Summary
Lipolysis is the biochemical pathway responsible for the catabolism of triacylglycerol (TAG) stored in cellular lipid droplets. The hydrolytic cleavage of TAG generates non-esterified fatty acids, which are subsequently used as energy substrates, essential precursors for lipid and membrane synthesis, or mediators in cell signaling processes. Consistent with its central importance in lipid and energy homeostasis, lipolysis occurs in essentially all tissues and cell types, it is most abundant, however, in white and brown adipose tissue. Over the last 5 years, important enzymes and regulatory protein factors involved in lipolysis have been identified. These include an essential TAG hydrolase named adipose triglyceride lipase (ATGL) [annotated as patatin-like phospholipase domain-containing protein A2], the ATGL activator comparative gene identification-58 [annotated as α/β hydrolase containing protein 5], and the ATGL inhibitor G0/G1 switch gene 2. Together with the established hormone-sensitive lipase [annotated as lipase E] and monoglyceride lipase, these proteins constitute the basic “lipolytic machinery”. Additionally, a large number of hormonal signaling pathways and lipid droplet-associated protein factors regulate substrate access and the activity of the “lipolysome”. This review summarizes the current knowledge concerning the enzymes and regulatory processes governing lipolysis of fat stores in adipose and non-adipose tissues. Special emphasis will be given to ATGL, its regulation, and physiological function.
Abbreviations: 2-AG, 2-arachidonoyl glycerol; ABHD1-15, α/β hydrolase domain containing protein 1–15; ARF1, ADP-ribosylation factor 1; ATGL, adipose triglyceride lipase; BAT, brown adipose tissue; BiFC, bimolecular fluorescence complementation; CDS, Chanarin-Dorfman syndrome; CE, cholesterylester; CGI-58, comparative-gene-identification 58; COPI, coat protein complex-I; cPLA2, cytosolic phospholipase A2; DAG, diacylglycerol; ER, endoplasmic reticulum; FoxO1, forkhead box O1; G0S2, G0/G1 Switch Protein 2; GS2, gene sequence 2; HSL, hormone-sensitive lipase; LD, lipid droplet; LPAAT, lysophosphatidic acid acyltransferase; MAG, monoacylglycerol; MGL, monoglyceride lipase; mTor, mammalian target of rapamycin; NEFA, non-esterified fatty acid; NLSD, neutral lipid storage disease; NLSDI, NLSD with ichthyosis; NLSDM, NLSD with myopathy; PKA, protein kinase A; PNPLA1-5, patatin-like phospholipase domain containing protein 1–5; PPARα/γ, peroxisome proliferator-activated receptor-alpha/gamma; PPRE, PPAR-response element; RBP4, retinol-binding protein 4; RE, retinylester; STS, steroid sulfatase; TAG, triacylglycerol; TGH, triglyceride hydrolase; TNF-α, tumor necrosis factor alpha; WAT, white adipose tissue
Keywords: Lipolysis, Fat stores, Triacylglycerol, Lipase, Neutral lipid storage disease
1. Introduction and overview
Fat stores of white adipose tissue (WAT) represent the major energy reserves in mammals. During food intake, excess of dietary non-esterified fatty acids (NEFAs) are esterified to chemically relative inert triacylglycerols (TAGs), which are subsequently stored in cytosolic lipid droplets (LDs) of adipocytes. Upon increased energy demand, TAG stores are mobilized by their hydrolytic cleavage and the resulting NEFAs are delivered via the circulation to peripheral tissues for β-oxidation and ATP production. Additionally, also non-adipose tissues are able to esterify NEFAs into TAGs and re-hydrolyze them upon demand. Accordingly, TAG storage and mobilization is a general biological process in essentially all cells of the body and not restricted to adipose tissue. However, whereas adipocytes are able to secrete NEFAs and provide them as systemic energy substrate, non-adipose cells do not secrete NEFAs but utilize TAG-derived NEFAs in a cell autonomous manner for energy production or lipid synthesis. Consistent with this local utilization, the TAG storage capacity of non-adipose tissues and cells is relatively minor compared to adipose tissue providing NEFAs for the whole organism. In fact, excessive ectopic lipid deposition in non-adipose tissues leads to lipotoxicity and is associated with prevalent metabolic diseases, such as type-2 diabetes [1–4].
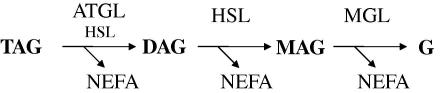
The cellular concentration of NEFAs is tightly controlled by the balance between TAG hydrolysis and NEFA esterification [5–7]. The hydrolysis of the primary and secondary ester bonds between long chain fatty acids and the glycerol backbone in TAG is called “lipolysis” and depends on specific hydrolases commonly designated lipases [8,9]. To date, three enzymes have been implicated in the complete hydrolysis of TAG molecules in cellular lipid stores (Fig. 1): adipose triglyceride lipase (ATGL) selectively performs the first and rate-limiting step hydrolyzing TAGs to generate diacylglycerols (DAGs) and NEFAs [10]. Hormone-sensitive lipase (HSL) is a multifunctional enzyme capable of hydrolyzing a variety of acylesters including TAG, DAG, and monoacylglycerol (MAG). Within the TAG hydrolysis cascade this enzyme is rate-limiting for DAG catabolism [11,12]. Finally, monoglyceride lipase (MGL) efficiently cleaves MAG into glycerol and NEFAs [13].
