWF to 175 for L-VWF. XS-VWF(AA) is identical to XS-VWF except that the Y1605-M1606 amino acids in the ADAMTS13 cleavage site are replaced by A1605-A1606. B. Relative positions of Cerulean (cyan) and Venus (yellow) in the family of FRET proteins are shown annotated in the VWF-A2 crystal (3GBX). Distances between Cerulean and Venus insertion sites in the crystal structure are listed. C. Silver stain showing the purity of VWF-A2 FRET proteins.
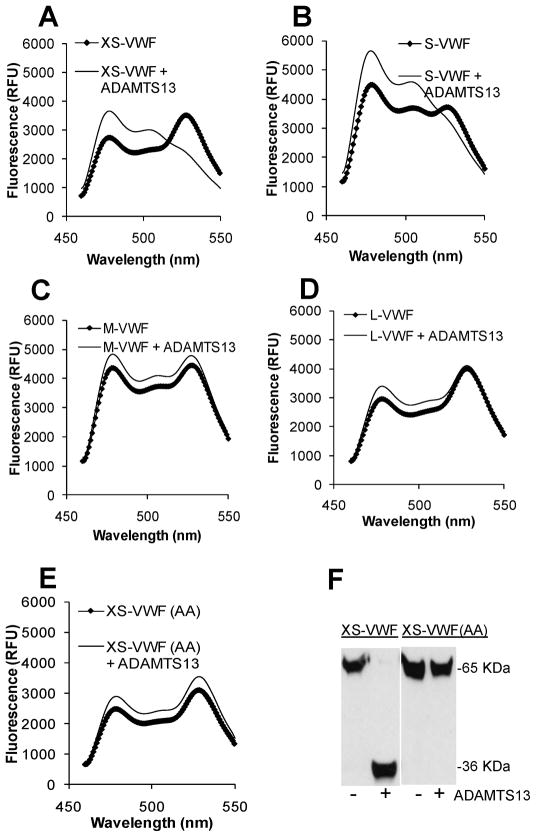
All proteins exhibited FRET properties (Fig. 2). FRET ratio varied from 0.74 for L-VWF to 1.24 for S-VWF. This corresponds to FRET efficiencies of 31% and 19% respectively. Such variation is consistent with the notion that in addition to the spatial distance separating the GFP-variant insertion sites, the distance between these fluorophores in the fusion protein and their relative orientation also determine the energy transfer efficiency [21]. Upon addition of ADAMTS13, FRET ratio increased from 0.78 to 1.73 for XS-VWF (Fig. 2A) and from 1.24 to 1.84 for S-VWF (Fig. 2B) within 1h. Complete cleavage corresponds to a FRET ratio of ~2.2. There was very little change in the FRET ratio of M-VWF (Fig. 2C), L-VWF (Fig. 2D), and XS-VWF(AA) (Fig. 2E) upon addition of ADAMTS13. Western blot analysis of XS-VWF and XS-VWF(AA) (Fig. 2F) is consistent with the fluorescence measurements, and it confirms that changes in FRET signal of XS-VWF is primarily due to the cleavage of the Y1605-M1606 scissile bond by ADAMTS13.
Fig. 2. Fluorescence spectra and ADAMTS13 cleavage.
1μM of each of the VWF-A2 FRET proteins was incubated for 1h at 37ºC either in the presence or absence of concentrated recombinant ADAMTS13. No denaturant was added. A–E. Samples diluted 60-fold were analyzed using the fluorescence spectrophotometer using 435nm excitation wavelength. All spectra were normalized by arbitrarily setting YFP fluorescence intensity to 10,000 (excitation=485nm; emission=530nm). F. Selected samples at 1h were subjected to western blot analysis, using anti-his pAb for detection. XS-VWF but not XS-VWF(AA) was cleaved by recombinant ADAMTS13. Data are representative of >3 experiments.
The KM and kcat values of XS-VWF and S-VWF were quantified based on the Lineweaver-Burk plot in the presence of 20% human plasma (Supplemental Fig. S2). These results show that both parameters were lower for S-VWF in comparison to XS-VWF (Table 2). Further, the measured KM and kcat values were similar to previously published values for other proteins constructed based on the VWF-A2 domain.
Table 2.
