Abstract
The histone deacetylase (HDAC) inhibitor suberoylanilide hydroxamic acid (SAHA) was used to turn on the biosynthesis of EGM-556, a new cyclodepsipeptide of hybrid biosynthetic origin, isolated from the Floridian marine sediment-derived fungus Microascus sp. The absolute configurations of 3 chiral centers were determined by Marfey’s derivatization. EGM-556 represents one of the few examples in which silent biosynthetic genes, encoding a new secondary metabolite, were activated by means of epigenetic manipulation of the fungal metabolome.
Marine-derived fungi1 continue to be a source of novel bioactive metabolites, even in genera as well studied as Aspergillus2 and Penicillium.3 Continuing attention on these taxa is, in part, being motivated by advances in genome sequencing,4 especially through genome mining.5 A core intent of these studies usually involves identifying biosynthetic gene clusters producing novel structures6,7 to test the hypothesis that such approaches will allow expression of ‘silent’ metabolite pathways.8,6a Another related strategy involves unraveling the regulatory mechanisms controlling the biosynthetic genes9 using insights gained from chemical genetics10 and epigenetics11 experiments.
The use of small molecule HDAC (Histone Deacetylase) inhibitors12 to perturb the fungal secondary biosynthetic machinery10c has been rewarding and represents a proof-of-principle of ideas summarized above. Commercially available compounds such as 5-azacytidine (Aza)13 and suberoylanilide hydroxamic acid (SAHA),12a, explored by several labs, have provided valuable outcomes. For example, the culturing of terrestrial Aspergillus niger in the presence of SAHA yielded the novel secondary metabolite nygerone, carrying a highly unusual 1-phenylpyridin-4(1H)-one core.11c New oxylipins plus the unique pigments, the cladochromes, were produced by C. cladosporioides cultured in the presence of either Aza or SAHA. As another parallel result, the lunalides, octadeca-dienoic acids, were produced by cultures of Diatrype sp. grown in the presence of Aza.11d
We used a test bed of 12 marine sediment-derived fungi (strain numbers and properties shown in Table S1, Supporting Information) to assess changes in their biosynthetic products induced by adding 10−4 M SAHA to the saltwater cultures. All fungi were isolated from shallow water sediments collected in Florida, and cultured for 18 days to reach the stationary growth phase. The group of 24 ethyl acetate crude extracts (from broths + or − SAHA), were profiled by analytical RP C18 LC-ELSD-MS, and significant changes were observed in the scans between the matched pairs. One member of this set, Microascus sp. (strain number 098059A, identified by morphological and molecular methods14), showed noteworthy changes and was chosen for further study. Shown for this sample in Figure 1, are the primary data consisting of LC-ELSD-MS traces for control (C) and SAHA-spiked (SP) cultures. Significantly, the SP sample contained a new peak at retention time 21.5 min with an m/z of 557 ([M+H]+) that was not present in the control culture. In addition, the ethyl acetate crude extract yields were very different between the two samples: SP = 114.6 mg vs. C = 30.6 mg. Of further note was our literature search that revealed that only ~10 secondary metabolites have been reported for Microascus and its asexual anamorph Scopulariopsis species, described as human pathogens in immuno-compromised patients.15
Figure 1.
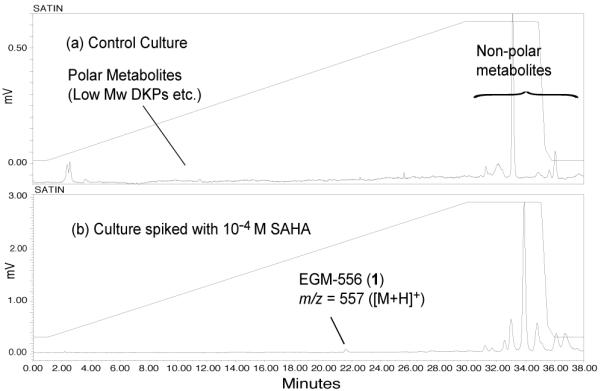
RP LC-ELSD-MS traces showing the absence (a) and presence (b) of EGM-556 (1) in cultures of Microascus sp.