Fig. 1.
Schematic delineation of the coordinate breakdown of triacylglycerols. Abbreviations: ATGL, adipose triacylglycerol lipase; DAG, diacylglycerol; G, glycerol; HSL, hormone-sensitive lipase; MAG, monoacylglycerol; MGL, monoacylglycerol lipase; NEFA, non-esterified fatty acid; TAG, triacylglycerol.
The important role of ATGL for TAG catabolism became evident from the analysis and examination of ATGL-deficient mice and human patients with mutations in the gene for ATGL [14–16]. ATGL deficiency in mice is associated with severely reduced lipolysis resulting in increased fat deposition in virtually all tissues of the body, most notably in highly oxidative tissues, such as muscle, testis, and the tubular system of the kidney. The massive fat deposition in the heart is causative for cardiac dysfunction and premature death of the animals [16]. Similarly, ATGL mutations in humans are associated with systemic TAG accumulation and cardiac myopathy [14,15]. The group of Fischer [14] elucidated the molecular basis of this rare inherited disease annotated as “neutral lipid storage disease with myopathy (NLSDM)”. Importantly, the deficiency or dysfunction of a potent coactivator of ATGL, CGI-58 (annotated as α/β hydrolase domain containing 5), also results in a severe systemic TAG accumulation in mice and human patients. Patients with mutations in CGI-58 additionally develop severe ichthyosis, a condition not observed in patients with mutations in the ATGL gene. Accordingly, this disorder was named “neutral lipid storage disease with ichthyosis (NLSDI)”. More recently, G0S2, a protein specifically inhibiting ATGL, has been identified [17,18]. However, the physiological relevance of this protein in the regulation of lipolysis has not been demonstrated so far. One would expect that a constitutive over-expression of this protein would result in a phenotype similar to that of ATGL deficiency.
In contrast to ATGL deficiency, hormone-sensitive lipase (HSL)-deficient mice do not show increased fat deposition, are not overweight or obese, and lose WAT mass with increasing age [11,12,19,20]. Adipocytes of HSL-deficient mice exhibit only a moderate decrease in stimulated lipolysis [11,21]. Notably, these mice accumulate DAG in several tissues indicating that HSL is rate-limiting for DAG hydrolysis [11]. Mutations in the HSL gene of humans leading to enzyme dysfunction or deficiency have not been reported. The physiological role of MGL in lipolysis has not been evaluated so far. Yet, the physiological importance of MGL in the breakdown of the endocannabinoid 2-arachidonoylglycerol has recently been elucidated using inhibitor studies and a MGL-deficient mouse model [22–30], confirming the pivotal role of MGL in the endocannabinoid system.
The recently characterized genetic mouse models and human disorders suggest that ATGL, HSL, and MGL are the main lipases involved in the catabolism of TAG. The activities of these enzymes are, however, delicately regulated. In addition to the factors and processes involved in lipolysis discussed in this review, many more remain to be elucidated.
2. Lipolytic enzymes
2.1. Adipose triglyceride lipase
2.1.1. Enzymatic properties: ATGL is a selective TAG hydrolase
In 2004, three groups [10,19,31] independently identified an enzyme capable of hydrolyzing TAG. This enzyme was named adipose triglyceride lipase [10], desnutrin [31], and phospholipase A2ξ [19] (now annotated as patatin-like phospholipase domain containing protein 2, PNPLA2). The enzyme selectively performs the first step in TAG hydrolysis generating DAGs and NEFAs [10]. The substrate- and stereo-selectivity of this reaction has not been studied in detail. Since most animal and microbial lipases preferentially cleave the primary ester bond at the sn-1(3) position of TAG [32], this may also be assumed for the positional priority of ATGL. Yet, the phylogenetic ancestry of the patatin domain of ATGL would rather suggest a sn-2 preference because various known patatin domain containing glycoproteins in plants [33,34] as well as the cytosolic phospholipase A2 (cPLA2) [35] preferentially cleave the sn-2-acyl ester bond of phospholipids to release arachidonic acid. Compared to the hydrolytic activity towards TAG, ATGL exhibits only minor or no activity when other lipids such as DAG, MAG, cholesteryl- (CE), or retinylesters (RE) are provided as substrates [10]. The enzyme was reported, however, to exhibit measurable phospholipase [36] and DAG transacylase activities, the latter generating TAG and MAG from two DAG molecules in an acyl-CoA independent manner [19].
2.1.2. Gene and protein structure: ATGL contains a patatin domain
The human and mouse genes for ATGL (PNPLA2 and Pnpla2, respectively) encode proteins with 504 and 486 residues, respectively. Orthologues of ATGL have also been described in other species including rat, cow, pig, chicken, fly, plants, and yeast [37–42]. The human gene for ATGL is located on chromosome 11p15.5 and comprises 10 exons [14]. Promoter sequences of the ATGL gene (PNPLA2) that regulate its tissue-specific and hormonally controlled expression have not been characterized so far. The only exception relates to the characterization of a peroxisome proliferator-activated receptor-gamma (PPARγ) responsive element within the promoter of the murine Atgl gene [43]. The human gene encodes for a 2.4 kb mRNA (the murine mRNA is 2.6 kb long) with relatively short 5′ and 3′ untranslated regions.