Kinetic constants for the cleavage of different ADAMTS13 substrates
| Substrates | KM | kcat (min−1) | Reference |
|---|---|---|---|
| XS-VWF | 4.6 ± 0.8μM | 44.8 ± 4.3 | (this paper) |
| S-VWF | 1.8 ± 0.2μM | 17.7 ± 0.5 | (this paper) |
| FRETS-VWF73* | 3.2 ± 1.1μM | 58 | [39] |
| VWF115** | 1.6 ± 0.5μM | 8.4 ± 3.6 | [40] |
| Multimeric VWF | 15nM | 0.83 | [39] |
Substrate similar to XS-VWF;
Substrate similar to S-VWF
Comparison of XS-VWF with FRETS-VWF73
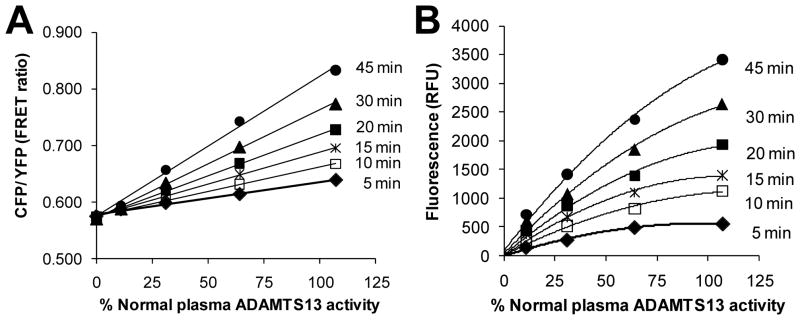
We tested the ability of one of the VWF-A2 FRET proteins, XS-VWF, to detect ADAMTS13 activity in human plasma samples. Upon comparing cleavage rates of XS-VWF with FRETS-VWF73 using samples having a range of plasma ADAMTS13 activity and measured over time, we noted a linear change in the FRET ratio of XS-VWF with ADAMTS13 activity (Fig. 3A). Under identical conditions, FRETS-VWF73 fluorescence readout exhibited a non-linear relationship with plasma ADAMTS13 activity (Fig. 3B).
Fig. 3. Detection of plasma ADAMTS13 levels by XS-VWF.
Various dilutions of human plasma were incubated with 1.5μM XS-VWF or FRETS-VWF73under conditions optimized for the latter reagent. A. FRET ratio varied linearly with plasma ADAMTS13 activity for XS-VWF. B. FRETS-VWF73 fluorescence readout varied non-linearly with plasma ADAMTS13 activity. Data are representative of >3 experiments.
When using normal human plasma, XS-VWF could be used to detect ADAMTS13 activity within 5 min, the earliest measured time point. In the presence of lower ADAMTS13 activity, more time was required for reliable detection of enzyme activity. Since auto-fluorescence due to plasma proteins plays only a minor role in assays utilizing XS-VWF (Supplemental Fig. S3), higher plasma concentrations can be used for this substrate compared to FRETS-VWF73 which exhibits higher auto-fluorescence. When assays were performed by diluting 20μL of a plasma mixture containing various ratios/amounts of normal plasma (100% ADAMTS13 activity) and heat-inactivated plasma (0% ADAMTS13 activity) in 100μL reaction volume, 10% of normal plasma protease activity could be detected within 30 min (Supplemental Fig. S4).
In order to further test our reagent, we compared substrate proteolysis with a panel of human plasma samples that were collected in sodium citrate (Table 3). The test samples include: i) Five calibration standards that had ADAMTS13 activity values (AVs) assigned based on the dilutions of a pool of normal plasma collected by the ISTH [International Society on Thrombosis and Haemostasis]; ii) Proficiency specimens obtained from individuals deficient in ADAMTS13 activity (Plasma 1), individuals with anti-ADAMTS13 antibodies (Plasma 2), and normal control (Plasma 3). For these three samples, in some cases, ADAMTS13 activity was heat inactivated by incubating plasma at 56ºC for 30min (specimens identified as “HI” in Table 3). In other runs (identified as ‘mixed’), heat inactivated plasma was mixed with an equal volume of normal plasma in order to check for anti-ADAMTS13 inhibitory antibodies [22]. iii) Six Factor Assay ConTrol plasma from George King Biomedical (Overland Park, Kansas): two FACT (plasma 11, 12), two A-FACT (plasma 5, 6) and two B-FACT (plasma 8, 9) samples that exhibit normal, abnormal and borderline activity in a wide variety of coagulation tests. iv) Additional human plasma specimen exhibiting a range of ADAMTS13 activity levels (plasma 4, 7, 10). As seen (Table 3), the results from XS-VWF activity measurements were largely consistent with findings noted using FRETS-VWF73. XS-VWF consistently gave lower values in the case of patients with <20% ADAMTS13 levels while FRETS-VWF73 appears to over predict this. This is particularly noted in case of heat inactivated plasma, where XS-VWF reported values close to zero while FRETS-VWF73 reported values ~14%. Differences were also noted in some plasma samples, namely 5,6 and 8,9. The reason for this has not been currently identified though it may be attributed to inherent structural differences in the two substrates and the manner in which ADAMTS13 binds them.
