Abstract
Francisella tularensis is a category A select agent based on its infectivity and virulence but disease mechanisms in Francisella tularensis infection remain poorly understood. Murine pulmonary models of infection were therefore employed to assess and compare dissemination and pathology and to elucidate the host immune response to infection with the highly virulent Type A F. tularensis strain Schu4 versus the less virulent Type B live vaccine strain (LVS). We found that dissemination and pathology in the spleen was significantly greater in mice infected with F. tularensis Schu4 compared to mice infected with F. tularensis LVS. Using gene expression profiling to compare the response to infection with the two F. tularensis strains, we found that there were significant differences in the expression of genes involved in the apoptosis pathway, antigen processing and presentation pathways, and inflammatory response pathways in mice infected with Schu4 when compared to LVS. These transcriptional differences coincided with marked differences in dissemination and severity of organ lesions in mice infected with the Schu4 and LVS strains. Therefore, these findings indicate that altered apoptosis, antigen presentation and production of inflammatory mediators explain the differences in pathogenicity of F. tularensis Schu4 and LVS.
Keywords: Francisella tularensis, immune, bacterial, microarray, pathology
1. Introduction
Francisella tularensis causes a fatal disseminated infection upon inhalation. While the majority of clinically diagnosed disease is due to arthropod bites or handling of infectious material, interest in Francisella pneumonic infection has been renewed due to the fear of bioterrorism and its history of weaponization [1, 2]. Pneumonic tularemia is the most severe form of the disease and can result in mortality if treatment is not initiated early in infection. In addition, relapse after the completion of treatment is a primary concern with bacteria of such high virulence [3–5]. Despite a substantial revival in F. tularensis research, the mechanisms of pathogenesis and dissemination remain to be elucidated. Greater knowledge of the host-pathogen interaction will aid in development of protective vaccines and effective chemotherapeutics.
F. tularensis Schu4 and LVS were selected for these studies because of the known difference in virulence associated with these strains, which provides a model with which to assess differences in host interaction and response genes [6–10]. Schu4 and LVS belong to the F. tularensis subspecies tularensis and holarctica respectively [11, 12]. Subspecies tularensis, referred to as type A, represents the most virulent of the Francisella subgroups whereas subspecies holarctica, referred to as type B, tends to be less deadly in humans [1]. LVS is the vaccine strain of F. tularensis, which was derived from a less virulent type B isolate [12]. While F. tularensis LVS retains its virulence in mice, lethal infection requires challenge with greater than103 CFU by the pulmonary route, whereas challenge with Schu4 causes a consistent lethal infection with fewer than 102 CFU [9, 7]. In addition time-to-death in the murine pulmonary infection model differs, with Schu4 infection typically resulting in death by 120 hours post infection, while LVS infected mice survive up to 14 days following infection [13, 7, 9, 14]
To understand how infection with virulent F. tularensis leads to a rapidly disseminating and lethal infection, studies have been performed in a variety of different infection models. In vitro studies aimed at characterizing the transcriptional response to F. tularensis using multiple cell types have revealed some insights into the host pathogen interaction [15–18]. Andersson et al. examined the whole lung transcriptional response to infection with type A Francisella isolate FSC033, and found limited host gene expression in the first 4 days of infection, suggesting a subversion of host recognition and delayed immune responses until immediately before death [15]. Schu4 mutant strains have also been used to assess host-pathogen interactions [19–22]. The mutant bacterial strains are generally less virulent in mouse models of infection. However, studies with mutant strains of F. tularensis have not yet been employed to study the overall host response to infection. Rather, they have been more instrumental in assessing the role of specific bacterial components in establishing infection leading to pathology in the lung.
Monitoring bacterial growth and dissemination along with pathology and in vivo transcriptional profiling of the host response to infection has provided important advances in understanding the host-pathogen interaction for organisms such as Listeria [23], Mycobacteria [24], and Yersinia [25]. Therefore, we believe this is also an appropriate technique for assessing the host response to infection in the F. tularensis mouse model of infection. The present work is to our knowledge the first comprehensive comparative study to define the host transcriptional response to F. tularensis infection following dissemination from the lungs to secondary sites of infection. In the present study, bacterial burden was monitored, pathology was assessed, and global gene expression was examined throughout the course of infection with F. tularensis, comparing infection with the Schu4 and LVS strains in a murine model. Here we report significant differences in pathology and regulation of expression of host immune response genes following infection with the Schu4 and LVS strains of F. tularensis.
2. Materials and Methods
2.1 Bacterial strains
F. tularensis Schu4 and LVS were provided by Dr. J. Peterson (Centers for Disease Control, Fort Collins, CO). Schu4 and LVS were cultured in modified Mueller-Hinton broth at 37 °C with constant shaking overnight, supplemented with 10% glycerol and aliquoted into 1 ml samples, frozen at −80 °C, and thawed just before use. Frozen stocks were titered by enumerating viable bacteria from serial dilutions plated on modified Mueller-Hinton agar as previously described [26]. The number of viable bacteria in frozen stock vials varied <5% over a 10-month period.
2.2 Mice
Six week-old female C57BL/6 mice were purchased from Jackson Laboratories, Bar Harbor, Maine. All mice were housed in sterile micro-isolator cages in the laboratory animal resources facility or in the Rocky Mountain Regional Biocontainment Laboratory BSL-3 facility at Colorado State University (Fort Collins, CO) and provided water and food ad libitum. All research involving animals was conducted in accordance with the Animal Care and Use Committee approved animal guidelines and protocols.
2.2.1 Murine models of infection
Mice were infected with either F. tularensis Schu4 or F. tularensis LVS via intranasal (i.n.) or aerosol routes as described previously [27, 28] depending on the objective of the study. For pathology, qRT-PCR, and bacterial burden studies mice were infected via intranasal route. Mice were anesthetized with ketamine-xylazine and 10 μL inocula was administered to each of the nares in sequential droplets allowing mice to inhale the fluid (20 μL total). Infected mice were monitored for morbidity twice daily and were euthanized at pre-determined endpoints. For global transcriptional profiling, mice were exposed to F. tularensis Schu4 or F. tularensis LVS by exposure in a Glas-Col Inhalation Exposure System (Glas-Col, Inc, Terre Haute, IN). Exposure was conducted by aerosolizing approximately 3.5 × 107 CFU in a volume of 5 cubic feet over a period of 30 min, followed by a 20 minute period of cloud decay.
2.3. Histopathology
C57BL/6 mice (n = 4 per group per time point) were infected i.n. with the F. tularensis Schu4 strain (102 CFU) or the F. tularensis LVS strain (104 CFU) and then sacrificed at 48 and 120 hours after exposure. Lung and spleen tissues were removed, divided and placed in 10% neutral buffered formalin for histopathology or in sterile PBS for bacterial quantification. Organs for histopathological examination were fixed, imbedded in paraffin, sectioned, and stained with hematoxylin and eosin.
2.4 Bacterial quantification
Samples of lung and spleen tissues were homogenized in 5 mL sterile PBS using a stomacher (Teledyne Tekmar, Mason, OH). Bacterial CFU per mL of organ homogenate were determined by plating serial 10-fold dilutions of organ homogenates on modified Mueller-Hinton agar and incubating at 37 °C for 72 hours. RT-PCR was carried out on RNA samples from the lungs using a 16s primer set and approach adapted from Cole et al. [29]. Relative detection of 16s molecules was determined using the ΔCT method.
2.5 RNA Isolation and amplification
RNA was stabilized and recovered from mouse organs by the addition of TRIzol reagent and organic partitioning. Total RNA was extracted from the TRIzol by the addition of chloroform (1:1) to achieve a bi-phase separation, then precipitated by the addition of isopropanol and subjected to Dnase treatment, and purified using a Qiagen RNeasy kit (Valencia, CA). Messenger RNA was converted to cDNA using poly(T) primers and amplified in the presence of modified dUTPs using the AminoAllyl Message Amplification Kit (Ambion, Foster City, CA). Indirect labeling of cDNA for hybridization was conducted by conjugating Cy3 dye with modified dUTPs in a subsequent reaction.
2.6 Microarray Scanning and Analysis
Full mouse genome version 4.0.3 (Operon Biotechnology, Huntsville, AL) cDNA spotted microarrays were obtained from the Genomics Proteomics Core of the Rocky Mountain Regional Center of Excellence (http://www.rmrce.colostate.edu/). The 70mer oligonucleotide cDNAs were printed on poly-amine coated slides (ArrayIt Corporation, Sunnyvale, CA) and post-processed by UV cross linking and blocking with 10% BSA and 3X SSC at 42 °C. Dye coupled cDNA, was combined with yeast tRNA (10 mg/mL), and hybridization buffer (formamide, 20XSSC and 10%SDS) and heated. Single channel (Cy3) hybridization was carried out in triplicate for each sample. Slides were scanned using the Genepix 4000B (Molecular Devices, Sunnyvale, CA) fluorescent scanner and analyzed using Genepix Pro 6.0 software. Background fluorescence was corrected for by subtracting background from foreground intensity values. Technical replicates were averaged before normalizing to the global mean intensity values from the entire data set. Log transformation, t-test, ANOVA, principal component analysis and Benjamini and Hochberg false discovery correction were applied to the data using the Genesifter software (Geospiza, Seattle, WA). Genes considered to be differentially expressed were induced or repressed by 1.5 fold or higher and had a p-value of 0.01 or lower. Clustering was conducted using Cluster software [30] (http://rana.lbl.gov/eisen/?page_id=42). Functional enrichment analysis was conducted using the DAVID Bioinformatics Database [31, 32] (http://david.abcc.ncifcrf.gov/). Response to each strain was then compared to controls to examine changes in expression of genes during the progression of the infections. The complete dataset is available through the Gene Expression Omnibus (GEO) database using accession # GSE22203.
2.7 qRT-PCR
Quantitative real time PCR was used to assess bacterial burden in infected tissues, validate microarray data, and monitor molecular markers of disease. Briefly, cDNA synthesis from total RNA was carried out using First Strand cDNA Synthesis Kit (Invitrogen, Carlsbad, CA). Briefly, 1 mg of total RNA was combined with random hexamer and oligo (dT) primers and heated in 10ml total volume for 5 minutes. 10 μL of buffered enzyme mix (2 μL 10X buffer, 4 μL MgCl2 (6 mM), 2 μL DTT (0.1 M), 1 μL RNAse out, and 1 μL Superscripttm) was added and incubated at 25 °C for 10 minutes, 50 °C for 50 minutes, and 85 °C for 5 minutes. Platinum SYBR Green qPCR Supermix-UDG (Invitrogen, Carlsbad, CA) was combined with gene specific primers (5nmol) and 50ng of template (cDNA) and run in triplicate on an IQ5 thermocycler (Bio-Rad, Hercules, CA). The transcripts encoding 18S rRNA, GapDH, and β-actin were used to monitor consistency in biological replicates. Other genes described in the text where employed to confirm the expression trends identified by microarray analysis. Resulting data from each condition was compared to controls in an independent fashion using the ΔCT method.
