Abstract
The deubiquitinating enzyme A20 (also known as TNFAIP3) is essential for maintaining immune homeostasis. A20 is a key regulator of inflammatory, antiviral and apoptotic signaling pathways. Here, we review recent advances illustrating the role of A20 as an essential negative regulator of inflammatory and antiviral signaling.
Keywords: A20, NF-κB, IKK, ubiquitin, antiviral signaling, inflammation
1) Introduction
The innate arm of the immune system plays a critical role as a first line of defense against invading pathogens including bacteria, fungi, and viruses. This immune response is initiated upon the recognition of highly invariant microbial components common to pathogens (aptly termed microbial-associated molecular patterns (MAMPs)) by germline-encoded sensors called pattern recognition receptors (PRRs) [1]. Receptor ligation yields the activation of key anti-microbial programs and further initiates a secondary adaptive immune response which ultimately results in pathogen elimination. Because anti-microbial programs typically utilize pro-inflammatory cytokines to serve as mediators of host defense, tight regulation is critical upon pathogen clearance as excessive inflammation is associated with numerous autoimmune pathologies and cancers. A20 (also known as TNFAIP3) is a key negative regulator of the inflammatory response and here we review the role of A20 in regulating three essential innate immune functions: 1) the inflammatory response, 2) the antiviral program and 3) programmed cell death. We further provide insight into potential mechanisms by which A20 functions in the context of a multi-protein ubiquitin-editing complex to attenuate inflammatory signaling.
2) Innate Signaling via Polyubiquitination
Ubiquitination is an enzymatic process that results in the covalent linkage of substrate proteins with a ubiquitin moiety-a small peptide containing 76 amino acids, in a post-translational manner. The process of protein ubiquitination is carried out in a three-step enzymatic cascade where a ubiquitin activating protein (E1) initially forms a high energy thioester linkage with a ubiquitin molecule, which is then transferred to a ubiquitin conjugating enzyme (E2) in a second step of activation [2]. Lastly, in conjunction with a ubiquitin ligase (E3), the ubiquitin molecule is then targeted to the epsilon amino group of a lysine residue on the substrate protein forming an isopeptide bond, resulting in monoubiquitination of the target protein [2]. Further addition of ubiquitin molecules onto the previously conjugated ubiquitin via the above enzymatic cascade results in the formation of a polyubiquitin chain. As the ubiquitin peptide itself contains seven lysine residues, the manner by which each ubiquitin moiety is linked to each other within a polyubiquitin chain is critical because the topological structure attributed to various polyubiquitin chain types results in disparate outcomes. Accordingly, ubiquitin molecules linked via lysine at position 48 (K48) within the ubiquitin chain typically target the substrate for proteasomal-mediated degradation [3]. Alternatively, ubiquitin molecules linked via lysine at position 63 (K63) yield proteasome-independent outcomes and often potentiate signaling by serving as signaling scaffolds that recruit proteins bearing ubiquitin-binding domains [4]. Like many other post-translational modifications, ubiquitination is a reversible process, therefore editing of ubiquitin chains provides an additional layer of regulation for protein function. Indeed, K63-linked polyubiquitination of substrate proteins has pervaded many biological programs including innate immune pathways [4], and are subject to additional regulation by isopeptidases or deubiquitinases (DUBs) that disassemble ubiquitin chains [5].
3) Ubiquitin-Editing Function of A20
In most cell types, A20 functions as a negative feedback regulator of inflammatory signaling pathways. As such, A20 is transcriptionally upregulated by Nuclear Factor-kappa B (NF-κB), a family of evolutionarily conserved transcription factors critical in mediating the pro-inflammatory response. A20 is a 90 kDa protein that belongs to the ovarian tumor family of DUBs where it contains an N-terminal cysteine protease/DUB domain [6]. Because A20 also harbors seven carboxy-terminal zinc fingers, of which zinc finger 4 (ZNF4) confers E3 ubiquitin ligase activity, it can be classified as a dual function ubiquitin-editing enzyme [7]. A20 has been shown to cleave K63-linked ubiquitin residues via its DUB domain, as well as to conjugate K48-linked ubiquitin chains via its zinc finger domain [7]. However, two recent reports have described an additional mechanism for substrate inactivation whereby A20 disrupts the interactions between essential components of the ubiquitination cascade [8, 9]. Specifically, A20 targets E3 ubiquitin ligase-substrate and/or E2 ubiquitin conjugating enzyme-E3 ubiquitin ligase interactions, and thus attenuates the activation of specific E3 ubiquitin ligases such as TNFα Receptor Associated Factor 6 (TRAF6) [8, 9]. It was additionally shown that A20 could further promote K48-linked ubiquitination and proteasomal degradation of the E2 enzyme responsible for facilitating K63-linked ubiquitin conjugation as a measure to fully terminate the assembly and formation of K63-linked ubiquitin chains [9]. Nonetheless, this dual nature of A20 with a capacity to disassociate K63-linked ubiquitin chains to initially block signal transduction, and then assemble K48-linked ubiquitin chains to promote degradation of key signaling molecules, establishes a novel mechanism to modulate innate immune pathways.
4) A20-Mediated Inhibition of Inflammation
The induction of pro-inflammatory cytokines is controlled in part by the NF-κB family of transcription factors. NF-κB dimers are sequestered in the cytoplasm as inactive subunits by IκB proteins that contain ankyrin repeat domains [10]. In response to specific stimuli, including pro-inflammatory cytokines or microbial infection, the IκB kinase (IKK) signalosome, consisting of IKKα, IKKβ and IKKγ (also known as NEMO), phosphorylates IκB proteins thus triggering their ubiquitination and proteasomal degradation allowing NF-κB to enter the nucleus and activate target genes [11]. In feedback regulation, A20 associates with the IKK complex and further undergoes IKKβ-dependent phosphorylation resulting in optimal effector function [12]. A20 targets key upstream components such as RIP1 and TRAF6 that are critical in signaling to the IKK complex, ultimately resulting in the inhibition of inflammatory pathways [7, 13]. Indeed, mice lacking A20 die prematurely due to severe multi-organ inflammation and cachexia [14], underscoring an essential role of this gene in limiting inflammatory signaling.