Table 3.
XS-VWF versus FRETS-VWF73
| % normal plasma ADAMTS13 activity | ||
|---|---|---|
| Sample | XS-VWF | FRETS-VWF73 |
| CalA [AV=0%] | −0.1±0.14 | −3.0±0.7 |
| CalB [AV=11%] | 8.3±1 | 15.2±1.4 |
| CalC [AV=31%] | 32.9±0.3 | 31.6±0.7 |
| CalD [AV=64%] | 66.7±0.2 | 60.9±0 |
| CalE [AV=107%] | 104.8±2.5 | 108.9±0.3 |
| Plasma 1 | 5.8±0.2 | 22.9±0.6 |
| Plasma 1 (HI) | 0.6±1 | 18.6±0.3 |
| Plasma 1 mixed | 46.7±0.1 | 46.9±0.1 |
| Plasma 2 | −3.2±1.1 | 14.2±1 |
| Plasma 2 (HI) | −1.4±1.2 | 13.4±0.8 |
| Plasma 2 mixed | −1.6±1.2 | 16.2±2.1 |
| Plasma 3 | 103.2±2.2 | 105.2±3 |
| Plasma 3 (HI) | −3.0±1.4 | 9.6±7.8 |
| Plasma 3 mixed | 48.9±0.4 | 46.5±1.1 |
| Plasma 4 | 7.3±1.8 | 12.4±0.2 |
| Plasma 5 | 25.8±6.3 | −1.6±1.6 |
| Plasma 6 | 21.0±5.4 | −4.6±3.4 |
| Plasma 7 | 24.9±0.6 | 36.9±1.6 |
| Plasma 8 | 70.4±14.9 | 32.1±1.41 |
| Plasma 9 | 67.4±14.35 | 32.9±1.3 |
| Plasma 10 | 114.3±23.4 | 80.7±3.8 |
| Plasma 11 | 116.0±23.7 | 88.7±2.1 |
| Plasma 12 | 93.7±0.4 | 80.9±1.2 |
ADAMTS13 activity was measured using either XS-VWF or FRETS-VWF73 as substrate for up to 45 min. Measured fluorescence value was converted to %ADAMTS13 activity using calibration curve in Fig. 3. Data from 30 and 45 min were averaged, and are presented as Mean ± standard deviation.
Effect of urea on VWF-A2 folding and access to proteolysis site
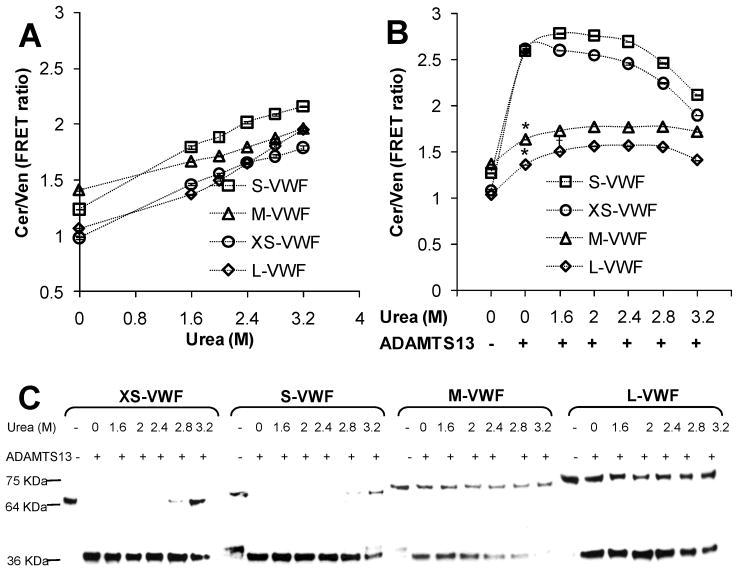
Since ADAMTS13 alone could not cleave M- and L-VWF (Fig. 2), studies were performed where urea concentration in the cleavage buffer was varied from 0 to 3.2M. Experiments were performed both in the absence (Fig. 4A) and presence (Fig. 4B–C) of ADAMTS13. In Fig. 4A, FRET ratio increased approximately linearly with urea concentration even in the absence of ADAMTS13. Over this range of urea concentration, there was negligible change in the fluorescence properties of Venus or Cerulean alone (data not shown). Significant structural changes occur at urea concentration down to 1.6M.