Isolation of the RT = 21.5 min substance using RP HPLC (see General Experimental Procedures, Supporting Information), was straightforward and yielded 3.5 mg of a white solid. This compound, named EGM-556 (1) with [α]23D = +2.60 (c 0.60, MeOH), possessed a molecular formula of C30H44N4O6, (11 unsaturation units) assigned by HR ESITOFMS data for the [M+H]+ ion at m/z 557.33064 (calcd. 557.33336, Δ = −4.8 ppm, see General Experimental Procedures, Supporting Information). The DEPT NMR spectrum (Table 1, and Figs S2, S3, Table S2, Supporting Information) showed a partial molecular formula of C30H40 and the remaining H4N4O6 atoms were indirectly accounted for by four amides (NH protons at δH 7.60, 7.74, 8.10, 8.56, see Fig S1, Supporting Information) and one ester (C=O)-Z (see five peaks δC 168.7 - 173.0). These functionalities comprised five unsaturation units with five more from an mono-substituted benzene ring (δH 7.44 (2H, d), 7.36 (2H, dd), δ 7.28 (1H, t)) and an E-disubstituted conjugated double bond (δH 6.28 (dd, J = 15.9, 7.1 Hz, 1H), δH 6.68 (d, J = 15.9 Hz, 1H)). Collectively, these observations meant that an additional ring must also be present as a macrocyclic depsipeptide. Using all of the preceding structural constraints and formulas as input, extensive dereplication searches were carried out but yielded no structural matches.
Table 1.
NMR Assignments for EGM-556 (1) in DMSO-d6
| structural unit |
position | 13Ca δ, typeb | 1H δ (m, J(Hz), int)c |
|---|---|---|---|
| Val-1 | CO | 168.7, qC | |
| α | 58.2, CH | 4.24 (dd, 9.6, 6.0, 1H) | |
| β | 28.5, CH | 2.25 (ds, 6.0, 6.6, 1H ) | |
| β-CH3 | 18.9, CH3 | 0.89 (d, 6.6, 3H) | |
| β-CH3 | 19.3, CH3 | 0.89 (d, 6.6, 3H) | |
| NH | 7.60 (d, 9.6, 1H) | ||
| Ile-2 | CO | 170.4, qC | |
| α | 57.2, CH | 4.26 (d, 9.6, 1H) | |
| β | 35.6, CH | 2.05 (m, 1H) | |
| γ | 23.6, CH2 | 1.26 (m, 2H) | |
| β-CH3 | 15.5, CH3 | 0.81 (d, 6.6, 3H) | |
| γ-CH3 | 11.9 CH3 | 0.81 (t, 7.5, 3H) | |
| NH | 8.10 (d, 9.6, 1H) | ||
| Ala-3 | CO | 173.0, qC | |
| α | 48.8, CH | 4.32 (dq, 6.3, 6.4, 1H) | |
| α-CH3 | 16.3, CH3 | 1.18 (d, 7.2, 3H) | |
| NH | 8.56 (d, 6.3, 1H) | ||
| Val-4 | CO | 172.3, qC | |
| α | 57.5, CH | 4.14 (dd, 8.7, 7.2, 1H) | |
| β | 29.8, CH | 1.96 (ds, 6.9, 6.8, 1H) | |
| β-CH3 | 19.6, CH3 | 0.89 (d, 6.6, 3H) | |
| β-CH3 | 18.2, CH3 | 0.89 (d, 6.6, 3H) | |
| NH | 7.74 (d, 8.7, 1H) | ||
| Hyppa-5 | 1 | 168.7, qC | |
| 2 | 40.1, CH2* | 2.42 (dd. 14.4, 1.8, 1H) | |
| 2.88 (dd, 14.4, 10.5, 1H) | |||
| 3 | 72.9, CH | 5.50 (ddd, 10.5, 7.1, 1.8, 1H) | |
| 4 | 126.7, CH | 6.28 (dd, 15.9, 7.1, 1H) | |
| 5 | 132.4, CH | 6.68 (d, 15.9, 1H) | |
| 6 | 135.7, qC | ||
| 7 | 126.5, CH | 7.44 (d, 7.2, 2H) | |
| 8 | 128.7, CH | 7.36 (dd, 7.5, 7.2, 2H) | |
| 9 | 128.1, CH | 7.28 (t, 7.5, 1H) |
Measured at 150 MHz.