Virtually all tissues examined express measurable amounts of ATGL mRNA [10,31,44,45]. WAT and brown adipose tissue (BAT) exhibit the highest expression levels, an observation which was eponymous for the protein. Much lower expression is detectable in other tissues such as testis, skeletal and cardiac muscle [10]. Systemic TAG accumulation in humans and mice lacking ATGL activity argues for a critical physiological function of the enzyme also in non-adipose tissues (see below).
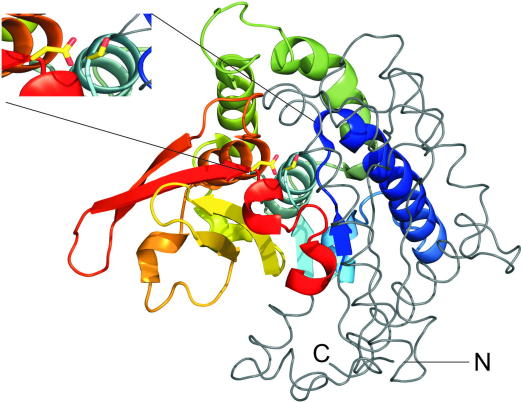
Human and murine ATGL share 84% amino acid identity. Notable regions of low sequence conservation are clustered around residue 260 and the C-terminal end of the protein, outside of conserved domain areas (Fig. 2). Sequence similarities predict that the N-terminal half of ATGL is an α/β-fold protein, belonging to the superfamily of patatin-like phospholipases. The name patatin derives from a plant protein, which is a non-specific lipid acyl hydrolase present in high amounts in potato tubers [33]. The 3-dimensional (3D) structure for two members of the superfamily have been determined, Pat17 [34] and human cPLA2 [35]. In these proteins, the hydrolytic reaction is mediated through a catalytic serine-aspartate dyad (Ser47–Asp215 in Pat17, Ser228–Asp549 in cPLA2), with the nucleophilic serine located within a GXSXG motif typically found in lipases of the α/β hydrolase fold family [34]. Although no 3D structures for ATGL is available to date, it is assumed that the enzyme also acts through a catalytic dyad. Identification of a GXSXG sequence with the presumed active site serine and the presence of a DXG/A sequence, all located within the patatin domain, lead to the conclusion that the catalytic dyad is composed of Ser47 and Asp166. Fig. 3 displays the 3D structure of Pat17 highlighting the catalytic area and further regions with sequence similarities to human ATGL. The critical role of Ser47 and Asp166 for ATGL enzyme activity was proven by mutation studies. Replacement of Ser47 of the murine protein or Asp166 of the human protein with an alanine in both cases led to catalytically inactive proteins [8,46]. The enzyme–substrate transition state is stabilized via a glycine-rich oxyanion hole in patatin domain containing proteins. The C-terminal part of ATGL is expected to consist mostly of α-helical and loop regions. It is assumed that a hydrophobic stretch (amino acid 315–360) represents a lipid binding region and is involved in the regulation of enzyme activity [47,48] (see Fig. 2).
Fig. 2.
Conserved areas and domain organization of ATGL. Protein sequences of human and mouse ATGL were aligned using web-based “T-Coffee Multiple Sequence Alignments” tool [203,204]. Identical amino acids in the protein sequences are depicted as black, differences in the sequence as white bars. Human ATGL as the longer orthologue (504 amino acids) was used as template. Domain organization is shown for the human protein.
Fig. 3.
3D structure of Pat17 depicting sequence similarities with human ATGL. A region of human ATGL, commonly annotated as the patatin-domain of ATGL (Ile10–Lys179), shares sequence similarities with Pat17 (Leu32–Ser228). The 3D structure of this sequence area in Pat17 is depicted as colored cartoon [34]. The remainder of the Pat17 3D structure is displayed in grey ribbon style. N- and C-terminal ends are indicated with capital letters. The insert shows the catalytic dyad of Pat17 with the catalytic residues Ser77 and Asp215 (corresponding to Ser47 and Asp166 in human ATGL) highlighted as yellow sticks. The figure was prepared using PyMol (The PyMOL Molecular Graphics System, Version 1.2, Schrödinger, LLC).
2.2. Hormone-sensitive lipase: enzymatic properties – gene and protein structure
HSL was initially discovered in WAT of mammals as an enzyme which is induced by fasting and stimulated by catabolic hormones [49–52]. Since then, numerous studies have addressed the biochemistry, cell biology, and physiology of HSL [53–59]. They found that HSL hydrolyzes a variety of substrates including TAG, DAG, MAG, CE and RE as well as short chain carbonic esters and artificial substrates, such as p-nitrophenyl butyrate. In fact, the relative maximal hydrolysis rates for TAG:DAG:MAG:CE:RE are in the range of 1:10:1:4:2. These in vitro data were indicative that HSL may be more important as a DAG than TAG hydrolase.
The tissue-specific expression pattern of HSL resembles the one for ATGL. Highest expression is observed in WAT and BAT. Low HSL expression is found in many other tissues and cells particularly in steroidogenic cells, muscle, pancreatic β-cells, and macrophages [55,60,61].
The human Hsl gene (designated Lipe) is located on chromosome 19q13.2. It spans a genomic region of 25.9 kb comprising 10 exons. Alternative exon usage results in a significant variation in the 5′ region of HSL transcripts [62] leading to different tissue-specific mRNA and protein sizes in adipose tissue, pancreatic β-cells, ovaries, and testis. Multiple potential transcription factor-binding elements upstream of each transcriptional start site suggest the possibility of differential transcriptional regulation of HSL in different tissues and under various physiological conditions [63].