Upon engagement of TNFα receptor 1 (TNFR1) by TNFα, Receptor Interacting Protein 1 (RIP1) undergoes K63-linked polyubiquitination in lipid rafts, where its ubiquitin modification is an essential prerequisite for IKK complex activation and downstream NF-κB signaling [15, 16]. To blunt signaling in a negative feedback loop, A20 is recruited to RIP1 and promotes RIP1 deubiquitination via its OTU domain [7]. In addition, A20 targets the RIP1 E3 ligase complex consisting of cellular inhibitor of apoptosis 1/2 (cIAP1/2), TRAF2 and possibly TRAF5 [9]. A20 antagonizes E3 ligase activity of this complex by disrupting the associations between cIAP1/2, TRAF2 and the E2 enzyme Ubc13 [9]. A20 sequentially assembles K48-linked polyubiquitin chains onto Ubc13 resulting in its proteasomal degradation and further inhibition of NF-κB signaling [9]. In an additional step to ultimately terminate TNFR1 signaling to NF-κB, RIP1 also undergoes K48-linked polyubiquitination and proteasomal-mediated degradation [7]. However, it is currently unclear if A20 is the E3 ligase that conjugates K48-linked polyubiquitin chains onto RIP1 although the zinc finger domain of A20 is required for RIP1 degradation [7]. In addition, the E3 ubiquitin ligases Itch (also known as AIP4) and RNF11 have also been shown to facilitate RIP1 degradation in an E3 ligase-dependent manner [17, 18]. Furthermore, the E3 ubiquitin ligase TRIAD3A inhibits NF-κB signaling by degrading RIP1 [19], however it remains to be determined if TRIAD3A is a component of the A20 ubiquitin-editing complex. Interestingly, both Itch and RNF11 are dispensable in TNFα-mediated degradation of Ubc13 [9]. Because the above E3 ligases appear to have nonredundant functions, it will be of great interest to fully elucidate their precise roles in controlling the inflammatory response.
The IL-1®R and Toll-like receptors (TLRs) share structurally similar intracellular domains and consequently signal downstream to the IKK complex utilizing common adaptor molecules [20]. Ligation of these receptors results in oligomerization and K63-linked auto-ubiquitination of the E3 ubiquitin ligase TRAF6 and subsequent activation of the IKK signalosome [21]. Similar to RIP1 targeting in the TNFR pathway, A20 is recruited to TRAF6 and disassembles K63-linked polyubiquitin chains [13]. In addition, A20 attenuates TRAF6 activation by disrupting its interactions with its cognate E2 conjugating enzymes Ubc13 and UbcH5 (Fig. 1) [9]. Likewise, in a second step of inhibition, A20 further catalyzes K48-linked polyubiquitin chains onto Ubc13 and UbcH5 (but not TRAF6) yielding their degradation to completely terminate signaling [9]. Because Itch and RNF11 also play critical roles in mediating TRAF6 deubiquitination and have dispensable roles in Ubc13 and UbcH5 degradation, it remains to be determined exactly how these E3 ubiquitin ligases contribute towards A20-mediated inhibition of TRAF6-dependent signaling. One possibility is that the Itch and RNF11 E3 ligases may not be true feedback inhibitors, but rather ligand-triggered molecules that nucleate substrates to rapidly drive feedback inhibition by initiating the recruitment of A20.
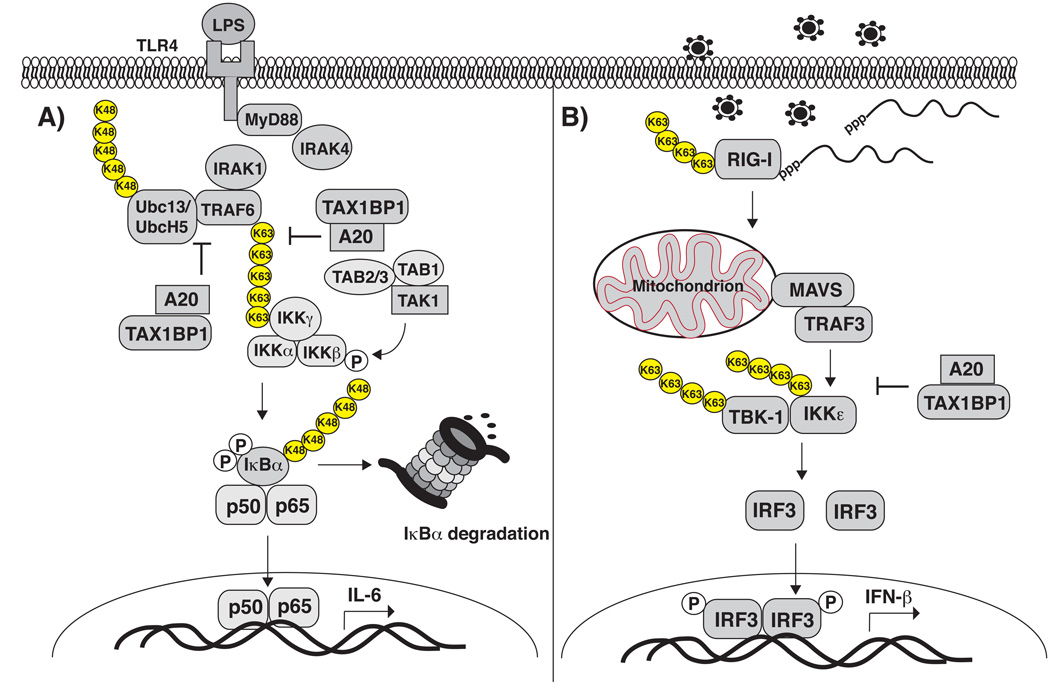
Figure 1. The role of A20 in the inhibition of NF-κB and antiviral signaling pathways.
