Abstract
The post-translational modification of proteins by sugars has been demonstrated in diabetes and classical galactosemia. In diabetes, the glycation process occurs as a result of D-glucose nonenzymatically reacting with proteins such as albumin and hemoglobin, used today as important tools to monitor the efficiency of dietary control and therapy during treatment of diabetes. In classical galactosemia, D-galactose contributes to the formation of glycated proteins also, suggesting that akin to diabetes with glucated proteins, the monitoring of galactated proteins may facilitate management of patients with galactosemia. The objectives of this project were: 1) galactate HSA, in vitro; 2) determine, by a sodium borohydride dependent mass peptide mapping method, the galactation sites in HSA; and 3) compare HSA’s galactation sites with the protein’s reported glucation sites. Treatment of galactated HSA with sodium borohydride stabilized the condensed sugars on the protein and yielded discrete fragmentation patterns by tandem MS, allowing reliable identification of HSA’s galactation sites. LC/ESI/MS, in combination with tandem MS, revealed the principal sites of galactation in HSA were the amino groups of lysine 12, 233, 281/276, 414 and 525. Lysyl residues 12, 233, 276, and 525 were previously reported as privileged sites for the nonenzymatic binding of D-glucose with HSA.
Keywords: Nonenzymatic glycation, Galactated HSA, Amadori adducts, Mass spectrometry, MALDI-TOF/MS, Tandem MS, Peptide mapping
Introduction
The nonenzymatic glucation of human serum albumin (HSA) and human hemoglobin has been extensively studied and has been the subject of several reviews [1–5]. What has made the glucation of these proteins clinically significant is that they both provide for an assessment of glycemic control in diabetes, with glucated HSA and glucated hemoglobin serving as sensitive indicators of short term and long term hyperglycemic control, respectively [6–8]. Studies in our laboratory and those of others have demonstrated that the in vivo glycation of HSA and hemoglobin does not occur with D-glucose only and that in certain conditions such as galactosemia, the high levels of D-galactose in blood can also promote the galactation of HSA and hemoglobin [9–13].
The glucation and galactation of proteins begins with the formation of an unstable Schiff base that occurs between the carbonyl group of the reducing sugars D-glucose or D-galactose and the free amino groups on proteins. This Schiff base then rearranges to form a more stable Amadori product that with time forms a heterogeneous group of compounds referred to Advanced Glycation End Products (AGEs) [14–16]. The mechanistic similarity in the formation of nonenzymatically glucated and galactated proteins [14, 15] suggests that akin to diabetes with glucated proteins, the monitoring of galactated proteins may provide a valuable tool for the management of patients with classical galactosemia, a rare genetic disorder characterized with increased D-galactose and galactitol levels in tissues and body fluids [9,17–20]. To explore this possibility and develop an assay specific for galactated proteins, we focused our attention to HSA and to its amino acid sites of galactation. Emphasis was placed on the vulnerable sites of galactation for two reasons: 1) to design genuine galactated peptides similar to those found in HSA for developing monoclonal antibodies to the protein, and 2) to determine if the sites of galactation coincided with HSA’s reported glucation sites.
Mass spectrometry using ESI and MALDI-MS has been successfully applied to the study of protein glucation in diabetes, particularly, when mapping for glycated peptides, characterizing advanced glucation end products and determining the number of D-glucose residues condensed with proteins [21, 22].
This paper describes the application of a sodium borohydride dependent mass peptide mapping method for identifying HSA’s galactation sites. In this procedure, in vitro prepared galactated HSA was first reduced with sodium borohydride, and the resulting protein was then digested with trypsin. The tryptic digests were analyzed by LC/ESI/MS and tandem MS which yielded quality spectra with minimal neutral water losses.
Neutral water loss behavior is a phenomenon commonly observed in collision induced dissociation (CID) of glycated peptide ions. During CID, the preferential release of water from the labile Amadori adducts results in poor production of sequence specific ions from the peptide backbone. Consequently, this yields a CID spectrum with little or no useful information impeding the identification of peptide sequences and sites of glycation [23–25].
The reduction of galactated HSA with sodium borohydride appeared to not only stabilize the linkages between the nonenzymatically reacted sugars and the peptides, but also yielded discrete fragmentation patterns by tandem MS allowing for the easy and reliable identification of HSA’s galactation sites.
In this report, we describe the galactation sites in HSA and compare our findings with previously reported glucation sites for the protein.
Materials and Methods
Reagents
Sterile filtered normal human serum screened and tested for and found non-reactive for Hepatitis B & C and non-reactive for Human Immunodeficiency Virus (HIV) antibody was purchased from the BioBank of Sera Care Life Sciences, Inc. (Oceanside, CA). D-galactose, aminophenylboronic acid resins, silica C18 resins, trypsin and solvents for HPLC were purchased from Sigma Aldrich Chemical Co. (St. Louis, MO). AffiGel Blue, Bio-Gel-P-150, standard chromatographic resins and Coomassie G 250 reagent were obtained from Bio-Rad lnc. (Richmond, CA). Unless otherwise indicated, all other reagents and supplies were obtained from Pierce Chemical Company (Rockford, IL).
Assessment of Protein Concentration
Protein levels were determined by Coomassie G 250 reagent using human albumin as the standard protein.
Purification of Albumin from Human Serum
HSA was isolated and purified from normal human serum by affinity chromatography on Affi-Gel Blue followed by gel filtration on Bio-Gel-P-150 as described previously [14]. The purified albumin fractions from the gel filtration step were pooled, dialyzed at 4°C against distilled water and then lyophilized. The purity of HSA was assessed by SDS-PAGE and Coomassie Blue staining.
Separation of Non-Glycated Albumin from Purified Albumin
Non-glycated HSA (hereinafter also referred to as ‘native’ HSA) was isolated from the purified HSA preparation by aminophenylboronic acid resins as described previously [26]. An aliquot of the resulting sample was then assayed by the thiobarbituric method of Fluckinger and Winterhalter [27] to confirm absence of glycated albumin species. Once confirmed free of glycated albumin, the protein preparation was dialyzed overnight at 4 °C against distilled water and was lyophilized.
In vitro Preparation of Galactated HSA
Unless otherwise indicated, galactated HSA was prepared by incubating 240 mg of the nonglycated protein with 1g of D-galactose in 3ml of 50mM phosphate buffer, pH 7.4 at 56°C for 2 hrs. Following incubation, samples were dialyzed overnight at 4°C against two changes of the phosphate buffer (2L each), and then applied to separate aminophenylboronate columns (1.5x10 cm) previously equilibrated with 50mM phosphate buffer, pH 7.4. Non bound material was eluted with the phosphate buffer, and bound material was eluted with a 0.1 M sorbitol solution prepared in 50mM phosphate buffer, pH 7.4.