Carbon type established from 13C DEPT and/or gHMQC experiments.
Measured at 600 MHz.
Overlapping with DMSO-d6
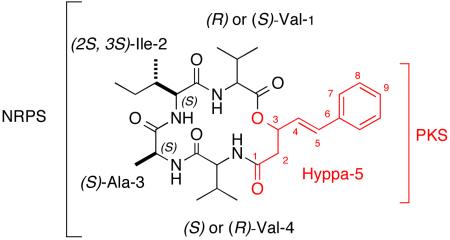
The 2D COSY NMR results clearly showed five separate proton spin systems consistent with the substructures I-V drawn in Scheme 1. The presence of seven methyl groups plus the 1H-1H COSY NMR data accounted for two valines (Val-1, Val-4), alanine (Ala-3), and isoleucine (Ile-2). The remaining substructure V, 3-hydroxy-5-phenyl-4(E)-pentenoic acid, Hyppa, was visualized from the 2D NMR correlations shown in Scheme 1. The inter-substructural gHMBC correlations, also shown in Scheme 1, unambiguously indicated the sequence Val-Ile-Ala-Val. The insertion of the Hyppa (V) substructure was complicated by the accidental isochrony of two carbonyl groups at δC 168.7 (due to Val and Hyppa arrays, indicated in red and blue in Scheme 1). The gHMBC correlations between protons at δH 4.24, 2.25 (Val-1), 4.14, 7.74 (Val-4), 5.50, 2.88 and 2.42 (Hyppa-5) and these carbonyls were consistent with two possible ways to dock V into the tetrapeptide as shown by structural candidates 1 and 1′ (Scheme 1). The gray carbonyl group in substructure IV is common to a carbonyl group in V, either in red, resulting in alternative structure 1 or in blue resulting in alternative structure 1′. Fortunately, the latter structure was quickly ruled out because it contains a carbamate and a ketone whose diagnostic 13C shifts of δC ~157 and δC ~200 respectively, were not observed.
Scheme 1.
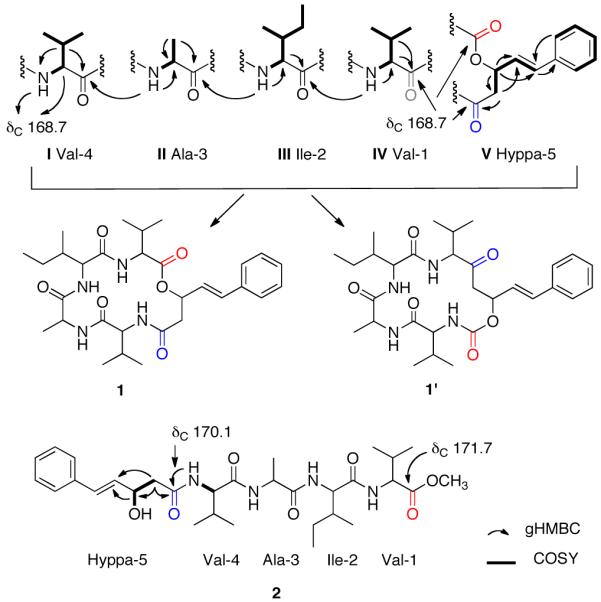
Substructures, alternative final structures, and key 2D NMR correlations. Accidentally isochronous carbonyl groups occurred in 1 at δC 168.7, indicated in red, gray and blue.
The pathway to further confirm the structure proposed of 1 along with efforts to resolv the configurations at the six chiral centers was approached in the following way. Hydrolysis of 1 (1.5 mg, 1N NaOH) followed by esterification with diazomethane16 afforded the acyclic methyl ester 2, [α]23D = −1.36 (c 1.30, MeOH) shown in Scheme 1. Its molecular formula of C31H48N4O7 was confirmed by HR ESITOFMS, based on the [M+Na]+ ion m/z 611.34268 (calcd. 611.34152, Δ = 1.9 ppm, see General Experimental Procedures, Supporting Information) and as expected was one unsaturation unit less than that of 1. The NMR data for 2 (See Table S4, Supporting Information) were consistent with the expected sequence and all five carbonyl resonances were anisochronous, which made it possible to unequivocally confirm the juxtaposition of Hyppa-5 and Val-4 through an amide bond.