A) A20 inhibits TLR4-mediated NF-κB activation by targeting TRAF6 for inactivation. A20 may directly disassemble K63-linked polyubiquitin chains from TRAF6 via its OTU domain. In addition, A20 disrupts the interactions between TRAF6 and its E2 enzymes Ubc13 and UbcH5, and later triggers the K48-linked polyubiquitination and degradation of the E2 enzymes. A20 is dependent on the ubiquitin-binding adaptor molecule TAX1BP1 to disrupt interactions between TRAF6 and its E2 enzymes Ubc13 and UbcH5. B) A20 inhibits RIG-I-dependent antiviral signaling by disrupting TRAF3 binding to TBK1 and IKKε, resulting in the inhibition of K63-linked polyubiquitination of TBK1 and IKKε. A20 also requires TAX1BP1 to inhibit antiviral signaling.
Although TRAF6 clearly undergoes K63-linked polyubiquitination in the IL-1/TLR4 pathway, the functional importance of TRAF6 auto-ubiquitination remains unclear. Lamothe et al. mapped the auto-ubiquitination site on TRAF6 to Lys124, and demonstrated that this lysine residue is critical for IKK activation and osteoclast differentiation [22]. However, an independent study revealed that a lysine-deficient TRAF6 mutant remains fully competent to activate TAK1 and NF-κB suggesting that TRAF6 auto-ubiquitination may not be essential for NF-κB activation in the IL-1R/TLR pathway [23]. If this is indeed the case, it is puzzling why TRAF6 is subject to ubiquitination and deubiquitination after receptor stimulation. It is plausible that K63-linked polyubiquitination of TRAF6 serves as a signal or homing marker for recruiting A20 to TRAF6. Indeed, A20 requires the ubiquitin sensor and adaptor molecule TAX1BP1 to target to K63-linked polyubiquitinated substrates [24, 25]. This can be described in a model whereby A20 is recruited to Ubc13 and UbcH5 indirectly via TRAF6 as part of a TRAF6/Ubc13/UbcH5 complex (where only TRAF6 is K63-linked polyubiquitinated) via TAX1BP1 thereby allowing A20 to disrupt TRAF6-E2 binding and consequently disable TRAF6 E3 ligase function. Because TRAF6 E3 ligase activity is dependent on Ubc13 and UbcH5 for generating free polyubiquitin chains or ubiquitinating the regulatory kinase IRAK1 [26, 27], A20 targeting to ubiquitinated TRAF6 may be essential to disrupt TRAF6/Ubc13/UbcH5 interactions and thereby terminate NF-κB signaling. Interestingly, A20 also requires TAX1BP1 to target Ubc13 and UbcH5 although these E2s do not appear to be conjugated by K63-linked ubiquitin [9]. Because A20 facilitates the disruption between TRAF6 and Ubc13/UbcH5 in a manner dependent on TAX1BP1, it will be of interest to determine exactly how these E2s are targeted by A20 for K48-linked polyubiquitination since they are displaced from the TRAF6/TAX1BP1/A20 complex upon A20 recruitment.
Nucleotide Oligomerization Domain 2 (NOD2) is an intracellular receptor of the bacterial cell wall component muramyl dipeptide, which upon ligand binding also signals to NF-κB via the IKK complex [28, 29]. In NOD2-mediated signaling to IKK, the adaptor protein RIP2 (also known as RICK or Cardiak) is subject to K63-linked polyubiquitination via cIAP1/2 to drive NF-κB signaling [30, 31]. Polyubiquitinated RIP2 is then targeted by A20 for deubiquitination to terminate NF-κB activation [32]. In vitro studies suggested that A20 required its cysteine protease activity to directly cleave K63-linked polyubiquitin chains from RIP2, however a stable RIP2-A20 association was not detected, consistent with the observation that A20 does not further promote RIP2 degradation [32]. Because A20 can inhibit K63-linked polyubiquitination by targeting E2–E3 enzyme complexes, it is possible that the cIAP1/2-Ubc13/UbcH5 complex is first disrupted by A20, and then Ubc13/UbcH5 are further conjugated via K48-linked polyubiquitin chains to attenuate RIP2 K63-linked polyubiquitination and activation of NF-κB upon NOD2 stimulation. Interestingly, it was recently shown that the E3 ligase Itch served as a positive regulator of RIP2-mediated induction of Map kinase (MAPK) activation, as well as a negative regulator of RIP2-mediated activation of NF-κB [33]. Mechanistically, it was demonstrated that Itch triggered the K63-linked polyubiquitination of RIP2 to drive MAPK activation [33]. In this scenario, Itch may initially serve as an E3 ligase that conjugates K63-linked polyubiquitin chains onto RIP2 after receptor stimulation to activate MAPKs, including c-Jun N-terminal kinase (JNK). Because JNK phosphorylates Itch, in part to activate its E3 ligase function for assembling K48-linked polyubiquitin chains [34], a phosphorylation-dependent conformational change on Itch may facilitate its association with A20 and further mediate NF-κB inhibitory function. Indeed, the kinetics of Itch targeting to its substrates, at least in TNFR and TLR signaling, is more rapid than that of A20 targeting to the same complex [9], in accordance with the above model whereby Itch mediates disparate A20-independent and dependent roles. As in IL-1R/TLR signaling, substrates for K48-linked polyubiquitination by Itch in NOD2-dependent signaling however, have yet to be identified.
In inflammatory signaling pathways, A20 clearly serves as a negative feedback inhibitor that is induced by NF-κB transcription factors. In T cells however, A20 is constitutively expressed thus allowing for A20 to act as both a steady-state inhibitor as well as a feedback inhibitor during T-cell receptor (TCR) signaling to NF-κB [35]. Upon TCR ligation, a CARMA1-BCL10-MALT1 (CBM) signaling complex is assembled via PKCθ-dependent phosphorylation of CARMA1 [36]. The CBM complex then recruits the E3 ligase TRAF6 (and possibly TRAF2) which triggers the K63-linked polyubiquitination of MALT1, BCL10 and IKKγ [36–39]. Polyubiquitinated MALT1 likely serves as a scaffold to recruit the IKK complex via ubiquitin-binding domains present within IKKγ [40]. In early signaling, MALT1 functions as a paracaspase to cleave A20, thus compromising the effector function of A20 and allowing for downstream signal transmission to the IKK complex [41]. A20 also undergoes proteasomal degradation at early times upon antigen receptor signaling, but is induced at later times in a negative feedback loop and targets polyubiquitinated MALT1 for deubiquitination which attenuates IKK and NF-κB activation [42]. Because TRAF6 likely serves as the E3 ligase for MALT1 and Ubc13 has been demonstrated to be essential for TCR signaling to NF-κB [43], it is plausible that in addition to deubiquitinating MALT1, A20 may disrupt a MALT1-TRAF6/Ubc13 complex and/or the TRAF6/2-Ubc13 complex to further block MALT1 K63-linked polyubiquitination and promote Ubc13 degradation at later times. Like TRAF6 and RIP2, A20 does not target MALT1 for degradation during feedback inhibition, however CARMA1 has been demonstrated to undergo K48-linked polyubiquitination followed by proteasomal degradation in response to TCR stimulation, although A20 has not been directly implicated in this process [44]. Although the roles of Itch, RNF11 and TAX1BP1 have not been formally demonstrated in regulating TCR-mediated signaling to NF-κB, it is likely that these molecules contribute towards modulating A20 effector function in this pathway.