Fig. 4. Effect of urea.
A. 2μM each of the VWF-A2 FRET proteins was incubated with varying urea concentrations in the absence of ADAMTS13. FRET ratio, measured at 1h, increased with urea concentration. B. Concentrated recombinant ADAMTS13 was added to 1μM VWF-FRET proteins in the presence of varying urea concentrations for 2.5h. Samples were then diluted in 20-fold excess buffer for additional 5 min prior to FRET measurement. XS-VWF and S-VWF were cleaved efficiently in the absence of urea. Cleavage rate decreased at urea > 2.4M. The extent of L- and M-VWF cleavage increased with urea concentration though neither substrate was fully cleaved in the available time. *p<0.05 with respect to sample without ADAMTS13. †p<0.05 with respect to both samples that lack urea. C. Western blot analysis of 3h samples performed using the protocol in Fig. 2F, are consistent with fluorescence data.
ADAMTS13 was added to the VWF-A2 FRET proteins in the presence of varying urea concentrations. FRET ratio measurement (Fig. 4B) and western blot analysis (Fig. 4C) of selected samples was performed. Complete cleavage of XS-VWF and S-VWF was observed at urea concentrations <2.8M. The data confirm that urea is not required for the cleavage of these proteins and that ADAMTS13 activity is diminished at high urea concentrations. In the absence of urea, the extent of cleavage of both L- and M-VWF was 20%. Increasing urea concentration to 1.6M resulted in 30% proteolysis. Complete proteolysis was not observed under any condition. Western blot analysis results (Fig. 4C) are consistent with the fluorescence measurements.
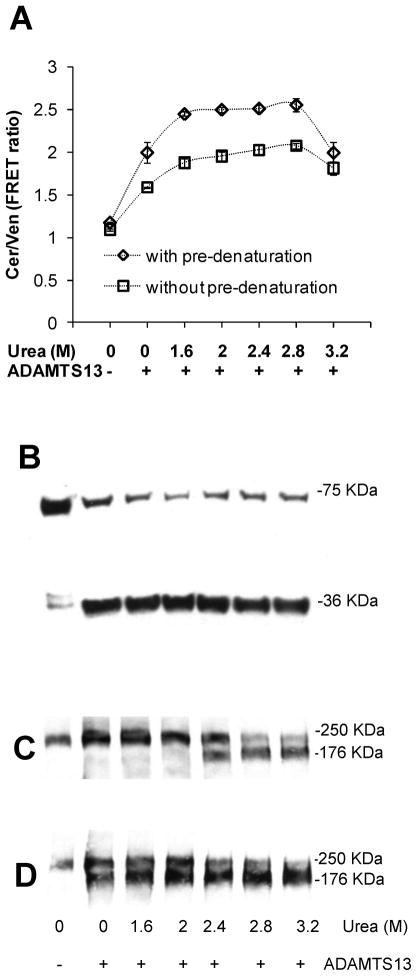
Since complete cleavage of L-VWF was not observed under any condition, we further increased the urea concentration by incubating L-VWF and multimeric human plasma VWF with 4M urea for 1h prior to dilution into a solution which contained ADAMTS13 along with lower urea concentrations (Fig. 5). Here, we noted substantial cleavage of L-VWF even when the protein was diluted into buffer that lacked urea. >90% cleavage of L-VWF was observed at 2M urea upon inclusion of the pre-denaturation step (Fig. 5A, 5B). Similar to L-VWF, multimeric VWF was also not fully cleaved when the protein was incubated with varying urea concentrations and ADAMTS13 (Fig. 5C). Proteolysis was however complete at urea >2.4M, provided a pre-denaturation step was included (Fig. 5D). The observation suggests that the proteolysis of L-VWF proceeds under similar denaturation conditions as the plasma protein.
Fig. 5. Pre-denaturation with urea.
10μM L-VWF or 40μg/mL multimeric plasma VWF was incubated either in the presence or absence of 4M urea at 37°C for 1h. Following this, VWF samples were diluted 10-fold into cleavage buffer containing concentrated ADAMTS13 and varying urea amounts for 24h. A. FRET ratio was measured similar to Fig. 4B. Pre-denaturation of L-VWF with 4M urea enhanced proteolysis rates. B. Western blot of L-VWF samples pre-denatured with urea prior to ADAMTS13 addition show >90% proteolysis. C and D. Western blot of plasma VWF that was either not subjected to (panel C) or was subjected to urea pre-denaturation (panel D). Pre-denaturation is required for the efficient cleavage of both L-VWF and plasma VWF.