3. Results
3.1 Dissemination, and lung and spleen pathology following infection
To assess possible differences in dissemination to the spleen, and pathology between F. tularensis strains Schu4 and LVS, mice were infected by the intranasal route. The intranasal route of infection was chosen for the dissemination and pathology studies in order to facilitate equalizing the bacterial burden in the lungs at the 48h time point. To accomplish this, it was necessary to accurately administer higher challenge doses of F. tularensis LVS than for F. tularensis Schu4. In addition, we also employed a higher intranasal challenge dose of F. tularensis LVS because there was minimal lung pathology noted when mice were subjected to low-dose aerosol challenge with F. tularensis LVS (data not shown). Accordingly, by assuring that mice had equivalent bacterial burdens at the appropriate time points after infection, we were able to directly compare the efficiency of bacterial dissemination from the lungs and the associated organ pathology.
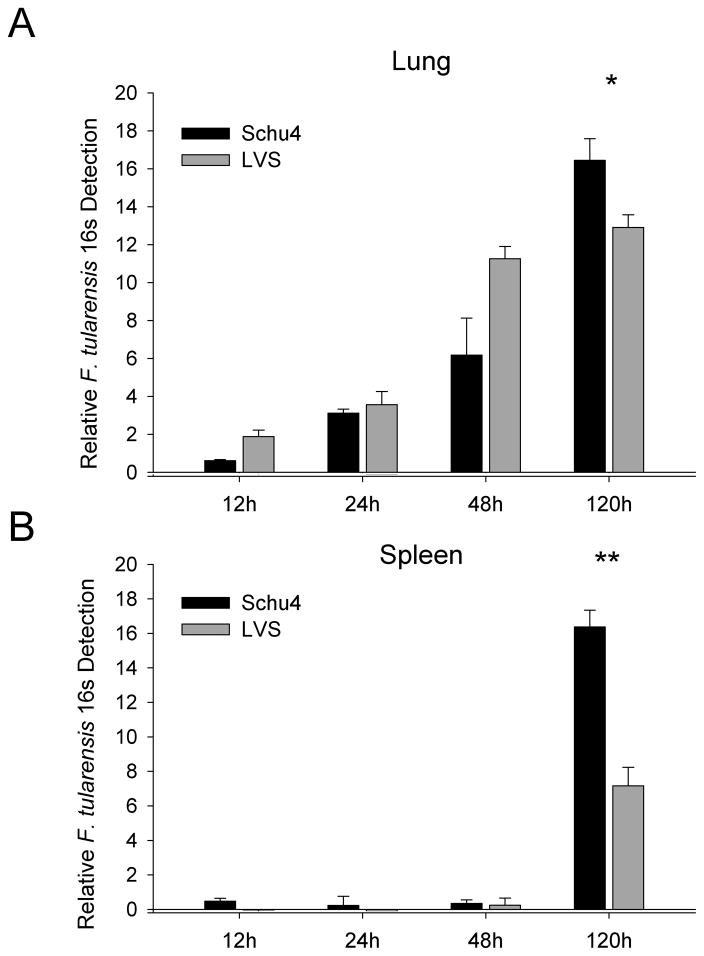
Bacterial load in lung and spleen tissue at different times of infection was determined by molecular detection of F tularensis 16S RNA and confirmed by direct plating of organ homogenates for F. tularensis colony detection. At 24 hours after inoculation, the bacterial load of Schu4 and LVS in the lungs was similar based on 16S RNA (Fig. 1A) and colony counting. For example, the lungs contained 4.4 ± 0.30 Log10 CFU Schu4 and 4.9 ± 0.30 Log10 CFU LVS at 24 hours of infection. In contrast, by 120 hours of infection, there was a significantly higher bacterial load in the lungs of mice infected with Schu4 (8.6 ± 0.09 Log10 CFU Schu4 versus 7.3 ± 0.49 Log10 CFU LVS). Although the bacterial load in the spleen was relatively low, Schu4 was detectable by 16S RNA as early as 24 hours after infection, while LVS was not detectable by 16S RNA until 48 hours after infection (Fig. 1B). At 120 hours after infection, Schu4 and LVS were detected in the spleen, though the splenic bacterial burden was significantly (p<0.001) higher in Schu4 infected mice (8.3 ± 0.26 Log10 CFU Schu4 versus 5.8 ± 0.28 Log10 CFU LVS). Overall, Schu4 demonstrated greater growth in the lungs, quicker dissemination to the spleen, and more rapid growth in the spleen compared to LVS. This observation is consistent with the known, more rapid disease progression and virulence of Schu4 compared to LVS [9, 7]. Further these data suggest that rapidity of dissemination to secondary sites is related to the extent of infection in the lungs.
Figure 1. Time-course of lung and spleen bacterial burden in mice infected with F. tularensis LVS and Schu4.
C57BL/6 mice (n = 4 per group) were inoculated i.n. with lethal doses of F. tularensis LVS (104 CFU) or Schu4 (102 CFU), as described in Methods. Lung and spleen tissues were collected 12, 24, 48 and 120 hours after infection and homogenized in TRiZol or PBS for isolation or total RNA or CFU enumeration, error bars represent standard deviation of all 4 samples. (A) F. tularensis 16s rRNA detection in the lungs of mice infected with Schu4 and LVS. (B) F. tularensis 16s rRNA detection in the spleens of mice infected with Schu4 and LVS. Data show similar growth trends through 48 hours in the lung, whereas 120 hours post infection Schu4 shows statistically significantly higher numbers in both the lung and spleen. Data from each time point was subjected to students T-test, (*)=p<0.01, (**)=p<0.001.
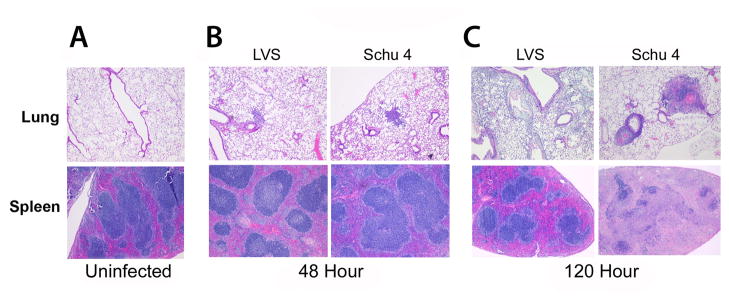
In lung tissues collected from mice 48 hours after infection with either Schu4 or LVS, there was mild to moderate perivascular edema, extravasation of erythrocytes and mild leukocyte margination in pulmonary vessels (Fig. 2A). In addition, there were perivascular interstitial accumulations of granulocytes, monocytes and macrophages, which were accompanied by minimal cell degeneration and necrosis. At 120 hours of infection both F. tularensis Schu4 and LVS infected lungs had pathologic changes consisting of multifocal interstitial edema and increased infiltration of granulocytes and monocytes and macrophages along with intra-alveolar accumulations of macrophages (Fig. 2B). While infection with both Schu4 and LVS induced lung pathology, the lungs from F. tularensis LVS-infected mice lacked abscesses and had less well-defined lesions with minimal necrosis compared to lungs from F. tularensis Schu4-infected mice, which were characterized by prominent abscesses and well-defined lesions with an increased amount of necrosis (Figs. 2B, C).
Figure 2. Time-course of lung and spleen pathology in mice infected with F. tularensis LVS and Schu4.
C57BL/6 mice (n = 4 per group) were inoculated i.n. with lethal doses of F. tularensis LVS (104 CFU) or Schu4 (102 CFU), as described in Methods. Lung and spleen tissues were collected 48 hours or 120 hours after infection and processed with hematoxylin and eosin staining for histopathological examination. (A) Histology from the lung and spleen of control (uninfected) mice. (B) Histology from the lung and spleen 48 hours post-infection with Schu4 or LVS. (C) Histology from the lung and spleen 120 hours post-infection with Schu4 or LVS. Pathological changes at 48 hours after infection were mild in both the lungs and spleen and indistinguishable between F. tularensis LVS and Schu4 infected mice. At 120 hours after infection, more severe lesions were noted in the lungs and especially the spleens of F. tularensis Schu4 infected mice, compared to LVS infected mice. Image magnification was 40X for all images displayed.
Spleen tissues from F. tularensis Schu4 and LVS infected mice collected 48 hours after infection were histologically unremarkable and indistinguishable from spleens of uninfected animals (Figs. 2A, B). By 120 hours after infection (Fig. 2C), spleens from F. tularensis LVS infected mice had mild lymphocyte depletion of the white pulp and multifocal accumulations of granulocytes and macrophages in the red pulp, with little evidence of necrosis. In contrast, spleens from F. tularensis Schu4-infected mice had almost complete destruction of parenchymal structures due to diffuse severe necrosis, fibrin-deposition and massive lymphocyte depletion. The marked increase in spleen pathology in F. tularensis Schu4-infected mice was the most notable histological difference between infections caused by the two strains of bacteria.
3.2 Common trends in the host response to F. tularensis Schu4 and LVS infection
Whole genome transcriptional profiling of lungs and spleen tissues collected at 12, 24, 48, and 120 hours of infection from mice infected via aerosol with Schu4 or LVS was conducted to investigate the global host response to infection with each bacterium. Low dose aerosol inoculation was used for the transcriptional studies in mice because this route is believed to more closely approximate human infection by inhalation of F. tularensis than other routes of infection. Genes that were considered to be differentially regulated had a variance <0.01 (ANOVA) and were up or down-regulated > 1.5 fold compared to uninfected mice. The complete dataset is available through the Gene Expression Omnibus (GEO) Accession # GSE22203.
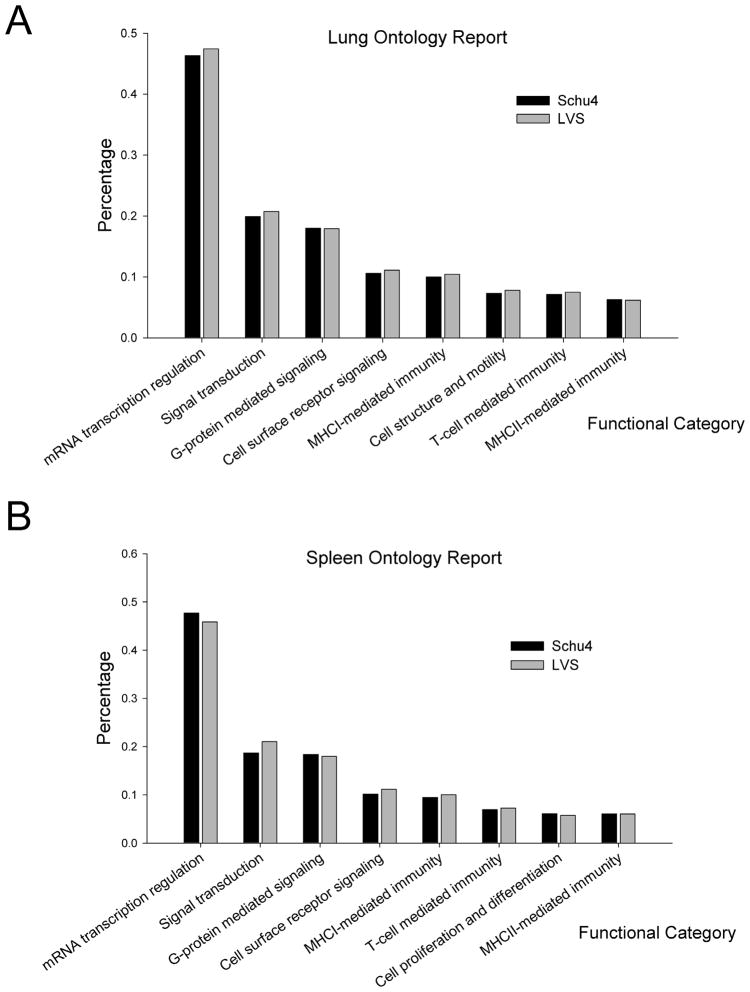
The total number of differentially expressed genes in the lung and spleen paralleled the bacterial burden. Infection with Schu4 resulted in differential regulation of 3,958 and 5,442 genes in lungs and spleen respectively, compared to uninfected mice. A similar range of differences in global responses was also observed in LVS infected mice, which resulted in 2,230 differentially regulated genes in the lungs and 9,388 differentially modulated open reading frames in the spleen. Global gene expression response data from all time points of infection with LVS and Schu4 were interrogated to identify ontologies and pathways that were over-represented in the host response to infection (Fig. 3A and 3B). Genes associated with inflammation, host-pathogen interactions, cellular activation/differentiation, host antimicrobial activity, and leukocyte receptor signaling constituted the majority of the host response to infection with both strains of F. tularensis (Table 1).