Acid mediated hydrolysis of 1 (1 mg, 6M HCl) followed by derivatization with Marfey’s reagent17 and subsequent C18 LC-ELSD-MS analysis of the adducts, was the approach taken to deduce the absolute configurations of I-IV. The overall results indicated the constituent amino acids were: (R)-Val, (S)-Val, (2S, 3S)-Ile, and (S)-Ala. Retention times of the standard amino acids under our analysis conditions were as follows: (R)-Ala 59.7 min., (S)-Ala 64.4 min., (S)-Val 68.9 min., (R)-Val 75.4 min., (2S, 3S)-Ile 74.7 min., (2R, 3R)-Ile 81.3 min (See also Fig S13, Supplemental Information). Since both (R)-Val and (S)-Val were observed, EGM-556 (1) is one of four possible structures: cyclic ((R)-Hyppa-5)-((R)-Val-4)-((S)-Ala-3)-((2S, 3S)-Ile-2)-((S)-Val-1), cyclic ((R)-Hyppa-5)-((S)-Val-4)-((S)-Ala-3)-((2S,3S)-Ile-2)-((R)-Val-1), cyclic ((S)-Hyppa-5)-((R)-Val-4)-((S)-Ala-3)-((2S, 3S)-Ile-2)-((S)-Val-1) or cyclic ((S)-Hyppa-5)-((S)-Val-4)-((S)-Ala-3)-((2S, 3S)-Ile-2)-((R)-Val-1).
Reverse phase chromatography did not differentiate between (2S, 3S)-isoleucine and (2S, 3R)-isoleucine and these amino acids were separated using a chiral column (Chirex 3010, Phenomenex, 4.1 × 150 mm).18 Solvent A was 0.1 M aqueous ammonium acetate, pH = 3 with TFA and solvent B acetonitrile. The gradient used was 20 – 75 % solvent B in A over 50 min, 1 mL/min). Retention times of amino acids under these conditions were: (2S, 3S)-Ile 23.0 min., (2R, 3R)-Ile 24.3 min., (2S, 3R)-Ile 22.3 min., (2R, 3S)-Ile 23.5 min. (See also Fig. S14, Supporting Information). Our results showed that an (2S, 3S)-Ile is present.
Another approach considered, to assign the unresolved configuration of 1 at C3 of Hyppa-5, involved attempts to make a Mosher ester derivative19 of the alcohol moiety of 2. However, all of our experiments were unsuccessful. The configuration is therefore left unassigned since there is precedent for both (R) and (S) in similar compounds in the literature, as discussed below.
As an important additional note, we utilized MSn data to also address the subunit sequence for both 1 and 2. The complex array of fragmentations observed for 1 was consistent with principles outlined in the literature for cyclic depsipeptides.20 By contrast, the pattern observed for 2 was quite distinct. These contrasting outcomes will be fully described elsewhere.
The EGM-556 (1) substructural units are likely derived from a HPN (Hybrid PKS/NRPS) biosynthetic pathway. Moreover, the 16 atom peptolide core of EGM-556 (1) is rare; the only other examples are: the antimicrobial unnarmicins (3) from a marine-derived Photobacterium sp. MBIC0648521 and the histone deacetylase inhibitory/antitumor active FK228 (FR901228, 4) from Chromobacterium violaceum No. 968.22 Side-by-side comparison of 1, 3, and 4 (see Figure 2) reveals some additional similarities and contrasts. The ketide portions of all three contain β-hydroxy amides; and, analogous to the situation of 1, FK228 (4) possess (R)-Val and (S)-Val units in close proximity to the PKS side chain. The mono substituted benzene ring-containing side chain of 1 is analogous to similar functionality present in the cryptophycins, especially -26, isolated from Nostoc strains of cyanobacteria.23 Rigorous biosynthetic analysis24 has shown the phenyl-octenoic polyketide unit of cryptophycin-26 is likely derived from a phenylalanine (via trans-cinnamic acid), which is clearly present in 1. Finally, the 3(R)-hydroxy-5-phenyl-4(E)-pentenoic acid (5) coded as Hyppa in 1, as shown in Figure 3, has one precedent. The free acid was first isolated from the Chinese plant Avicennia marina together with five new iridoid glucosides, and one marinoid E (6) also contained the (3R)-Hyppa unit.25
Figure 2.
