Abstract
Cleft palate, including submucous cleft palate, is among the most common birth defects in humans. While overt cleft palate results from defects in growth or fusion of the developing palatal shelves, submucous cleft palate is characterized by defects in palatal bones. In this report, we show that the Bmpr1a gene, encoding a type I receptor for bone morphogenetic proteins (Bmp), is preferentially expressed in the primary palate and anterior secondary palate during palatal outgrowth. Following palatal fusion, Bmpr1a mRNA expression was upregulated in the condensed mesenchyme progenitors of palatal bone. Tissue-specific inactivation of Bmpr1a in the developing palatal mesenchyme in mice caused reduced cell proliferation in the primary and anterior secondary palate, resulting in partial cleft of the anterior palate at birth. Expression of Msx1 and Fgf10 was downregulated in the anterior palate mesenchyme and expression of Shh was downregulated in the anterior palatal epithelium in the Bmpr1a conditional mutant embryos, indicating that Bmp signaling regulates mesenchymal-epithelial interactions during palatal outgrowth. In addition, formation of the palatal processes of the maxilla was blocked while formation of the palatal processes of the palatine was significantly delayed, resulting in submucous cleft of the hard palate in the mutant mice. Our data indicate that Bmp signaling plays critical roles in the regulation of palatal mesenchyme condensation and osteoblast differentiation during palatal bone formation.
Keywords: Bmp signaling, Bmpr1a, submucous cleft palate, palate development, palatal bone
Introduction
During mammalian embryonic development, the secondary palate initiates from the oral sides of the maxillary processes and grow initially vertically down the sides of the developing tongue. At a precise developmental stage the bilateral palatal shelves elevate to a horizontal position above the dorsum of the tongue and fuse with each other at the midline. In addition, the bilateral palatal shelves fuse anteriorly with the primary palate, derived from medial nasal processes, to form the intact roof of the oral cavity. Any disturbance of the growth, elevation or fusion of the palatal shelves could result in cleft palate, one of the most common birth defects in humans (reviewed by Ferguson, 1988; Gritli-Linde, 2007).
The secondary palate is anatomically divided into the anterior bony region (hard palate) and posterior muscular region (soft palate) (Sperber, 2002). Cleft palate defects affecting the entire secondary palate or either the anterior or posterior regions have been documented (reviewed by Hilliard et al., 2005). Consistent with the morphological and pathological differences in the anterior and posterior palate, recent studies have clearly demonstrated that there is molecular heterogeneity along the anterior-posterior axis of the developing secondary palate in mice (reviewed by Hilliard et al., 2005; Li and Ding, 2007; Welsh and O’Brien, 2009). During early palate development, expression of several transcription factors, including Barx1, Mn1, Msx1, Meox2, Shox2, and Tbx22, is highly restricted along the anterior-posterior axis. Expression of Msx1 and Shox2 mRNAs is restricted to the anterior region whereas Meox2 and Tbx22 mRNAs are restricted to the posterior region, with the anterior-posterior gene expression boundary coinciding with a morphological landmark, the first formed palatal rugae, a thickened palatal epithelial structure perpendicular to the anterior-posterior axis on the oral side of the developing palatal shelves (Zhang et al., 2002; Yu et al., 2005; Hilliard et al., 2005; Li and Ding, 2007, Pantalacci et al., 2008; Welsh and O’Brien, 2009). Barx1 and Mn1 mRNAs are also preferentially expressed in the posterior regions of the developing palatal mesenchyme during palatal outgrowth (Liu et al., 2008; Welsh and O’Brien, 2009). Although mice lacking either Msx1 or Mn1 exhibited complete cleft palate, the Msx1−/− mutant mice exhibited specific cell proliferation defects in the anterior region whereas Mn1−/− mutant mice showed preferential growth deficit in the posterior regions of the developing palatal shelves (Zhang et al., 2002; Liu et al., 2008). Shox2 is required for growth of the anterior palate and mice lacking Shox2 exhibited incomplete cleft within the anterior palate while the mutant posterior palate fused normally (Yu et al., 2005). In contrast, mice lacking Tbx22 showed submucous cleft palate, with delayed palatal bone development (Pauws et al., 2009).
In addition to findings of distinct transcription factors involved in the growth of the anterior and posterior regions of the developing palatal shelves, recent studies showed that the growth and elongation of palatal shelves along the anterior-posterior axis are associated with periodic addition of new palatal rugae just anterior to the first formed rugae (Pantalacci et al., 2008; Welsh and O’Brien, 2009). Palatal rugae express high levels of Shh, which has been shown to play critical roles in regulating cell proliferation and expression of the fibroblast growth factor Fgf10 in the developing palatal mesenchyme (Pantalacci et al., 2008; Lan and Jiang, 2009; Welsh and O’Brien, 2009). Fgf10 function in the developing palate is required for palatal ruga formation and maintenance of Shh mRNA expression in the palatal epithelium (Rice et al., 2004; Welsh and O’Brien, 2009). Thus, the periodic formation of palatal rugae is believed to play critical roles in palatal outgrowth (Welsh and O’Brien, 2009).
Previous studies have also suggested important roles for bone morphogenetic protein (Bmp) signaling in secondary palate development. Zhang et al. (2002) reported that Bmp4 expression in the anterior palate was lost in the Msx1−/− mutant mouse embryos and that transgenic expression of Bmp4 under the control of an Msx1 promoter rescued anterior palatal growth defects of the Msx1−/− mutant mice, suggesting that Bmp4 acts downstream of Msx1 to regulate anterior palatal growth. However, whereas Bmp4 null mutant mouse embryos died before palate development, tissue-specific inactivation of the Bmp4 gene in the early developing maxillary mesenchyme did not disrupt secondary palate development (Winnier et al., 1995; Liu et al., 2005). In contrast, inactivation of the mouse Bmpr1a gene, encoding one of the type-I Bmp receptors, in the early maxillary mesenchyme caused significant reduction in maxillary mesenchyme proliferation prior to the onset of secondary palate outgrowth and resulted in smaller palatal shelves and subsequently cleft palate at birth (Liu et al., 2005). It is not known, however, whether Bmpr1a signaling plays a critical role in the developing secondary palate. Moreover, while it is well known that Bmp signaling plays critical roles in bone development, and although submucous cleft palate with defects in the palatal bones is as common as overt cleft palate (Garcia et al., 1988; Weather-White et al., 1972), little is known about the molecular mechanisms regulating palatal bone formation. Here we report the generation of mice with tissue-specific inactivation of Bmpr1a in the developing palatal mesenchyme and show that Bmpr1a is required for palatal shelf growth as well as palatal bone formation.
Materials and Methods
Mouse strains
The Bmpr1af/f conditional mice and the Osr2-IresCre mice have been described previously (Mishina et al., 2002; Lan et al., 2007). Both mouse strains were maintained as homozygotes in their own strain background. Osr2IresCre/IresCre homozygous male mice were crossed to Bmpr1af/f female mice to generate the Osr2-IresCre;Bmpr1af/+ male mice, which were subsequently crossed to Bmpr1af/f female mice to generate Osr2-IresCre;Bmpr1af/f embryos for analysis. Although the IresCre cassette in the Osr2-IresCre allele did not disrupt Osr2 gene function (Lan et al., 2007), all experimental analysis of Osr2-IresCre;Bmpr1af/f embryos used Osr2-IresCre;Bmpr1af/+ littermates as controls. Genotyping of mice and embryos were carried out by allele-specific PCR as previously described (Mishina et al., 2002; Lan et al., 2007).
Histology, in situ hybridization analysis, and skeletal preparations
For histology, embryos were harvested from euthanized timed pregnant mice, fixed in Bouin’s fixative, dehydrated through graded alcohols, embedded in paraffin wax, sectioned at 7-μm thickness. The sections were stained with hematoxylin and eosin or the trichrome reagents for morphological observations. Sections from late-term and newborn pups were stained with alcian blue and van Gieson’s picrofuchsin to visualize bone (red) and cartilage (blue).
For whole mount in situ hybridization, embryos were dissected from euthanized timed pregnant mice, fixed overnight in 4% paraformaldehyde in PBS at 4°C. The fixed embryos were washed in cold PBS, the embryonic head separated from the body using a sharp razor, and the mandible was removed from the upper jaw to expose the secondary palate. The fixed embryonic heads were washed with PBS, dehydrated through 25%, 50%, 75%, and 100% methanol, and stored in 100% methanol at −20°C. Following identification of genotypes, the samples of the same genotype were pooled and whole mount in situ hybridization carried out using previously described protocol (Jiang et al., 1998).
For section in situ hybridization, embryos were fixed overnight at 4°C in 4% paraformaldehyde in PBS, dehydrated through graded alcohols, embedded in paraffin and sectioned at 5-μm thickness. In situ hybridization of tissue sections were performed as described previously (Zhang et al., 1999).
Skeletal preparations of newborn pups and of E18.5 embryos were made according to the protocol of Martin et al. (1995).