The bound HSA fractions from each column were separately pooled and dialyzed overnight at 4°C against two changes of 50 mM phosphate buffer, pH 7.4 (2L each). The unbound fractions were separately pooled also, but were not dialyzed. The resulting preparations were then each tested for glycation with thiobarbituric assay which measures ketoamine bound carbohydrate as the amount of 5- hydroxymethyl furfuraldehyde released by dehydration of hexose sugars in boiling oxalic acid [24]. Samples bound to aminophenylboronate resin were each found to be glycated and positive by the thiobarbituric assay. Preparations not bound to the aminophenylboronate resin were each found to be nonglycated and to yield a negative thiobarbituric reaction.
Sodium Borohydride Reduction
A freshly prepared 100mM solution of sodium borohydride in 0.02N NaOH was mixed 1:4 with water, and then immediately reacted 1:1 with each of the HSA preparations. The mixing of the protein samples (i.e., dialyzed glycated HSA and non glycated HSA) with sodium borohydride solution was performed in such a manner as to ensure that each tube contained a final protein content of 5mg/ml. Incubations were allowed to proceed at room temperature for 1h and reactions were then terminated by 1N HCl (1:12.5 vol/vol).
Following the addition of HCl to samples, mixtures were incubated at room temperature for 5 minutes. The pH of samples was then adjusted to physiological pH by a 1:1 dilution with a 100 mM Tris buffer, pH 7.5.
Preparation & Analysis of Tryptic Digests of HSA by MALDI-TOF Mass Spectrometry
Unless otherwise indicated, borohydride treated HSA samples adjusted to pH 7.5 were digested with trypsin (1:10 w/w) overnight at room temperature. The tryptic digests of HSA were purified with C18 ZipTips (Millipore, Billerica MA) and analyzed on a Bruker Autoflex MALDI-TOF mass spectrometer (Billerica, MA). Prior to analysis, the peptide preparations (0.6 μL) were each mixed with a 50% aqueous acetonitrile solution (0.6 μl) of saturated α-cyano-4-hydroxy-cinnamic acid containing 0.05% TFA as matrix, spotted onto a stainless steel sample plate and allowed to air dry. The mass spectra were recorded in positive ion mode with a 60 nsec delay in the m/z range from 500 to 3500. Typically, 300 spectra were accumulated with 50 laser shots for each sample spot analyzed.
Preparation and Analysis of Intact HSA by MALDI-TOF Mass Spectrometry
Intact HSA preparations were processed similar to the HSA tryptic digests, but with minor modifications in the above protocol: i) instead of C18 Zip Tips, C4 ZipTips were employed, and ii) instead of α-cyano-4-hydroxy-cinnamic acid, sinapinic acid was used for the spotting of the samples.
Samples were analyzed as before on a Bruker Autoflex MALDI-TOF mass spectrometer, but with instrument settings optimized for intact protein analysis. Spectra were acquired in linear TOF mode with a 550 nsec delay in the m/z range from 40,000 to 90,000. Each spectrum was the sum of 500 single laser shots randomized over 10 positions within the same spot (500/50). Analysis of data was performed using FlexAnalysis and ClinProTools (Bruker Daltonics Inc., Billerica, MA)
Determining the Sites of Galactation in HSA by LC/ESI/MS
Samples adjusted to pH 7.5 were digested with trypsin (1:10 w/w) overnight at room temperature. The tryptic peptides were fractionated on 10μm spherical C18 resins (YMC, Inc. Milford, MA) and were eluted directly into the electrospray source of a ThermoFinnigan LCQ mass spectrometer (ThermoScientific, Waltham, MA) equipped with a quadrupole ion trap mass analyzer. The reverse phase microcapillary columns were constructed according to a procedure modified from Kennedy and Jorgensin [28]. The loading of peptides onto the columns was performed using a helium bomb. Once applied to the columns, the peptides were first washed with 0.1% acetic acid for 3 minutes and were then collectively eluted from the columns with 60% acetonitrile in 0.1% acetic acid at a flow rate of 2 μL/min using the helium bomb. The column eluates directed into the electrospray source were analyzed by MS and tandem mass spectrometry (CID fragmentation with He) favoring isolation and fragmentation of doubly charged ions.
The ESI/MS system was operated with the Xcalibur software (version 2.0, ThermoFinnigan), with the same software used also for data analysis. The mass spectrometer was set to function in the positive ion mode with parameters optimized during direct infusion of peptide standards with solvent. The solvent, consisting of 50/50 (v/v) 0.1% acetic acid in water/acetonitrile, was applied at a flow rate of 3 μL/min. Nitrogen was used as the sheath gas (setting at 60), and ultrapure helium was used as the collision gas. The ion spray voltage was set as 4.5 kV and the capillary temperature was 210°C.
Results
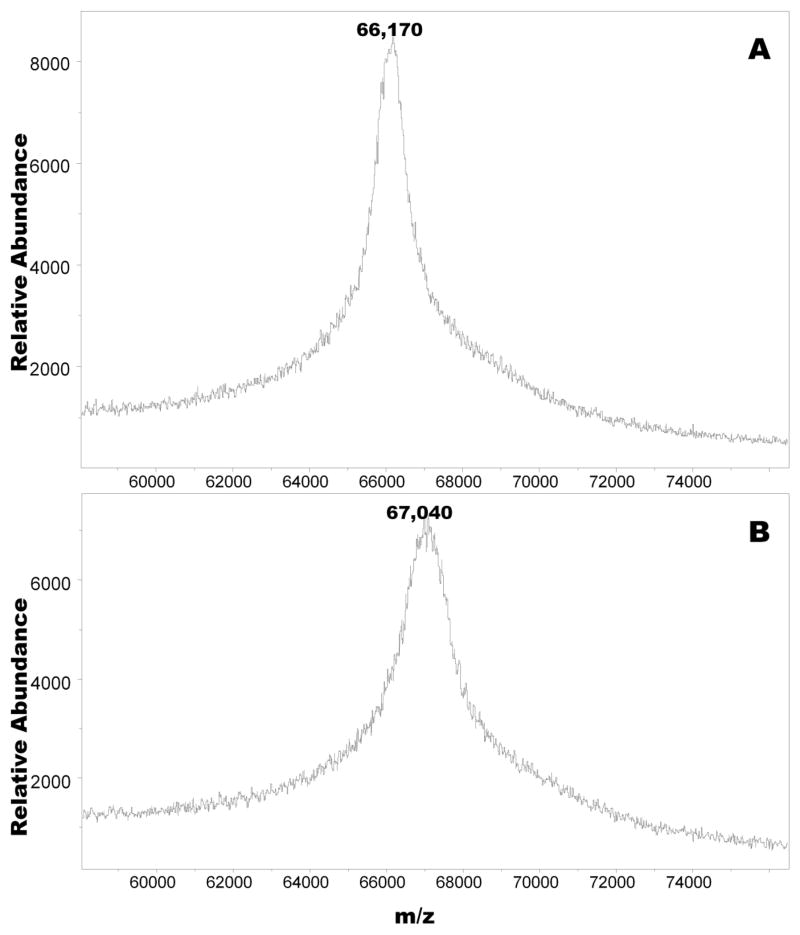
Figure 1 shows the MALDI-TOF mass spectra of nonglycated HSA and of a galactated HSA sample prepared at 56°C for 2h (see Materials and Methods). The spectrum of nonglycated HSA exhibited a narrow band centered at m/z=66170 Da, whereas that of the galactated protein yielded a wide band, commonly seen with ‘modified proteins’ [29], centered at m/z=67040 Da. Repeat analysis of the samples by MALDI-TOF mass spectrometry yielded similar results confirming that the shift in peaks corresponded to the condensation of about five reduced D-galactose (+164) molecules to the protein.