Figure 3. Functional enrichment of global transcriptional response data.
C57BL/6 mice (n = 2 per group) were inoculated via aerosol with lethal doses of F. tularensis LVS or Schu4 (104 CFU), as described in Methods. Total RNA from the lung and spleen tissues was collected 12, 24, 48 and 120 hours post infection, converted to cDNA, labeled and hybridized on full mouse genome microarrays. (A) Ontology analysis showing select functional categories relevant to infection in the lung in response to infection with Schu4 or LVS. (B) Ontology analysis showing select functional categories relevant to infection in the spleen in response to infection with Schu4 or LVS. Genes with a p-value < 0.01 and differentially regulated > 1.5 fold were used for clustering and ontology analysis.
Table 1. Genes with similar expression patterns in F. tularensis Schu4 and LVS infection.
C57BL/6 mice (n = 2 per group) were inoculated via aerosol with lethal doses of F. tularensis LVS or Schu4 (104 CFU), as described in Methods. Total RNA Lung and spleen tissues were collected 12, 24, 48 and 120 hours post infection, converted to cDNA, labeled and hybridized on full mouse genome microarrays. Genes with a p-value < 0.01 and differentially regulated > 1.5 fold were mined for genes common to each infection that fell into the categories of inflammatory response, cellular activation/differentiation, antimicrobial activity, leukocyte receptors, and cell signaling.
|
Lung | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Gene ID | Annotation | Accession |
Schu4 Infection |
LVS Infection |
||||||
| 12h | 24h | 48h | 120h | 12h | 24h | 48h | 120h | |||
| I. Inflammatory Response | ||||||||||
| Ccl25 | Chemokine (C-C motif) ligand 25 | NM_009138 | - | 1.94 | - | - | - | 2.47 | - | - |
| Chi3l1 | Chitinase 3-like 1 | NM_007695 | 3.92 | 3.84 | 4.03 | - | 2.54 | - | - | - |
| Chi3l4 | Chitinase 3-like 4 | NM_145126 | - | - | - | 2.33 | - | 2.87 | - | - |
| Csf2 | Colony stimulating factor 2 (granulocyte-macrophage) | NM_009969 | - | - | −1.57 | - | - | - | −1.60 | - |
| Cxcl14 | Chemokine (C-X-C motif) ligand 14 | NM_019568 | - | - | −1.97 | −1.58 | - | - | - | −2.48 |
| Cxcr7 | Chemokine (C-X-C motif) receptor 7 | NM_007722 | −1.50 | −2.18 | - | - | - | −2.59 | - | - |
| Il10ra | Interleukin 10 receptor, alpha | NM_008348 | - | −1.73 | - | - | - | −1.98 | - | - |
| Il10rb | Interleukin 10 receptor, beta | NM_008349 | - | - | - | −3.35 | - | −1.81 | - | - |
| Il18bp | Interleukin 18 binding protein | NM_010531 | - | - | - | 3.34 | - | - | - | 3.07 |
| Il1b | Interleukin 1 beta | NM_008361 | −3.22 | −2.32 | - | −2.18 | - | −2.95 | −2.46 | - |
| Il33 | Interleukin 33 | NM_133775 | −2.13 | −1.80 | - | −3.36 | - | −2.01 | - | - |
| Il9r | Interleukin 9 receptor | NM_008374 | - | - | - | −2.46 | - | −1.85 | - | - |
| II. Cellular Activation/Differentiation | ||||||||||
| Cd109 | CD109 antigen | NM_153098 | - | - | −2.24 | - | - | - | - | −2.03 |
| Cd2 | CD2 antigen | NM_013486 | - | - | 2.16 | - | - | 2.06 | - | - |
| Cd55 | CD55 antigen | NM_010016 | - | −2.67 | - | - | - | −2.20 | - | - |
| Cd63 | Cd63 antigen | NM_007653 | - | 3.09 | 2.72 | - | - | - | 2.16 | - |
| III. Antimicrobial Activity | ||||||||||
| Mmp8 | Matrix metallopeptidase 8 | NM_008611 | - | - | - | 3.60 | - | - | - | 1.64 |
| Timp1 | Tissue inhibitor of metalloproteinase 1 | NM_011593 | - | - | - | 2.61 | - | - | - | 3.86 |
| IV. Leukocyte Receptors | ||||||||||
| Klra22 | Killer cell lectin-like receptor subfamily A, member 22 | NM_053152 | - | - | −1.36 | −2.38 | - | - | - | 1.94 |
| V. Cell Signaling | ||||||||||
| Ptger1 | Prostaglandin E receptor 1 (subtype EP1) | NM_013641 | - | - | - | 2.59 | - | - | 2.43 | - |
| Spleen | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Gene ID | Annotation | Accession |
Schu4 Infection |
LVS Infection |
||||||
| 12 | 24 | 48 | 120 | 12 | 24 | 48 | 120 | |||
| I. Apoptosis | ||||||||||
| Aifm1 | Apoptosis-inducing factor, mitochondrion-associated 1 | NM_012019 | - | - | −1.82 | −3.97 | - | −1.78 | −1.82 | −3.08 |
| Bnip2 | BCL2/adenovirus E1B interacting protein 1, NIP2 | NM_016787 | −1.74 | −2.28 | −1.83 | −3.01 | - | −1.75 | −1.66 | −2.55 |
| Bnip3l | BCL2/adenovirus E1B interacting protein 3-like | NM_009761 | - | - | - | −3.11 | - | - | - | −3.58 |
| Casp7 | Caspase 7 | NM_007611 | 1.70 | - | - | - | - | - | 1.77 | - |
| Pdcd2 | Programmed cell death 2 | NM_008799 | - | - | - | −2.36 | - | - | - | −2.37 |
| II. Inflammatory Response | ||||||||||
| Ccl21b | Chemokine (C-C motif) ligand 21b | NM_011124 | - | - | - | −3.11 | - | - | - | −2.00 |
| Ccr2 | Chemokine (C-C motif) receptor 2 | NM_009915 | - | −2.47 | - | −3.87 | - | −1.69 | - | −2.51 |
| Ccr6 | Chemokine (C-C motif) receptor 6 | NM_009835 | - | - | - | −3.67 | - | - | - | −2.34 |
| Ccr8 | Chemokine (C-C motif) receptor 8 | NM_007720 | - | 2.22 | - | 1.76 | - | - | - | 1.79 |
| Cx3cr1 | Chemokine (C-X3-C) receptor 1 | NM_009987 | - | - | - | −4.28 | - | - | - | −2.39 |
| Cxcl3 | Chemokine (C-X-C motif) ligand 3 | NM_203320 | - | - | - | 2.38 | - | - | - | 2.21 |
| Il10 | Interleukin 10 | NM_010548 | - | - | 2.65 | - | - | - | 1.72 | - |
| Il10rb | Interleukin 10 receptor, beta | NM_008349 | −1.83 | - | - | −3.44 | - | - | - | −3.69 |
| Il17a | Interleukin 17A | NM_010552 | - | - | - | −2.68 | - | −1.97 | - | −1.96 |
| Il18bp | Interleukin 18 binding protein | NM_010531 | - | - | - | 2.84 | - | - | - | 1.58 |
| Il18rap | Interleukin 18 receptor accessory protein | NM_010553 | - | −1.61 | - | −1.54 | - | - | −1.74 | - |
| Il1b | Interleukin 1 beta | NM_008361 | - | - | - | 1.75 | 2.03 | - | - | −2.10 |
| Il22 | Interleukin 22 | NM_016971 | - | - | - | 3.24 | - | - | - | 2.78 |
| Il3 | Interleukin 3 | NM_010556 | - | - | - | 3.41 | - | - | - | 2.37 |
| Tgfb1 | Transforming growth factor, beta 1 | NM_011577 | - | - | 1.83 | - | - | - | - | 1.75 |
| III. Cellular Activation/Differentiation | ||||||||||
| Cd163 | CD163 antigen | NM_053094 | - | - | - | −2.27 | - | - | - | −1.74 |
| Cd300a | CD300A antigen | NM_170758 | - | - | - | −3.31 | - | - | - | −2.12 |
| Cd34 | CD34 antigen | NM_133654 | 1.87 | - | 2.43 | 2.72 | - | - | 2.42 | 2.46 |
| Cd37 | CD37 antigen | NM_007645 | - | 2.01 | - | - | - | - | 2.99 | - |
| Cd48 | CD48 antigen | NM_007649 | - | - | - | −1.73 | - | - | - | −1.74 |
| Cd63 | Cd63 antigen | NM_007653 | - | - | - | 3.61 | - | - | 1.84 | - |
| Cd74 | CD74 antigen | NM_010545 | - | 2.84 | - | - | - | - | 2.66 | - |
| Cd79b | CD79B antigen | NM_008339 | - | 1.64 | - | - | - | - | - | −2.01 |
| Cd83 | CD83 antigen | NM_009856 | - | 1.67 | - | - | - | 1.77 | 1.61 | - |
| Cd86 | CD86 antigen | NM_019388 | - | −1.74 | - | −2.58 | - | - | - | −3.04 |
| Cd97 | CD97 antigen | NM_011925 | - | - | - | −1.74 | - | - | - | −1.97 |
| IV. Antimicrobial Activity | ||||||||||
| Adamts1 | A disintegrin-like and metallopeptidase thrombospondin type 1 motif, 1 | NM_009621 | - | - | - | 3.58 | - | - | - | 2.22 |
| C9 | Complement component 9 | NM_013485 | −1.55 | - | - | −2.65 | - | −1.68 | −1.52 | −1.99 |
| F5 | Coagulation factor V | NM_007976 | - | −1.74 | - | −3.14 | - | - | −2.05 | - |
| V. Leukocyte Receptors | ||||||||||
| H2-Ab1 | Histocompatibility 2, class II antigen A, beta 1 | NM_207105 | - | 3.02 | - | - | - | - | 4.07 | - |
| H2-Bl | Histocompatibility 2, blastocyst | NM_008199 | - | - | - | 2.55 | - | - | 2.03 | - |
| H2-M10.2 | Histocompatibility 2, M region locus 10.2 | NM_177923 | - | - | - | −1.82 | - | −2.39 | - | - |
| H2-M3 | Histocompatibility 2, M region locus 3 | NM_013819 | - | - | - | 3.81 | - | - | 2.84 | 3.31 |
| H2-Q8 | Histocompatibility 2, Q region locus 8 | NM_207648 | - | - | - | 2.75 | - | - | 1.98 | 2.30 |
| Klra1 | Killer cell lectin-like receptor, subfamily A, member 1 | NM_013793 | −1.90 | −3.63 | - | −3.82 | −2.53 | −2.45 | −1.67 | −3.69 |
| Klra10 | Killer cell lectin-like receptor subfamily A, member 10 | NM_008459 | - | −2.88 | - | −2.77 | −2.48 | −2.07 | - | - |