Quantitative real-time reverse transcription-polymerase chain reaction (RT-PCR)
For quantitative analysis of gene expression in the developing primary palate, the primary palate tissues were dissected from E14.0 embryos in cold DEPC-treated PBS, quickly frozen in liquid nitrogen and stored individually at −80 °C. Following identification of the genotypes of the embryos by allele-specific PCR, tissues were pooled by genotype and total RNA extracted using Trizol reagents (Invitrogen). Following total RNA extraction and quantification, first-strand cDNA was synthesized using SuperScript™ First-Strand Synthesis System for RT-PCR (Invitrogen). Quantitative PCR amplifications were performed in an iCycler real-time PCR machine (Bio-Rad) using the SYBR GreenER™ qPCR Supermix (Invitrogen). The reaction was run in a PCR program of 50 °C for 2 minutes, 95 °C for 8.5 minutes, followed by 40 cycles of 95 °C for 15 seconds and 60 °C for 60 seconds, with a melt curve generation cycle at the end. For each gene, the PCR reaction was carried out in triplicates and the relative levels of mRNAs were normalized to that of Hprt using the standard curve method. Student’s t-test was used to analyze the significance of difference and a P value less than 0.05 was considered statistically significant.
Assay of osteoblast differentiation by alkaline phosphatase staining
Embryos were harvested from euthanized timed pregnant mice, fixed in 4% paraformaldehyde overnight at 4°C. After washing with PBS, the embryos were equilibrated in 10%, 20%, and then 30% sucrose in PBS, embedded in Negative-50 cryomedium, frozen sectioned at 10-μm thickness. The sections were incubated in the N™T buffer (100 mM NaCl, 100 mM TrisHCl pH 9.5, 50 mM MgCl2, 0.1% Tween-20) containing 4.5 μl/ml nitroblue tetrazolium (NBT) and 3.5 μl/ml 5-bromo-4-chloro-3-indolyl phosphate (BCIP). When staining was complete, the slides were washed in PBT containing 20mM EDTA. Sections were counterstained with Nuclear Fast Red, dehydrated through graded alcohols and mounted with Permount.
Detection of cell proliferation and apoptosis
For detection of cell proliferation in the palatal shelves, timed pregnant female mice were injected once intraperitoneally at gestational day 13.5, 14.5, 15.5, or 16.5 (with 12 pm on the day of plug was observed counted as 0.5 day of gestation) with the BrdU Labeling Reagent (Roche, 45 μg/g body weight). One hour after injection, embryos were dissected, fixed with Carnoy’s fixatives at 4°C overnight, embedded in paraffin and sectioned at 5 μm thickness in the coronal plane for immunodetection of BrdU using the BrdU Labeling and Detection kit (Roche) as described previously (Lan et al., 2004). Following BrdU immunostaining, the embryonic sections were counterstained with Nuclear Fast Red to visualize all cellular nuclei. Cell counts were recorded separately for the primary palate and each of the bilateral palatal shelves from five continuous sections of matching areas of the primary palate, anterior secondary palate (from section between the palatal rugae 2 and 3), and of the mid-palate (from sections through the middle of the maxillary first molar tooth germs) regions in the control and mutant samples. The cell proliferation rate was calculated as a percentage of the cell nuclei with BrdU labeling. Data were collected from at least three pairs of mutant and control littermates at each developmental stage. Students’ t-test was used to analyze the significance of difference and a P-value less than 0.05 was considered statistically significant.
Apoptotic cell death in the developing palate was assessed by staining paraffin sections using the DeadEnd™ Fluorometric TUNEL System reagents (Promega) following the manufacturer’s instructions. Following TUNEL staining, sections were counterstained with 4′,6-diamidino-2-phenylindole (DAPI).
Results
Expression pattern of Bmpr1a in the developing palate
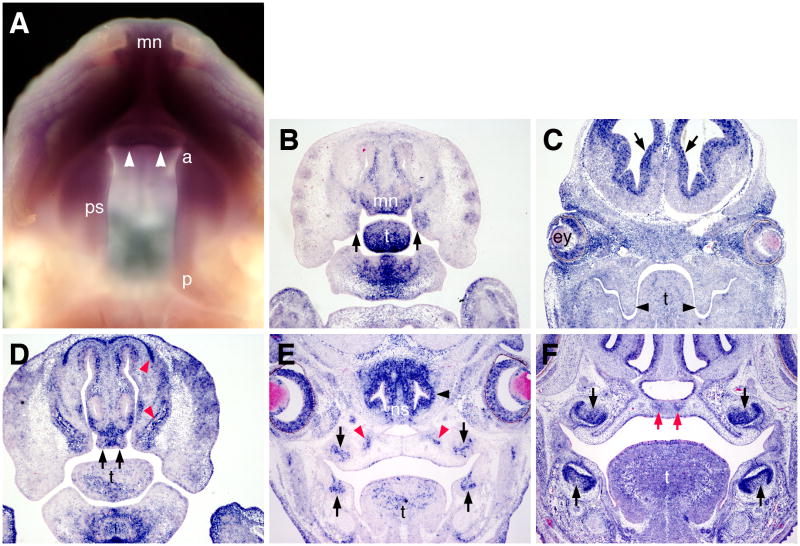
We used whole mount and section in situ hybridization analyses to examine the expression patterns of Bmpr1a mRNA during palate development. By whole mount in situ hybridization, strong Bmpr1a mRNA expression was detected in the medial nasal processes and in the anterior half of the developing palatal shelves while its expression in the posterior half of the palatal shelves was significantly weaker at E13.5 (Fig. 1A). Section in situ hybridization analyses detected strong Bmpr1a mRNA expression in the mesenchyme of the medial nasal processes and in the anterior secondary palate at this stage (Fig. 1B). Strong Bmpr1a mRNA expression was also detected in the mesenchyme of the distal mandible and tongue (Fig. 1B). Consistent with the whole mount data, expression of Bmpr1a in the posterior palate was much weaker although strong signals were detected in the ventricular zone cells of the brain and in the developing eyelid and retina tissues on the same sections (Fig. 1C). At E14.5, strong Bmpr1a mRNA expression was detected in the mesenchyme of the developing primary palate, in the olfactory and dental epithelia, and in the precursor cells of the facial and palatal bone (Fig. 1D, E). By E16.5, Bmpr1a mRNA is highly expressed in the dental epithelium and mesenchyme, and in the condensed mesenchyme precursor cells of the palatal processes of the maxilla (Fig. 1F). These results suggest that Bmpr1a signaling may be involved in the outgrowth of the primary and secondary palate as well as in palatal bone formation.
Fig. 1.
Expression of Bmpr1a mRNA in the developing palate. (A) Whole mount in situ hybridization detection of Bmpr1a mRNA expression at E13.5. Purple color indicates mRNA expression. Arrowheads point to the primary palate outgrowth. Strong Bmpr1a mRNA expression was detected in the medial nasal processes, the primary palate, and the anterior part of the secondary palate shelves. (B) A frontal section through the primary and anterior secondary palate of an E13.5 embryo. Strong Bmpr1a mRNA expression was detected in the medial nasal processes, including the primary palatal outgrowth, in the anterior secondary palate (arrows), in the distal tongue, and mandibular tissues. (C) A frontal section through the posterior secondary palate of an E13.5 embryo. Strong Bmpr1a mRNA expression was detected in the ventricular zones of the developing brain (arrows), in the retina and eyelids. The signal in the palatal shelves was not significantly above background. (D) A frontal section through the developing primary palate of an E14.5 embryo. High levels of Bmpr1a mRNAs were detected in the developing primary palate (arrows) and in the osteogenic mesenchyme (red arrowheads). (E) A frontal section through the midpalate and the developing first molar tooth germs. Strong Bmpr1a mRNA expression was detected in the developing dental epithelium (black arrows), olfactory epithelium (black arrowhead), and in the condensed mesenchyme at the osteogenic fronts (red arrowheads). (F) A frontal section through the first molar tooth germs of an E16.5 embryo. Strong Bmpr1a mRNA expression was detected in developing dental epithelium and mesenchyme (black arrows) and in the condensed mesenchyme forming the osteogenic centers of the palatal processes of the maxilla (red arrows). a, anterior end of the developing secondary palate; ey, eye; mn, medial nasal processes; ns, nasal septum; p, posterior end of the secondary palate; ps, palatal shelf; t, tongue.
Bmpr1a is required in the palatal mesenchyme for normal palatogenesis
To specifically investigate the roles of Bmpr1a signaling in palate development, we used the Osr2-IresCre mouse strain (Lan et al., 2007) in combination with the Bmpr1af/f mouse strain (Mishina et al., 2002) to inactivate Bmpr1a in the developing palatal mesenchyme. In contrast to the Wnt1Cre and Nestin-cre transgenic mouse lines, in which Cre activity was present in the premigratory neural crest cells and in the early developing facial tissues, respectively (Chai et al., 2000; Liu et al., 2005), Cre expression was activated in the nascent palatal mesenchyme at E10.5 and Cre/loxP-mediated recombination occurred highly specifically throughout the early developing palatal mesenchyme by E12.5 in the Osr2-IresCre mice (Lan et al., 2007; Lan and Jiang, 2009). Thus, whereas Wnt1Cre;Bmpr1af/− mouse embryos died at midgestation and Nestin-cre;Bmpr1af/− mouse embryos had significantly reduced maxillary mesenchyme cell proliferation prior to the onset of palatal outgrowth (Stottmann et al., 2002; Liu et al., 2005), we expected that using the Osr2-IresCre mouse strain in combination with the Bmpr1af/f mice would reveal specific roles of Bmpr1a signaling within the developing palate.