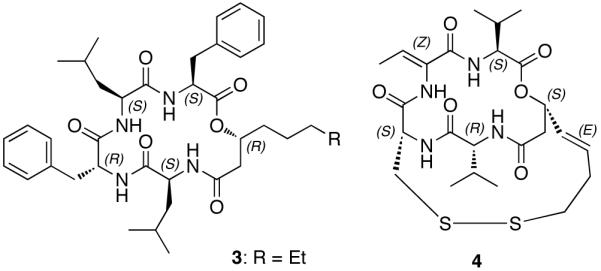
Unnarmicin C (3) from marine Photobacterium sp.21 and FK228 (4) from C. violaceum.22
Figure 3.
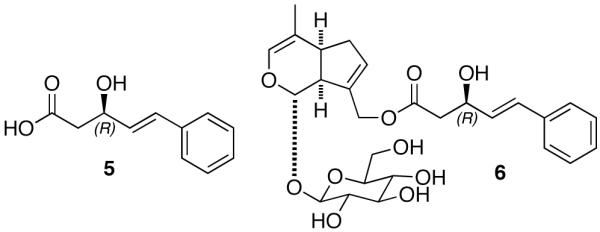
3(R)-hydroxy-5-phenyl-4(E)-pentenoic acid (Hyppa) (5) and marinoid E (6) isolated from A. marina.25
Our successful initial experiments with the 12 marine-derived fungi explored here, extend previous parallel results where spiking cultures with the potent actin inhibitor Jasplakinolide8b upregulated different biosynthetic products. The discovery of EGM-556 (1) represents an exciting outcome of expressing hitherto silent HPN biosynthetic genes. In the future, we will report on parallel outcomes based on additional HDAC spiking experiments to extend the biosynthetic products from well-studied fungi including Aspergillus and Penicillium, but isolated from marine sources.
Supplementary Material
Acknowledgement
This work was supported by a supplement to NIH grant U19 CA 052955 to PC, a grant to HV and PC under the American Recovery and Reinvestment Act (ARRA). We wish to thank William Fenical, Scripps Institute of Oceanography, UC San Diego, for collection of sediments from which M. trigonosporus was isolated.
Footnotes
Supporting Information Available: 14 Figures and 4 Tables are provided including the experimental procedures, 1D and 2D NMR data of 1 and 2 and chromatograms obtained in determining the absolute configuration of the amino acids. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- (1).(a) Saleem M, Ali MS, Hussain S, Jabbar A, Ashraf M, Lee YS. Nat. Prod. Rep. 2007;24:1142–1152. doi: 10.1039/b607254m. [DOI] [PubMed] [Google Scholar]; (b) Ebel R. In: Comprehensive Natural Products II: Chemistry and Biology. 1 ed. Moore BS, Crews P, editors. Vol. 2. Elsevier; 2010. pp. 223–262. Ch. 2. [Google Scholar]
- (2).(a) Tsukamoto S, Umaoka H, Yoshikawa K, Ikeda T, Hirota H. J. Nat. Prod. 2010;73:1438–1440. doi: 10.1021/np1002498. [DOI] [PubMed] [Google Scholar]; (b) Zhao WY, Zhu TJ, Fan GT, Liu HB, Fang YC, Gu QQ, Zhu WM. Nat. Prod. Res. 2010;24:953–957. doi: 10.1080/14786410902726134. [DOI] [PubMed] [Google Scholar]; (c) Tsukamoto S, Kawabata T, Kato H, Greshock TJ, Hirota H, Ohta T, Williams RM. Org. Lett. 2009;11:1297–1300. doi: 10.1021/ol900071c. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Wu Q-X, Crews MS, Drašković M, Sohn J, Johnson TA, Tenney K, Valeriote FA, Yao X-J, Bjeldanes LF, Crews P. Org. Lett. 2010;12:4458–4461. doi: 10.1021/ol101396n. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Fenical W, Jensen PR, Cheng XC. Halimide, a cytotoxic marine natural product, and derivatives thereof. US 6,069,146. 2000; (f) Abbas HK, editor. Aflatoxin and food safety. CRC Press; 2005. [Google Scholar]