| Klra21 | Killer cell lectin-like receptor subfamily A, member 21 | NM_010650 | - | −2.18 | - | −2.16 | −2.75 | - | - | −1.87 |
| Klra22 | Killer cell lectin-like receptor subfamily A, member 22 | NM_053152 | - | −2.10 | - | −3.82 | - | - | - | −3.72 |
| Klra18 | Killer cell lectin-like receptor subfamily A, member 18 | NM_053153 | - | - | 2.99 | 3.12 | - | - | - | 2.14 |
| Klre1 | Killer cell lectin-like receptor family E member 1 | NM_153590 | - | −2.83 | - | - | −1.82 | −2.91 | −1.90 | −3.95 |
| Pecam1 | Platelet/endothelial cell adhesion molecule 1 | NM_008816 | - | - | - | 1.82 | - | - | - | 1.54 |
| VI. Signaling | ||||||||||
| Irf2 | Interferon regulatory factor 2 | NM_008391 | - | - | 1.66 | 3.12 | - | - | - | 2.79 |
| Lck | Lymphocyte protein tyrosine kinase | NM_010693 | - | - | - | −3.93 | - | - | - | −4.10 |
| Ltc4s | Leukotriene C4 synthase | NM_008521 | - | - | - | 3.72 | - | - | - | 3.87 |
| Ptger1 | Prostaglandin E receptor 1 (subtype EP1) | NM_013641 | - | - | 2.37 | - | - | - | - | 3.94 |
Infection with either strain resulted in the down-regulation of Il-1β expression immediately following infection in the lungs. Il-1β is a potent inflammatory cytokine and its suppression may be a key mechanism in Francisella infection. Upregrulation of Tgfβ1 and Ptger1 expression was noted 48 hours post infection in Schu4 infected mice, and expression of these immunosuppressive cytokines may be key to the rapid dissemination of Schu4. For example, Tgfβ1 and Ptger1 have both been shown to play a role in the suppression of host defenses in the lungs of LVS infected mice and in human dendritic cells infected with Schu4 [33, 34],. There was also altered expression of several MHC genes and the killer cell lectin-like receptor family genes, including (Ly49/Klra) and H2-Ab1, H2-Bl, H2-M10.2, H2-M3, H2-Q8. The Ly49/Kllra killer cell lectin-like receptors have been shown to be vital for recognition and activation/inhibition of natural killer cells [35, 36]. The fact that Schu4 and LVS infection both decreased the expression of these receptors adds further evidence to the notion that F. tularensis evades the host innate immune response by suppressing key mediators of this response.
3.3 Differences in host response to infection with Schu4 and LVS
Although the overall host response to infection with F. tularensis Schu4 and LVS is similar, unique host transcriptional responses to infection with Schu4 or infection with LVS infection were identified. Further inspection of the transcriptional response to Schu4 revealed notable differences in the transcription of immunologically important genes relative to their expression in LVS-infected mice. These differentially expressed genes included genes encoding components involved in apoptosis, antimicrobial activity, inflammatory response, cellular activation and differentiation, leukocyte receptors, and cell signaling (Table 2).
Table 2. Unique genes differentially expressed in response to F.tularensis Schu4 discussed in the text.
C57BL/6 mice (n = 2 per group) were inoculated via aerosol with lethal doses of F. tularensis LVS or Schu4 (104 CFU), as described in Methods. Total RNA Lung and spleen tissues were collected 12, 24, 48 and 120 hours post infection, converted to cDNA, labeled and hybridized on full mouse genome microarrays. Genes with a p-value < 0.01 and differentially regulated > 1.5 fold were mined for genes unique to Schu4 infection that fell into the categories of inflammatory response, cellular activation/differentiation, antimicrobial activity, leukocyte receptors, and cell signaling.
|
Lung | ||||||
|---|---|---|---|---|---|---|
| Gene ID | Annotation | Accession |
Hours Post-Infection |
|||
| 12 | 24 | 48 | 120 | |||
| I. Apoptosis | ||||||
| Anxa5 | Annexin A5 | NM_009673 | - | 3.49 | 3.79 | - |
| Bad | Bcl-associated death promoter | NM_007522 | - | - | - | −2.55 |
| Bbc3 | Bcl-2 binding component 3 | NM_133234 | 2.34 | - | - | 2.69 |
| Bcl2 | B-cell leukemia/lymphoma 2 | NM_177410 | - | −1.96 | −2.34 | - |
| Bclaf1 | BCL2-associated transcription factor 1 | NM_153787 | 1.93 | - | - | - |
| Bnip1 | BCL2/adenovirus E1B interacting protein 1, NIP1 | NM_172149 | - | 1.58 | 1.77 | - |
| Bnip2 | BCL2/adenovirus E1B interacting protein 1, NIP2 | NM_016787 | −2.20 | - | - | −4.24 |
| Bnip3l | BCL2/adenovirus E1B interacting protein 3-like | NM_009761 | - | - | - | −3.56 |
| Pdcd2 | Programmed cell death 2 | NM_008799 | −1.85 | −2.07 | −1.94 | - |
| Pdcd4 | Programmed cell death 4 | NM_011050 | - | - | - | −3.34 |
| Pdcd6 | Programmed cell death 6 | NM_011051 | - | - | - | −2.76 |
| II. Inflammatory Response | ||||||
| Ccl2 | Chemokine (C-C motif) ligand 2 | NM_011333 | - | - | - | 3.67 |
| Ccl22 | Chemokine (C-C motif) ligand 22 | NM_009137 | - | 1.93 | 2.04 | - |
| Ccl6 | Chemokine (C-C motif) ligand 6 | NM_009139 | - | - | 1.86 | - |
| Ccr6 | Chemokine (C-C motif) receptor 6 | NM_009835 | - | - | - | −3.27 |
| Cxcl10 | Chemokine (C-X-C motif) ligand 10 | NM_021274 | - | - | - | 1.84 |
| Ifna1 | Interferon alpha 1 | NM_010502 | - | - | - | −3.15 |
| Il13 | Interleukin 13 | NM_008355 | - | 1.80 | - | - |
| Il13ra2 | Interleukin 13 receptor, alpha 2 | NM_008356 | 2.11 | - | - | - |
| Il18 | Interleukin 18 | NM_008360 | - | −1.56 | - | - |
| Il1r2 | Interleukin 1 receptor, type II | NM_010555 | - | - | - | 2.28 |
| Sdf2 | Stromal cell derived factor 2 | NM_009143 | - | - | - | −1.96 |
| Tgfbr2 | Transforming growth factor, beta receptor II | NM_009371 | - | - | - | −3.14 |
| Tnfrsf8 | Tumor necrosis factor receptor superfamily, member 8 | NM_009401 | - | - | - | −2.79 |
| III. Cellular Activation/Differentiation | ||||||
| Cd164 | CD164 antigen | NM_016898 | - | - | - | −2.94 |
| Cd209a | CD209a antigen | NM_133238 | - | - | - | −3.66 |
| Cd37 | CD37 antigen | NM_007645 | - | 2.47 | 3.92 | - |
| Cd4 | CD4 antigen | NM_013488 | - | - | - | 3.93 |
| Cd52 | CD52 antigen | NM_013706 | - | - | 2.73 | - |
| Cd74 | CD74 antigen | NM_010545 | - | 3.47 | - | - |
| Cd79b | CD79B antigen | NM_008339 | - | - | 2.99 | - |
| Cd99l2 | Cd99 antigen-like 2 | NM_138309 | - | - | 2.01 | - |
| IV. Antimicrobial Activity | ||||||
| - | complement component 8, gamma subunit | XM_130127 | - | - | - | 3.12 |
| Adam2 | A disintegrin and metallopeptidase domain 2 | NM_009618 | −2.06 | - | −3.09 | −3.05 |
| Adam9 | A disintegrin and metallopeptidase domain 9 (meltrin gamma) | NM_007404 | - | - | - | −2.99 |
| Arg1 | Arginase 1, liver | NM_007482 | - | - | - | 1.69 |
| C1qc | Complement component 1, q subcomponent, C chain | NM_007574 | - | 2.12 | 2.60 | - |
| C9 | Complement component 9 | NM_013485 | - | - | - | −2.26 |
| Ctsd | Cathepsin D | NM_009983 | - | −2.41 | −1.85 | - |
| Ctsl | Cathepsin L | NM_009984 | - | - | - | −2.92 |
| Ctss | Cathepsin S | NM_021281 | - | 2.36 | 2.51 | - |
| Ctsz | Cathepsin Z | NM_022325 | - | 1.52 | 1.58 | - |
| F11r | F11 receptor | NM_172647 | - | - | - | −2.21 |
| F2r | Coagulation factor II (thrombin) receptor | NM_010169 | 1.60 | - | - | - |
| F2rl2 | Coagulation factor II (thrombin) receptor-like 2 | NM_010170 | - | −1.72 | - | - |
| F5 | Coagulation factor V | NM_007976 | −1.63 | - | - | −2.29 |
| Oasl1 | 2–5 oligoadenylate synthetase-like 1 | NM_145209 | - | - | - | 2.07 |
| Thbd | Thrombomodulin | NM_009378 | - | - | - | −3.04 |
| Thbs1 | Thrombospondin 1 | NM_011580 | - | - | - | 1.84 |
| Thbs2 | Thrombospondin 2 | NM_011581 | - | - | - | −2.36 |
| Timp3 | Tissue inhibitor of metalloproteinase 3 | NM_011595 | - | 2.22 | 1.86 | - |
| V. Leukocyte Receptors | ||||||
| Fcer1a | Fc receptor, IgE, high affinity I, alpha polypeptide | NM_010184 | - | - | - | 2.71 |
| Fcgrt | Fc receptor, IgG, alpha chain transporter | NM_010189 | - | 2.61 | 2.90 | - |
| H2-Ab1 | Histocompatibility 2, class II antigen A, beta 1 | NM_207105 | - | - | 2.86 | - |
| H2-D1 | Histocompatibility 2, T region locus 23 | NM_010398 | - | 2.59 | 3.30 | - |
| H2-DMa | Histocompatibility 2, class II, locus DMa | NM_010386 | - | - | 2.70 | - |