5) A20-Mediated Inhibition of Innate Antiviral Pathways
The Toll-like receptors and Rig-I like helicases are pattern recognition receptors that recognize MAMPs derived from viral components to drive the activation of antiviral programs during viral infection. This antiviral response is mediated in part by the production of both antiviral and pro-inflammatory cytokines regulated by the Interferon Regulatory Factor 3/7 (IRF3/7) and NF-κB transcription factors, respectively [45]. As activation of both transcription factors in the context of virus infection requires the K63-linked protein polyubiquitination of key signaling components, it is not surprising that A20 plays an important role in terminating the antiviral immune response by inhibiting ubiquitin-dependent signal transduction. NF-κB activation during antiviral signaling is mediated by TRAF6 and is likely targeted by A20 in negative feedback regulation analogous to IL-1®R and TLR4 signaling inhibition. The mechanism by which A20 inhibits IRF3 activation however, has remained elusive until recently, although several groups had reported that A20 was a potent inhibitor of innate antiviral signaling [46–48]. Interestingly, it was shown that A20 did not require its DUB domain, but rather its carboxy-terminal zinc finger domains to inhibit signaling to IRF3 [47]. This result suggested that A20 inhibited antiviral signaling independently of protein deubiquitination. Consistent with these results, we found that A20 (also in a manner independent of its DUB function), in conjunction with TAX1BP1, disrupted TRAF3-dependent K63-linked polyubiquitination of TBK1 and IKKi kinases to inhibit signaling to IRF3 (Fig. 1) [8]. A20 also required TAX1BP1 to target to TBK1/IKKi and block TBK1/IKKi-dependent activation of IRF3 [8]. Because overexpression of TAX1BP1 was shown to be sufficient to block signaling to IRF3 by disrupting the TRAF3-TBK1/IKKi complex, it is plausible that TAX1BP1 serves as an early effector in blocking the antiviral response, whereas A20 functions at later times by targeting UbcH5 for proteasomal degradation, since the role of this E2 has been recently demonstrated in facilitating signaling to IRF3 [49]. The Itch and TRIAD3A E3 ubiquitin ligases have also been shown to play essential roles in terminating IRF3 activation by targeting the mitochondrial antiviral signaling adaptor (MAVS; also known as IPS-1, Cardif or VISA) and the E3 ligase TRAF3, respectively, for degradation [50, 51]. Because TRIAD3A had previously been shown to target and degrade RIP1 [19], which functions as a key signaling adaptor in TLR-independent signaling to IRF3, it would be interesting to determine if TRIAD3A also targets RIP1 in the context of antiviral signaling. It is currently unknown if TAX1BP1 and A20 additionally cooperate with either Itch or TRIAD3A to modulate their effector functions in antiviral signaling.
An additional report further identified the IRF7 transcription factor as a substrate for A20-mediated inhibition of antiviral signaling [52]. However, similar to the effects of A20 on TBK1 and IKKi, A20 was found not to directly deubiquitinate IRF7 [52]. Because it was previously reported that TRAF6 is the E3 ligase that assembles K63-linked polyubiquitin chains onto IRF7 [53], it may be possible that A20 disrupts TRAF6-dependent ubiquitination of IRF7 to inhibit its activation. Unlike A20-mediated inhibition of IRF3 however, A20 was instead shown to require its DUB domain but not its carboxy-terminal zinc fingers to block IRF7 activity [52]. Indeed, the DUB domain of A20 was determined to be important for IRF7 association, thus providing evidence for its requirement in blocking IRF7 [52]. Furthermore, since the catalytically inactive DUB mutant of A20 is defective in its capacity to interact with UbcH5/Ubc13 (likely due to a change in conformation) [9], and thus fails to disrupt its association with TRAF6 (and consequently TRAF6 E3 ligase capability), it is in good agreement that A20 would require a catalytically intact DUB domain to block TRAF6-dependent polyubiquitination and activation of IRF7. However, since the A20 zinc fingers and in particular ZnF4, which has E3 ligase acticity, was shown to be dispensable for IRF7 inhibition, it remains to be determined if A20 has a physiological substrate for K48-linked polyubiquitin targeting in antiviral signaling. Moreover, since ZnF4 of A20 is essential for interacting with TAX1BP1, the requirement of TAX1BP1 in regulating A20 effector function in IRF7 inhibition is currently unclear and further suggests a TAX1BP1-independent role for A20 in blocking IRF7.
6) A20-Mediated Inhibition of Cell Death
Programmed cell death or apoptosis is a common mechanism used by the host to eliminate virus-infected cells or to limit the number of activated lymphoid cells to maintain immune homeostasis. The initiation of apoptotic programs involves the activation of effector caspases that execute cell death. In apoptotic signaling via death receptors, upstream initiator caspases (i.e. caspase 8) form a death inducing signaling complex (DISC) and are activated in a manner dependent on the ubiquitin conjugation machinery [54]. Incidentally, A20 was initially identified as an inhibitor of programmed cell death however, whether A20 modulates ubiquitin in death receptor signaling as a sole mechanism of inhibition is not entirely clear. In this context, K63-linked polyubiquitination of caspase 8 has been shown to be antagonized by A20 to block death receptor-induced apoptosis [55]. Like RIP1, caspase 8 is also subject to K48-linked polyubiquitination, however it is currently unknown if A20 has a direct role in targeting caspase 8 for degradative ubiquitination. In the TNFR1 pathway, A20 prevents cell death by disrupting the recruitment of TRADD and RIP1 to the receptor signaling complex [56]. It was recently shown that the A20 binding protein ABIN1 functions as a ubiquitin sensor to block apoptosis, but interestingly both A20 and ABIN1 were demonstrated to operate independently of each other in inhibiting cell death [57]. Although ABIN1 was shown to require its ubiquitin binding domain to block apoptosis, the mechanism of inhibition was shown to be via disruption of the DISC complex [57]. Because a disruption-based mechanism of the DISC complex was also attributed to A20 in mediating the anti-apoptotic response, it is currently unclear whether A20 utilizes multiple mechanisms to achieve effector function.