Figure 1.
MALDI-TOF mass spectrum of non-glycated HSA treated with sodium borohydride (A), and of sodium borohydride reduced galactated HSA derived from incubation mixtures of non-glycated protein (240 mg) with D-galactose (1 g) in 50 mM phosphate buffer (3ml), pH 7.4 at 56ºC for 2h (B).
To determine the amino acid sites of galactation, the non glycated and galactated HSA samples were separately reacted with trypsin and the tryptic digests from each was evaluated by MALDI-TOF mass spectrometry. The findings from this analysis were then compared with the molecular weights for a theoretical HSA tryptic digest.
Of the resulting peptides, only one peptide with m/z=1651 was found to exhibit a mass shift. This peptide was found to condense with one reduced D-galactose molecule (+164 Da) yielding a band with m/z=1815 (spectrum not shown). Relative to its counterpart non-modified peptide, the glycated peptide was seen at about 70 % abundance. Based on the primary sequence of HSA, the site of galactation was localized around peptideaa 226–240 (AEFAEVSKLVTDLTK). The finding of no additional galactated peptides in the tryptic digests of HSA was surprising, especially, since prior to the digestion of the protein, HSA was shown condensed with five D-galactose residues.
We speculate that this anomaly occurred due to one of several causes. One possibility could be that the glycated peptides might have exhibited a mass overlapping those of the unmodified peptides in the tryptic digest. In a complex mixture such as our tryptic digest where the majority of the peptides were unmodified, it is also likely that competitive ionization in the gas phase might have caused certain species to preferentially ionize. This phenomenon, known as ‘ion suppression’, might have also contributed to the obscurity in the detection of galactated peptides depending on the extent complexity of the tryptic mixture [30]. A third possibility could be that during the ionization process some sugars were detached from the peptides.
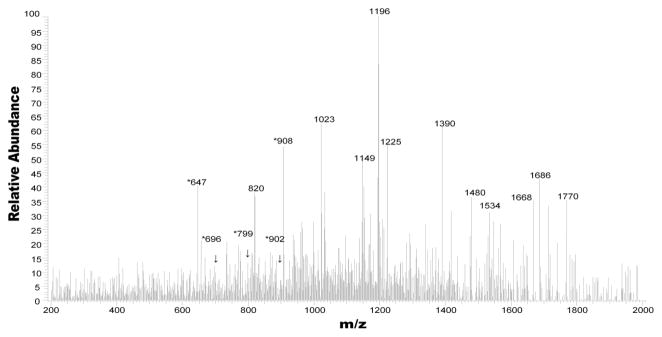
To improve the analysis of galactated peptides, we shifted focus from MALDI-TOF mass spectrometry to LC/ESI/MS. Figure 2 shows the LC/ESI mass spectrum of the tryptic digests of galactated HSA evaluated earlier by MALDI-TOF mass spectrometry. Analysis of the spectrum identified five peptides (m/z of 647, m/z 696, m/z 798, m/z 901, and m/z 908) as having masses corresponding to galactated peptides in HSA. As several of these galactated peptides were in low abundance relative to the non glycated peptides from the protein, the identification of the galactation sites remained challenging without further analysis by tandem mass spectrometry. A follow up evaluation of the five peptides by CID fragmentation confirmed that each was glycated with D-galactose and that the sugar in each of the peptides was condensed with lysine residues only.
Figure 2.
The tryptic mass fingerprint spectrum of HSA incubated with D-galactose for 2 hours at 37ºC. The protein was reduced with sodium borohydride and digested with trypsin. *Denotes the +2 ions of glycated peptides.