To date no 3D protein structure for HSL is available. Yet, HSL protein domain architecture suggests that the enzyme comprises of three functional regions [64]: the N-terminal domain (amino acids 1–300) is believed to mediate lipid binding, enzyme dimerization, and interaction with the fatty acid binding protein 4 (FABP4) [64–67]. These features are thought to modulate the in vivo activity of HSL; in fact, the interaction with FABP4 is known to enhance HSL catalytic activity also in vitro [68]. The C-terminal domain (amino acids 301–768) contains a common structural fold found in many lipases and esterases, called the α/β hydrolase fold, which harbors the classical catalytic triad of Ser424, Asp693, and His723 of the human protein [64,69,70]. The third region represents the regulatory module of the enzyme. This region (amino acids 521–669) is located within the catalytic domain and contains all five known phosphorylation sites of HSL [54,71–73].
Besides the catalytic site motif (GXSXG) commonly found in lipases, HSL shows no homology with other known lipases or proteins; however, 61 of the last 283 amino acids of the lipase 2 of Moraxella TA144, an antarctic psychotrophic bacterium, are identical to that of rat HSL in the catalytic domain. Since in lipase 2 also respective amino acids of the triad are conserved it is believed that it forms a α/β hydrolase fold with an active site [62,64]. In the C-terminal region of HSL, DNA sequence of HSL shows identity rates of around 60% with corresponding stretches of acetylcholinesterase, bile salt-stimulated lipase, and two fungal lipases from Geotrichum candidum and Candida rugosa [69,74].
2.3. Monoglyceride lipase (MGL): enzymatic properties – gene and protein structure
MGL was first isolated from rat adipose tissue and was shown to specifically hydrolyze MAG but not TAG or DAG [75]. This led to the assumption that MGL is responsible for the hydrolysis of MAG in the lipolytic cascade. Interestingly, recent studies with an MGL-deficient mouse model and an MGL specific inhibitor have established a pivotal role in endocannabinoid signaling but have not addressed the role of MGL in the breakdown of lipolytic MAG [23–30]. Yet, in MGL-deficient mice elevated levels of various MAG species have been described in the brain [23]. Thus, it appears likely that MGL might also be indispensable for the breakdown of MAG in other tissues including WAT. Whether other enzymes with known hydrolytic activities for MAG in in vitro assays such as HSL or α/β-hydrolase domain-containing 6 (ABHD6) are relevant for MAG hydrolysis in vivo remains to be determined.
The human gene for MGL (annotated as MGLL) is located on chromosome 3q21.3. It spans over 134 kb, contains eight exons, with a transcript length of 4.6 kb. It codes for a protein of 303 residues with a molecular weight of ∼33 kDa. The 3D structure of MGL has been determined very recently by two independent groups [76,77]. MGL also belongs to the large superfamily of α/β hydrolase fold proteins with a GXSXG motif. The catalytic triad is composed by Ser122, Asp239, and His269 [13]. MGL is ubiquitously expressed among tissues. MGL mRNA levels are particularly high in adipose tissue, kidney, and testis [13]. More recently, MGL has also been implicated in the degradation of the bioactive MAG 2-arachidonoyl glycerol, which is known to be a potent endogenous agonist of cannabinoid receptors [78]. With the availability of mouse models that lack or overexpress MGL it will be interesting to investigate the physiological role of MGL in endocannabinoid metabolism and the regulation of appetite, pain sensation, mood control, and other endocannabinoid-affected physiological and psychological conditions.
2.4. Other lipases
Assessment of lipolytic activities in ATGL-deficient mice using a specific inhibitor for HSL revealed that ATGL and HSL are responsible for more than 90% of the lipolytic capacity in murine adipose tissue [79]. Although this finding suggests that these two enzymes are the major lipases for TAG catabolism in WAT, it does not exclude a significant contribution of other enzymes in non-adipose tissues or under specific physiological conditions. In recent years, a number of alternative TAG hydrolases has been identified and their physiological relevance in the lipolytic process is currently being elaborated [80]. Four of these proteins are members of the PNPLA family with high sequence homology to ATGL. The PNPLA family of genes consists of 9 and 10 members in the murine and human genomes, respectively [80]. The structurally closest relative to ATGL (annotated as PNPLA2) within this family is PNPLA3 (also called adiponutrin). Similarly as described for ATGL, this protein acts as TAG hydrolase and transacylase, however, the specific TAG hydrolase activity is at least two orders of magnitudes smaller than for ATGL [19]. PNPLA3 has recently gained major interest because of a strong association of specific variants of the enzyme (I148M) with non-alcoholic fatty liver disease, hepatosteatosis, steato-hepatitis, and liver cirrhosis [81–84]. Initially, it was assumed that a loss of function in the lipase activity of the protein causes the higher susceptibility to develop fatty liver [81]. However, the recent finding that adiponutrin-deficient mice have no increased liver TAG content questions the proposed mechanism [85].