- (3).(a) Drawz SM, Bonomo RA. Clin. Microbiol. Rev. 2010;23:160–201. doi: 10.1128/CMR.00037-09. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Barrios-González J, Miranda R. Appl. Microbiol. Biotechnol. 2010;85:869–883. doi: 10.1007/s00253-009-2239-6. [DOI] [PubMed] [Google Scholar]; (c) Gautschi JT, Amagata T, Amagata A, Valeriote FA, Mooberry SL, Crews P. J. Nat. Prod. 2004;67:362–367. doi: 10.1021/np030388m. [DOI] [PubMed] [Google Scholar]; (d) de Silva ED, Geiermann A-S, Mitova MI, Kuegler P, Blunt JW, Cole ALJ, Munro MHG. J. Nat. Prod. 2009;72:477–479. doi: 10.1021/np800627f. [DOI] [PubMed] [Google Scholar]
- (4).Ansorge WJ. New Biotechnol. 2009;25:195–203. doi: 10.1016/j.nbt.2008.12.009. [DOI] [PubMed] [Google Scholar]
- (5).Challis GL. J. Med. Chem. 2008;51:2618–2628. doi: 10.1021/jm700948z. [DOI] [PubMed] [Google Scholar]
- (6).(a) Chiang Y-M, Szewczyk E, Davidson AD, Keller N, Oakley BR, Wang CCC. J. Am. Chem. Soc. 2009;131:2965–2970. doi: 10.1021/ja8088185. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Ding Y, de Wet JR, Cavalcoli J, Li S, Greshock TJ, Miller KA, Finefield JM, Sunderhaus JD, McAfoos TJ, Tsukamoto S, Williams RM, Sherman DH. J. Am. Chem. Soc. 2010;132:12733–12740. doi: 10.1021/ja1049302. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Cuomo CA, Birren BW. In: Methods Enzymol. Jonathan W, Christine G, Gerald RF, editors. Vol. 470. Academic Press; 2010. pp. 833–855. [DOI] [PubMed] [Google Scholar]; (d) Schroeckh V, Scherlach K, Nützmann H-W, Shelest E, Schmidt-Heck W, Schuemann J, Martin K, Hertweck C, Brakhage AA. Proc. Nat. Acad. Sci. 2009;106:14558–14563. doi: 10.1073/pnas.0901870106. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Wang M, Liu S, Li Y, Xu R, Lu C, Shen Y. Curr. Microbiol. 2010;61:254–260. doi: 10.1007/s00284-010-9604-7. [DOI] [PubMed] [Google Scholar]; (f) Yin W-B, Grundmann A, Cheng J, Li S-M. J. Biol. Chem. 2009;284:100–109. doi: 10.1074/jbc.M807606200. [DOI] [PubMed] [Google Scholar]; (g) Crawford JM, Vagstad AL, Ehrlich KC, Townsend CA. Bioorg. Chem. 2008;36:16–22. doi: 10.1016/j.bioorg.2007.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).(a) Ricke DO, Wang S, Cai R, Cohen D. Curr. Opin. Chem. Biol. 2006;10:303–308. doi: 10.1016/j.cbpa.2006.06.024. [DOI] [PubMed] [Google Scholar]; (b) Van Lanen SG, Shen B. Curr. Opin. Microbiol. 2006;9:252–260. doi: 10.1016/j.mib.2006.04.002. [DOI] [PubMed] [Google Scholar]; (c) Bergmann S, Schumann J, Scherlach K, Lange C, Brakhage AA, Hertweck C. Nat. Chem. Biol. 2007;3:213–217. doi: 10.1038/nchembio869. [DOI] [PubMed] [Google Scholar]