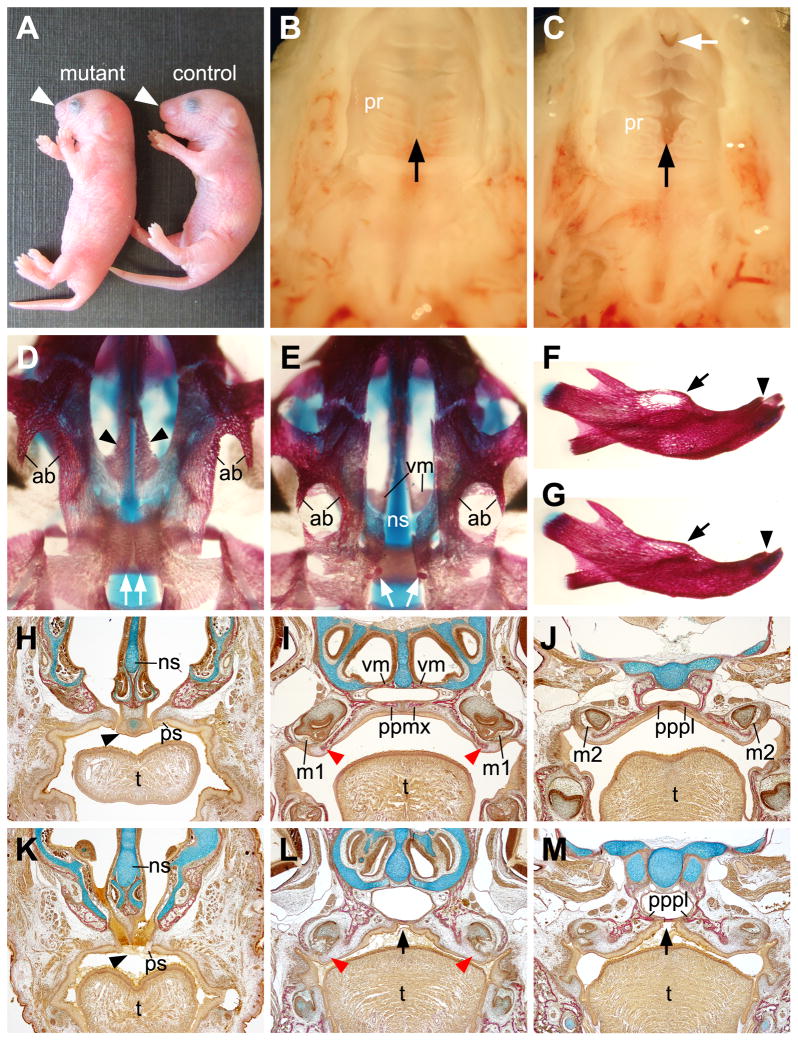
Osr2-IresCre;Bmpr1af/f mice were born alive but died shortly after birth (n = 10). In comparison with their control littermates, Osr2-IresCre;Bmpr1af/f pups exhibited shortened snout (Fig. 2A). Anatomical analysis showed that these mutants had an open gap between the primary and secondary palate (Fig. 2B, C). In addition, the mutant pups exhibited disorganization of the palatal rugae and submucous cleft of the anterior palate (Fig. 2C). Examination of skeletal preparations and histological sections of newborn pups showed that the Osr2-IresCre;Bmpr1af/f mice lacked the palatal processes of the maxilla and had severely retarded palatal processes of the palatine (Fig. 2, D–M). The mutant primary palate was also significantly retarded compared with control littermates (Fig. 2H, K). In addition, the Osr2-IresCre;Bmpr1af/f mutants had deficiency in the alveolar bones on the lingual side of the incisor and molar teeth (Fig. 2, D–M), indicating a requirement for Bmpr1a signaling in alveolar bone development. The lingual bias of the alveolar bone defect seen in these mutants is most likely due to the lingual bias of Cre expression from the Osr2-IresCre allele in the mesenchyme surrounding the developing tooth germs (Lan et al., 2007; Chen et al., 2009).
Fig. 2.
Osr2-IresCre;Bmpr1af/f mutant mice exhibited shortened snout, anterior palatal cleft, and defects in palatal and alveolar bone formation. (A) Whole mount pictures of newborn Osr2-IresCre;Bmpr1af/f (mutant) and Osr2-IresCre;Bmpr1af/+ (control) pups. Arrowhead points to the snout. (B,C) Oral view of the secondary palate of the control (B) and mutant (C) pups. Black arrow points to the midline of the hard palate. White arrow in C points to the cleft at the anterior end of the secondary palate. (D,E) Ventral view of the cranial skeleton of the newborn control (D) and mutant (E) pups. Black arrowheads point to the palatal processes of the maxilla, while the white arrows point to the palatal processes of the palatal processes of the palatine bones. The mutant lacked the palatal processes of the maxilla, allowing direct visualization of the vomer bones, which are normally hidden above. (F,G) Comparison of the mandibular skeletons of the newborn control (F) and mutant (G) samples. Arrows point to the molar alveolar bones and arrowheads point to the incisor alveolar bones. (H–M) Van Gieson stained frontal sections of newborn control (H–J) and mutant (K–M) littermates. Bone was stained red while cartilage blue. H and K show sections from the anterior end of the secondary palate, I and L show sections through the middle of the maxillary first molars, J and M show sections through the maxillary second molars. Black arrowheads in H and K point to the junction of primary and secondary palate. Red arrowheads in I and L point to the lingual alveolar crest of the maxillary first molars. Black arrows in L and M point to the arched midline mucosa without underlying bone in the mutant palate. ab, alveolar bone; m1, first molar; m2, second molar; ns, nasal septum; ppmx, palatal process of the maxilla; pppl, palatal process of the palatine; pr, palatal ruga; ps, palatal shelf; vm, vomer; t, tongue.
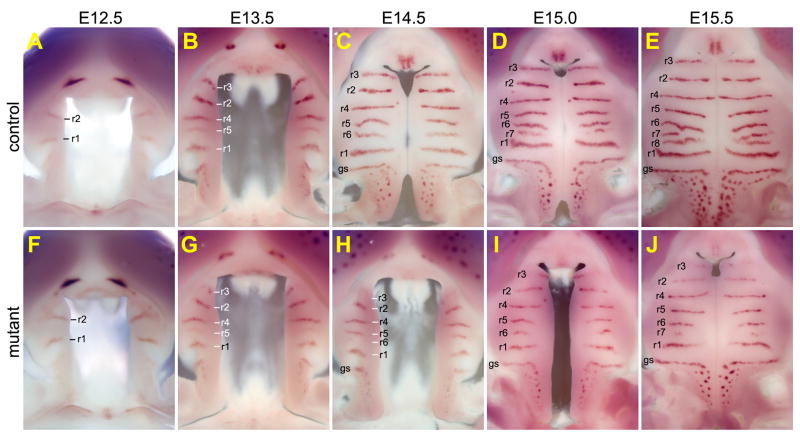
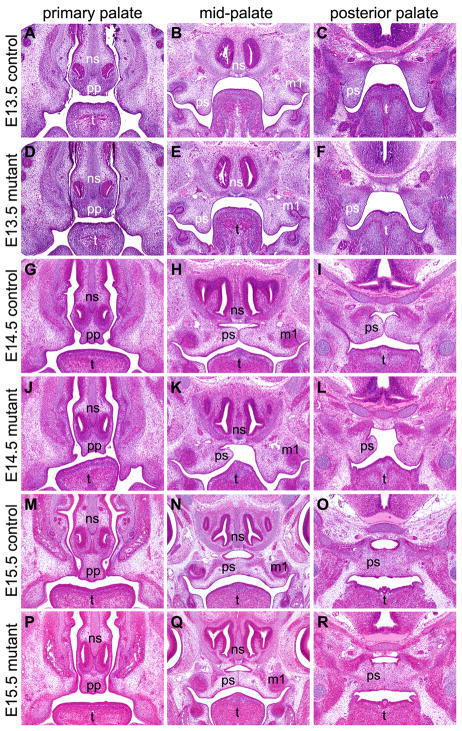
To investigate the pathogenic processes of the palatal defects in Osr2-IresCre;Bmpr1af/f mutant mice, we carried out detailed morphological and histological analyses of embryos collected at various stages from the beginning of palatal outgrowth to the completion of palate fusion. Since the secondary palate develop at the time of significant rostral expansion of the jaws, we used whole mount detection of Shh mRNA expression in the developing palatal rugae to reveal possible differences in growth patterns along the anterior-posterior axis and used histology to visualize differences in palatal shelf growth, elevation, and fusion in the Osr2-IresCre;Bmpr1af/f mutant and control littermates. At E12.5, palatal shelves are comparable in size and two Shh-expressing palatal rugae had formed on each palatal shelf in the control and mutant embryos (Fig. 3, A and F). At E13.5, while the palatal shelves in the control and mutant embryos were still comparable in shape and size (Fig. 3, B and G; Fig. 4, A–F), Shh mRNA expression in ruga-3, the most anterior palatal ruga, was reduced in the mutant embryos in comparison with the control littermates (Fig. 3, B and G). By E14.5, the bilateral palatal shelves had elevated to the horizontal position above the developing tongue and initiated contact at the midline in the mid-palatal region in the control embryos (Fig. 3C; Fig. 4, G–I). In contrast, the mutant palatal shelves exhibited delay in elevation (Fig. 3H; Fig. 4, J–L). The rostral expansion of the mutant palatal shelves was also slower than that in the control littermate, as detected by the delay in formation of ruga-6 and by the shorter distance between ruga-1 and ruga-4 (Fig. 3, compare C and H). In addition, expression of Shh mRNA in the mutant ruga-3 was significantly weaker than that in the control littermates. Expression of Shh mRNA was also significantly weaker in the primary palate in the mutant embryos than that in the control littermates (Fig. 3, compare C and H; Supplementary Fig. 1). By E15.0, while the palatal shelves had expanded further rostrally in both control and mutant embryos (Fig. 3, D and I), the delay in palatal fusion and rostral expansion in the mutant embryos was consistently detected, with the control embryo forming the 7th palatal rugae while the mutant embryo was still forming the 6th rugae. By E15.5, fusion between the bilateral palatal shelves was nearly complete in the control embryos, with disintegration of the midline epithelial seam and mesenchymal confluence through much of the secondary palate and with the anterior ends of the secondary palatal shelves making contact with the primary palate (Fig. 3E; Fig. 4, M–R). In contrast, the bilateral palatal shelves failed to contact each other in the region anterior to ruga-2 in the mutant embryos although fusion occurred in the middle and posterior regions of the secondary palate (Fig. 3J; Fig. 4, P–R). In addition, Shh mRNA expression in the primary palate and in rugae 2 and 3 was dramatically downregulated in the mutant embryos in comparison with the control littermates (Fig. 3, E and J). Together, these data indicate that development of both the primary palate and anterior secondary palate was impaired in the Osr2-IresCre;Bmpr1af/f mutant mice.