Other members of close ATGL relatives are less well characterized (PNPLA1, 4, and 5) [44]. PNPLA4 (alternative name: gene sequence 2; GS2) was shown to hydrolyze TAG and act as transacylase, transferring an acyl chain of one DAG molecule to another, thereby forming one TAG and one MAG molecule [19]. In keratinocytes, this protein hydrolyzes REs at neutral pH, whereas in acidic pH conditions the reverse reaction, the esterification of free retinol, is favored [86,87]. These GS2-driven activities are consistent with the loss of triolein and accumulation of REs during the differentiation process of keratinocytes and suggest that GS2 may be involved in the maturation of the epidermal skin barrier. The GS2 gene is closely linked to the gene coding for steroid sulfatase (STS) on the distal short arm of the human X chromosome [88]. Notably, the phenotype of patients with X-linked ichthyosis who have deletions of both GS2 and STS is indistinguishable from those with mutations within the STS gene alone [89]. This suggests that the deletion of GS2 may not cause any apparent phenotype (so far no GS2 homologue has been identified in the mouse genome) or it may cause defective epidermal development and ichthyosis similar to X-linked ichthyosis. Whether the two remaining proteins of the family with demonstrated lipase activity, PNPLA1 and PNPLA5 (also called GS2-like) [19,44] have any physiological role in lipolysis is currently not known.
Finally, two proteins of the carboxylesterase family, carboxylesterase 3 (also called triglyceride hydrolase (TGH) or TGH-1) and carboxylesterase ML1 (also called TGH-2, now annotated as carboxylesterase B-1) have been shown to hydrolyze a triolein substrate and to be expressed in both liver and adipose tissue [90]. These enzymes show a strong preference for short chain esters and, accordingly, are weak hydrolases for long chain fatty acid TAGs. Moreover, these enzymes localize to the endoplasmic reticulum (ER) membrane and are thought to face with the active site to the lumen of the ER suggesting that TGH and TGH-2 may be more important for the hydrolysis of the microsomal TAG pool than for TAG in cytosolic LDs [90,91].
3. Regulation of lipolysis
3.1. Regulation of ATGL
3.1.1. Comparative gene identification-58 (CGI-58) activates ATGL
Extracellular TAG lipases, such as lipoprotein lipase or pancreatic lipase are catalytically much more active in the presence of coactivators (apolipoprotein CII and colipase, respectively). Similarly, ATGL activity can be drastically increased by an activator protein [8] called CGI-58 [also annotated as α/β hydrolase domain containing protein 5 and as 1-acylglycerol-3-phosphate O-acyltransferase] [8,17,47,92,93].
3.1.1.1. CGI-58: gene and protein structure
CGI-58 is highly conserved among species. The name comparative gene identification-58 derives from a proteomic approach to identify genes conserved between Caenorhabditis elegans (C. elegans) and humans. The human gene for CGI-58 is located on chromosome 3p21 and comprises seven exons. The gene encodes for a 5.4 kb mRNA (the murine mRNA is 3.1 kb long), with a short 5′ and a 4.2 kb long 3′ untranslated region. Human and murine CGI-58 display 94% sequence identity and consist of 349 and 351 amino acids, respectively. In Fig. 4 the conserved areas in the protein sequences of human and mouse CGI-58 are depicted showing that the N-terminal region, the area outside of the predicted alpha/beta hydrolase domain, harbors the largest number of sequence variations. Recently, an alternative splice isoform has been described for murine CGI-58 which lacks the second and third exon and exhibits altered cellular function (see below) [94].
Fig. 4.
Conserved areas and domain organization of CGI-58. Protein sequences of human and mouse CGI-58 were aligned using web-based “T-Coffee Multiple Sequence Alignments” tool [202,203]. Identical amino acids in the protein sequences are depicted as black, differences in the sequence as white bars. Mouse CGI-58 as the longer orthologue (351 amino acids) was used as template. Domain organization is shown for the murine protein.
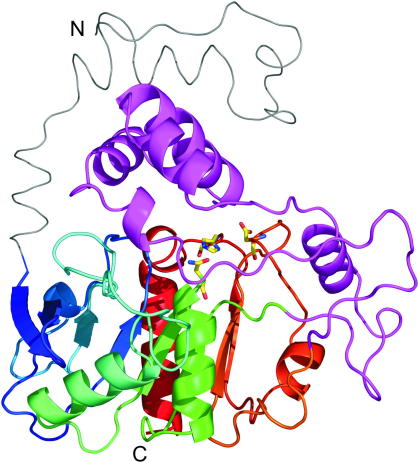
CGI-58 belongs to an α/β hydrolase-fold containing sub-family with 15 members annotated as α/β hydrolase domain containing proteins 1–15 (ABHD1–15). A 3D homology model of mouse CGI-58 was calculated and shows that the compact core constitutes a three-layer (αβα) sandwich typical for α/β hydrolases, starting around Cys50 and ranging to Val350 (Fig. 5) [92]. A large, primarily helical lid region (Pro180–Leu280) is inserted after strand β6 and covers the potential “active site.” An additional feature of the model is a long partly helical, partly unstructured region ranging from the N-terminus to Cys47, which seems to cap the lid. Truncations of this N-terminal region led to CGI-58 fragments incapable of localizing to the LD or stimulating ATGL lipolytic activity [92]. Typically, α/β-hydrolases exert their catalytic activity via a catalytic triad [95]. In murine CGI-58, this triad is located within the α/β hydrolase domain, however, the nucleophilic serine within the canonical GXSXG motif of the active site is replaced by an asparagine residue (Asn155) (Fig. 4). The loss of the nucleophilic serine residue resulting from this amino acid exchange provides a rational explanation for the observation that the protein does not possess intrinsic hydrolase activity [8]. Recent mutation studies in our laboratory exchanging Asn155 by Ser and thus reconstituting the canonical GXSXG motif failed, however, to create a catalytically active esterase/lipase (unpublished data).