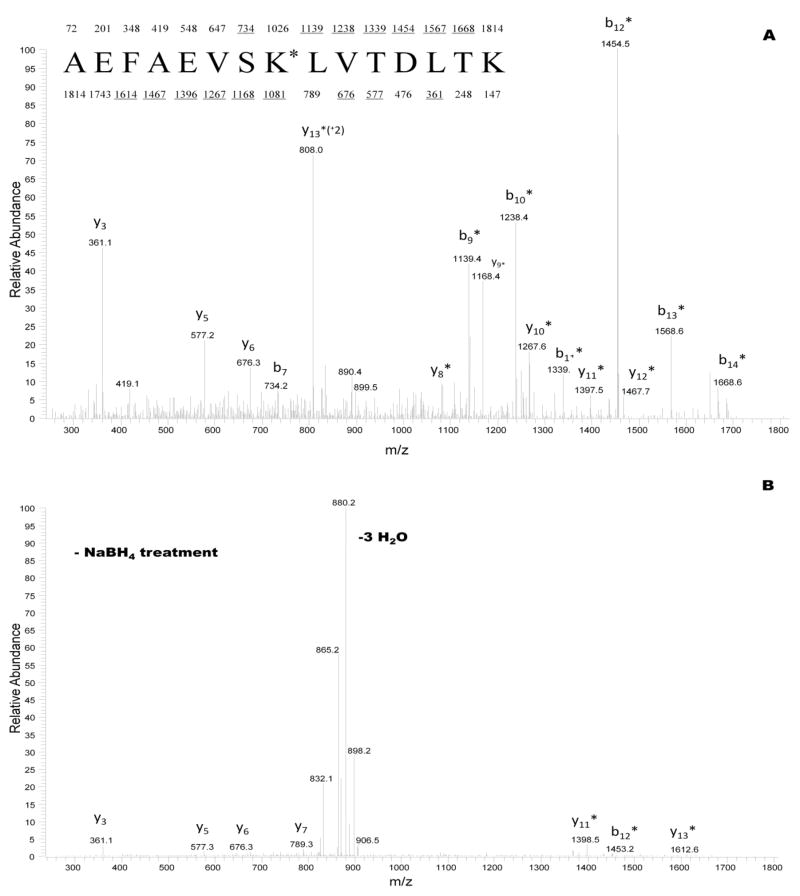
A dramatic improvement in the MS/MS spectral quality was obtained when the glycated peptides were reduced with sodium borohydride. Figure 3A shows the CID fragmentation for the +2 ion corresponding to the reduced (+ NaBH4 treatment) glycated peptide at m/z 908. Figure 3B displays the CID spectrum for the counterpart galactated peptide control not reduced with sodium borohydride. Analysis of the CID mass spectra revealed that the peptide (aa 226-240) had a sequence consisting of AEFAEVSK*LVTDLTK, with K* representing a lysine residue derivatized with D-galactose. The y and b ions in figure 3A demonstrated mass shifts of + 164 Da corresponding to reduced glycated peptide fragments. These fragments localized the galactation site at lysine 233 (233K*). Other findings revealed that trypsin failed to cleave C-terminal to lysine residues modified with reduced galactose, and that characteristic water loss peaks at m/z 890 and m/z 899 appeared in the reduced peptide CID spectra. These water loss peaks would allow the identification of glycated peptides in neutral water loss screens.
Figure 3.
CID mass spectra recorded on the (M+2H)+2 ion for sodium borohydride pretreated glycated peptide at m/z 908 (A), and for counterpart glycated peptide control not treated with sodium borohydride (B). The sequence of the peptide in (A) is shown with the predicted y and b fragments. Ions observed in spectrum (A) are underlined. K* represents a galactated lysine residue of 292 Da at position 233 in the primary structure of HSA.
The major fragment ions in figure 3B corresponded to: i) the loss of three water molecules from the peptide, and ii) the development of few informative y and b fragment ions. Treatment with sodium borohydride facilitated the identification of galactation sites in the HSA peptide, and yielded a spectrum with minimal neutral water losses. This technique may also prove versatile in the mass peptide mapping of other galactated proteins.
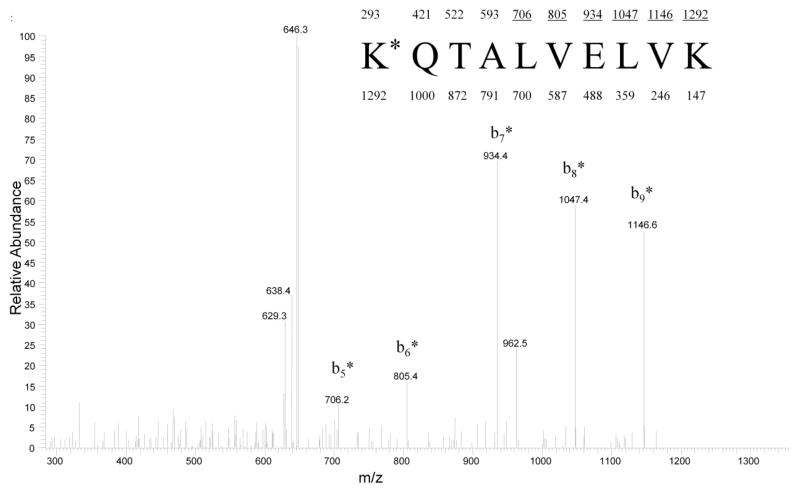
Figure 4 shows the CID mass spectrum for the reduced peptide aa 525-534 at m/z 647. Analysis of the spectrum revealed a strong b-ion series indicating mass shifts of +164 Da corresponding to derivatized galactated peptide fragments. The sequence for this peptide was identified as K*QTALVELVK , with K* representing a galactated lysine residue (525K*). The glycated peptides with m/z’s at 696, 798, and 901 were similarly subjected to CID fragmentation and analyzed by tandem MS as described previously (data not shown). Table 1 summarizes the data and shows the sequences and the location of D-galactose in each of the glycated peptides.
Figure 4.
CID mass spectrum recorded on the (M+2H)+2 ion at m/z 647. The sequence of the peptide is shown with the predicted y and b fragments. Ions observed in the spectrum are underlined. K* represents a galactated lysine residue of 292 Da (i,e., 525K in HSA). The water loss series at m/z 638 and m/z 629 are characteristic of glycated peptides.
Table I.
Glycated peptides identified by LC/ESI/MS and LC/ESI/MS/MS
| Tryptic Digest Fragments | Peptide aa | Theoretical Mass of Fragments in Da | LC/ESI/MS Observed Mass in Da (charge state) | Δ m/z | Analysis by Tandem MS Incorporating CID |
|---|---|---|---|---|---|
| FK*DLGEENFK | 11–20 | 1226.6 | 1391(+2) | 164 | 12K |
| AEFAEVSK*LVTDLTK | 226–240 | 1650.9 | 1815(+2) | 164 | 233K |
| LK*ECCEK*PLLEK | 275–286 | 1432.8 | 1597(+2) | 164 | 276 K or 281K |
| K*VPQVSTPTLVEVSR | 414–428 | 1639.9 | 1804(+2) | 164 | 414K |
| K*QTALVELVK | 525–534 | 1128.7 | 1293(+2) | 164 | 525K |
Galactated HSA was prepared by incubating the native protein (240 mg) with D-galactose (1.85 M) in 3 ml of 50 mM phosphate buffer, pH 7.4, for 2h at 56°C.
Denotes that these sites represent galactated lysine residues
To determine the effect of incubation time on the galactation of HSA, the ‘native’ protein was reacted with 1 g of D-galactose as before, but for variable periods of time from 2 to 5 h. The resulting HSA preparations were then applied to aminophenylboronate columns (1.5x10 cm) and processed as previously described, with the exception that samples were not treated with sodium borohydride. The control sample included native HSA (240 mg) in 3ml of 50 mM phosphate buffer, pH 7.4, to which no D-galactose was added. Unless otherwise indicated, the control HSA preparation was incubated at 56 °C for 5h and processed under equivalent conditions as the glycated HSA samples.