Fig. 3.
Osr2-IresCre;Bmpr1af/f mutant mouse embryos exhibited slower palatal growth and delayed palatal elevation. (A–E) Oral views of the developing secondary palate showing pattern of formation of Shh-expressing palatal rugae from E12.5 (A), E13.5 (B), E14.5 (C), E15.0 (D), and E15.5 (E) control embryos. Each ruga is marked with “r” followed by the number according to the temporal sequence of formation, with the first ruga labeled as “r1”. gs, geschmacksstreifen. (F–J) Oral views of the developing secondary palate showing pattern of Shh-expressing palatal rugae from E12.5 (F), E13.5 (G), E14.5 (H), E15.0 (I), and E15.5 (J) Osr2-IresCre;Bmpr1af/f mutant littermates of those shown in A–E, respectively.
Fig. 4.
Histology of frontal sections of the developing palate in Osr2-IresCre;Bmpr1af/f mutant and control mouse embryos. (A–F) Selected position matched sections of E13.5 control (A–C) and mutant (D–F) littermates. (G–L) Selected position matched sections of E14.5 control (G–I) and mutant (J–L) littermates. (M–R) Selected position matched sections of E15.5 control (M–O) and mutant (P–R) littermates. Sections shown in the left column (A, D, G, J, M, P) are from the anterior end of the developing secondary palate. Sections in the middle column (B, E, H, K, N, Q) were cut through the middle of the maxillary first molar tooth germs. Sections shown in the right column (C, F, I, L, O, R) are from the posterior soft palate region. m1, first molar; ns, nasal septum; pp, primary palate; ps, palatal shelf: t, tongue.
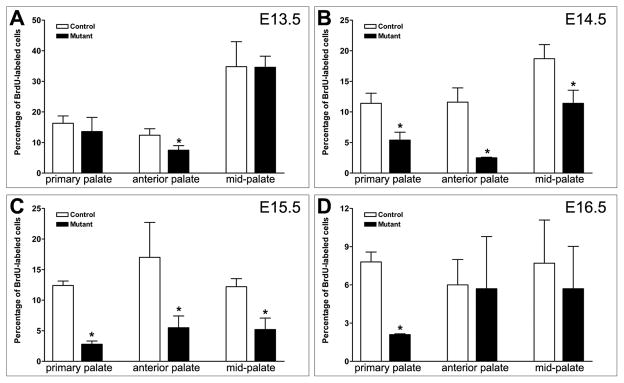
To examine further whether inactivation of Bmpr1a impaired palatal shelf growth, we compared palatal mesenchyme cell proliferation in the control and mutant embryos from E13.5 through E16.5. We found that the cell proliferation rate was significantly reduced in the anterior secondary palate at E13.5 in the mutant embryos in comparison with control littermates (Fig. 5A). At E14.5 and E15.5, we detected significantly reduced cell proliferation in both the primary palate and the anterior secondary palate in the mutant embryos compared with control littermates (Fig. 5, B and C). At E16.5, the cell proliferation rate was still reduced in the primary palate of the mutant in comparison with control littermates (Fig. 5D). These data indicate that Bmpr1a signaling plays a critical role in the outgrowth of the primary and anterior secondary palate and that defects in both the primary and anterior secondary palate contributed to the partial anterior cleft palate in the newborn mutant mice.
Fig. 5.
Osr2-IresCre;Bmpr1af/f mutant embryos had defects in palatal growth. The percentage of BrdU-labeled palatal mesenchyme cells were recorded and compared between Osr2-IresCre;Bmpr1af/f mutant and control littermates at E13.5 (A), E14.5 (B), E15.5 (C), and E16.5 (D). Standard deviation values were used for the error bars. Asteroid marks statistically significant reduction in palatal mesenchyme proliferation in the mutant in comparison with the control littermates.
Defects in mesenchymal-epithelial signaling in the developing primary and secondary palate in Osr2-IresCre;Bmpr1af/f mutant embryos
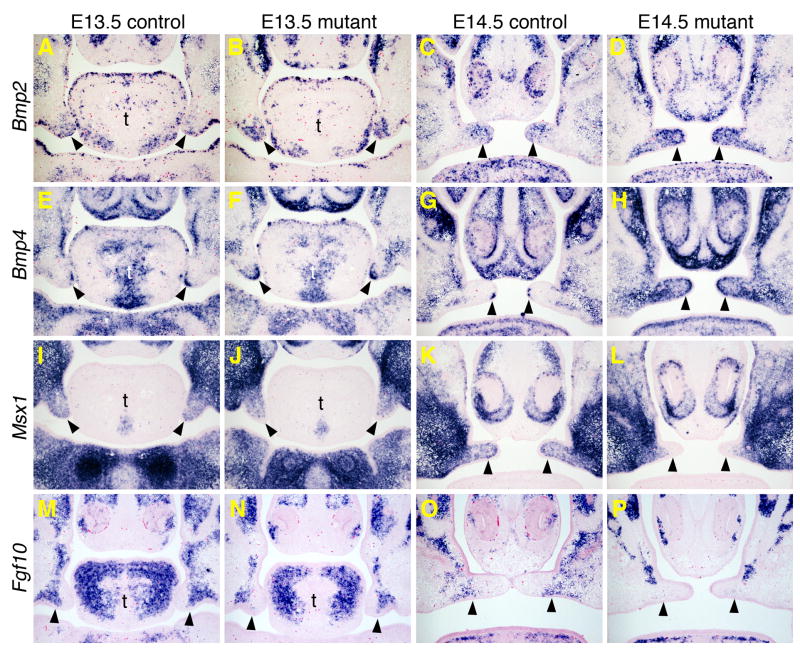
Since Shh is expressed in the palatal epithelium whereas Cre expression is highly specific in the developing palatal mesenchyme (Lan et al., 2007; Lan and Jiang, 2009), the reduction in Shh mRNA expression in the developing primary and anterior secondary palate in the Osr2-IresCre;Bmpr1af/f mutant embryos suggests defective mesenchymal-epithelial signaling in the these tissues. Previous studies identified Bmp4 and Fgf10, both expressed in the anterior palate mesenchyme, as important factors for the maintenance of Shh mRNA expression in the palatal epithelium (Zhang et al., 2002; Rice et al., 2004). Zhang et al. (2002) suggested that Bmp4 and Msx1 function in a positive regulatory loop in the anterior palatal mesenchyme. We compared expression of Bmp2, Bmp4, Msx1, and Fgf10 in the developing anterior secondary palate in control and mutant embryos by section in situ hybridization. At E13.5, expression of Bmp2 and Bmp4 mRNAs was upregulated while expression of Msx1 appeared moderately decreased in the anterior palate mesenchyme in the Osr2-IresCre;Bmpr1af/f mutant embryos in comparison with the control littermates (Fig. 6, A, B, E, F, I, J). At E14.5, changes in the expression of these genes were more apparent, with significant increases in expression of Bmp2 and Bmp4 and apparent downregulation of expression of Msx1 mRNAs in the anterior palate mesenchyme in the mutant embryos (Fig. 6, C, D, G, H, K, L). While the decrease in Msx1 mRNA expression is likely a direct effect of inactivation of Bmpr1a in the palatal mesenchyme, the increase in Bmp2 and Bmp4 mRNA expression suggests that the reduction in Shh expression is not direct effect of decreased Bmp signaling in the palatal epithelium. Similar to the changes in Msx1 mRNA expression, expression of Fgf10 mRNA was moderately decreased at E13.5 and more significantly downregulated by E14.5 in the anterior palatal mesenchyme in the Osr2-IresCre;Bmpr1af/f mutant embryos (Fig. 6, M–P). Quantitative RT-PCR analyses also revealed increased levels of Bmp2 and Bmp4 mRNAs and decreased levels of Fgf10 mRNAs in the developing primary palate in the Osr2-IresCre;Bmpr1af/f mutant embryos (Supplementary Fig. 1). These results suggest that Fgf10 acts downstream of Bmpr1a signaling to regulate the mesenchymal-epithelial interactions during palatal outgrowth.