Fig. 5.
3D model and domain organization of CGI-58. A homology model of CGI-58 was built using Swiss-Model based on Aspergillus niger epoxide hydrolase as template [204,205]. The compact αβα sandwich containing the α/β-hydrolase core structure is depicted as cartoon in rainbow colors; residues of the putative catalytic triad in corresponding hydrolases are highlighted as yellow sticks (Asn155, His329, Asp303). The cap region covering the potential active site is depicted in magenta, whereas the N-terminal extension is depicted as a grey ribbon. N- and C-termini are indicated with capital letters. The figure was prepared using PyMol (The PyMOL Molecular Graphics System, Version 1.2, Schrödinger, LLC).
3.1.1.2. CGI-58: activation of ATGL
An important regulatory mechanism for ATGL involves the activation of the enzyme upon interaction with CGI-58. Maximal stimulation is achieved at approximately equimolar concentrations of enzyme and activator protein [8]. Interestingly, a species difference exists in the activation potential between the mouse and human proteins. Both human and murine CGI-58 activate mouse ATGL much better than human ATGL. Mutation studies [17,47,92] revealed that ATGL activation by CGI-58 depends on the amino acid sequence present within the patatin domain in the N-terminal half of ATGL. The C-terminal half of ATGL appears to have an inhibitory effect on CGI-58 mediated enzyme activation [47]. This inhibitory effect is more pronounced in human ATGL than in murine ATGL because, compared to human ATGL, CGI-58 mediated enzyme activation was drastically improved when a hybrid enzyme consisting of the human N-terminus (the first 266 amino acids containing the active site) and the murine C-terminus (amino acids 267–486) was used in enzyme assays. Taken together, these studies are consistent with the concept that the patatin domain within the αβα sandwich fold is responsible for enzyme activity and CGI-58 interaction, whereas the C-terminal part of the enzyme has a regulatory function and mediates LD interaction of the enzyme.
Currently, it is unknown whether CGI-58 binding affects ATGL conformation, facilitates substrate presentation, or enhances ATGL’s lipolytic activity by removing reaction products from the active site. In vitro experiments and studies in living cells using bimolecular fluorescence complementation (BiFC) revealed that CGI-58-mediated activation of ATGL requires direct protein–protein interaction [8,96]. However, this interaction by itself is not sufficient for ATGL activation because CGI-58 variants, which were capable of binding ATGL, failed to stimulate enzyme activity [92]. ATGL activation in living cells additionally requires the binding of CGI-58 to the LD. Truncated variants of CGI-58, which fail to localize to the LD, but bind to ATGL, are unable to stimulate ATGL activity [92]. Very recently, a splice variant of CGI-58 was described lacking 149 amino acids in the N-terminal region (full length CGI-58 is 351 amino acids long). The shorter version of CGI-58 is unable to bind to LDs, lacks the GXNXG sequence, and fails to activate ATGL [94].
3.1.1.3. CGI-58: ATGL unrelated function
Several studies have reported that CGI-58 possesses acyl-CoA dependent lysophosphatidic acid acyltransferase (LPAAT) activity [97,98]. The acyltransferase activity of CGI-58 is thought to depend on the structural motif HX4D near the C-terminus, however, this assumption has not been tested by mutation studies. In this respect, it is interesting to note that the shorter splice variant of CGI-58 retains its enzymatic properties as LPAAT despite the fact that it cannot bind to LDs and does not activate ATGL. Although the physiological relevance of the LPAAT activity still needs to be demonstrated in vivo, the in vitro findings support the speculation that CGI-58 could affect both lipid synthesis and lipid hydrolysis depending on the metabolic status of the cell.
3.1.2. G0/G1 Switch Protein 2 (G0S2) inhibits ATGL
Recently, a protein called G0S2 was identified as a selective inhibitor of ATGL [17]. G0S2 was originally found to be expressed during re-entry of blood mononuclear cells from G0 into G1 phase of the cell cycle [99]. However, its mechanistic involvement in cell-cycling remained unclear. Yang et al. [17] have now demonstrated that G0S2 is predominantly expressed in adipose tissue and liver and that overexpression of G0S2 in cells causes massive lipid accumulation. These findings are indicative for a role for G0S2 in lipid/energy metabolism.
3.1.2.1. G0S2: gene and protein structure
The human gene for G0S2 is located on chromosome 1q32.2 and comprises two exons. The first exon is non-coding, the second exon contains the complete coding region for the protein. The respective mRNA consists of 978 nucleotides, with a shorter 5′ and a 409 nucleotide long 3′ untranslated region. Both human and murine G0S2 proteins consist of 103 amino acids and display 77% sequence identity. G0S2 is unique in that no homologous proteins could be identified in lower organisms, including C. elegans and Drosophila melanogater. In addition, G0S2 does not share any sequence similarities with proteins of known 3D structure. A hydrophobic region locates between amino acids 27–42. Deletion of this region disables the interaction of G0S2 with ATGL [17].