DISCUSSION
The manuscript describes the family of VWF-A2 FRET proteins’. Multiple lines of evidence suggest that the introduction of Venus and Cerulean in these molecules does not dramatically affect the function of these proteins. First, we observed that addition of β-sheets at the N-terminus increases the urea requirement for A2 cleavage. In this regard, while S- and XS-VWF are separated by α-helices and these were cleaved in the absence of urea, M-VWF includes the β2- β3 sheets while L-VWF includes an additional β1 sheet. This observation is consistent with the report by Kokame [15], who applied western blot analysis to study various truncated VWF fragments that lacked GFP-variants. While some protein had to be refolded from inclusion bodies in this previous study [15] and this complicated interpretation, the molecules we produced were soluble. In addition to differences in the protein constructs used in the two studies, differences in the E. coli culture conditions may also contribute to enhanced protein solubility in our work. Second, the urea requirement for L-VWF proteolysis was similar to multimeric plasma VWF. In both cases, while some proteolysis was observed at urea>1.6M, complete VWF-A2 proteolysis required denaturation using 4M urea.
XS-VWF reports on plasma ADAMTS13 activity
XS-VWF can be used as a rapid, reliable, sensitive and cost-effective reagent to monitor plasma ADAMTS13 activity. It can detect ADAMTS13 activity below 10% normal levels within 30 min. Such a reagent is important since it provides a measure of the hemostatic potential of blood. In addition to TTP, decreased ADAMTS13 activity is also associated with poor prognosis during sepsis-induced organ failure and with increased risk of non-fatal heart attack [11]. Reagents that detect ADAMTS13 activity can also be applied to distinguish TTP from other unrelated disorders like hemolytic uremic syndrome (HUS), which present similar clinical symptoms [23; 24].
While several methodologies have been previously developed to assay for ADAMTS13 activity [25], XS-VWF provides some advantages. In comparison to traditional assays that monitor ULVWF multimer distribution using western blot analysis, results from XS-VWF are obtained more rapidly and they are quantitative. As opposed to indirect measures of ADAMTS13 activity that monitor the binding of proteolytically cleaved patient/sample VWF to collagen in ELISA type assays [26], XS-VWF provides a more direct functional readout of enzyme activity. The experimental protocol is also less technically difficult to perform and it is rapid. Thus, XS-VWF shares many of the advantages of the fluorogenic substrate VWF FRETS-73 [15; 16]. In addition, XS-VWF is a recombinant protein that can likely be produced in a cost-effective manner. Under our assay conditions, it also exhibits a more linear relationship with plasma ADAMTS13 activity compared to VWF FRETS-73 and it more reliably detected low levels of ADAMTS13 activity. Finally, XS-VWF utilizes higher excitation and emission wavelengths compared to FRETS-VWF73. This reduces the effects of auto-fluorescence due to plasma proteins. The use of higher wavelengths may also partially improve substrate performance in plasma samples having high levels of bilirubin since this yellow product quenches light with peak absorbance at 450nm [27].
While XS-VWF is likely to detect many instances of ADAMTS13 defect, the reagent has limitations since it lacks the additional domains of VWF. It is established that extensive molecular interaction between VWF and ADAMTS13 enable the proteolysis of the VWF-A2 domain. Exosites of ADAMTS13 bind sites in VWF are located both within and outside the VWF-A2 domain. In this regard, based on X-ray crystallography and enzymatic investigations of the non-catalytic domains of ADAMTS13, it is suggested that multiple exosites located in the dis-integrin-like (D), cysteine-rich (C) and spacer (S) domains of ADAMTS13 engage VWF-A2 [28]. The amino acids engaged by these exosites are located in the A2-domain between Pro1645-Arg1668 [29]. In addition, Zanardelli et al. [30] suggest that VWF amino acids between the D4-CK domains interact with the TSP5-CUB region of ADAMTS13. Mice lacking the TSP7-CUB at the C-terminal end of ADAMTS13 also exhibit reduced ADAMTS13 enzyme activity [31], and increased thrombus volume in regions of high shear stress [32]. Thus, while XS-VWF is likely to detect reduced activity of ADAMTS13 in most instances, it may not assay alterations in ADAMTS13 activity that are caused by mutations in enzyme exosite regions that engage domains other than the C-terminal section of VWF-A2. In addition, XS-VWF cannot be used in shear based assays since the molecule is too small to ‘feel’ the effect of hydrodynamic shear [9]. It may also not be effective in instances where plasma is contaminated with hemoglobin since this is reported to inhibit ADAMTS13 activity [33].