Fig. 6.
Changes in gene expression in the developing anterior secondary palate in the Osr2-IresCre;Bmpr1af/f mutant embryos. Frontal sections through the anterior secondary palate of E13.5 and E14.5 embryos are shown. mRNA signals are shown in blue/purple color. Control embryos (A, C, E, G, I, K, M, O) are of Osr2-IresCre;Bmpr1af/+ genotype and mutant embryos (B, D, F, H, J, L, N, P) are of Osr2-IresCre;Bmpr1af/f genotype. Arrowheads point to the palatal shelves. (A–D) Bmp2 mRNA expression. (E–H) Bmp4 mRNA expression. (I–L) Msx1 mRNA expression. (M–P) Fgf10 mRNA expression. t, tongue.
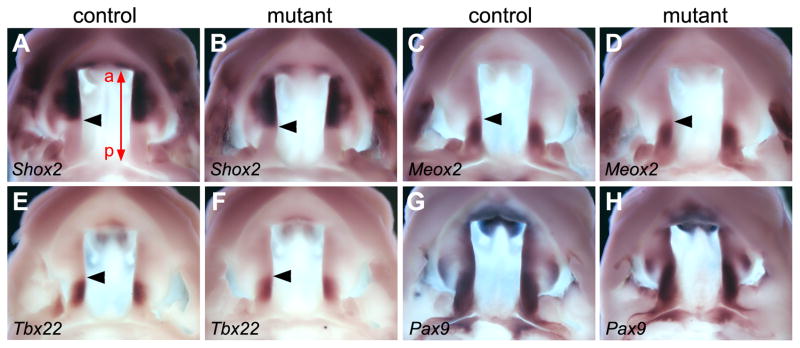
Previous studies suggested that Bmp signaling patterns gene expression along the anterior-posterior axis of the secondary palate (Zhang et al., 2002; Hilliard et al., 2005; Liu et al., 2005, Yu et al., 2005). To examine whether defects in the rostral expansion of the developing secondary palate was accompanied by defects in anterior-posterior patterning, we compared expression of molecular markers of anterior and posterior regions of the developing secondary palate, respectively, in the Osr2-IresCre;Bmpr1af/f mutant and control littermates by whole mount in situ hybridization. As shown in Fig. 7, the restricted expression patterns of Shox2 mRNA in the anterior palate and of Meox2 and Tbx22 in the posterior palate were not significantly altered in the Osr2-IresCre;Bmpr1af/f mutant embryos in comparison with control littermates (Fig. 7, A–F). A similar pattern of expression of Pax9 mRNA, which is differentially expressed along the anterior-posterior axis, was also detected in the developing palatal shelves in the Osr2-IresCre;Bmpr1af/f mutant and control littermates (Fig. 7, G and H). These data indicate that the defects in rostral expansion of the developing secondary palate is likely due to the decreased palatal mesenchyme proliferation, as described above, and not defects in anterior-posterior patterning.
Fig. 7.
Anterior and posterior regional identities of the developing secondary palate are maintained in the Osr2-IresCre;Bmpr1af/f mutant mice. (A,B) Shox2 mRNA expression was restricted to the anterior half of the palatal shelves in E13.5 control (A) and mutant (B) littermates. The red line with arrows at both ends in A mark the length of the palatal shelf, with “a” marking the anterior end and “p” marking the posterior end. Black arrowhead point to the posterior boundary of Shox2 mRNA expression domain. (C–F) Expression of Meox2 (C and D) and Tbx22 (E and F) mRNAs was restricted to the posterior side of the E13.5 palatal shelves in both control (C and E) and mutant (D and F) embryos. Arrowheads point to the anterior boundary of mRNA expression domain in these samples. (G,H) Expression of Pax9 mRNAs exhibited similar patterns along the anterior-posterior axis of the developing palatal shelves at E13.5 in the control (G) and mutant (H) littermates.
Bmpr1a signaling in the palatal mesenchyme is required for palatal bone formation
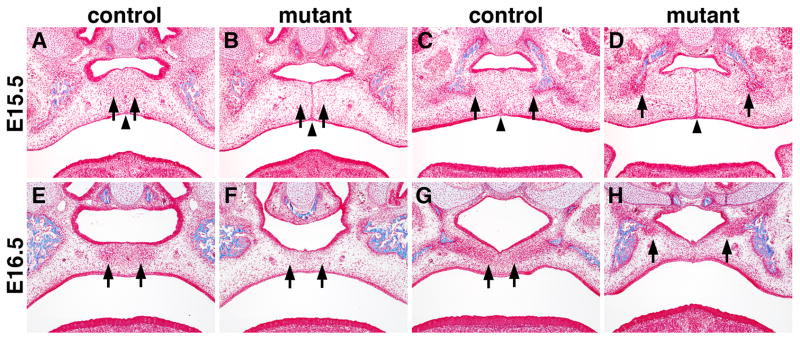
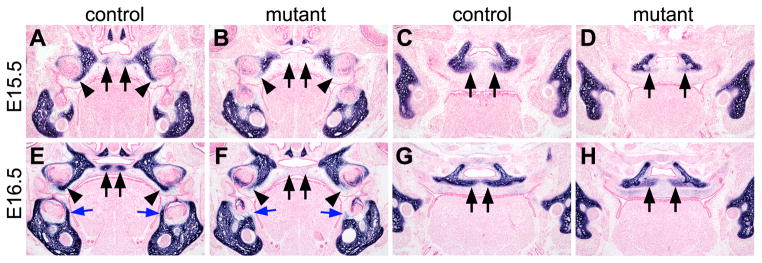
The palatal bones include the palatal processes of the maxilla in the anterior secondary palate and palatal processes of the palatine in the middle section of the secondary palate, both of which are deficient in the newborn Osr2-IresCre;Bmpr1af/f mutant mice (Fig. 2). Like most craniofacial bones, the palatal bones form by intramembranous ossification, which involves condensation of the neural crest derived mesenchyme cells that differentiate directly into osteoblasts. We carried out trichrome staining of serial paraffin sections to compare the cell condensation and ossification processes during palatal bone formation in the control and Osr2-IresCre;Bmpr1af/f mutant embryos. The palatal mesenchyme in the anterior secondary palate started to condense near the midline shortly after palatal fusion at E15.5 in the control embryos (Fig. 8A). In the middle section of the secondary palate, formation of the palatal processes of the palatine starts from mesenchymal condensations in close association with the osteogenic fronts of the palatine bone in the lateral regions (Fig. 8C). In the E15.5 Osr2-IresCre;Bmpr1af/f mutant embryos, no obvious mesenchymal condensations were detected in the anterior secondary palate and formation of the palatal processes of the palatine was also delayed (Fig. 8B, D). At E16.5, extensive mesenchymal condensations forming the primordia of the palatal processes of both the maxilla and palatine are clearly visible in the control embryos (Fig. 8E, G). In the E16.5 Osr2-IresCre;Bmpr1af/f mutant embryos, while the midline epithelial seam had completely disintegrated in most parts of the secondary palate, no mesenchyml condensations comparable to the primordia of the palatal processes of the maxillary bone were detected and formation of the palatal processes of the palatine was significantly delayed (Fig. 8F, H).
Fig. 8.
Defects in palatal bone formation in the Osr2-IresCre;Bmpr1af/f mutant embryos. (A, B) At E15.5, anterior palatal mesenchyme started to condense adjacent to the midline in the control embryo (A) while the corresponding cells (arrows) in the mutant (B) are loosely distributed. (C, D) At E15.5, palatal mesenchyme condensed at the bilateral osteogenic fronts (arrows) of the palatine in the control (C) and mutant (D) embryos. Arrowheads point to the midline of the palate, which is occupied by the midline epithelial seam in the mutant (B, D) while the midline epithelial seam had almost completely disappeared in the control embryo (A, C). (E, F) At E16.5, condensed palatal mesenchyme in the anterior secondary palate formed the osteogenic centers of the palatal processes of the maxilla in the control embryo (E) but the corresponding region of the secondary palate in the mutant embryo still contain loose mesenchyme. Arrows point to corresponding regions of the palatal mesenchyme in the control and mutant samples. (G, H) At E16.5, palatal mesenchyme condensation in the future palatal bone region was significantly delayed in the mutant embryo (H) in comparison with the control embryo (G). Arrows point to the medial edge of the condensed palatal mesenchyme.