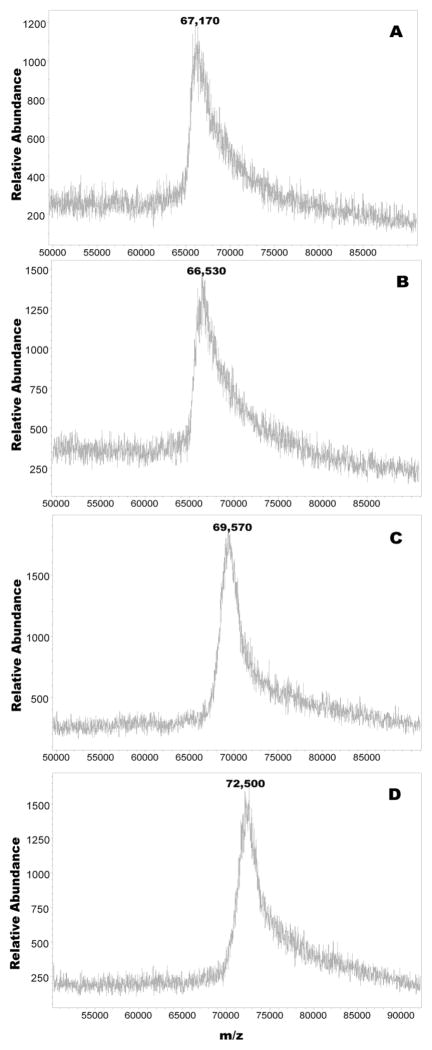
Figure 5 shows the MALDI-TOF mass spectra of the non glycated HSA control sample and of the HSA preparations incubated at 56 °C with 1g D-galactose (1.85 M) for 2, 3 and 5 h. A close scrutiny of the spectra revealed that the control HSA sample yielded a band centered at m/z=66,170 Da, whereas the two hour glycated preparation yielded a similar looking peak with a band centered at m/z=66,530 Da. The shift in mass between the two samples corresponded to the addition of about two D-galactose molecules to the protein.
Figure 5.
MALDI-TOF mass spectra of HSA control incubated in the absence of sugar (A), and in the presence of 5mM D-galactose at 56°C for 2 h (B), 3 h (C) and 5 h (D).
Under equivalent incubation conditions, but with the resulting glycated protein reduced with sodium borohydride, intact HSA was earlier found to condense with roughly five D-galactose molecules (see figure 1). This difference we believe may have occurred due to sodium borohydride stabilizing the nonenzymatically reacted D-galactose residues on HSA, diminishing the likelihood of the sugar dissociating from the protein. In the case of the HSA not treated with sodium borohydride, the opposite might have occurred where, for example, some of the labile Schiff base structures including reversible Amadori adducts could have split from the protein resulting in the lower number of D-galactose residues observed (figure 5) by MALDI-TOF mass spectrometry.
Other observations revealed that increases in incubation time increased the mass of the glycated proteins, with the 5h incubated HSA sample exhibiting the highest shift in m/z. In this instance, the shift in m/z was consistent with the addition of approximately 39 D-galactose residues to the protein.
Next, focus was placed on determining the preferred sites of galactation in HSA samples whose glycation was performed in vitro at a low concentration of sugar (5mM) and at physiological temperature and pH. To conduct this study, 240 mg of ‘native’ HSA was mixed with 2.7mg D-galactose in 3ml of 50mM phosphate buffer, pH 7.4 and incubated at 37°C for 7 days. After incubation, the resulting protein was isolated on aminophenylboronate column (1.5x10 cm) and processed with sodium borohydride as described previously. The control HSA sample consisted of ‘native HSA’ (240mg) incubated in 50mM phosphate buffer, pH 7.4 in the absence of D-galactose at 37°C for 7 days. Like its glycated counterpart, the control HSA sample was treated with sodium borohydride.
A close scrutiny of the MALDI-TOF mass spectra revealed two important findings. First, that under physiological temperature and pH, HSA was susceptible to glycation by 5mM D-galactose. Second that, under the aforementioned incubation conditions, only one sugar residue condensed with HSA (data not shown).
ESI/MS analysis of the tryptic digests of the protein yielded four peptides with mass to charge ratios corresponding to various galactated peptides in HSA (m/z of 647, m/z of 696, m/z of 901, and m/z of 907). These same peptides were found in their non glycated forms in the tryptic digests also (spectrum not shown). Since the intact HSA had condensed with only one D-galactose residue, these results suggested that i) there was no preferred site of galactation where the sugar was solely found to react with on the protein, and ii) that the resulting HSA preparation was actually a heterogeneous mixture of singly glycated HSA proteins.
A further analysis of the peptides by CID fragmentation revealed that each was condensed with D-galactose at lysine residues only, and that the glycated lysine residues were located at positions observed earlier when HSA was incubated at 56 °C with 1.85 M D-galactose (see Table II). The reactivity of D-galactose with lysine was not surprising, as earlier studies with HSA showed that lysine was the major amino acid residue condensing with D-glucose [32]. Glucose-lysine adducts have been also demonstrated in bovine collagen [33], basic myelin protein [34, 35], lens crystallin [36], and in proteins of the human erythrocyte membrane [37].
Table II.
Glycated peptides identified by LC/ESI/MS and LC/ESI/MS/MS
| Tryptic Digest Fragments | Peptide aa | Theoretical Mass of Fragments in Da | LC/ESI/MS Observed Mass in Da (charge state) | Δ m/z | Analysis by Tandem MS Incorporating CID |
|---|---|---|---|---|---|
| FK*DLGEENFK | 11–20 | 1226.6 | 1391(+2) | 164 | 12K |
| AEFAEVSK*LVTDLTK | 226–240 | 1650.9 | 1815(+2) | 164 | 233K |
| K*VPQVSTPTLVEVSR | 414–428 | 1639.9 | 1804(+2) | 164 | 414K |
| K*QTALVELVK | 525–534 | 1128.7 | 1293(+2) | 164 | 525K |
Galactated HSA was prepared by incubating the native protein (240 mg) with D-galactose (5 mM) in 3 ml of 50 mM phosphate buffer, pH 7.4, for 7 days at 37°C.