The TAX1BP1 adaptor, in addition to regulating A20 function in both anti-inflammatory and antiviral pathways has also been shown to coordinate A20 anti-apoptotic activities [58]. Indeed, it was shown that antisense TAX1BP1 blocks the ability of A20 to inhibit cell death in certain cell types, suggesting that TAX1BP1 and A20 function cooperatively to inhibit apoptosis [58]. Because RIP1 K63-linked polyubiquitination is enhanced during TNFR1 stimulation in both A20 and TAX1BP1-deficient cells, and K63-linked polyubiquitination of RIP1 has been shown to be a critical step in blocking the formation of the DISC complex [59], it is not yet clear why cells lacking either A20 or TAX1BP1 are exquisitely sensitive to cell death. Adding further complexity into the role of A20 in regulating cell survival is the E3 ligase Itch. Although Itch cooperates with A20 and TAX1BP1 to terminate NF-κB signaling [17], and was further shown to inhibit antiviral signaling [51], Itch does not inhibit cell death but rather promotes the apoptotic response during death receptor signaling by targeting the pseudo-caspase/DISC inhibitor c-FLIP [34]. Because Itch is important in targeting RIP1 for proteasomal degradation following TNF receptor engagement, and RIP1 is also a component of the DISC complex, the connection between Itch, RIP1 and apoptosis still requires further investigation.
A20 has also been recently linked to blocking autophagy, a form of programmed cell death that operates independently of death receptors under nutrient deprivation or innate immune activation [60]. During TLR4 or IL-1R activation, beclin-1 is subject to TRAF6-dependent K63-linked polyubiquitination, thought to facilitate beclin-1 disassociation from the anti-apoptotic protein Bcl-2, and further allow for self-association via its ubiquitin binding domain, ultimately promoting the autophagic process [61]. In feedback inhibition, A20 targets beclin-1 for deubiquitination and consequently inhibits autophagosome formation [61]. Although in vitro studies have suggested that A20 targets beclin-1 rather than TRAF6 to inhibit autophagy, the exact physiological substrate or target for A20 will require further investigation since TRAF6, and likely Ubc13/UbcH5, are part of a beclin-1 containing complex. Because TAX1BP1, Itch and RNF11 have been demonstrated to modulate A20 effector function in other signaling pathways, dissecting their roles in the autophagic process may provide further insight into how A20 regulates this and other innate immune pathways.
7) Conclusions and Perspective
The important role of A20 in restricting inflammation is underscored by numerous polymorphisms found within the A20 gene locus in nearly every human autoimmune disease [62]. A20 is also an important tumor suppressor in both Hodgkin and non-Hodgkin subtypes of B-cell lymphomas [63]. Although most studies have focused on the role of A20 in the inhibition of NF-κB and inflammatory signaling, little is known about how A20 inhibits cell death. This may be particularly relevant in certain cancers such as breast cancer or glioma where A20 is overexpressed and exerts an anti-apoptotic effect [64, 65]. In future studies, determining the precise role and mechanisms by which A20 inhibits cell death promises to be an exciting avenue of research.
Acknowledgements
The laboratory of E.W.H. is supported by NIH grants PO1CA128115, RO1CA135362 and RO1GM083143.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kawai T, Akira S. The roles of TLRs, RLRs and NLRs in pathogen recognition. Int Immunol. 2009;21:317–337. doi: 10.1093/intimm/dxp017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kerscher O, Felberbaum R, Hochstrasser M. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu Rev Cell Dev Biol. 2006;22:159–180. doi: 10.1146/annurev.cellbio.22.010605.093503. [DOI] [PubMed] [Google Scholar]
- 3.Varshavsky A. The ubiquitin system. Trends Biochem Sci. 1997;22:383–387. doi: 10.1016/s0968-0004(97)01122-5. [DOI] [PubMed] [Google Scholar]
- 4.Bhoj VG, Chen ZJ. Ubiquitylation in innate and adaptive immunity. Nature. 2009;458:430–437. doi: 10.1038/nature07959. [DOI] [PubMed] [Google Scholar]
- 5.Reyes-Turcu FE, Ventii KH, Wilkinson KD. Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu Rev Biochem. 2009;78:363–397. doi: 10.1146/annurev.biochem.78.082307.091526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Evans PC, Ovaa H, Hamon M, Kilshaw PJ, Hamm S, Bauer S, Ploegh HL, Smith TS. Zinc-finger protein A20, a regulator of inflammation and cell survival, has de-ubiquitinating activity. Biochem J. 2004;378:727–734. doi: 10.1042/BJ20031377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wertz IE, O'Rourke KM, Zhou H, Eby M, Aravind L, Seshagiri S, Wu P, Wiesmann C, Baker R, Boone DL, Ma A, Koonin EV, Dixit VM. De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-κB signalling. Nature. 2004;430:694–699. doi: 10.1038/nature02794. [DOI] [PubMed] [Google Scholar]