Consistent with the morphological defects observed, we found that osteoblast differentiation in the secondary palate was significantly impaired in Osr2-IresCre;Bmpr1af/f mutant embryos. Expression of alkaline phosphatase, an early osteoblast differentiation marker, was activated in the progenitor cells of the palatal bones by E15.5 and significantly upregulated in the condensed palatal mesenchyme by E16.5 in the control embryos (Fig. 9A, C, E, G). No alkaline phosphatase expression was detected in the anterior palatal mesenchyme in the Osr2-IresCre;Bmpr1af/f mutant embryos at these stages (Fig. 11, A, B, E, F). In the mid-palate region, alkaline phosphatase expression in the palatal mesenchyme at the osteogenic fronts of the palatal processes of the palatine bones was significantly reduced in the Osr2-IresCre;Bmpr1af/f mutant embryos in comparison with the control littermates at E15.5 and E16.5, respectively (Fig. 9, C, D, G, H).
Fig. 9.
Detection of alkaline phosphatase expression in the palatal mesenchyme in the Osr2-IresCre;Bmpr1af/f mutant and control littermates. Alkaline phosphatase staining is shown in blue color. (A–D) Position-matched frontal sections from the E15.5 control (A and C) and mutant (B and D) littermates through either the middle of the first molar tooth germs (A and B) or the developing palatal process of the palatine bone (C and D). Arrows point to corresponding regions of the palatal mesenchyme. Arrowheads in A and B point to the developing alveolar bone. Arrows in C and D point to alkaline phosphatase staining in the prospective palatal processes of the palatine bone. (E–H) Position-matched frontal sections from the E16.5 control (E and G) and mutant (F and H) littermates through either the middle of the first molar tooth germs (E and F) or the developing palatal process of the palatine bone (G and H). Black arrows in E point to the alkaline phosphatase-positive osteogenic centers of the developing palatal processes of the maxilla. Black arrows in F point to the absence of the palatal processes of the maxilla in the mutant. Black arrowheads in E and F point to the developing alveolar bone of the maxillary first molars. Blue arrows in E and F point to the developing alveolar bone of the mandibular first molars. Arrows in G and H point to alkaline phosphatase staining in the developing palatal processes of the palatine bone.
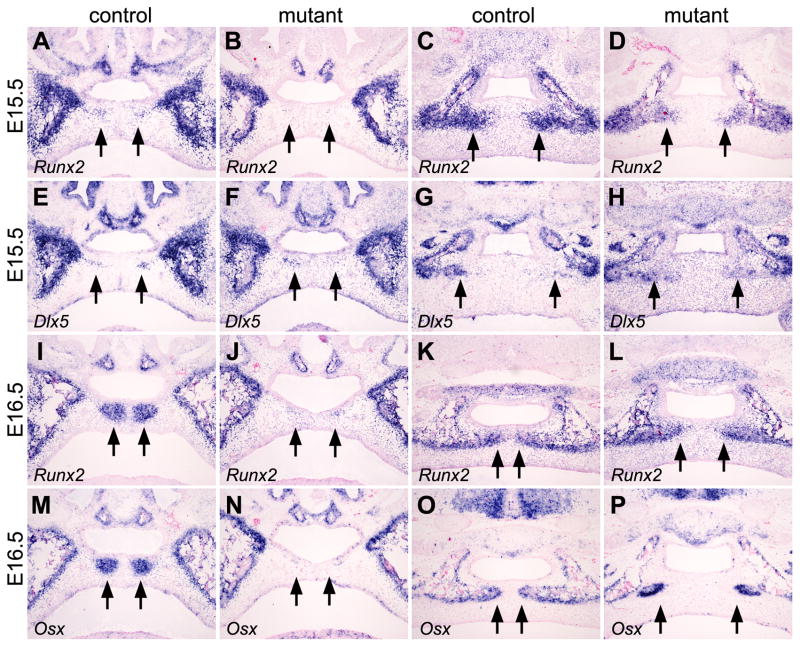
Expression of several transcription factors critical for osteoblast differentiation in vivo, including Runx2, Osterix (Osx), and Dlx5, have been shown to respond to Bmp induction of osteoblast differentiation from mesenchymal progenitor cells (Komori et al., 1997; Otto et al., 1997; Miyama et al., 1999; Lee et al., 2000; Nakashima et al., 2002; Holleville et al., 2007; Matsubara et al., 2008). Osx acts downstream of Runx2 and both Osx and Runx2 are essential for osteoblast differentiation in vivo (Komori et al., 1997; Otto et al., 1997; Nakashima et al., 2002). Dlx5−/− mutant mice had delayed ossification during craniofacial skeletogenesis, including defects in the palatal processes of both the maxilla and palatine (Acampora et al., 1999; Depew et al., 1999; Depew et al., 2005). Thus, we examined whether the expression of these transcription factors was affected in the developing palate in Osr2-IresCre;Bmpr1af/f mutant embryos. At E15.5, similar to alkaline phosphatase expression, Runx2 mRNA expression was activated in a subset of progenitor cells of the palatal processes of the maxilla in the control embryos but not in the corresponding regions of the secondary palate in the mutant embryos (Fig. 10A, B). In addition, Runx2 mRNA expression was significantly reduced in the progenitor cells of the palatal processes of the palatine in the mutant embryos compared with that in the control littermates (Fig. 10C, D). In contrast, expression of Dlx5 mRNA was not significantly altered in the Osr2-IresCre;Bmpr1af/f mutant embryos compared with control littermates (Fig. 10, E–H). At E16.5, expression of both Runx2 and Osx mRNAs was strongly upregulated in the condensed mesenchyme progenitor cells of the palatal processes of the maxilla in control embryos (Fig. 10, I and M). Little expression of Runx2 or Osx mRNAs was detected in the corresponding region of the anterior secondary palate in the Osr2-IresCre;Bmpr1af/f mutant embryos (Fig. 10, J and N). In the developing palatal processes of the palatine bones, the Osr2-IresCre;Bmpr1af/f mutant embryos exhibited delayed ossification, but high levels of expression of both Runx2 and Osx mRNAs were detected in the condensed mesenchyme progenitor cells at the osteogenic fronts at this stage (Fig. 10, K, L, O, P). Taken together, these data indicate that Bmpr1a signaling in the palatal mesenchyme is required for the initiation of the palatal processes of the maxilla and plays critical roles in the osteogenesis of the palatal processes of the palatine.
Fig. 10.
Comparison of expression of osteogenic transcription factor genes during palatal bone formation in the Osr2-IresCre;Bmpr1af/f mutant and control littermates. (A–D) Compared with the control littermates (A, C), the Osr2-IresCre;Bmpr1af/f mutant embryos (C and D) exhibited reduced expression of Runx2 mRNA in the mesenchymal precursors of the palatal processes of the maxilla (A and B) and palatine (C and D) at E15.5. Arrows point to the palatal mesenchyme in the prospective palatal processes of the maxilla (A and B) and palatine (C and D). (E–H) Dlx5 mRNA was expressed in the mesenchymal progenitor cells of the palatal processes of the maxilla (E and F) and palatine (G and H) in both the control (E and G) and mutant (F and H) littermates at E15.5. (I, J) In contrast to the strong Runx2 mRNA expression in the developing palatal processes of the maxilla in the control embryo (I), little Runx2 mRNA expression was detected in the corresponding regions of the secondary palate in the mutant littermate (J) at E16.5. (K,L) Although strong Runx2 mRNA expression was detected in the mesenchymal cells at the osteogenic fronts of the developing palatal processes of the palatine bone in both the control (K) and mutant (L) littermates at E16.5, the delay in osteogenesis of the palatal processes of the palatine was apparent in the mutant (L), with the bilateral osteogenic fronts separated by a much wider midline area devoid of Runx2 mRNA expression, compared with the control littermate (K). (M,N) Strong expression of Osx mRNA was detected in the developing palatal processes of the maxilla in the E16.5 control embryo (M) but no Osx mRNA expression was detected in the palatal mesenchyme in the corresponding regions of the mutant embryo (N). (O,P) While strong Osx mRNA expression was detected in the osteoblasts at the osteogenic fronts of the developing palatal processes of the palatine in both the control (O) and mutant (P) littermates at E16.5, the bilateral osteogenic fronts failed to expand toward the midline of the secondary palate in the mutant embryo (P) compared with the control (O).
In the newborn mice, the midline of the Osr2-IresCre;Bmpr1af/f mutant secondary palate appeared to contain little mesenchyme (Fig. 2L, M). At E16.5, the mutant secondary palate was visibly thinner than that in the control littermates, especially in the anterior secondary palate (Fig. 8E, F; Fig. 10, J and N). We investigated whether there was increased palatal mesenchyme cell death in the Osr2-IresCre;Bmpr1af/f mutant secondary palate by using TUNEL assays. While we clearly detected apoptosis of the midline epithelial seam during palatal fusion in both the control and mutant embryos, we did not find any increase in cell death in the palatal mesenchyme in the mutant embryos (Supplementary Fig. 2, and data not shown). Taken together, our data indicate that the submucous cleft phenotype of the Osr2-IresCre;Bmpr1af/f mutant mice resulted from the combination of significantly reduced palatal mesenchyme proliferation from E13.5 to E15.5 and lack of palatal bone support in late gestation.
Discussion
By using Cre/loxP-mediated palate mesenchyme-specific inactivation of the Bmpr1a gene in mice, this study has gained new insights into the roles of Bmp signaling in mammalian palatogenesis. We found that Bmpr1a mRNA is preferentially expressed in the primary and anterior secondary palate during palatal outgrowth and is upregulated during palatal mesenchyme differentiation into osteoblasts. Inactivation of Bmpr1a in the palatal mesenchyme caused partial anterior cleft palate and disruption of palatal bone formation.