| H2-K1 | Histocompatibility 2, Q region locus 1 | NM_010390 | - | - | −2.54 | - |
| H2-Ke2 | H2-K region expressed gene 2 | NM_010385 | - | - | 1.61 | - |
| H2-Q7 | Histocompatibility 2, Q region locus 7 | NM_010394 | - | 2.45 | - | - |
| Icam2 | Intercellular adhesion molecule 2 | NM_010494 | - | 3.68 | 3.88 | - |
| Klra17 | Killer cell lectin-like receptor, subfamily A, member 17 | NM_133203 | - | - | −1.51 | - |
| Pecam1 | Platelet/endothelial cell adhesion molecule 1 | NM_008816 | - | - | 2.64 | - |
| Tlr11 | Toll-like receptor 11 | NM_205819 | - | −1.64 | −2.23 | - |
| Tlr5 | Toll-like receptor 5 | NM_016928 | 1.90 | - | - | - |
| Tlr9 | Toll-like receptor 9 | NM_031178 | - | - | −2.75 | - |
| VI. Cell Signaling | ||||||
| Ifi204 | Interferon activated gene 204 | NM_008329 | −2.10 | - | - | - |
| Il1rap | Interleukin 1 receptor accessory protein | NM_134103 | - | - | - | −1.54 |
| Irak3 | Interleukin-1 receptor-associated kinase 3 | NM_028679 | - | - | −1.99 | - |
| Irak4 | Interleukin-1 receptor-associated kinase 4 | NM_029926 | - | - | - | 2.66 |
| Irf2 | Interferon regulatory factor 2 | NM_008391 | - | - | - | 1.63 |
| Irf4 | Interferon regulatory factor 4 | NM_013674 | - | - | 2.08 | - |
| Irf9 | Interferon regulatory factor 9 | NM_008394 | - | - | 1.89 | - |
| Ptger3 | Prostaglandin E receptor 3 (subtype EP3) | NM_011196 | - | - | - | −1.51 |
| Ptgfr | Prostaglandin F receptor | NM_008966 | - | - | - | −2.43 |
| Ptgis | Prostaglandin I2 (prostacyclin) synthase | NM_008968 | 2.60 | - | - | - |
| Ptgr2 | Prostaglandin reductase 2 | NM_029880 | - | - | - | −2.25 |
| Tbrg1 | Transforming growth factor beta regulated gene 1 | NM_025289 | - | - | 2.86 | - |
| Traf5 | Tnf receptor-associated factor 5 | NM_011633 | - | - | - | −3.10 |
| Traf7 | Tnf receptor-associated factor 7 | NM_153792 | - | - | 2.16 | - |
| Trap1 | TNF receptor-associated protein 1 | NM_026508 | - | 2.12 | 2.19 | - |
|
Spleen | ||||||
|---|---|---|---|---|---|---|
| Gene ID | Annotation | Accession |
Hours Post Infection |
|||
| 12 | 24 | 48 | 120 | |||
| I. Apoptosis | ||||||
| Apitd1 | Apoptosis-inducing, TAF9-like domain 1 | NM_027263 | - | - | - | −1.99 |
| Bclaf1 | BCL2-associated transcription factor 1 | NM_153787 | - | - | - | −2.43 |
| Casp6 | Caspase 6 | NM_009811 | - | - | - | −1.54 |
| Fadd | Fas (TNFRSF6)-associated via death domain | NM_010175 | 2.45 | 1.95 | - | 2.25 |
| Faim | Fas apoptotic inhibitory molecule | NM_011810 | - | - | - | −3.18 |
| Pdcd4 | Programmed cell death 4 | NM_011050 | - | - | - | −2.69 |
| II. Inflammatory Response | ||||||
| Ccl3 | Chemokine (C-C motif) ligand 3 | NM_011337 | - | - | - | 3.01 |
| Ccr1 | Chemokine (C-C motif) receptor 1 | NM_009912 | - | - | - | −1.50 |
| Ccr1l1 | Chemokine (C-C motif) receptor 1-like 1 | NM_007718 | - | - | - | −4.21 |
| Ccr3 | Chemokine (C-C motif) receptor 3 | NM_009914 | - | - | 2.94 | - |
| Ccr5 | Chemokine (C-C motif) receptor 5 | NM_009917 | - | - | 1.63 | - |
| Cx3cl1 | Chemokine (C-X3-C motif) ligand 1 | NM_009142 | - | - | - | −1.94 |
| Cxcl11 | Chemokine (C-X-C motif) ligand 11 | NM_019494 | - | - | - | 4.13 |
| Cxcl13 | Chemokine (C-X-C motif) ligand 13 | NM_018866 | - | - | - | 3.09 |
| Cxcl14 | Chemokine (C-X-C motif) ligand 14 | NM_019568 | - | - | - | −2.98 |
| Cxcr6 | Chemokine (C-X-C motif) receptor 6 | NM_030712 | - | - | - | −2.69 |
| Ifnb1 | Interferon beta 1, fibroblast | NM_010510 | - | - | - | 2.20 |
| Il10ra | Interleukin 10 receptor, alpha | NM_008348 | - | - | - | −1.57 |
| Il13 | Interleukin 13 | NM_008355 | - | - | - | 2.07 |
| Il13ra2 | Interleukin 13 receptor, alpha 2 | NM_008356 | - | - | - | −1.96 |
| Il18r1 | Interleukin 18 receptor 1 | NM_008365 | - | - | - | −2.09 |
| Il1f9 | Interleukin 1 family, member 9 | NM_153511 | - | - | - | 2.60 |
| Il1r2 | Interleukin 1 receptor, type II | NM_010555 | - | - | 1.53 | - |
| Il2rb | Interleukin 2 receptor, beta chain | NM_008368 | - | - | - | −2.21 |
| Il9r | Interleukin 9 receptor | NM_008374 | - | - | - | −1.69 |
| Lta | Lymphotoxin A | NM_010735 | - | - | 1.50 | - |
| Ltbp3 | Latent transforming growth factor beta binding protein 3 | NM_008520 | - | - | - | 2.70 |
| Tgfbr2 | Transforming growth factor, beta receptor II | NM_009371 | - | −1.69 | - | −3.55 |
| Tnfrsf1a | Tumor necrosis factor receptor superfamily, member 1a | NM_011609 | - | - | 2.38 | - |
| Vegfc | Vascular endothelial growth factor C | NM_009506 | - | - | 1.55 | - |
| Xcl1 | Chemokine (C motif) ligand 1 | NM_008510 | - | - | - | −2.54 |
| III. Cellular Activation/Differentiation | ||||||
| Cd247 | CD247 antigen | NM_031162 | - | - | - | −2.18 |
| Cd274 | CD274 antigen | NM_021893 | - | - | - | 1.82 |
| Cd300c | CD300C antigen | NM_199225 | - | - | - | −2.79 |
| Cd300e | CD300e antigen | NM_172050 | - | - | - | −3.91 |
| Cd300lb | CD300 antigen like family member B | NM_199221 | - | - | - | 3.72 |
| Cd320 | CD320 antigen | NM_019421 | - | - | - | −2.10 |
| Cd3d | CD3 antigen, delta polypeptide | NM_013487 | - | - | - | −3.05 |
| Cd3eap | CD3E antigen, epsilon polypeptide associated protein | NM_145822 | - | - | - | 2.54 |
| Cd3g | CD3 antigen, gamma polypeptide | NM_009850 | - | - | - | −1.78 |
| Cd44 | CD44 antigen | NM_009851 | - | - | - | −3.15 |
| IV. Antimicrobial Activity | ||||||
| - | complement factor properdin | XM_135820 | - | - | - | −2.88 |
| Adam15 | A disintegrin and metallopeptidase domain 15 | NM_009614 | - | - | - | 1.96 |
| Arg1 | Arginase 1, liver | NM_007482 | - | - | - | 3.09 |
| C2 | Complement component 2 (within H-2S) | NM_013484 | - | - | - | 1.98 |
| C6 | Complement component 6 | NM_016704 | - | - | - | −3.02 |
| Ctsb | Cathepsin B | NM_007798 | - | - | - | −2.63 |
| Ctsd | Cathepsin D | NM_009983 | - | - | - | −1.86 |
| Ctse | Cathepsin E | NM_007799 | - | - | - | −2.16 |
| Ctsw | Cathepsin W | NM_009985 | - | - | - | −3.14 |
| Defb1 | Defensin beta 1 | NM_007843 | - | - | - | 2.64 |
| Defb21 | Defensin beta 21 | NM_207276 | - | 1.51 | - | - |
| Gzmb | Granzyme B | NM_013542 | - | −1.71 | - | - |
| Igj | Immunoglobulin joining chain | NM_152839 | - | - | - | −3.23 |
| Mmp13 | Matrix metallopeptidase 13 | NM_008607 | - | - | - | 1.96 |
| Mmp14 | Matrix metallopeptidase 14 (membrane-inserted) | NM_008608 | - | - | - | 2.49 |
| Ncf1 | Neutrophil cytosolic factor 1 | NM_010876 | - | - | - | −1.99 |
| Nos2 | Nitric oxide synthase 2, inducible, macrophage | NM_010927 | - | - | −1.52 | −2.44 |
| Oas1d | 2–5 oligoadenylate synthetase 1D | NM_133893 | - | - | - | 1.62 |
| Oas2 | 2–5 oligoadenylate synthetase 2 | NM_145227 | - | - | - | 2.41 |
| Oasl2 | 2–5 oligoadenylate synthetase-like 2 | NM_011854 | - | - | - | −1.86 |
| Socs1 | Suppressor of cytokine signaling 1 | NM_009896 | 1.58 | - | 1.84 | - |
| Timp3 | Tissue inhibitor of metalloproteinase 3 | NM_011595 | - | - | - | 2.53 |
| Tslp | Thymic stromal lymphopoietin | NM_021367 | - | - | - | −3.34 |
| V. Leukocyte Receptors | ||||||
| H2-D1 | Histocompatibility 2, T region locus 23 | NM_010398 | - | - | - | 2.18 |
| H2-Ke2 | H2-K region expressed gene 2 | NM_010385 | - | - | - | 3.53 |
| H2-Ke6 | H2-K region expressed gene 6 | NM_013543 | - | - | - | −2.88 |
| H2-M11 | Histocompatibility 2, M region locus 11 | NM_177635 | - | - | - | −2.20 |
| H2-T22 | Histocompatibility 2, T region locus 10 | NM_010399 | - | - | - | 1.62 |
| H2-T22 | Histocompatibility 2, T region locus 10 | NM_010397 | - | - | - | 3.20 |
| Itgav | Integrin alpha V | NM_008402 | - | - | - | 2.45 |
| Jam3 | Junction adhesion molecule 3 | NM_023277 | - | - | 2.36 | - |
| Klra16 | Killer cell lectin-like receptor, subfamily A, member 16 | NM_013794 | - | −3.20 | - | −3.30 |
| Klrd1 | Killer cell lectin-like receptor, subfamily D, member 1 | NM_010654 | - | −2.15 | - | −3.42 |
| Ltb4r1 | Leukotriene B4 receptor 1 | NM_008519 | - | - | 1.69 | - |
| Ly6a | Lymphocyte antigen 6 complex, locus A | NM_010738 | - | - | - | 2.35 |
| Ly6e | Lymphocyte antigen 6 complex, locus E | NM_008529 | - | 2.77 | - | 3.19 |
| Ly6f | Lymphocyte antigen 6 complex, locus F | NM_008530 | - | - | - | 2.64 |