G0S2 mRNA is expressed ubiquitously. The highest expression levels are detected in adipose tissues and liver followed by muscle, ovary, and kidney [17,100]. In differentiating adipocyte cell lines, such as Simpson-Golabi-Behmel syndrome cells or 3T3-L1 adipocytes, G0S2 mRNA is detectable 2–3 days after the initiation of differentiation and remains high during the entire differentiation period [17,100]. In adipocytes, its expression is induced by insulin and inhibited by TNF-α or isoproterenol, both factors that stimulate lipolysis [17]. Importantly, Zandbergen et al. [100] identified G0S2 as a PPARγ target gene containing a PPAR-response element (PPRE) in its promoter sequence. In contrast to PPARγ, PPARα down-regulates G0S2 mRNA expression [100].
3.1.2.2. G0S2: mode of function
G0S2 directly interacts with the N-terminal patatin domain of ATGL [17]. Recent evidence [17] suggests that this interaction does not directly compete with the binding of CGI-58, which also interacts with the N-terminal region of ATGL [47]. Interestingly, in 3T3-L1 adipocytes both ATGL and G0S2 translocate to LDs upon stimulation of lipolysis, while this is not the case when ATGL expression was down-regulated by small interfering RNA [17]. This suggests that G0S2 by itself may be unable to bind to LDs but requires ATGL as its binding partner. Interestingly, Welch et al. [101] identified G0S2 as a mitochondrial protein, which interacts with Bcl-2, an anti-apoptotic factor, altering mitochondrial membrane permeability and promoting apoptosis. Thus, G0S2 interacts with proteins involved in different cellular processes and may link cell cycle, cell survival, and cell death with lipolysis. The existence of such a link has recently been demonstrated in yeast, where the entry of the S phase is mechanistically connected with an induction of TGL4 activity, the yeast orthologue of ATGL [102]. Future studies are required to focus on the interplay of CGI-58 and G0S2 in the regulation of lipolysis and its role in the pathophysiology of obesity, type II diabetes, and cancer.
3.1.3. Hormonal regulation – ATGL is a hormone-sensitive lipase
Lipolysis in adipocytes is predominantly regulated post-translationally involving phosphorylation and translocation processes of the participating enzymes. The molecular mechanism leading to the activation of HSL is well established (see below). Additionally, early experiments in HSL-deficient adipose tissue showed that the non-HSL lipolytic activity can also be activated by β-adrenergic stimulation [12,21,103,104]. This suggested the existence of another “hormone-sensitive” enzyme besides HSL. With the discovery of ATGL and the availability of ATGL-deficient mice it became evident that β-adrenergic activation of ATGL activity is required for full hormone-activated lipolysis in WAT [10,16,79]. In the absence of ATGL, NEFA and glycerol mobilization in response to β-adrenergic stimulation were decreased by ∼70%.
3.1.4. Regulation of ATGL by LD associated proteins
The molecular mechanisms that regulate ATGL activity in response to β-adrenergic stimulation are incompletely understood. Recent evidence suggests an indirect mechanism involving perilipin-1 and CGI-58. Perilipin-1 is a member of the PAT family (for review see [105]). The name “PAT” derives from the most noticed members of the family, perilipin (now annotated as perilipin-1), adipophilin (also called adipocyte differentiation-related protein; now annotated as perilipin-2), and tail-interacting protein 47 kDa (Tip-47; now annotated as perilipin-3). Other members include perilipin 4 (initially named S3-12) and perilipin-5 (initially named: myocyte LD protein, PAT protein in oxidative tissues, and LD specific protein-5) [106–108].
Perilipin-1 expression is restricted to β-adrenergic stimulatable cells, such as adipocytes and steroidogenic cells, and is essential for β-adrenergic stimulatable lipolysis. The protein governs the ATGL- and HSL-mediated breakdown of fat in WAT in multiple ways (see below). ATGL activity is regulated by the availability of its coactivator CGI-58. In non-stimulated adipocytes, CGI-58 is located at the surface of LDs and is mostly bound to perilipin-1 [93,96,109]. In the activated state, perilipin-1 is phosphorylated by cAMP-dependent protein kinase A (PKA). This causes the dissociation of CGI-58 from perilipin-1, which is now available for the activation of ATGL [96,109]. Granneman et al. [93] recently pinpointed the perilipin-1 phosphorylation sites responsible for the reversible binding of CGI-58 (Ser492 or Ser517) [96]. These observations indicate that perilipin-1 controls the activation of ATGL in WAT by interacting with its coactivator CGI-58 in a cAMP-dependent fashion.
In non-adipose tissues, the activation of lipolysis is less well characterized because these tissues express little or no perilipin-1. Yet, ATGL and CGI-58 play an essential role in TAG hydrolysis particularly in tissues with high NEFA demand, such as skeletal muscle, heart, and liver [16]. Thus, alternative, perilipin-1 independent mechanisms must exist to regulate ATGL activity in non-adipose tissues. Listenberger et al. [110] and Bell et al. [111] demonstrated that another PAT protein, the ubiquitously expressed perilipin-2, regulates the access of ATGL to LDs in various cell lines, such as AML-12 hepatocytes, Hek293 kidney cells, and 3T3-L1 adipocytes. In perilipin-1-deficient mice, perilipin-2 is the major LD coating protein in adipocytes [112]. Whether the regulation of ATGL activity by perilipin-2 in adipose and non-adipose tissues involves the reversible binding of CGI-58 by perilipin-2 is currently unknown. Perilipin-3 is also expressed in virtually all cell types and tissues. In murine hepatocytes downregulation of perilipin-3 expression leads to a dramatic increase of LD size and a decrease in LD number. In addition, these cells show increased lipolysis associated with increased ATGL localization on LD [111]. Thus, perilipin-3 exerts a protective role for LD via preventing ATGL access to the LD. Perilipin-4 is found primarily in WAT and to a lesser degree in skeletal muscle and heart [113] but nothing is known about its involvement in the lipolytic pathway [114]. Finally, perilipin-5 was identified as a LD binding protein by three independent groups [106–108]. Perilipin-5 is expressed predominantly in oxidative tissues, including heart, skeletal muscle, and fasted liver. In fasted tissues, perilipin-5 induces LD fragmentation and lipolysis [107]. Importantly, perilipin-5 has been shown to interact with CGI-58 and to colocalize on LDs. The expression of perilipin-5 greatly increases LD localization of both CGI-58 and ATGL and, thus, may promote their interaction [115]. These observations may indicate that perilipin-5 regulates fatty acid mobilization and oxidation in tissues with high oxidative capacity such as liver and muscle.