Progressive denaturation of VWF-A2 with urea concentration
While the smallest FRET protein is well suited for measuring ADAMTS13 activity, the largest molecule (L-VWF) provides insight into the dynamics of VWF conformation change upon addition of denaturant. In this regard, others have used urea as a surrogate for fluid shear since this denaturant exposes the cryptic Tyr1605-Met1606 cleavage site in VWF-A2 [34]. Our studies show that protein conformation changes progressively with urea concentration which is consistent with the notion that multiple salt bridges stabilize the A2 domain. Alternatively, as shown for cold-shock protein CspTm, chymotrypsin inhibitor 2 and other biomolecules [35; 36], VWF-A2 FRET may exist in only a limited number of states with urea regulating the distribution of the protein in these states. Additional single molecule FRET (smFRET) studies over a wider range of denaturation conditions are necessary in order to determine the number of intermediate states that exist between fully-folded and fully-denatured VWF-A2.
In conclusion, the current study introduces new CFP/YFP based FRET biosensors to measure VWF conformation change. While the current proteins are expressed in E. coli, similar constructs can also be expressed in mammalian systems. CFP/YFP may also be incorporated into multimeric VWF to quantify structural changes in VWF due to hydrodynamic shear [10; 37; 38]. Such FRET based proteins can be used to study the dynamics of VWF structural change under hydrodynamic shear, including the effects of protein glycosylation, trans-cis proline isomerization and vicinal cysteines [14].
Supplementary Material
Acknowledgments
This work was supported by NIH grant HL 77258. We thank Elizabeth Wuitschick for technical assistance. XS-VWF is available from the corresponding author.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Fujikawa K, Suzuki H, McMullen B, Chung D. Purification of human von Willebrand factor-cleaving protease and its identification as a new member of the metalloproteinase family. Blood. 2001;98:1662–6. doi: 10.1182/blood.v98.6.1662. [DOI] [PubMed] [Google Scholar]
- 2.Gerritsen HE, Robles R, Lammle B, Furlan M. Partial amino acid sequence of purified von Willebrand factor-cleaving protease. Blood. 2001;98:1654–61. doi: 10.1182/blood.v98.6.1654. [DOI] [PubMed] [Google Scholar]
- 3.Soejima K, Mimura N, Hirashima M, Maeda H, Hamamoto T, Nakagaki T, Nozaki C. A novel human metalloprotease synthesized in the liver and secreted into the blood: possibly, the von Willebrand factor-cleaving protease? J Biochem. 2001;130:475–80. doi: 10.1093/oxfordjournals.jbchem.a003009. [DOI] [PubMed] [Google Scholar]
- 4.Tsai HM, Sussman, Nagel RL. Shear stress enhances the proteolysis of von Willebrand factor in normal plasma. Blood. 1994;83:2171–9. [PubMed] [Google Scholar]
- 5.Shim K, Anderson PJ, Tuley EA, Wiswall E, Sadler JE. Platelet-VWF complexes are preferred substrates of ADAMTS13 under fluid shear stress. Blood. 2008;111:651–7. doi: 10.1182/blood-2007-05-093021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang J, Roth R, Heuser JE, Sadler JE. Integrin alpha(v)beta(3) on human endothelial cells binds von Willebrand factor strings under fluid shear stress. Blood. 2009;113:1589–97. doi: 10.1182/blood-2008-05-158584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Donadelli R, Orje JN, Capoferri C, Remuzzi G, Ruggeri ZM. Size regulation of von Willebrand factor-mediated platelet thrombi by ADAMTS13 in flowing blood. Blood. 2006;107:1943–50. doi: 10.1182/blood-2005-07-2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shida Y, Nishio K, Sugimoto M, Mizuno T, Hamada M, Kato S, Matsumoto M, Okuchi K, Fujimura Y, Yoshioka A. Functional imaging of shear-dependent activity of ADAMTS13 in regulating mural thrombus growth under whole blood flow conditions. Blood. 2008;111:1295–8. doi: 10.1182/blood-2007-09-110700. [DOI] [PubMed] [Google Scholar]