| Ly6g6e | Lymphocyte antigen 6 complex, locus G6E | NM_027366 | - | - | - | −3.92 |
| Ly6i | Lymphocyte antigen 6 complex, locus I | NM_020498 | - | - | - | 2.67 |
| Ly6k | Lymphocyte antigen 6 complex, locus K | NM_029627 | - | - | - | 3.09 |
| Lyve1 | Lymphatic vessel endothelial hyaluronan receptor 1 | NM_053247 | - | - | - | 2.52 |
| Marco | Macrophage receptor with collagenous structure | NM_010766 | - | −2.87 | - | - |
| Mrc1 | Mannose receptor, C type 1 | NM_008625 | - | - | - | −1.67 |
| Mrcl | Mannose receptor-like precursor | NM_181549 | - | - | - | 2.38 |
| Scarb2 | Scavenger receptor class B, member 2 | NM_007644 | - | - | - | −1.70 |
| Tlr11 | Toll-like receptor 11 | NM_205819 | - | - | - | −2.16 |
| VI. Signaling | ||||||
| Cd2bp2 | CD2 antigen (cytoplasmic tail) binding protein 2 | NM_027353 | - | - | - | 4.22 |
| Ifi202b | Interferon activated gene 202B | NM_008327 | - | - | - | 4.16 |
| Ifi204 | Interferon activated gene 204 | NM_008329 | −1.69 | −1.86 | −2.24 | - |
| Ifi205 | Interferon activated gene 205 | NM_172648 | - | - | - | 3.51 |
| Ifi27 | Interferon, alpha-inducible protein 27 | NM_029803 | - | - | - | 2.57 |
| Ifi35 | Interferon-induced protein 35 | NM_027320 | - | - | - | 4.15 |
| Ifitm2 | Interferon induced transmembrane protein 2 | NM_030694 | - | - | 1.58 | 4.09 |
| Ifitm3 | Interferon induced transmembrane protein 3 | NM_025378 | - | - | - | 3.32 |
| Il6st | Interleukin 6 signal transducer | NM_010560 | - | −1.51 | - | −2.24 |
| Irf2bp1 | Interferon regulatory factor 2 binding protein 1 | NM_178757 | - | - | - | −1.95 |
| Isg20 | Interferon-stimulated protein | NM_020583 | - | - | - | 3.59 |
| Prnd | Prion protein dublet | NM_023043 | - | - | - | −3.13 |
| Ptgds2 | Prostaglandin D2 synthase 2, hematopoietic | NM_019455 | - | - | - | −1.90 |
| Ptgis | Prostaglandin I2 (prostacyclin) synthase | NM_008968 | - | - | - | −4.02 |
| Ptgr2 | Prostaglandin reductase 2 | NM_029880 | - | - | - | −2.72 |
| Tnfaip1 | Tumor necrosis factor, alpha-induced protein 1 (endothelial) | NM_009395 | - | 1.65 | - | 1.60 |
| Tnfaip8l1 | Tumor necrosis factor, alpha-induced protein 8-like 1 | NM_025566 | - | - | - | 3.05 |
| Tnfaip8l2 | Tumor necrosis factor, alpha-induced protein 8-like 2 | NM_027206 | - | - | - | −1.89 |
| Tnfrsf13c | Tumor necrosis factor receptor superfamily, member 13c | NM_028075 | - | - | - | −2.43 |
| Traf3 | Tnf receptor-associated factor 3 | NM_011632 | - | - | - | 3.31 |
| Traf3ip3 | TRAF3 interacting protein 3 | NM_153137 | - | - | - | −2.20 |
| Vezf1 | Vascular endothelial zinc finger 1 | NM_016686 | - | - | - | −2.46 |
Genes associated with apoptosis and antimicrobial activity had different expression patterns in Schu4 as compared to LVS and uninfected mice. For example, expression of the pro-apoptotic genes Bad, Bnip2, Bnip3l, Pdcd2, Pdcd4 and Pdcd6, and the anti-apoptotic genes Bcl2 was repressed in Schu4 infected lungs compared to LVS-infected lungs. Similarly, in the spleen there was also repression of apitd1, Bclaf1 and Casp6 expression. Inhibition of apoptosis has been shown to be an important mechanism for replication and survival during infection of other bacteria such as C. burnetii [37–39].
The antimicrobial activity response in the lungs of F. tularensis Schu4 infected mice was dominated by altered expression of Adam2 and Adam9, cathepsin D, L, S and Z, thrombomodulin, thrombospondin 1 and 2, and Timp3. In the spleen, Adam15, Defb1, and Defb21 showed increased expression, while cathepsin B, D and E had significantly reduced expression. The transcriptional response of these genes indicated a reduction in tissue remodeling and breakdown, intracellular protein metabolism, and breakdown of antigenic proteins for MHC-II presentation. Defensins are intrinsically antimicrobial but the isoforms induced during infection have been shown to have little effect on Francisella using human alveolar cells in vitro [40].
The transcription of CD4, CD52, CD74 (Ii, Invariant chain), and B lymphocyte markers CD37 and CD79B (Igβ) involved in cellular activation and differentiation were uniquely upregulated in Schu4 infection. The increased expression of these particular components involved in MHC-II antigen presentation is consistent with augmented cell-mediated immunity. As antigen presentation is a tightly regulated process [41], these data in addition to the cathepsin data above may implicate F. tularensis induced alterations in processing and presentation of antigens during infection with F. tularensis Schu4.
There were also important differences in the molecular mediators of the inflammatory response in mice infected with Schu4 compared to LVS infected mice. Expression of the genes for IL-13, IL-13Ra2, CCL2, CCL6, CCL22, and CXCL10 were only induced in the lungs of Schu4 infected mice. Upregulated expression of IL-13 is important because of its role as a Th2 related cytokine, which can be associated with downregulation of Th1 immunity. Upregulation of the chemokine genes suggests that Schu4 infection may lead to increased recruitment of monocytes. A similar trend of altered expression of cytokines and chemokines, specifically IL-13, CCL3, CCR3, CCR5, interferon activated genes, prostaglandin D2 synthase 2, prostacyclin I2, prostaglandin reductase 2, and several Ly6-family genes was observed in the spleen of Schu4 infected mice, albeit later time points in infection. Interferon activated gene families as well as prostaglandin signaling has been shown to be involved in the response to virulent Francisella [13, 42]. We found similar involvement of these pathways in response to Schu4 infection in the mouse spleen. Interestingly, the expression of the T-helper 2 type interleukin IL-13, anti-inflammatory cytokines IL-10 and TGFβ, and the down regulation of the proinflammatory cytokines IL-18 and interferon alpha suggest a disruption in the activation of the protective defenses in Schu4 infection compared to LVS infection.
3.4 Validation of transcriptional trends by qRT-PCR
To confirm the transcriptional response of select immunological genes during F. tularensis infection, quantitative real-time PCR (qRT) was performed on lung and spleen tissue from independent infections (Table 3). Analysis revealed that the trends identified by global microarray analysis were 85% and 62% concordant with qRT data in the lung and spleen, respectively. The lower concordance noted in the spleen is attributed to temporal differences in dissemination. The expression of the 12 key pro-inflammatory and anti-inflammatory markers in the lung and spleen was limited in the initial 24 hours of infection. However, 48 hours post infection with LVS shows activation of cytokine and chemokine expression not seen until 120 hours post infection with Schu4. The genes significantly up-regulated as determined by qRT in the lungs during Schu4 infection included the pro-inflammatory chemokines CCL4, CXCL1 and CXCL10; the pro-inflammatory cytokines IL-6 and IL-12a; the gene for inducible nitric oxide, Nos2; and the gene for a type I interferon, IFN-β. A similar trend in the differential expression of these genes was observed in the spleen of Schu4-infected mice at 120 hours. This trend in transcriptional activity indicates a delayed and reduced host response to infection with Schu4 and is consistent with a lack of host recognition or active mechanism of host-response suppression by the Schu4, consistent with previous reports [43, 26, 34].
Table 3. Relative expression values of inflammatory markers from lung and spleen of mice infected with F. tularensis Schu4 and LVS.
C57BL/6 mice (n = 4 per group) were inoculated i.n. with lethal doses of F. tularensis LVS (104 CFU) or Schu4 (102 CFU), as described in Methods. Quantitative real time PCR was used to validate microarray data, and monitor molecular markers of disease. Data was monitored for consistency by the housekeeping genes 18S rRNA, GapDH, and β-actin. Data from each condition was compared to controls using the ΔCT method.