- 8.Parvatiyar K, Barber GN, Harhaj EW. TAX1BP1 and A20 inhibit antiviral signaling by targeting TBK1-IKKi kinases. J Biol Chem. 2010;285:14999–15009. doi: 10.1074/jbc.M110.109819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shembade N, Ma A, Harhaj EW. Inhibition of NF-κB signaling by A20 through disruption of ubiquitin enzyme complexes. Science. 2010;327:1135–1139. doi: 10.1126/science.1182364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karin M, Ben-Neriah Y. Phosphorylation meets ubiquitination: the control of NF-κB activity. Annu Rev Immunol. 2000;18:621–663. doi: 10.1146/annurev.immunol.18.1.621. [DOI] [PubMed] [Google Scholar]
- 11.Hayden MS, Ghosh S. Signaling to NF-κB. Genes Dev. 2004;18:2195–2224. doi: 10.1101/gad.1228704. [DOI] [PubMed] [Google Scholar]
- 12.Hutti JE, Turk BE, Asara JM, Ma A, Cantley LC, Abbott DW. IκB Kinase β Phosphorylates the K63 Deubiquitinase A20 To Cause Feedback Inhibition of the NF-κB Pathway. Mol Cell Biol. 2007;27:7451–7461. doi: 10.1128/MCB.01101-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boone DL, Turer EE, Lee EG, Ahmad RC, Wheeler MT, Tsui C, Hurley P, Chien M, Chai S, Hitotsumatsu O, McNally E, Pickart C, Ma A. The ubiquitin-modifying enzyme A20 is required for termination of Toll-like receptor responses. Nat Immunol. 2004;5:1052–1060. doi: 10.1038/ni1110. [DOI] [PubMed] [Google Scholar]
- 14.Lee EG, Boone DL, Chai S, Libby SL, Chien M, Lodolce JP, Ma A. Failure to regulate TNF-induced NF-κB and cell death responses in A20-deficient mice. Science. 2000;289:2350–2354. doi: 10.1126/science.289.5488.2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ea CK, Deng L, Xia ZP, Pineda G, Chen ZJ. Activation of IKK by TNFα requires site-specific ubiquitination of RIP1 and polyubiquitin binding by NEMO. Mol Cell. 2006;22:245–257. doi: 10.1016/j.molcel.2006.03.026. [DOI] [PubMed] [Google Scholar]
- 16.Legler DF, Micheau O, Doucey MA, Tschopp J, Bron C. Recruitment of TNF receptor 1 to lipid rafts is essential for TNFα-mediated NF-κB activation. Immunity. 2003;18:655–664. doi: 10.1016/s1074-7613(03)00092-x. [DOI] [PubMed] [Google Scholar]
- 17.Shembade N, Harhaj NS, Parvatiyar K, Copeland NG, Jenkins NA, Matesic LE, Harhaj EW. The E3 ligase Itch negatively regulates inflammatory signaling pathways by controlling the function of the ubiquitin-editing enzyme A20. Nat Immunol. 2008;9:254–262. doi: 10.1038/ni1563. [DOI] [PubMed] [Google Scholar]
- 18.Shembade N, Parvatiyar K, Harhaj NS, Harhaj EW. The ubiquitin-editing enzyme A20 requires RNF11 to downregulate NF-κB signalling. EMBO J. 2009;28:513–522. doi: 10.1038/emboj.2008.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fearns C, Pan Q, Mathison JC, Chuang TH. Triad3A regulates ubiquitination and proteasomal degradation of RIP1 following disruption of Hsp90 binding. J Biol Chem. 2006;281:34592–34600. doi: 10.1074/jbc.M604019200. [DOI] [PubMed] [Google Scholar]
- 20.Verstrepen L, Bekaert T, Chau TL, Tavernier J, Chariot A, Beyaert R. TLR-4, IL-1R and TNF-R signaling to NF-κB: variations on a common theme. Cell Mol Life Sci. 2008;65:2964–2978. doi: 10.1007/s00018-008-8064-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deng L, Wang C, Spencer E, Yang L, Braun A, You J, Slaughter C, Pickart C, Chen ZJ. Activation of the IκB kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain. Cell. 2000;103:351–361. doi: 10.1016/s0092-8674(00)00126-4. [DOI] [PubMed] [Google Scholar]
- 22.Lamothe B, Besse A, Campos AD, Webster WK, Wu H, Darnay BG. Site-specific Lys-63-linked tumor necrosis factor receptor-associated Factor 6 autoubiquitination is a critical determinant of IκB kinase activation. J Biol Chem. 2007;282:4102–4112. doi: 10.1074/jbc.M609503200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Walsh MC, Kim GK, Maurizio PL, Molnar EE, Choi Y. TRAF6 autoubiquitination-independent activation of the NFκB and MAPK pathways in response to IL-1 and RANKL. PLoS ONE. 2008;3:e4064. doi: 10.1371/journal.pone.0004064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iha H, Peloponese JM, Verstrepen L, Zapart G, Ikeda F, Smith CD, Starost MF, Yedavalli V, Heyninck K, Dikic I, Beyaert R, Jeang KT. Inflammatory cardiac valvulitis in TAX1BP1-deficient mice through selective NF-κB activation. EMBO J. 2008;27:629–641. doi: 10.1038/emboj.2008.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shembade N, Harhaj NS, Liebl DJ, Harhaj EW. Essential role for TAX1BP1 in the termination of TNF-α-, IL-1- and LPS-mediated NF-κB and JNK signaling. EMBO J. 2007;26:3910–3922. doi: 10.1038/sj.emboj.7601823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Conze DB, Wu CJ, Thomas JA, Landstrom A, Ashwell JD. Lys63-linked polyubiquitination of IRAK-1 is required for interleukin-1 receptor- and toll-like receptor-mediated NF-κB activation. Mol Cell Biol. 2008;28:3538–3547. doi: 10.1128/MCB.02098-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xia ZP, Sun L, Chen X, Pineda G, Jiang X, Adhikari A, Zeng W, Chen ZJ. Direct activation of protein kinases by unanchored polyubiquitin chains. Nature. 2009;461:114–119. doi: 10.1038/nature08247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Inohara N, Ogura Y, Fontalba A, Gutierrez O, Pons F, Crespo J, Fukase K, Inamura S, Kusumoto S, Hashimoto M, Foster SJ, Moran AP, Fernandez-Luna JL, Nunez G. Host recognition of bacterial muramyl dipeptide mediated through NOD2. Implications for Crohn's disease. J Biol Chem. 2003;278:5509–5512. doi: 10.1074/jbc.C200673200. [DOI] [PubMed] [Google Scholar]