Denotes that these sites represent galactated lysine residues
Discussion
Ever since the description of hemoglobin A1c, there has been increased interest in the peptide mapping of glycated proteins. Hemoglobin A1c is a glucated form of hemoglobin, used for the long term management of patients with diabetes [8]. Peptide mapping of hemoglobin A1c has played a significant role in our understanding of the glycation sites of the protein, allowing for the design of genuine peptides that facilitated the development of monoclonal antibodies to the protein [38–40]. Like hemoglobin A1c, the peptide mapping of glucated albumin has provided valuable information for the development of immune probes to the protein [41,42], aiding the development of assays for its quantification in human plasma. The levels of glucated albumin provide an index of the prevailing blood glucose concentrations over the preceding 2–3 weeks, making it an ideal candidate for the short term monitoring of patients with diabetes [6–8, 42].
Studies have demonstrated that, similar to D-glucose, D-galactose can nonenzymatically react with proteins in vivo, and in diseases such as classical galactosemia, contribute to the post- translational modification of hemoglobin and albumin [9-11, 13]. We have focused on the identification of HSA’s galactation sites, reasoning that knowledge of these may aid in the design of immunogens for developing antibody probes to the protein.
Mass Peptide mapping of glycated proteins has been performed by a number of mass spectrometric techniques. Some of these techniques have relied on MALDI-TOF MS [43], liquid chromatography coupled to MS via electrospray ionization [23], tandem MS incorporating CID (25), or electron transfer dissociation (ETD) [44,45]. In this study, we identified the sites of galactation in HSA, primarily relying on LC/ESI/MS and tandem MS incorporating CID. Focus was placed on in vitro prepared galactated HSA, and on early glycation products of HSA with D-galactose. Galactated HSA, treated with sodium borohydride, yielded tryptic digests readily resolvable by MS/MS spectra, as compared to galactated controls not treated with sodium borohydride.
There have been only a few reports on the mass spectrometry of proteins nonenzymatically glycated with D-galactose. One group assessed the galactation sites of a bovine serum albumin preparation heat-dried in the presence of D-galactose at 60° C for 2 hours [31]. Their results revealed that, under these conditions, D-galactose nonenzymatically reacted with ten sites on the protein, with each of the sites involving a lysyl residue (117K, 140K, 256K, 285K, 346K, 374K, 420K, 523K, 297K, and 597K). These galactation sites were identified by LC/MS with no reported follow up tandem mass spectrometry to confirm the findings.
A comparison of HSA’s galactation sites, with reported HSA’s glucation sites, revealed several shared sites of glycation, where both D-glucose and D-galactose were found condensed with the protein. Galactated HSA prepared under physiological temperature and pH showed that the protein was vulnerable to nonenzymatic attack by D-galactose at four lysyl sites. These sites were 12K, 233K, 414K, and 525K. A further analysis of these positions indicated that 525K is the major galactation site in HSA. Incubation of the protein at 56°C for 2h yielded one additional site of glycation localized at 276K or 281K. The uncertainty in the assignment of these lysyl residues was due to minimal production of y and b fragment ions in this region of the peptide.
Iberg and Flückiger [46] working with HSA from diabetic patients found 199K, 281K, 439K, and 525K as the privileged glycation sites in the protein, with 525K being the site most vulnerable to nonenzymatic attack by sugar. Working with freshly isolated HSA, Garlick and Mazer [47] also identified 525K as the site most receptive to Maillard reaction in vivo. In both of these studies, peptide mapping was performed by first labeling the glycated protein with tritiated sodium borohydride, and finally analyzing the hydrolysates from radioactively labeled peptides.
In addition to the in vivo findings, several glucation sites were reported in HSA minimally glycated with D-glucose in vitro. Using MALDI-TOF mass spectrometry as the basis for analysis, one group reported 12K, 51K, 199K,159K, 205K, 286 K, 378 K,439K, 538K, 160R, 222R, and 742R as the sites of glycation in HSA (48). Of these residues, only six sites were found to be modified through the formation of fructosyl-lysine (12K, 51K, 199K, 205K, 439K, and 538K). While several of these glucation sites were reported for the first time (K51, K159, K205, K286, K538, R160, R222 and R472), it is not clear from the study how the analyzed glycated HSA was prepared. Considering that the sugar modified HSA was obtained from a commercial source, no details were provided as to the incubation temperature and period of time the protein was exposed to D-glucose in vitro. Consequently, some of these new sites of glucation, could be due to forced glycation. In another study where HSA was glucated under physiological conditions, Lapolla et al [23] reported the condensation of the sugar primarily with five sites. These sites included 233K, 276K, 378K, 525K, and 545K, respectively.
As mentioned before, we found 414K as one of the privileged glycation sites of HSA with D-galactose. To our knowledge, this residue was never reported as a site of glucation in HSA. One reason this residue was overlooked earlier, may be due to 414K being more receptive to forming a labile Schiff base structure than an Amadori adduct with sugars. The rationale for this speculation comes from: 1) the presence of 410R and 413K in proximity to 414K, which could enhance the nucleophilicity of the epsilon amino group of the lysine, encouraging it to form a Schiff base with sugars; and 2) the absence of an acidic amino acid nearby, diminishing the likelihood of sugars condensed at this site to undergo an Amadori rearrangement. Therefore, treatment of the glycated HSA with sodium borohydride must have stabilized the linkage between D-galactose and 414K, allowing its identification by mass spectrometry.
It is interesting to note that, in terms of its location in HSA, 414K exhibits several similarities with 525K, the most predominant site of galactation in the protein. For example, like 414K, 525K is part of a α-helical structure in HSA and is positioned in proximity of arginine. Moreover, it exhibits an analogous sequence motif represented by R--KK; the presence of di-lysine motifs has been suggested to accelerate glycation due to local acid-base catalysis [49].
This study revealed two important findings: 1) that sodium borohydride treatment, coupled with LC/ESI /MS/MS using CID as the fragmentation mode, is an efficient approach for analysis of galactated proteins, and 2) that out of the five galactation sites in HSA, four sites were the same as those reported previously with HSA nonenzymatically reacted with D-glucose.