| Lung |
||||||||
|---|---|---|---|---|---|---|---|---|
| Schu4 | LVS | |||||||
| 12 hours | 24 hours | 48 hours | 120 hours | 12 hours | 24 hours | 48 hours | 120 hours | |
| Tnfα | −0.88±1.09 | −0.25±0.45 | −0.29±0.25 | 2.51±1.53 | 0.44±0.30 | −0.69±0.68 | 0.51±0.30 | −0.95±0.19 |
| Ifn-γ | −7.72±0.09 | −7.01±1.72 | −0.81±0.45 | 1.79±0.39 | −7.31±0.29 | −7.32±0.11 | 1.25±0.38 | 4.22±0.47 |
| Ifn-β | −3.80±0.99 | −1.92±1.07 | −2.69±1.00 | 4.61±0.58 | −4.18±0.95 | −2.66±0.80 | 2.45±0.25 | 3.03±0.44 |
| TgfB1 | 0.97±0.33 | 1.38±0.57 | 1.02±0.13 | 1.84±0.63 | 0.40±0.66 | 2.09±0.40 | 1.39±0.21 | 1.72±0.34 |
| Cxcl1 | 0.69±0.98 | 2.35±0.43 | 1.70±0.88 | 2.85±0.94 | 2.19±0.69 | 1.11±1.30 | 4.12±0.47 | 5.29±0.21 |
| Cxcl10 | −0.90±0.55 | −0.81±0.50 | 0.74±0.87 | 7.74±0.21 | −0.34±0.38 | −0.75±0.88 | 5.78±1.03 | 9.07±0.40 |
| Ccl4 | −1.20±1.17 | 0.00±0.52 | −0.51±0.52 | 1.78±0.55 | −1.17±0.43 | −1.57±0.87 | 2.21±0.45 | 4.38±0.29 |
| II-1β | −1.64±0.63 | −0.55±1.14 | 1.00±0.71 | −0.24±0.51 | −2.42±0.43 | −2.48±0.65 | 2.93±0.81 | 3.8±0.26 |
| II-6 | −2.17±0.81 | −0.23±1.05 | 1.84±1.05 | 4.30±0.43 | −2.32±0.83 | −1.00±1.17 | 4.64±1.16 | 5.50±0.71 |
| II-10 | 1.21±1.08 | −2.46±0.82 | −0.67±0.84 | 4.28±0.89 | −1.06±1.10 | −1.94±0.89 | 1.60±0.35 | 4.36±0.30 |
| II-12a | 0.68±1.91 | 1.57±0.63 | 1.51±0.16 | 4.67±0.88 | 0.72±0.75 | 0.91±1.65 | 3.89±0.22 | 5.15±0.35 |
| Nos2 | 0.57±1.16 | 0.61±0.16 | 1.94±0.27 | 3.78±0.43 | 0.04±0.76 | 0.77±0.47 | 2.73±0.33 | 7.58±0.51 |
| Spleen |
||||||||
|---|---|---|---|---|---|---|---|---|
| Schu4 | LVS | |||||||
| 12 hours | 24 hours | 48 hours | 120 hours | 12 hours | 24 hours | 48 hours | 120 hours | |
| Tnfa | 0.55±0.41 | 0.26±0.44 | −1.81±0.33 | 3.23±0.18 | 2.03±0.42 | 0.79±0.45 | −0.06±0.88 | −2.52±0.16 |
| Ifn-γ | −6.93±0.83 | −7.89±0.86 | 0.47±0.43 | −1.19–0.55 | −5.72±0.62 | −6.69±0.25 | 1.76±0.38 | 3.28±0.30 |
| Ifn-β | −3.94±0.46 | −4.25±1.22 | −4.53±1.10 | 3.36±0.24 | −3.51±1.75 | −3.16±1.22 | 0.69±0.61 | −1.98±0.73 |
| TgfB1 | −1.27±0.25 | −0.26±1.37 | −1.50±0.51 | 3.68±0.81 | 1.53±0.54 | −0.94±0.49 | 0.36±0.21 | 1.77±0.23 |
| Cxcl1 | −1.27±0.67 | −0.26±0.39 | −1.50±0.31 | 3.68±0.45 | 0.35±0.52 | −0.94±0.98 | 0.36±0.93 | 1.77±0.84 |
| Ccl4 | −0.44±0.79 | −1.04±1.03 | −0.27±0.80 | 2.69±0.34 | 0.33±0.57 | −0.19±0.59 | 0.61±0.94 | 1.63±0.47 |
| II-1β | −0.20±0.73 | −0.05±0.36 | 1.29±0.65 | 1.40±0.28 | −0.28±0.57 | −0.81±0.43 | 2.00±0.62 | 2.53±0.82 |
| II-6 | −6.04±2.26 | −4.68±1.96 | −4.79±1.40 | 2.23±0.43 | −4.02±1.57 | −3.21±1.18 | −1.51±1.74 | 0.81±1.23 |
| II-10 | 0.03±0.44 | −0.10±0.98 | −1.05±0.87 | 3.63±0.41 | 2.08±0.51 | 0.32±0.53 | 0.98±0.35 | 2.97±0.35 |
| II-12a | −1.28±0.74 | 1.39±0.58 | −1.48±0.76 | 0.46±0.40 | −1.28±0.74 | −1.39±0.58 | −0.13±0.20 | 0.22±0.24 |
| Nos2 | 0.34±0.50 | 0.50±0.37 | −0.10±0.39 | 4.85±0.48 | 0.78±0.86 | 0.27±0.67 | 0.21±0.40 | 4.76±0.57 |
4. Discussion
A critical question in understanding F. tularensis pathobiology is to determine which critical host responses are altered during the first 4–5 days following infection. Whole genome microarrays are an established post-genomic approach that allow the assessment of global host responses in an unbiased fashion. In the present study, we coupled whole genome microarray analysis with analysis of tissue pathology and organ bacterial burden to gain a more complete understanding of disease progression and host response to infection with a fully virulent and a less virulent strain of F. tularensis. By means of this combined approx we were able to identify important host response differences to infection with the two strains of F. tularensis.
Quantification of bacterial burden in the lungs revealed that Schu4 had increased growth compared to LVS, such that by 120 hours the bacterial load of Schu4 in the lungs significantly exceeded that of mice infected with the LVS strain. In addition, F. tularensis Schu4 demonstrated increased dissemination to the spleen, as indicated by detection within 48 hours of infection and significantly increased bacterial burden in the spleen at later time points following infection. Tissue damage was markedly more severe in the spleen following infection with Schu4, particularly at later time points of infection. Notably, both Schu4 and LVS established similar levels of infection in the lung, but eventually the Schu4 infection progressed to more severe pulmonary pathology, presumably due to more rapid replication and avoidance of host immune responses. Efficient dissemination appears to be an important distinction and hallmark of infection with highly virulent strains of F. tularensis [14, 44, 45]. Importantly, the correlation between controlled dissemination and survival has been observed in drug development studies that indicate that drug efficacy is related to control of dissemination to secondary organs such as the spleen [45].
Rapid dissemination is an important determinant of disease outcome and likely relies on the initial recognition and control of pathogen replication at the site of infection. A study conducted by Chiavolini et. al showed the importance of the initial inflammatory response in determining survival following F. tularensis infection. For example, survival was predicted by the induction of several inflammatory genes before day 7 of infection with LVS in mice [44]. Since replication of Schu4 was actually higher in the spleens of infected mice than replication of LVS, it is likely that the decrease in cytokine gene expression in the lungs of Schu4 infected mice reflects either failure to activate immune responses, or active immune suppression.
The results of the global analyses of the host response to infection with Francisella Schu4 or LVS strains indicate highly virulent strains are capable of subverting the host innate immune response and cell mediated immunity. In the present study, these altered responses included apoptosis, antigen processing and presentation, the inflammatory response, and leukocyte receptor signaling. The down regulation of multiple host defense mechanisms by F. tularensis is consistent with results reported in previous studies [15, 13, 18, 17, 16][34, 43, 26]. In addition, the transcriptional response to Schu4 and F. tularensis subspecies novicida in human monocytes found that reported that there was less inflammatory gene activation by Schu4 as compared to the less virulent F. novicida strain [17].
In addition, we found F. tularensis Schu4 versus LVS induced changes in novel gene subsets, particularly IL-13, cathepsins, and most strikingly, the killer cell lectin-like receptor family (Ly49/Klre1). Studies have shown interferon activated macrophages treated with IL-13 have a reduced capacity to inhibit the growth of intracellular bacteria. A previous transcriptional profiling study showed the increased expression of interferon activated genes four days after infection in the lungs of mice infected with Type A FSC033 [13]. Moreover, the killer cell lectin-like receptors have been shown to be vital for recognition and activation/inhibition of natural killer cells [35, 36]. Evidence that infection with F. tularensis Schu4 decreased the expression of these receptors further highlights the immuno-evasive activity of Francisella Schu4 compared to the less virulent F. tularensis LVS. Furthermore, expression of the prostaglandin E1 receptor (Ptger1) confirms recent reports implicating prostaglandin signaling as an important mechanism of Francisella manipulation of the host-response to infection [46, 33, 42].
Our studies also indicate confirm previous studies and indicate that dissemination to secondary sites of infection leading to multi-organ damage and failure are key contributing factors to mortality from F. tularensis infection. We have also identified gene expression patterns that may reflect immune responses to bacterial dissemination from the lung to spleen tissues. These data may also be useful for facilitating the development of diagnostics for monitoring treatment efficacy, the effectiveness of chemotherapeutic or vaccine strategies. For example, gene expression correlates of host evasion during early infection combined with gene expression signatures of dissemination provide a panel of genes that can be used to assess disease progression and severity that can be used as checkpoints of therapeutic efficacy. In addition, as high throughput RNA sequencing becomes more readily available, biomarkers can be correlated to in vivo transcriptional data from the pathogen in an attempt to decipher complex host-pathogen interactions. Importantly the expression of markers that are associated with differences infection with LVS and Schu4 will be useful for assessing immune response to immunotherapeutic drugs. These studies therefore provide a foundation for continued research in this area that will ultimately provide unique opportunities that can be exploited for the development of protective vaccines and effective chemotherapeutics with enhanced efficacy and that prevent relapse of disease.