- 29.Inohara N, Nunez G. NODs: intracellular proteins involved in inflammation and apoptosis. Nat Rev Immunol. 2003;3:371–382. doi: 10.1038/nri1086. [DOI] [PubMed] [Google Scholar]
- 30.Hasegawa M, Fujimoto Y, Lucas PC, Nakano H, Fukase K, Nunez G, Inohara N. A critical role of RICK/RIP2 polyubiquitination in Nod-induced NF-κB activation. EMBO J. 2008;27:373–383. doi: 10.1038/sj.emboj.7601962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bertrand MJ, Doiron K, Labbe K, Korneluk RG, Barker PA, Saleh M. Cellular inhibitors of apoptosis cIAP1 and cIAP2 are required for innate immunity signaling by the pattern recognition receptors NOD1 and NOD2. Immunity. 2009;30:789–801. doi: 10.1016/j.immuni.2009.04.011. [DOI] [PubMed] [Google Scholar]
- 32.Hitotsumatsu O, Ahmad RC, Tavares R, Wang M, Philpott D, Turer EE, Lee BL, Shiffin N, Advincula R, Malynn BA, Werts C, Ma A. The ubiquitin-editing enzyme A20 restricts nucleotide-binding oligomerization domain containing 2-triggered signals. Immunity. 2008;28:381–390. doi: 10.1016/j.immuni.2008.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tao M, Scacheri PC, Marinis JM, Harhaj EW, Matesic LE, Abbott DW. ITCH K63-ubiquitinates the NOD2 binding protein, RIP2, to influence inflammatory signaling pathways. Curr Biol. 2009;19:1255–1263. doi: 10.1016/j.cub.2009.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chang L, Kamata H, Solinas G, Luo JL, Maeda S, Venuprasad K, Liu YC, Karin M. The E3 ubiquitin ligase itch couples JNK activation to TNFα-induced cell death by inducing c-FLIP(L) turnover. Cell. 2006;124:601–613. doi: 10.1016/j.cell.2006.01.021. [DOI] [PubMed] [Google Scholar]
- 35.Tewari M, Wolf FW, Seldin MF, O'Shea KS, Dixit VM, Turka LA. Lymphoid expression and regulation of A20, an inhibitor of programmed cell death. J Immunol. 1995;154:1699–1706. [PubMed] [Google Scholar]
- 36.Matsumoto R, Wang D, Blonska M, Li H, Kobayashi M, Pappu B, Chen Y, Lin X. Phosphorylation of CARMA1 plays a critical role in T Cell receptor-mediated NF-κB activation. Immunity. 2005;23:575–585. doi: 10.1016/j.immuni.2005.10.007. [DOI] [PubMed] [Google Scholar]
- 37.Oeckinghaus A, Wegener E, Welteke V, Ferch U, Arslan SC, Ruland J, Scheidereit C, Krappmann D. Malt1 ubiquitination triggers NF-κB signaling upon T-cell activation. EMBO J. 2007;26:4634–4645. doi: 10.1038/sj.emboj.7601897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun L, Deng L, Ea CK, Xia ZP, Chen ZJ. The TRAF6 ubiquitin ligase and TAK1 kinase mediate IKK activation by BCL10 and MALT1 in T lymphocytes. Mol Cell. 2004;14:289–301. doi: 10.1016/s1097-2765(04)00236-9. [DOI] [PubMed] [Google Scholar]
- 39.Wu CJ, Ashwell JD. NEMO recognition of ubiquitinated Bcl10 is required for T cell receptor-mediated NF-κB activation. Proc Natl Acad Sci U S A. 2008;105:3023–3028. doi: 10.1073/pnas.0712313105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Laplantine E, Fontan E, Chiaravalli J, Lopez T, Lakisic G, Veron M, Agou F, Israel A. NEMO specifically recognizes K63-linked poly-ubiquitin chains through a new bipartite ubiquitin-binding domain. EMBO J. 2009;28:2885–2895. doi: 10.1038/emboj.2009.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Coornaert B, Baens M, Heyninck K, Bekaert T, Haegman M, Staal J, Sun L, Chen ZJ, Marynen P, Beyaert R. T cell antigen receptor stimulation induces MALT1 paracaspase-mediated cleavage of the NF-κB inhibitor A20. Nat Immunol. 2008;9:263–271. doi: 10.1038/ni1561. [DOI] [PubMed] [Google Scholar]
- 42.Duwel M, Welteke V, Oeckinghaus A, Baens M, Kloo B, Ferch U, Darnay BG, Ruland J, Marynen P, Krappmann D. A20 negatively regulates T cell receptor signaling to NF-κB by cleaving Malt1 ubiquitin chains. J Immunol. 2009;182:7718–7728. doi: 10.4049/jimmunol.0803313. [DOI] [PubMed] [Google Scholar]
- 43.Yamamoto M, Sato S, Saitoh T, Sakurai H, Uematsu S, Kawai T, Ishii KJ, Takeuchi O, Akira S. Cutting Edge: Pivotal function of Ubc13 in thymocyte TCR signaling. J Immunol. 2006;177:7520–7524. doi: 10.4049/jimmunol.177.11.7520. [DOI] [PubMed] [Google Scholar]
- 44.Moreno-Garcia ME, Sommer K, Shinohara H, Bandaranayake AD, Kurosaki T, Rawlings DJ. MAGUK-controlled ubiquitination of CARMA1 modulates lymphocyte NF-κB activity. Mol Cell Biol. 30:922–934. doi: 10.1128/MCB.01129-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hiscott J. Convergence of the NF-κB and IRF pathways in the regulation of the innate antiviral response. Cytokine Growth Factor Rev. 2007;18:483–490. doi: 10.1016/j.cytogfr.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 46.Saitoh T, Yamamoto M, Miyagishi M, Taira K, Nakanishi M, Fujita T, Akira S, Yamamoto N, Yamaoka S. A20 is a negative regulator of IFN regulatory factor 3 signaling. J Immunol. 2005;174:1507–1512. doi: 10.4049/jimmunol.174.3.1507. [DOI] [PubMed] [Google Scholar]