Acknowledgments
This work was supported by STTR Grant 1R41DK076481 from the National Institutes of Health awarded to Vandalia Research, Inc. Special thanks go to Jason Cohenford for his review of the manuscript and comments, and to Dr. Faith Hays for her valuable suggestions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Levenson D. Everyone should watch glycosated hemoglobin and blood sugar levels not just diabetics, research says. Rep Med Guidel Outcomes Res. 2004;15:1–2. 5–6. [PubMed] [Google Scholar]
- 2.Akazawa Y, Tsuji S, Tashiro S. Glycated serum albumin: An indicator of short term glycemic control in diabetic patients. J Med Pharm Sci. 1993;29:269–274. [Google Scholar]
- 3.Ono M, Ikegami H, Ogihara T. Glycated serum protein (GSP), glycated albumin (GA) and fructosamine. Nippon Rinsho. 2002;60(Suppl 8):415–9. (article in Japanese) [PubMed] [Google Scholar]
- 4.Cohen MP, Clements RS. Measuring glycated proteins: Clinical and methodological aspects. Diabetes Technol Ther. 1999;1:57–70. doi: 10.1089/152091599317585. [DOI] [PubMed] [Google Scholar]
- 5.Carter AW. An Analysis of the Assessment of Glycated Hemoglobin Using A1cNow+ Point-of-Care Device Compared to Central Laboratory Testing– an Important Addition to Pharmacist-Managed Diabetes Programs? J Diabetes Sci Technol. 2008;2:828–830. doi: 10.1177/193229680800200513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wieland OH. Protein glycation: measurement and clinical relevance. J Clin Chem Clin Biochem. 1989;27:577–87. [PubMed] [Google Scholar]
- 7.Ba JM, Dou JT, Zhang XQ, Ma FL, Zhai HW, Zou XM, Mu YM, Lu JM. Clinical study of glycated albumin measurement by enzymatic method in type 2 diabetes mellitus. Zhonghua Yi Xue Za Zhi. 2009;89:1570–2. (article in Chinese) [PubMed] [Google Scholar]
- 8.Takahashi S, Uchino H, Shimizu T, Kanazawa A, Tamura Y, Sakai K, Watada H, Hirose T, Kawamori R, Tanaka Y. Comparison of glycated albumin (GA) and glycated hemoglobin (HbA1c) in type 2 diabetic patients: usefulness of GA for evaluation of short-term changes in glycemic control. Endocr J. 2007;54:139–44. doi: 10.1507/endocrj.k06-103. [DOI] [PubMed] [Google Scholar]
- 9.Urbanowski JC, Cohenford MA, Levy HL, Crawford JD, Dain JA. Nonenzymatically galactosylated serum albumin in a galactosemic infant. N Engl J Med. 1982;14:306, 84–6. doi: 10.1056/NEJM198201143060207. [DOI] [PubMed] [Google Scholar]
- 10.Bohles H, Schadle J, Endres W, Shin YS, Kollmann F, Bender SW, Kruse K. Increased concentrations of HbAlab in hereditary fructose intolerance and galactosemia. Padiatr Padol. 1987;22:25–31. [PubMed] [Google Scholar]
- 11.Howard NJ, Monaghan H, Martin JM. Hemoglobin A1 in galactosemia, a possible role in monitoring dietary compliance. Acta Paediatr Scand. 1981;70:695–8. doi: 10.1111/j.1651-2227.1981.tb05770.x. [DOI] [PubMed] [Google Scholar]
- 12.Mohr S, Xi X, Tang J, Kern TS. Caspase Activation in Retinas of Diabetic and Galactosemic Mice and Diabetic Patients. Diabetes. 2002;51:1172–1179. doi: 10.2337/diabetes.51.4.1172. [DOI] [PubMed] [Google Scholar]
- 13.Dain JA, Urbanowski JC, Cohenford MA, Shepard DC, Hitz JB. Nonenzymatic galactosylation of proteins in galactosemia. In: Sun G, et al., editors. Neural Membranes. Humana Press Inc; New Jersey: 1983. pp. 415–424. [Google Scholar]
- 14.Urbanowski JC, Cohenford MA, Dain JA. Nonenzymatic galactation of human serum albumin in vitro preparation. J Biol Chem. 1982;257:111–115. [PubMed] [Google Scholar]
- 15.Armbruster DA. Fructosamine: structure, analysis, and clinical usefulness. Clin Chem. 1987;33:2153–63. [PubMed] [Google Scholar]
- 16.Bohlender JM, Sybille F, Stein G, Wolf G. Advanced glycation end products and the kidney, C. Am J Physiol Renal Physiol. 2005;289:F645–F659. doi: 10.1152/ajprenal.00398.2004. [DOI] [PubMed] [Google Scholar]
- 17.Lambert, Boneh A. The impact of galactosaemia on quality of life-A pilot study. J Inherit Metab Dis. 2004;27:601–8. doi: 10.1023/b:boli.0000042957.98782.e4. [DOI] [PubMed] [Google Scholar]
- 18.Bosch AM, Grootenhuis MA, Bakker HD, Heijmans HS, Wijburg FA, Last BF. Living with classical galactosemia: health-related quality of life consequences. Pediatrics. 2004;113:423–8. doi: 10.1542/peds.113.5.e423. [DOI] [PubMed] [Google Scholar]
- 19.Lyon IC, Chapman CJ, Houston IB, Veale AM. Galactosaemia: estimated live birth incidence in New Zealand. Humangenetik. 1975;28:79–82. doi: 10.1007/BF00272487. [DOI] [PubMed] [Google Scholar]
- 20.Schweitzer-Krantz S. Early diagnosis of inherited metabolic disorders towards improving outcome: the controversial issue of galactosaemia. Eur J Pediatr. 2003;162(Suppl 1):S50–53. doi: 10.1007/s00431-003-1352-2. [DOI] [PubMed] [Google Scholar]
- 21.Lapolla A, Fedele D, Seraglia R, Traldi P. The role of mass spectrometry in the study of non-enzymatic protein glycation in diabetes: an update. Mass Spectrom Rev. 2006;25:775–97. doi: 10.1002/mas.20090. [DOI] [PubMed] [Google Scholar]
- 22.Frolov A, Hoffmann R. Analysis of amadori peptides enriched by boronic acid affinity chromatography. Ann N Y Acad Sci. 2008;1126:253–6. doi: 10.1196/annals.1433.060. [DOI] [PubMed] [Google Scholar]
- 23.Lapolla A, Fedele D, Reitano R, Arico NC, Seraglia R, Traldi P, Marotta E, Tonani R. Enzymatic digestion and mass spectrometry in the study of advanced glycation end products/peptides. J Am Soc Mass Spectrom. 2004;15:496–509. doi: 10.1016/j.jasms.2003.11.014. [DOI] [PubMed] [Google Scholar]