Bmpr1a is required in the palatal mesenchyme for mesenchymal-epithelial interactions controlling palatal outgrowth
Previous analysis of the Msx1−/− mutant mice uncovered a genetic pathway in which Msx1 controls Bmp4 expression in the anterior palatal mesenchyme and Bmp4 signals to maintain Shh expression in the anterior palatal epithelium to control palatal shelf growth (Zhang et al., 2002). However, there is no direct evidence that Bmp signaling in the palatal epithelium is necessary for Shh expression during palate development. Our data show that inactivation of Bmpr1a in the palatal mesenchyme resulted in downregulation of Shh expression in the primary and anterior secondary palate during palatal outgrowth. It is likely that Bmp signaling activates other signaling molecules in the palatal mesenchyme, which in turn signals to the palatal epithelium to maintain Shh expression. Studies of Fgf10−/− and Fgfr2b−/− mutant mice have shown that both mesenchymally expressed Fgf10 and the epithelial Fgfr2b are required for maintenance of Shh mRNA expression in the palatal epithelium (Rice et al., 2004). We found that Fgf10 mRNA expression was also downregulated in the anterior palatal mesenchyme in Osr2-IresCre;Bmpr1af/f mutant embryos. Taken together, these data suggest that Bmp4, Fgf10, and Shh signaling pathways function in an integrated molecular network controlling anterior palatal growth.
In contrast to the loss of Bmp4 mRNA expression in the palatal mesenchyme in Msx1−/− mutant embryos (Zhang et al., 2002), we found that Msx1 mRNA expression was downregulated but Bmp4 mRNA expression was upregulated in the anterior palatal mesenchyme from E13.5 to E14.5 in the Osr2-IresCre;Bmpr1af/f mutant embryos. The increase of Bmp4 mRNA expression may be due to activation of a compensatory mechanism that is Msx1-independent in the Osr2-IresCre;Bmpr1af/f mutant embryos. Further studies are necessary to identify the molecular mechanism and to investigate whether the increase in expression of Bmp2 and Bmp4 may have contributed to the developmental defects in the anterior palate of Osr2-IresCre;Bmpr1af/f mutant embryos.
The role of Bmpr1a in palatal bone formation and submucous cleft palate pathogenesis
We found that the palatal processes of the maxilla and of the palatine are formed through distinct skeletogenic processes, with the formation of palatal process of the maxilla involving mesenchymal condensation to form de novo ossification centers separate from the maxillary bones while formation of palatal process of the palatine involves expansion of osteogenic fronts from the bilateral palatine toward the midline. In the Osr2-IresCre;Bmpr1af/f mutant mice, the palatal processes of the maxilla failed to form and the palatal processes of the palatine were reduced. We found that the palatal mesenchyme progenitor cells of the palatal processes of the maxilla in the Osr2-IresCre;Bmpr1af/f mutant embryos failed to condense and did not express the essential osteogenic transcription factor genes Runx2 and Osx. Since Runx2 induction occurred concurrently with palatal mesenchyme condensation, further studies will be necessary to investigate whether Bmpr1a signaling directly activates Runx2 expression to initiate palatal bone formation.
The differences in severity of defects in the palatal processes of the maxilla and of the palatine likely result from partial functional complementation by other type I Bmp receptors. Although mice lacking Bmpr1b did not have obvious craniofacial defects (Baur et al., 2000; Yi et al., 2000), mice with neural crest-specific inactivation of another type I Bmp receptor Alk2 had overt cleft palate (Dudas et al., 2004). Generation of mice with palate-specific inactivation of two or all three type I receptors will provide additional insights into the roles of Bmp signaling in palate morphogenesis.
Although submucous cleft palate forms a clinically important subgroup and has a reported incidence of 1:1250 to 1:5000 (Garcia et al., 1988; Weatherley-White et al., 1972), little is known about the pathogenic mechanisms. Only two other mutant mouse strains have been reported to exhibit submucous cleft palate phenotypes (Pauws et al., 2009; Xu et al., 2006). Mice lacking Tbx22 had reduced palatal bone formation at the posterior palatine region (Pauws et al., 2009). We examined expression of Tbx22 mRNA in the developing palate in the control and Osr2-IresCre;Bmpr1af/f mutant embryos at various developmental stages and did not find any significant differences (Fig. 7, E and F, and data not shown). Mice with epithelium-specific deletion of the type II receptor for the transforming growth factor-beta family ligands, K14-Cre;Tgfbr2fl/fl; exhibited submucous cleft palate in the hard palate region due to incomplete disintegration of the midline epithelial seam during palate fusion (Xu et al., 2006). While we found that Bmpr1a expression is upregulated in the condensing palatal mesenchyme and that the Osr2-IresCre;Bmpr1af/f mutant embryos had clear defects in palatal bone formation, most of these mutant embryos also had delayed palatal fusion. It is possible that the early defects in anterior palatal mesenchyme proliferation might have contributed to the delay in palatal shelf elevation and fusion, which in turn contributed to the delay in palatal bone formation. The failure of anterior palatal mesenchyme to form the ossification centers of the palatal processes of the maxilla in the Osr2-IresCre;Bmpr1af/f mutant embryos, however, identify a specific role for Bmpr1a signaling in palatal bone formation and distinguishes the submucous cleft palate phenotype in this mouse strain from those in the Tbx22 null and K14-Cre;Tgfbr2fl/fl mutant mice.
Research highlights.
In situ hybridization analysis showed differential expression of Bmpr1a mRNA along the anterior-posterior axis of the developing palate during palatal outgrowth and upregulation of Bmpr1a mRNA during palatal bone formation in mice;
Tissue-specific inactivation of Bmpr1a in the developing palatal mesenchyme caused defects in primary palate and anterior secondary palate outgrowth;
Bmpr1a signaling in the palatal mesenchyme is required for palatal bone formation;
In addition to providing new insights into the roles of Bmp signaling in palatogenesis, this study also revealed distinct processes in the formation of the palatal processes of the maxilla and palatine.
Supplementary Material
Acknowledgments
We thank Dr. Amel Gritli-Linde for sharing the protocol for van Gieson staining of paraffin sections. This work was supported by the NIH/NIDCR grants R01DE013681 and R01DE015207 to RJ.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Acampora D, Merlo GR, Paleari L, Zerega B, Postiglione MP, Mantero S, Bober E, Barbieri O, Simeone A, Levi G. Craniofacial, vestibular and bone defects in mice lacking the Distal-less-related gene Dlx5. Development. 1999;126:3795–3809. doi: 10.1242/dev.126.17.3795. [DOI] [PubMed] [Google Scholar]
- Baur ST, Mai JJ, Dymecki SM. Combinatorial signaling through BMP receptor IB and GDF5: shaping of the distal mouse limb and genetics of distal limb diversity. Development. 2000;127:605–619. doi: 10.1242/dev.127.3.605. [DOI] [PubMed] [Google Scholar]
- Chai Y, Jiang X, Ito Y, Bringas P, Han J, Rowitch DH, Soriano P, McMahon AP, Sucov HM. Fate of the mammalian cranial neural crest during tooth and mandibular morphogenesis. Development. 2000;127:1671–1679. doi: 10.1242/dev.127.8.1671. [DOI] [PubMed] [Google Scholar]
- Chen J, Lan Y, Baek JA, Gao Y, Jiang R. Wnt/beta-catenin signaling plays an essential role in activation of odontogenic mesenchyme during early tooth development. Dev Biol. 2009;334:174–185. doi: 10.1016/j.ydbio.2009.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depew MJ, Simpson CA, Morasso M, Rubenstein JLR. Reassessing the Dlx code: the genetic regulation of branchial arch skeletal pattern and development. J Anat. 2005;207:501–561. doi: 10.1111/j.1469-7580.2005.00487.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depew MJ, Liu JK, Long JE, Presley R, Meneses JJ, Pedersen RA, Rubenstein JL. Dlx5 regulates regional development of the branchial arches and sensory capsules. Development. 1999;126:3831–3846. doi: 10.1242/dev.126.17.3831. [DOI] [PubMed] [Google Scholar]
- Dudas M, Sridurongrit S, Nagy A, Okazaki K, Kaartinen V. Craniofacial defects in mice lacking BMP type I receptor Alk2 in neural crest cells. Mech Dev. 2004;121:173–182. doi: 10.1016/j.mod.2003.12.003. [DOI] [PubMed] [Google Scholar]
- Ferguson MWJ. Palate Development. Development [suppl] 1988;103:41–60. doi: 10.1242/dev.103.Supplement.41. [DOI] [PubMed] [Google Scholar]