- 47.Lin R, Yang L, Nakhaei P, Sun Q, Sharif-Askari E, Julkunen I, Hiscott J. Negative regulation of the retinoic acid-inducible gene I-induced antiviral state by the ubiquitin-editing protein A20. J Biol Chem. 2006;281:2095–2103. doi: 10.1074/jbc.M510326200. [DOI] [PubMed] [Google Scholar]
- 48.Wang YY, Li L, Han KJ, Zhai Z, Shu HB. A20 is a potent inhibitor of TLR3- and Sendai virus-induced activation of NF-κB and ISRE and IFN-b promoter. FEBS Lett. 2004;576:86–90. doi: 10.1016/j.febslet.2004.08.071. [DOI] [PubMed] [Google Scholar]
- 49.Zeng W, Xu M, Liu S, Sun L, Chen ZJ. Key role of Ubc5 and lysine-63 polyubiquitination in viral activation of IRF3. Mol Cell. 2009;36:315–325. doi: 10.1016/j.molcel.2009.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nakhaei P, Mesplede T, Solis M, Sun Q, Zhao T, Yang L, Chuang TH, Ware CF, Lin R, Hiscott J. The E3 ubiquitin ligase Triad3A negatively regulates the RIG-I/MAVS signaling pathway by targeting TRAF3 for degradation. PLoS Pathog. 2009;5:e1000650. doi: 10.1371/journal.ppat.1000650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.You F, Sun H, Zhou X, Sun W, Liang S, Zhai Z, Jiang Z. PCBP2 mediates degradation of the adaptor MAVS via the HECT ubiquitin ligase AIP4. Nat Immunol. 2009;10:1300–1308. doi: 10.1038/ni.1815. [DOI] [PubMed] [Google Scholar]
- 52.Ning S, Pagano JS. The A20 deubiquitinase activity negatively regulates LMP1 activation of IRF7. J Virol. 84:6130–6138. doi: 10.1128/JVI.00364-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ning S, Campos AD, Darnay B, Bentz GL, Pagano JS. TRAF6 and the Three C-Terminal Lysine Sites on IRF7 Are Required for Its Ubiquitination-mediated Transactivation by the TNFR Family Member LMP1. Mol Cell Biol. 2008 doi: 10.1128/MCB.00785-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wertz IE, Dixit VM. Ubiquitin-mediated regulation of TNFR1 signaling. Cytokine Growth Factor Rev. 2008;19:313–324. doi: 10.1016/j.cytogfr.2008.04.014. [DOI] [PubMed] [Google Scholar]
- 55.Jin Z, Li Y, Pitti R, Lawrence D, Pham VC, Lill JR, Ashkenazi A. Cullin3-based polyubiquitination and p62-dependent aggregation of caspase-8 mediate extrinsic apoptosis signaling. Cell. 2009;137:721–735. doi: 10.1016/j.cell.2009.03.015. [DOI] [PubMed] [Google Scholar]
- 56.He KL, Ting AT. A20 inhibits tumor necrosis factor (TNF) alpha-induced apoptosis by disrupting recruitment of TRADD and RIP to the TNF receptor 1 complex in Jurkat T cells. Mol Cell Biol. 2002;22:6034–6045. doi: 10.1128/MCB.22.17.6034-6045.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Oshima S, Turer EE, Callahan JA, Chai S, Advincula R, Barrera J, Shifrin N, Lee B, Benedict Yen TS, Woo T, Malynn BA, Ma A. ABIN-1 is a ubiquitin sensor that restricts cell death and sustains embryonic development. Nature. 2009;457:906–909. doi: 10.1038/nature07575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.De Valck D, Jin DY, Heyninck K, Van de Craen M, Contreras R, Fiers W, Jeang KT, Beyaert R. The zinc finger protein A20 interacts with a novel anti-apoptotic protein which is cleaved by specific caspases. Oncogene. 1999;18:4182–4190. doi: 10.1038/sj.onc.1202787. [DOI] [PubMed] [Google Scholar]
- 59.O'Donnell MA, Legarda-Addison D, Skountzos P, Yeh WC, Ting AT. Ubiquitination of RIP1 regulates an NF-κB-independent cell-death switch in TNF signaling. Curr Biol. 2007;17:418–424. doi: 10.1016/j.cub.2007.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kundu M, Thompson CB. Autophagy: basic principles and relevance to disease. Annu Rev Pathol. 2008;3:427–455. doi: 10.1146/annurev.pathmechdis.2.010506.091842. [DOI] [PubMed] [Google Scholar]
- 61.Shi CS, Kehrl JH. TRAF6 and A20 regulate lysine 63-linked ubiquitination of Beclin-1 to control TLR4-induced autophagy. Sci Signal. 2010;3:ra42. doi: 10.1126/scisignal.2000751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Coornaert B, Carpentier I, Beyaert R. Central gatekeeper in inflammation and immunity. J Biol Chem. 2008;A20 doi: 10.1074/jbc.R800032200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kato M, Sanada M, Kato I, Sato Y, Takita J, Takeuchi K, Niwa A, Chen Y, Nakazaki K, Nomoto J, Asakura Y, Muto S, Tamura A, Iio M, Akatsuka Y, Hayashi Y, Mori H, Igarashi T, Kurokawa M, Chiba S, Mori S, Ishikawa Y, Okamoto K, Tobinai K, Nakagama H, Nakahata T, Yoshino T, Kobayashi Y, Ogawa S. Frequent inactivation of A20 in B-cell lymphomas. Nature. 2009;459:712–716. doi: 10.1038/nature07969. [DOI] [PubMed] [Google Scholar]
- 64.Vendrell JA, Ghayad S, Ben-Larbi S, Dumontet C, Mechti N, Cohen PA. A20/TNFAIP3, a new estrogen-regulated gene that confers tamoxifen resistance in breast cancer cells. Oncogene. 2007;26:4656–4667. doi: 10.1038/sj.onc.1210269. [DOI] [PubMed] [Google Scholar]
- 65.Hjelmeland AB, Wu Q, Wickman S, Eyler C, Heddleston J, Shi Q, Lathia JD, Macswords J, Lee J, McLendon RE, Rich JN. Targeting A20 decreases glioma stem cell survival and tumor growth. PLoS Biol. 2010;8:e1000319. doi: 10.1371/journal.pbio.1000319. [DOI] [PMC free article] [PubMed] [Google Scholar]