- (8).(a) Chiang Y-M, Lee K-H, Sanchez JF, Keller NP, Wang CCC. Nat. Prod. Commun. 2009;4:1505–1510. [PMC free article] [PubMed] [Google Scholar]; (b) Christian OE, Compton J, Christian KR, Mooberry SL, Valeriote FA, Crews P. J. Nat. Prod. 2005;68:1592–1597. doi: 10.1021/np050293f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).(a) Keats Shwab E, Keller NP. Mycol. Res. 2008;112:225–230. doi: 10.1016/j.mycres.2007.08.021. [DOI] [PubMed] [Google Scholar]; (b) Bok JW, Keller NP. Eukaryot. Cell. 2004;3:527–535. doi: 10.1128/EC.3.2.527-535.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Stadler M, Keller NP. Mycol. Res. 2008;112:127–130. doi: 10.1016/j.mycres.2007.12.002. [DOI] [PubMed] [Google Scholar]
- (10).(a) Hertweck C. Nat. Chem. Biol. 2009;5:450–452. doi: 10.1038/nchembio0709-450. [DOI] [PubMed] [Google Scholar]; (b) Scherlach K, Hertweck C. Org. Biomol. Chem. 2009;7:1753–1760. doi: 10.1039/b821578b. [DOI] [PubMed] [Google Scholar]; (c) Keats Shwab E, Bok JW, Tribus M, Galehr J, Graessle S, Keller NP. Eukaryot. Cell. 2007;6:1656–1664. doi: 10.1128/EC.00186-07. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Bok JW, Chiang Y-M, Szewczyk E, Reyes-Dominguez Y, Davidson AD, Sanchez JF, Lo H-C, Watanabe K, Strauss J, Oakley BR, Wang CCC, Keller NP. Nat. Chem. Biol. 2009;5:462–464. doi: 10.1038/nchembio.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).(a) Cichewicz RH. Nat. Prod. Rep. 2010;27:11–22. doi: 10.1039/b920860g. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Fisch K, Gillaspy A, Gipson M, Henrikson J, Hoover A, Jackson L, Najar F, Wägele H, Cichewicz R. J. Ind. Microbiol. Biotechnol. 2009;36:1199–1213. doi: 10.1007/s10295-009-0601-4. [DOI] [PubMed] [Google Scholar]; (c) Henrikson JC, Hoover AR, Joyner PM, Cichewicz RH. Org. Biomol. Chem. 2009;7:435–438. doi: 10.1039/b819208a. [DOI] [PubMed] [Google Scholar]; (d) Williams RB, Henrikson JC, Hoover AR, Lee AE, Cichewicz RH. Org. Biomol. Chem. 2008;6:1895–1897. doi: 10.1039/b804701d. [DOI] [PubMed] [Google Scholar]
- (12).(a) Marks PA. Oncogene. 2007;26:1351–1356. doi: 10.1038/sj.onc.1210204. [DOI] [PubMed] [Google Scholar]; (b) Wiech NL, Fisher JF, Helquist P, Wiest O. Curr. Top. Med. Chem. 2009;9:257–271. doi: 10.2174/156802609788085241. [DOI] [PubMed] [Google Scholar]
- (13).Raj K, Mufti GJ. Ther. Clin. Risk Manag. 2006;2:377–388. doi: 10.2147/tcrm.2006.2.4.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Dr. E. H. Thompson and Dr. B. L.Wickes at the Department of Pathology UT Health Science Center at San Antonio, kindly provided taxonomic identification of fungal strain 098059A placing it within the Microascus/Scopulariopsis genera (details on the identification of the strain described in the Identification of Fungal strain 098059A and Table S2, Supporting Information)
- (15).(a) Mohammedi I, Piens MA, Audigier-Valette C, Gantier JC, Argaud L, Martin O, Robert D. Eur. J. Clin. Microbiol. Infect. Dis. 2004;23:215–217. doi: 10.1007/s10096-003-1096-y. [DOI] [PubMed] [Google Scholar]; (b) Baddley JW, Moser SA, Sutton DA, Pappas PG. J. Clin. Microbiol. 2000;38:395–397. doi: 10.1128/jcm.38.1.395-397.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Glastrup J. Journal of Chromatography A. 1998;827:133–136. [Google Scholar]