- Garcia VM, Ysunza A, Hernandez X, Marquez C. Diagnosis and treatment of submucous cleft palate: a review of 108 cases. Cleft Palate J. 1988;25:171–173. [PubMed] [Google Scholar]
- Gritli-Linde A. Molecular control of secondary palate development. Dev Biol. 2007;301:309–326. doi: 10.1016/j.ydbio.2006.07.042. [DOI] [PubMed] [Google Scholar]
- Hilliard SA, Yu L, Gu S, Zhang Z, Chen YP. Regional regulation of palatal growth and patterning along the anterior-posterior axis in mice. J Anat. 2005;207:655–667. doi: 10.1111/j.1469-7580.2005.00474.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holleville N, Mateos S, Bontoux M, Bollerot K, Monsoro-Burq AH. Dlx5 drives Runx2 expression and osteogenic differentiation in developing cranial suture mesenchyme. Dev Biol. 2007;304:860–874. doi: 10.1016/j.ydbio.2007.01.003. [DOI] [PubMed] [Google Scholar]
- Jiang R, Lan Y, Norton CR, Sundberg JP, Gridley T. The Slug gene is not essential for mesoderm or neural crest development in mice. Dev Biol. 1998;198:277–285. [PubMed] [Google Scholar]
- Komori T, Yagi H, Nomura S, Yamaguchi A, Sasaki K, Deguchi K, Shimizu Y, Bronson RT, Gao YH, Inada M, et al. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell. 1997;89:755–764. doi: 10.1016/s0092-8674(00)80258-5. [DOI] [PubMed] [Google Scholar]
- Lan Y, Jiang R. Sonic hedgehog signaling regulates reciprocal epithelial-mesenchymal interactions controlling palatal outgrowth. Development. 2009;136:1378–1396. doi: 10.1242/dev.028167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan Y, Ovitt CE, Cho ES, Maltby KM, Wang Q, Jiang R. Odd-skipped related 2 (Osr2) encodes a key intrinsic regulator of secondary palate growth and morphogenesis. Development. 2004;131:3207–3216. doi: 10.1242/dev.01175. [DOI] [PubMed] [Google Scholar]
- Lan Y, Wang Q, Ovitt CE, Jiang R. A unique mouse strain expressing Cre recombinase for tissue-specific analysis of gene function in palate and kidney development. Genesis. 2007;45:618–624. doi: 10.1002/dvg.20334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KS, Kim HJ, Li QL, Chi XZ, Ueta C, Komori T, Wozney JM, Kim EG, Choi JY, Ryoo HM, Bae SC. Runx2 is a common target of transforming growth factor beta1 and bone morphogenetic protein 2, and cooporation between Runx2 and Smad5 induces osteoblast-specific gene expression in the pluoripotent mesenchymal precursor cell line C2C12. Mol Cell Biol. 2000;20:8783–8792. doi: 10.1128/mcb.20.23.8783-8792.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Ding J. Gene expression analysis reveals that formation of the mouse anterior secondary palate involves recruitment of cells from the posterior side. Int J Dev Biol. 2007;51:167–172. doi: 10.1387/ijdb.062212ql. [DOI] [PubMed] [Google Scholar]
- Liu W, Lan Y, Pauws E, Meester-Smoor MA, Stanier P, Zwarthoff EC, Jiang R. The Mn1 transcription factor acts upstream of Tbx22 and preferentially regulates posterior palate growth in mice. Development. 2008;135:3959–68. doi: 10.1242/dev.025304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Sun X, Braut A, Mishina Y, Behringer RR, Mina M, Martin JF. Distinct functions for Bmp signaling in lip and palate fusion in mice. Development. 2005;132:1453–61. doi: 10.1242/dev.01676. [DOI] [PubMed] [Google Scholar]
- Martin JF, Bradley A, Olson EN. The paired-like homeobox gene Mhox is required for early events of skeletogenesis in multiple lineages. Genes Dev. 1995;9:1237–1249. doi: 10.1101/gad.9.10.1237. [DOI] [PubMed] [Google Scholar]
- Matsubara T, Kida K, Yamaguchi A, Hata K, Ichida F, Meguro H, Aburatani H, Nishimura R, Yoneda T. BMP2 regulates Osterix through Msx2 and Runx2 during osteoblast differentiation. J Biol Chem. 2008;283:29119–29125. doi: 10.1074/jbc.M801774200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishina Y, Hanks MC, Miura S, Tallquist MD, Behringer RR. Generation of Bmpr/Alk3 conditional knockout mice. Genesis. 2002;32:69–72. doi: 10.1002/gene.10038. [DOI] [PubMed] [Google Scholar]
- Mishina Y, Suzuki A, Ueno N, Behringer RR. Bmpr encodes a type I bone morphogenetic protein receptor that is essential for gastrulation during mouse embryogenesis. Genes Dev. 1995;9:3027–3037. doi: 10.1101/gad.9.24.3027. [DOI] [PubMed] [Google Scholar]
- Miyama K, Yamada G, Yamamoto TS, Takagi C, Miyado K, Sakai M, Ueno N, Shibuya H. A BMP-inducible gene, dlx5, regulates osteoblast differentiation and mesoderm induction. Dev Biol. 1999;208:123–133. doi: 10.1006/dbio.1998.9197. [DOI] [PubMed] [Google Scholar]
- Nakashima K, Zhou X, Kunkel G, Zhang Z, Deng JM, Behringer RR, de Crombrugghe B. The novel zinc finger-containing transcription factor Osterix is required for osteoblast differentiation and bone formation. Cell. 2002;108:17–29. doi: 10.1016/s0092-8674(01)00622-5. [DOI] [PubMed] [Google Scholar]
- Otto F, Thornell AP, Crompton T, Denzel A, Gilmour KC, Rosewell IR, Stamp GW, Beddington RS, Mundlos S, Olsen BR, et al. Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell. 1997;89:765–771. doi: 10.1016/s0092-8674(00)80259-7. [DOI] [PubMed] [Google Scholar]
- Pantalacci S, Prochazka J, Martin A, Rothova M, Lambert A, Bernard L, Charles C, Viriot L, Peterkova R, Laudet A. Patterning of palatal rugae through sequential addition reveals an anterior/posterior boundary in palatal development. BMC Dev Biol. 2008;8:116. doi: 10.1186/1471-213X-8-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauws E, Hoshino A, Bentley L, Prajapati S, Keller C, Hammond P, Martinez-Barbera J, Moore GE, Stanier P. Tbx22null mice have a submucous cleft palate due to reduced bone formation and also display ankyloglossia and choanal atresia phenotypes. Hum Mol Genet. 2009;18:4171–4179. doi: 10.1093/hmg/ddp368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice R, Spencer-Dene B, Connor EC, Gritli-Linde A, McMahon AP, Dickson C, Thesleff I, Rice DP. Disruption of Fgf10/Fgfr2b-coordinated epithelial-mesenchymal interactions causes cleft palate. J Clin Invest. 2004;113:1692–1700. doi: 10.1172/JCI20384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperber GH. Palatogenesis: closure of the secondary palate. In: Wyszynski DF, editor. Cleft Lip & Palate, From Origin To Treatment. Oxford University Press; 2002. pp. 14–24. [Google Scholar]
- Stottmann RW, Choi M, Mishina Y, Meyers EN, Klingensmith J. BMP receptor 1A is required in mammalian neural crest cells for development of the cardiac outflow tract and ventricular myocardium. Development. 2004;131:2205–2218. doi: 10.1242/dev.01086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weatherley-White RC, Sakura CY, Jr, Brenner LD, Stewart JM, Ott JE. Submucous cleft palate, its incidence, natural history, and indications for treatment. Plast Reconstr Surg. 1972;49:297–304. [PubMed] [Google Scholar]
- Welsh IC, O’Brien TB. Signaling integration in the rugae growth zone directs sequential SHH signaling center formation during the rostral outgrowth of the palate. Dev Biol. 2009;336:53–67. doi: 10.1016/j.ydbio.2009.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winnier G, Blessing M, Labosky PA, Hogan BL. Bone morphogenetic protein-4 is required for mesoderm formation and patterning in the mouse. Genes Dev. 1995;9:2105–2116. doi: 10.1101/gad.9.17.2105. [DOI] [PubMed] [Google Scholar]
- Xu X, Han J, Ito Y, Bringas P, Jr, Urata MM, Chai Y. Cell autonomous requirements for Tgfbr2 in the disappearance of medial edge epithelium during palatal fusion. Dev Biol. 2006;297:238–248. doi: 10.1016/j.ydbio.2006.05.014. [DOI] [PubMed] [Google Scholar]
- Yi SE, Daluiski A, Pederson R, Rosen V, Lyons KM. The type I BMP receptor BMPR1B is required for chondrogenesis in the mouse limb. Development. 2000;127:621–630. doi: 10.1242/dev.127.3.621. [DOI] [PubMed] [Google Scholar]
- Yu L, Alappat S, Song S, Yan Y, Zhang M, Zhang X, Jiang G, Zhang Y, Zhang Z, Chen YP. Shox2-deficient mice exhibit a rare type of incomplete clefting of the secondary palate. Development. 2005;132:4393–4406. doi: 10.1242/dev.02013. [DOI] [PubMed] [Google Scholar]
- Zhang YD, Zhao X, Hu Y, St Amand TR, Ramamurthy R, Qiu MS, Chen YP. Msx1 is required for the induction of Patched by Sonic hedgehog in the mammalian tooth germ. Dev Dyn. 1999;215:45–53. doi: 10.1002/(SICI)1097-0177(199905)215:1<45::AID-DVDY5>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Song Y, Zhao X, Zhang X, Fermin C, Chen Y. Rescue of cleft palate in Msx1-deficient mice by transgenic Bmp4 reveals a network of BMP and Shh signaling in the regulation of mammalian palatogenesis. Development. 2002;129:4135–4146. doi: 10.1242/dev.129.17.4135. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.