- 9.Shankaran H, Neelamegham S. Hydrodynamic forces applied on intercellular bonds, soluble molecules, and cell-surface receptors. Biophys J. 2004;86:576–88. doi: 10.1016/S0006-3495(04)74136-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dayananda KM, Singh I, Mondal N, Neelamegham S. von Willebrand factor self-association on platelet GpIb{alpha} under hydrodynamic shear: effect on shear-induced platelet activation. Blood. 2010;116:3990–8. doi: 10.1182/blood-2010-02-269266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tsai HM. Pathophysiology of thrombotic thrombocytopenic purpura. Int J Hematol. 2010;91:1–19. doi: 10.1007/s12185-009-0476-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rizzo MA, Springer GH, Granada B, Piston DW. An improved cyan fluorescent protein variant useful for FRET. Nat Biotechnol. 2004;22:445–9. doi: 10.1038/nbt945. [DOI] [PubMed] [Google Scholar]
- 13.Nagai T, Ibata K, Park ES, Kubota M, Mikoshiba K, Miyawaki A. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat Biotechnol. 2002;20:87–90. doi: 10.1038/nbt0102-87. [DOI] [PubMed] [Google Scholar]
- 14.Zhang Q, Zhou YF, Zhang CZ, Zhang X, Lu C, Springer TA. Structural specializations of A2, a force-sensing domain in the ultralarge vascular protein von Willebrand factor. Proc Natl Acad Sci U S A. 2009;106:9226–31. doi: 10.1073/pnas.0903679106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kokame K, Matsumoto M, Fujimura Y, Miyata T. VWF73, a region from D1596 to R1668 of von Willebrand factor, provides a minimal substrate for ADAMTS-13. Blood. 2004;103:607–12. doi: 10.1182/blood-2003-08-2861. [DOI] [PubMed] [Google Scholar]
- 16.Kokame K, Nobe Y, Kokubo Y, Okayama A, Miyata T. FRETS-VWF73, a first fluorogenic substrate for ADAMTS13 assay. Br J Haematol. 2005;129:93–100. doi: 10.1111/j.1365-2141.2005.05420.x. [DOI] [PubMed] [Google Scholar]
- 17.Peyvandi F, Palla R, Lotta LA, Mackie I, Scully MA, Machin SJ. ADAMTS-13 assays in thrombotic thrombocytopenic purpura. J Thromb Haemost. 2010;8:631–40. doi: 10.1111/j.1538-7836.2010.03761.x. [DOI] [PubMed] [Google Scholar]
- 18.Singh I, Shankaran H, Beauharnois ME, Xiao Z, Alexandridis P, Neelamegham S. Solution structure of human von Willebrand factor studied using small angle neutron scattering. J Biol Chem. 2006;281:38266–75. doi: 10.1074/jbc.M607123200. [DOI] [PubMed] [Google Scholar]
- 19.Shankaran H, Alexandridis P, Neelamegham S. Aspects of hydrodynamic shear regulating shear-induced platelet activation and self-association of von Willebrand factor in suspension. Blood. 2003;101:2637–45. doi: 10.1182/blood-2002-05-1550. [DOI] [PubMed] [Google Scholar]
- 20.Soejima K, Nakamura H, Hirashima M, Morikawa W, Nozaki C, Nakagaki T. Analysis on the molecular species and concentration of circulating ADAMTS13 in Blood. J Biochem. 2006;139:147–54. doi: 10.1093/jb/mvj013. [DOI] [PubMed] [Google Scholar]
- 21.Piston DW, Kremers GJ. Fluorescent protein FRET: the good, the bad and the ugly. Trends Biochem Sci. 2007;32:407–14. doi: 10.1016/j.tibs.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 22.Tsai HM, Lian EC. Antibodies to von Willebrand factor-cleaving protease in acute thrombotic thrombocytopenic purpura. N Engl J Med. 1998;339:1585–94. doi: 10.1056/NEJM199811263392203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Forzley BR, Clark WF. TTP/HUS and prognosis: the syndrome and the disease(s) Kidney Int Suppl. 2009:S59–61. doi: 10.1038/ki.2008.623. [DOI] [PubMed] [Google Scholar]
- 24.Marques MB. Thrombotic thrombocytopenic purpura and heparin-induced thrombocytopenia: two unique causes of life-threatening thrombocytopenia. Clin Lab Med. 2009;29:321–38. doi: 10.1016/j.cll.2009.03.003. [DOI] [PubMed] [Google Scholar]