- (17).(a) Bhushan R, Brückner H. Amino Acids. 2004;27:231–247. doi: 10.1007/s00726-004-0118-0. [DOI] [PubMed] [Google Scholar]; (b) Marfey P, Ottesen M. Carlsberg Res. Commun. 1984;49:585–590. [Google Scholar]
- (18).Capon RJ, Skene C, Stewart M, Ford J, O’Hair RAJ, Williams L, Lacey E, Gill JH, Heiland K, Friedel T. Org. Biomol. Chem. 2003;1:1856–1862. doi: 10.1039/b302306k. [DOI] [PubMed] [Google Scholar]
- (19).(a) Trost BM, Belletire JL, Godleski S, McDougal PG, Balkovec JM, Baldwin JJ, Christy ME, Ponticello GS, Varga SL, Springer JP. J. Org. Chem. 1986;51:2370–2374. [Google Scholar]; (b) Latypov SK, Seco JM, Quiñoá E, Riguera R. J. Am. Chem. Soc. 1998;120:877–882. [Google Scholar]
- (20).(a) Paizs B, Suhai S. Mass Spectrom. Rev. 2005;24:508–548. doi: 10.1002/mas.20024. [DOI] [PubMed] [Google Scholar]; (b) Jegorov A, Paizs B, Žabka M, Kuzma M, Havlíček V. Eur. J. Mass Spectrom. 2003;9:105–116. doi: 10.1255/ejms.531. [DOI] [PubMed] [Google Scholar]; (c) Ngoka LCM, Gross ML. J. Am. Soc. Mass Spectrom. 1999;10:732–746. doi: 10.1016/S1044-0305(99)00049-5. [DOI] [PubMed] [Google Scholar]; (d) Williams SM, Brodbelt JS. J. Am. Soc. Mass Spectrom. 2004;15:1039–1054. doi: 10.1016/j.jasms.2004.03.015. [DOI] [PubMed] [Google Scholar]
- (21).(a) Oku N, Kawabata K, Adachi K, Katsuta A, Shizuri Y. J. Antibiot. 2008;61:11–17. doi: 10.1038/ja.2008.103. [DOI] [PubMed] [Google Scholar]; (b) Tanabe K, Lamping E, Adachi K, Takano Y, Kawabata K, Shizuri Y, Niimi M, Uehara Y. Biochem. Biophys. Res. Commun. 2007;364:990–995. doi: 10.1016/j.bbrc.2007.10.110. [DOI] [PubMed] [Google Scholar]
- (22).(a) Ueda H, Nakajima H, Hori Y, Fujita T, Nishimura M, Goto T, Okuhara M. J. Antibiot. 1994;47:301–310. doi: 10.7164/antibiotics.47.301. [DOI] [PubMed] [Google Scholar]; (b) Sasakawa Y, Naoe Y, Inoue T, Sasakawa T, Matsuo M, Manda T, Mutoh S. Biochem. Pharmacol. 2002;64:1079–1090. doi: 10.1016/s0006-2952(02)01261-3. [DOI] [PubMed] [Google Scholar]
- (23).(a) Golakoti T, Ohtani I, Patterson GML, Moore RE, Corbett TH, Valeriote FA, Demchik L. J. Am. Chem. Soc. 1994;116:4729–4737. [Google Scholar]; (b) Golakoti T, Ogino J, Heltzel CE, Le Husebo T, Jensen CM, Larsen LK, Patterson GML, Moore RE, Mooberry SL. J. Am. Chem. Soc. 1995;117:12030–12049. [Google Scholar]
- (24).Magarvey NA, Beck ZQ, Golakoti T, Ding Y, Huber U, Hemscheidt TK, Abelson D, Moore RE, Sherman DH. ACS Chem. Biol. 2006;1:766–779. doi: 10.1021/cb6004307. [DOI] [PubMed] [Google Scholar]
- (25).Sun Y, Ouyang J, Deng Z, Li Q, Lin W. Magn. Reson. Chem. 2008;46:638–642. doi: 10.1002/mrc.2224. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.