Acknowledgments
We are thankful for post-genomics resources and instrumentation, and animal models expertise provided by the Genomics and Proteomics Core and the Animal Models Core in the Rocky Mountain Regional Center of Excellence (AI065357) respectively. We would like to thank Laurel Respicio for technical assistance and reviewing of the manuscript. Funding from the Rocky Mountain Regional Center of Excellence (AI065357) to R.A.S supported this work.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Dennis DT, Inglesby TV, Henderson DA, Bartlett JG, Ascher MS, Eitzen E, Fine AD, Friedlander AM, Hauer J, Layton M, Lillibridge SR, McDade JE, Osterholm MT, O’Toole T, Parker G, Perl TM, Russell PK, Tonat K. Tularemia as a biological weapon: medical and public health management. JAMA. 2001;285:2763–2773. doi: 10.1001/jama.285.21.2763. [DOI] [PubMed] [Google Scholar]
- 2.Petersen JM, Schriefer ME. Tularemia: emergence/re-emergence. Vet Res. 2005;36:455–467. doi: 10.1051/vetres:2005006. [DOI] [PubMed] [Google Scholar]
- 3.Edwin LO, Tigertt WD, Paul JK, Martha KW, Charkes ND, Robert MR, Theodore ES, Mallory S. An analysis of forty-two cases of laboratory-acquired tularemia: Treatment with broad spectrum antibiotics. The American journal of medicine. 1961;30:785–806. doi: 10.1016/0002-9343(61)90214-5. [DOI] [PubMed] [Google Scholar]
- 4.Chocarro A, Gonzalez A, Garcia I. Treatment of Tularemia with Ciprofloxacin. Clinical Infectious Diseases. 2000;31:623–623. doi: 10.1086/313946. [DOI] [PubMed] [Google Scholar]
- 5.Sawyer WD, Dangerfield HG, Hogge AL, Crozier D. Antibiotic prophylaxis and therapy of airborne tularemia. Bacteriol Rev. 1966;30:542–550. doi: 10.1128/br.30.3.542-550.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Metzger DW, Bakshi CS, Kirimanjeswara G. Mucosal immunopathogenesis of Francisella tularensis. Ann N Y Acad Sci. 2007;1105:266–283. doi: 10.1196/annals.1409.007. [DOI] [PubMed] [Google Scholar]
- 7.Twine SM, Shen H, Kelly JF, Chen W, Sjostedt A, Conlan JW. Virulence comparison in mice of distinct isolates of type A Francisella tularensis. Microb Pathog. 2006;40:133–138. doi: 10.1016/j.micpath.2005.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moe JB, Canonico PG, Stookey JL, Powanda MC, Cockerell GL. Pathogenesis of tularemia in immune and nonimmune rats. Am J Vet Res. 1975;36:1505–1510. [PubMed] [Google Scholar]
- 9.Fortier AH, Slayter MV, Ziemba R, Meltzer MS, Nacy CA. Live vaccine strain of Francisella tularensis: infection and immunity in mice. Infect Immun. 1991;59:2922–2928. doi: 10.1128/iai.59.9.2922-2928.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elkins KL, Cowley SC, Bosio CM. Innate and adaptive immune responses to an intracellular bacterium, Francisella tularensis live vaccine strain. Microbes Infect. 2003;5:135–142. doi: 10.1016/s1286-4579(02)00084-9. [DOI] [PubMed] [Google Scholar]
- 11.Keim P, Johansson A, Wagner DM. Molecular epidemiology, evolution, and ecology of Francisella. Ann N Y Acad Sci. 2007;1105:30–66. doi: 10.1196/annals.1409.011. [DOI] [PubMed] [Google Scholar]
- 12.Sjostedt A. Tularemia: history, epidemiology, pathogen physiology, and clinical manifestations. Ann N Y Acad Sci. 2007;1105:1–29. doi: 10.1196/annals.1409.009. [DOI] [PubMed] [Google Scholar]
- 13.Andersson H, Hartmanova B, Kuolee R, Ryden P, Conlan W, Chen W, Sjostedt A. Transcriptional profiling of host responses in mouse lungs following aerosol infection with type A Francisella tularensis. J Med Microbiol. 2006;55:263–271. doi: 10.1099/jmm.0.46313-0. [DOI] [PubMed] [Google Scholar]
- 14.Conlan JW, Chen W, Shen H, Webb A, KuoLee R. Experimental tularemia in mice challenged by aerosol or intradermally with virulent strains of Francisella tularensis: bacteriologic and histopathologic studies. Microb Pathog. 2003;34:239–248. doi: 10.1016/s0882-4010(03)00046-9. [DOI] [PubMed] [Google Scholar]
- 15.Andersson H, Hartmanova B, Back E, Eliasson H, Landfors M, Naslund L, Ryden P, Sjostedt A. Transcriptional profiling of the peripheral blood response during tularemia. Genes Immun. 2006;7:503–513. doi: 10.1038/sj.gene.6364321. [DOI] [PubMed] [Google Scholar]
- 16.Cremer TJ, Amer A, Tridandapani S, Butchar JP. Francisella tularensis regulates autophagy-related host cell signaling pathways. Autophagy. 2009;5:125–128. doi: 10.4161/auto.5.1.7305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Butchar JP, Cremer TJ, Clay CD, Gavrilin MA, Wewers MD, Marsh CB, Schlesinger LS, Tridandapani S. Microarray analysis of human monocytes infected with Francisella tularensis identifies new targets of host response subversion. PLoS ONE. 2008;3:e2924. doi: 10.1371/journal.pone.0002924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Andersson H, Hartmanova B, Ryden P, Noppa L, Naslund L, Sjostedt A. A microarray analysis of the murine macrophage response to infection with Francisella tularensis LVS. J Med Microbiol. 2006;55:1023–1033. doi: 10.1099/jmm.0.46553-0. [DOI] [PubMed] [Google Scholar]
- 19.Pechous RD, McCarthy TR, Mohapatra NP, Soni S, Penoske RM, Salzman NH, Frank DW, Gunn JS, Zahrt TC. A Francisella tularensis Schu S4 purine auxotroph is highly attenuated in mice but offers limited protection against homologous intranasal challenge. PLoS ONE. 2008;3:e2487. doi: 10.1371/journal.pone.0002487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Twine S, Bystrom M, Chen W, Forsman M, Golovliov I, Johansson A, Kelly J, Lindgren H, Svensson K, Zingmark C, Conlan W, Sjostedt A. A mutant of Francisella tularensis strain SCHU S4 lacking the ability to express a 58-kilodalton protein is attenuated for virulence and is an effective live vaccine. Infect Immun. 2005;73:8345–8352. doi: 10.1128/IAI.73.12.8345-8352.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qin A, Scott DW, Mann BJ. Francisella tularensis subsp. tularensis Schu S4 disulfide bond formation protein B, but not an RND-type efflux pump, is required for virulence. Infect Immun. 2008;76:3086–3092. doi: 10.1128/IAI.00363-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mahawar M, Kirimanjeswara GS, Metzger DW, Bakshi CS. Contribution of citrulline ureidase to Francisella tularensis strain Schu S4 pathogenesis. J Bacteriol. 2009;191:4798–4806. doi: 10.1128/JB.00212-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ng HH, Frantz CE, Rausch L, Fairchild DC, Shimon J, Riccio E, Smith S, Mirsalis JC. Gene expression profiling of mouse host response to Listeria monocytogenes infection. Genomics. 2005;86:657–667. doi: 10.1016/j.ygeno.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 24.Gonzalez-Juarrero M, Kingry LC, Ordway DJ, Henao-Tamayo M, Harton M, Basaraba RJ, Hanneman WH, Orme IM, Slayden RA. Immune response to Mycobacterium tuberculosis and identification of molecular markers of disease. Am J Respir Cell Mol Biol. 2009;40:398–409. doi: 10.1165/rcmb.2008-0248OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu H, Wang H, Qiu J, Wang X, Guo Z, Qiu Y, Zhou D, Han Y, Du Z, Li C, Song Y, Yang R. Transcriptional profiling of a mice plague model: insights into interaction between Yersinia pestis and its host. J Basic Microbiol. 2009;49:92–99. doi: 10.1002/jobm.200800027. [DOI] [PubMed] [Google Scholar]
- 26.Bosio CM, Dow SW. Francisella tularensis induces aberrant activation of pulmonary dendritic cells. J Immunol. 2005;175:6792–6801. doi: 10.4049/jimmunol.175.10.6792. [DOI] [PubMed] [Google Scholar]
- 27.Troyer RM, Propst KL, Fairman J, Bosio CM, Dow SW. Mucosal immunotherapy for protection from pneumonic infection with Francisella tularensis. Vaccine. 2009;27:4424–4433. doi: 10.1016/j.vaccine.2009.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jia Q, Lee BY, Clemens DL, Bowen RA, Horwitz MA. Recombinant attenuated Listeria monocytogenes vaccine expressing Francisella tularensis IglC induces protection in mice against aerosolized Type A F. tularensis. Vaccine. 2009;27:1216–1229. doi: 10.1016/j.vaccine.2008.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cole LE, Elkins KL, Michalek SM, Qureshi N, Eaton LJ, Rallabhandi P, Cuesta N, Vogel SN. Immunologic Consequences of Francisella tularensis Live Vaccine Strain Infection: Role of the Innate Immune Response in Infection and Immunity. J Immunol. 2006;176:6888–6899. doi: 10.4049/jimmunol.176.11.6888. [DOI] [PubMed] [Google Scholar]
- 30.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 32.Dennis G, Sherman B, Hosack D, Yang J, Gao W, Lane HC, Lempicki R. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biology. 2003;4:3. [PubMed] [Google Scholar]
- 33.Woolard MD, Hensley LL, Kawula TH, Frelinger JA. Respiratory Francisella tularensis Live Vaccine Strain Infection Induces Th17 Cells and Prostaglandin E2, Which Inhibits Generation of Gamma Interferon-Positive T Cells. Infect Immun. 2008;76:2651–2659. doi: 10.1128/IAI.01412-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chase JC, Celli J, Bosio CM. Direct and indirect impairment of human dendritic cell function by virulent Francisella tularensis Schu S4. Infect Immun. 2009;77:180–195. doi: 10.1128/IAI.00879-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ortaldo JR, Young HA. Mouse Ly49 NK receptors: balancing activation and inhibition. Molecular Immunology. 2005;42:445–450. doi: 10.1016/j.molimm.2004.07.024. [DOI] [PubMed] [Google Scholar]
- 36.Westgaard IH, Dissen E, Torgersen KM, Lazetic S, Lanier LL, Phillips JH, Fossum S. The Lectin-like Receptor KLRE1 Inhibits Natural Killer Cell Cytotoxicity. J Exp Med. 2003;197:1551–1561. doi: 10.1084/jem.20021253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Voth DE, Howe D, Heinzen RA. Coxiella burnetii Inhibits Apoptosis in Human THP-1 Cells and Monkey Primary Alveolar Macrophages. Infect Immun. 2007;75:4263–4271. doi: 10.1128/IAI.00594-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Benoit M, Ghigo E, Capo C, Raoult D, Mege JL. The Uptake of Apoptotic Cells Drives Coxiella burnetii Replication and Macrophage Polarization: A Model for Q Fever Endocarditis. PLoS Pathog. 2008;4:e1000066. doi: 10.1371/journal.ppat.1000066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Luhrmann A, Roy CR. Coxiella burnetii Inhibits Activation of Host Cell Apoptosis through a Mechanism That Involves Preventing Cytochrome c Release from Mitochondria. Infect Immun. 2007;75:5282–5289. doi: 10.1128/IAI.00863-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Han S, Bishop BM, van Hoek ML. Antimicrobial activity of human beta-defensins and induction by Francisella. Biochemical and Biophysical Research Communications. 2008;371:670–674. doi: 10.1016/j.bbrc.2008.04.092. [DOI] [PubMed] [Google Scholar]
- 41.Lennon-Duménil A-M, Bakker AH, Maehr R, Fiebiger E, Overkleeft HS, Rosemblatt M, Ploegh HL, Lagaudriére-Gesbert Cc. Analysis of Protease Activity in Live Antigen-presenting Cells Shows Regulation of the Phagosomal Proteolytic Contents During Dendritic Cell Activation. The Journal of Experimental Medicine. 2002;196:529–540. doi: 10.1084/jem.20020327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Woolard MD, Wilson JE, Hensley LL, Jania LA, Kawula TH, Drake JR, Frelinger JA. Francisella tularensis-Infected Macrophages Release Prostaglandin E2 that Blocks T Cell Proliferation and Promotes a Th2-Like Response. J Immunol. 2007;178:2065–2074. doi: 10.4049/jimmunol.178.4.2065. [DOI] [PubMed] [Google Scholar]
- 43.Bosio CM, Bielefeldt-Ohmann H, Belisle JT. Active suppression of the pulmonary immune response by Francisella tularensis Schu4. J Immunol. 2007;178:4538–4547. doi: 10.4049/jimmunol.178.7.4538. [DOI] [PubMed] [Google Scholar]
- 44.Chiavolini D, Alroy J, King CA, Jorth P, Weir S, Madico G, Murphy JR, Wetzler LM. Identification of immunologic and pathologic parameters of death versus survival in respiratory tularemia. Infect Immun. 2008;76:486–496. doi: 10.1128/IAI.00862-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.England K, am Ende C, Lu H, Sullivan TJ, Marlenee NL, Bowen RA, Knudson SE, Knudson DL, Tonge PJ, Slayden RA. Substituted diphenyl ethers as a broad-spectrum platform for the development of chemotherapeutics for the treatment of tularaemia. J Antimicrob Chemother. 2009;64:1052–1061. doi: 10.1093/jac/dkp307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wilson JE, Katkere B, Drake JR. Francisella tularensis Induces Ubiquitin-Dependent Major Histocompatibility Complex Class II Degradation in Activated Macrophages. Infect Immun. 2009;77:4953–4965. doi: 10.1128/IAI.00844-09. [DOI] [PMC free article] [PubMed] [Google Scholar]