- 25.Tripodi A, Peyvandi F, Chantarangkul V, Palla R, Afrasiabi A, Canciani MT, Chung DW, Ferrari S, Fujimura Y, Karimi M, Kokame K, Kremer Hovinga JA, Lammle B, de Meyer SF, Plaimauer B, Vanhoorelbeke K, Varadi K, Mannucci PM. Second international collaborative study evaluating performance characteristics of methods measuring the von Willebrand factor cleaving protease (ADAMTS-13) J Thromb Haemost. 2008;6:1534–41. doi: 10.1111/j.1538-7836.2008.03099.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gerritsen HE, Turecek PL, Schwarz HP, Lammle B, Furlan M. Assay of von Willebrand factor (vWF)-cleaving protease based on decreased collagen binding affinity of degraded vWF: a tool for the diagnosis of thrombotic thrombocytopenic purpura (TTP) Thromb Haemost. 1999;82:1386–9. [PubMed] [Google Scholar]
- 27.Meyer SISC, Lämmle B, Kremer Hovinga JA. Hyperbilirubinemia interferes with ADAMTS-13 activity measurement by FRETS-VWF73 assay: diagnostic relevance in patients suffering from acute thrombotic microangiopathies. J Thromb Haemost. 2007:866–867. doi: 10.1111/j.1538-7836.2007.02438.x. [DOI] [PubMed] [Google Scholar]
- 28.Akiyama M, Takeda S, Kokame K, Takagi J, Miyata T. Crystal structures of the noncatalytic domains of ADAMTS13 reveal multiple discontinuous exosites for von Willebrand factor. Proc Natl Acad Sci U S A. 2009;106:19274–9. doi: 10.1073/pnas.0909755106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu JJ, Fujikawa K, McMullen BA, Chung DW. Characterization of a core binding site for ADAMTS-13 in the A2 domain of von Willebrand factor. Proc Natl Acad Sci U S A. 2006;103:18470–4. doi: 10.1073/pnas.0609190103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zanardelli S, Chion AC, Groot E, Lenting PJ, McKinnon TA, Laffan MA, Tseng M, Lane DA. A novel binding site for ADAMTS13 constitutively exposed on the surface of globular VWF. Blood. 2009;114:2819–28. doi: 10.1182/blood-2009-05-224915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou W, Bouhassira EE, Tsai HM. An IAP retrotransposon in the mouse ADAMTS13 gene creates ADAMTS13 variant proteins that are less effective in cleaving von Willebrand factor multimers. Blood. 2007;110:886–93. doi: 10.1182/blood-2007-01-070953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Banno F, Chauhan AK, Kokame K, Yang J, Miyata S, Wagner DD, Miyata T. The distal carboxyl-terminal domains of ADAMTS13 are required for regulation of in vivo thrombus formation. Blood. 2009;113:5323–9. doi: 10.1182/blood-2008-07-169359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Studt JD, Hovinga JA, Antoine G, Hermann M, Rieger M, Scheiflinger F, Lammle B. Fatal congenital thrombotic thrombocytopenic purpura with apparent ADAMTS13 inhibitor: in vitro inhibition of ADAMTS13 activity by hemoglobin. Blood. 2005;105:542–4. doi: 10.1182/blood-2004-06-2096. [DOI] [PubMed] [Google Scholar]
- 34.Auton M, Cruz MA, Moake J. Conformational stability and domain unfolding of the Von Willebrand factor A domains. J Mol Biol. 2007;366:986–1000. doi: 10.1016/j.jmb.2006.10.067. [DOI] [PubMed] [Google Scholar]
- 35.Schuler B, Eaton WA. Protein folding studied by single-molecule FRET. Curr Opin Struct Biol. 2008;18:16–26. doi: 10.1016/j.sbi.2007.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Borgia A, Williams PM, Clarke J. Single-molecule studies of protein folding. Annu Rev Biochem. 2008;77:101–25. doi: 10.1146/annurev.biochem.77.060706.093102. [DOI] [PubMed] [Google Scholar]
- 37.Singh I, Themistou E, Porcar L, Neelamegham S. Fluid shear induces conformation change in human blood protein von Willebrand factor in solution. Biophys J. 2009;96:2313–20. doi: 10.1016/j.bpj.2008.12.3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Themistou E, Singh I, Shang C, Balu-Iyer SV, Alexandridis P, Neelamegham S. Application of fluorescence spectroscopy to quantify shear-induced protein conformation change. Biophys J. 2009;97:2567–76. doi: 10.1016/j.bpj.2009.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Anderson PJ, Kokame K, Sadler JE. Zinc and calcium ions cooperatively modulate ADAMTS13 activity. J Biol Chem. 2006;281:850–7. doi: 10.1074/jbc.M504540200. [DOI] [PubMed] [Google Scholar]
- 40.Zanardelli S, Crawley JT, Chion CK, Lam JK, Preston RJ, Lane DA. ADAMTS13 substrate recognition of von Willebrand factor A2 domain. J Biol Chem. 2006;281:1555–63. doi: 10.1074/jbc.M508316200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.