- 24.Zhang Q, Frolov A, Tang N, Hoffmann R, van de Goor T, Metz TO, Smith RD. Application of Electron Transfer Dissociation Mass Spectrometry in Analyses of Non-enzymatically Glycated Peptides. Rapid Commun Mass Spectrom. 2007;21:661–666. doi: 10.1002/rcm.2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Q, Petyuk VA, Schepmoes AA, Orton DJ, Monroe ME, Yang F, Smith RD, Metz TO. Analysis of non-enzymatically glycated peptides: neutral-loss-triggered MS3 versus multi-stage activation tandem mass spectrometry. Rapid Commun Mass Spectrom. 2008;19:3027–3034. doi: 10.1002/rcm.3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kazuyoshi I, Yuichiro S, Yukie K, Takehiro M, Kimikazu T, Takeyoshi U, Yoshiaki S, Shoichi U, Seikoh H. Determination of glycated albumin by enzyme-linked boronate immunoassay (ELBIA) Clinical Chemistry. 1998;44:256–263. [PubMed] [Google Scholar]
- 27.Fluckiger R, Winterhalter KH. Glycosylated hemoglobins. In: Caughey WS, editor. Biochemical and Clinical Aspects of Hemoglobin Abnormalities. Academic Press, Inc; New York: 1978. pp. 205–214. [Google Scholar]
- 28.Kennedy RT, Jorgensen JW. Preparation and evaluation of packed capillary liquid chromatography columns with inner diameters from 20 to 50 μm. Analyt Chem. 1989;61:1128–1135. [Google Scholar]
- 29.Kislinger T, Humney A, Seeber S, Becker CM, Pischetsrieder M. Qualitative determination of early Maillard-Products by MALDI-TOF mass spectrometry peptide mapping. Eur Food Res Technol. 2002;215:65–71. [Google Scholar]
- 30.Montgomery H, Tanaka K, Belgacem O. Glycation pattern of peptides condensed with maltose, lactose and glucose determined by ultraviolet matrix-assisted laser desorption/ionization tandem mass spectrometry. Rapid Commun Mass Spectrom. 2010;24:841–848. doi: 10.1002/rcm.4455. [DOI] [PubMed] [Google Scholar]
- 31.Ledesma-Osuna AI, Ramos-Clamont G, Vázquez-Moreno L. Characterization of bovine serum albumin glycated with glucose, galactose and lactose. Acta Biochim Pol. 2008;55:491–7. [PubMed] [Google Scholar]
- 32.Day JF, Thorpe SR, Baynes JW. Nonenzymatically glucosylated albumin. J Biol Chem. 1979;254:595–597. [PubMed] [Google Scholar]
- 33.Tanzer ML, Fairweather R, Gallop PM. Collagen crosslinks: isolation of reduced NEhexosyl-hydroxylysine from borohydride reduced calf skin insoluble collagen. Arch Biochemn Biophys. 1972;151:137–141. doi: 10.1016/0003-9861(72)90482-1. [DOI] [PubMed] [Google Scholar]
- 34.Persaud R, Fraser P, Wood DD, Moscarello MA. The glycosylation of human myelin basic protein at threonines 95 and 98 occurs sequentially. Biochim Biophys Acta. 1988;966:357–361. doi: 10.1016/0304-4165(88)90085-2. [DOI] [PubMed] [Google Scholar]
- 35.Cruz TF, Moscarello MA. Identification of the major sites of enzymic glycosylation of myelin basic protein. Biochim Biophys Acta. 1983;760:403–410. doi: 10.1016/0304-4165(83)90381-1. [DOI] [PubMed] [Google Scholar]
- 36.Stevens VJ, Rouzer CA, Monnier VM, Cerami A. Diabetic cataract formation: potential role of glycosylationand lens crystallins. Proc Natl ACID Sci U S A. 1978;75:2918–2922. doi: 10.1073/pnas.75.6.2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bailey AJ, Robins SD, Tanner MJA. Reducible components in the proteins of human erythrocyte membrane. Biochim Biophys Acta. 1976;434:51–57. doi: 10.1016/0005-2795(76)90034-9. [DOI] [PubMed] [Google Scholar]
- 38.Knowles WJ, Marches V, Haigh W. Peptides useful in preparing hemoglobin A.sub.1c immunogens. 4,647,654 United States Patent. 1987
- 39.Blackshear PJ. Glycated peptides and methods use of, United. States. 20090093066 Patent Application. 2009
- 40.Mezei LM, Chen JS, Huang JJ, Lovins RE. Method of obtaining antibodies. 4,478,744 United States Patent. 1984
- 41.Cohen MP. Monoclonal antibodies against glycated albumin, hybrid cell lines producing these antibodies, and use therefore. 5,223,392 United States Patent. 1993
- 42.Cohen MP, Hud E. Measurement of plasma glycoalbumin levels with a monoclonal antibody based ELISA. J Immunol Methods. 1989;122:279–283. doi: 10.1016/0022-1759(89)90275-5. [DOI] [PubMed] [Google Scholar]
- 43.Kislinger T, Humeny A, Peich CC, Becker CM, Pischetsrieder M. Analysis of protein glycation products by MALDI-TOF/MS. Ann N Y Acad Sci. 2005;1043:249–59. doi: 10.1196/annals.1333.030. [DOI] [PubMed] [Google Scholar]
- 44.Zhang Q, Tang N, Brock JW, Mottaz HM, Ames JM, Baynes JW, Smith RD, Metz TO. Enrichment and Analysis of Nonenzymatically Glycated Peptides: Boronate Affinity Chromatography Coupled with Electron-Transfer Dissociation Mass Spectrometry. J Proteome Res. 2007;6:2323–2330. doi: 10.1021/pr070112q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mikesh LM, Ueberheide B, Chi A, Coon JJ, Syka JE, Shabanowitz J, Hunt DF. The utility of ETD mass spectrometry in proteomic analysis. Biochim Biophys Acta. 2006;1764:18111822. doi: 10.1016/j.bbapap.2006.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Iberg N, Flückiger R. Nonenzymatic glycosylation of albumin in vivo. Identification of multiple glycosylated sites. J Biol Chem. 1986;261:13542–5. [PubMed] [Google Scholar]
- 47.Garlick RL, Mazer JS. The principal site of non-enzymatic glycosylation of human serum albumin in vivo. J Biol Chem. 1983;258:6142–6146. [PubMed] [Google Scholar]
- 48.Wa C, Cerny RL, Clarke WA, Hage DS. Characterization of glycation adducts on human serum albumin by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Clin Chim Acta. 2007;385:48–60. doi: 10.1016/j.cca.2007.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Howard MJ, Smales CM. NMR Analysis of Synthetic Human Serum Albumin -Helix 28 Identifies Structural Distortion upon Amadori Modification. J Biol Chem. 2005;280:22582–22589. doi: 10.1074/jbc.M501480200. [DOI] [PubMed] [Google Scholar]