Cidec (FSP27) is a recently identified LD protein that is unrelated to the PAT family on a sequence level. Cidec is predominantly expressed in adipose tissue and liver and has a strong inhibitory effect on lipolysis, thus promoting lipid storage [116,117]. It will be interesting to see whether this protein acts via inhibition of ATGL.
3.1.5. Regulation of ATGL by its targeting to LDs
Two independent genetic screens in D. melanogaster L2 cells for genes that affect LD size and morphology revealed that proteins of the ER-Golgi transport machinery are essential for normal LD number and size [118,119]. Silencing of either coat protein complex-I (COPI) or (ADP-ribosylation factor 1) ARF1, two essential members of the transport machinery, in L2 cells inhibited ATGL-mediated lipolysis and increased the cellular TAG content. Soni et al. [120] elegantly demonstrated in human Hela cells that the delivery of ATGL to LDs critically depends on the vesicular transport machinery and its protein components ARF1, SAR1 (small GTP binding proteins), guanine–nucleotide exchange factor, and the effector coatamer proteins COPI and II. In the absence of these proteins, ATGL stays strictly associated with the ER membrane.
3.1.6. ATGL regulation by enzyme phosphorylation
Human ATGL is phosphorylated at two serine residues (Ser404 and Ser428) [121]. However, the relevance of phosphorylation for the regulation of enzyme activity is unclear. In contrast to HSL (see below), phosphorylation of ATGL does not involve PKA [10]. Furthermore, the known phosphorylation sites are not critical for LD localization or in vitro TAG hydrolysis [46]. Interestingly, the ATGL orthologue in C. elegans ATGL-1 is phosphorylated at multiple sites by AMPK thereby inactivating enzyme activity [122]. ATGL-1 inhibition via enzyme phosphorylation prolongs the life span of C. elegans larvae during the dormant state of dauer [122]. Whether matching regulatory phosphorylation sites exist on mammalian ATGL orthologues and whether they are involved in enzyme inactivation during hibernation, long-term fasting, etc. is not known.
3.1.7. Transcriptional regulation of ATGL
ATGL expression and activity have been shown to be regulated by numerous effectors or conditions (summarized in Table 1). The enzyme is upregulated during adipose differentiation [10,31] and a target for transcription factors PPARγ [43,123,124] and insulin-responsive transcription factor forkhead box O1 (FoxO1) [125]. Furthermore, glucocorticoids such as dexamethasone [31], the PPARγ agonists thiazolidinediones [43,123,124,126], and fasting induce mRNA expression. In contrast, insulin [43,127,128], TNF-α [43,128], mTor complex 1 [129], and feeding [45] repress ATGL mRNA expression. Interestingly, the β-adrenergic agonist isoproterenol reduces ATGL (and HSL) mRNA levels in adipocytes [128] although the enzyme activity is induced at the same time. The role of leptin in the regulation of ATGL is controversial. Leptin is known to restrain energy intake and to promote lipolysis, a process involving upregulation of PPARγ expression [130–132]. Yet, a study on porcine adipocyte lipolysis found that leptin decreased ATGL protein expression while it increased mRNA expression [133]. Insulin resistance and obesity have also been correlated with changes in ATGL mRNA or protein levels [43,123,124,126–128].
Table 1.
Regulation of ATGL mRNA expression and enzyme activity.
| mRNA down | mRNA/protein up | Activity down | Activity up |
|---|---|---|---|
| Feeding [45] | Catecholamines [124,126] | Perilipin [144] | CGI-58 [8] |
| Insulin [43,127,128] | Dexamethasone [31] | Adipophilin [110] | Isoproterenol [16,79] |
| Isoproterenol [128] | Fasting [31,45] | G0S2 [17] | |
| mTOR1 [129] | FoxO1 [125] | ||
| TNF-α [43,128] | Thiazolidinediones [43,123,124,126] |
3.2. HSL – regulation of enzyme activity
The regulation of HSL activity has been intensively studied over nearly five decades [134–137]. The best-studied system is the regulation of HSL in adipocytes. Adipose HSL activity is controlled by two distinct mechanisms in response to β-adrenergic stimulation: first, the enzyme is phosphorylated by cAMP-dependent PKA. This leads to an increase of the intrinsic enzyme activity (∼two-fold). HSL harbors at least five distinct serine residues that can be phosphorylated. Recent results demonstrated that phosphorylation of Ser650 (human HSL) or Ser660 (rat HSL) are particularly important for enzyme activity [138]. Besides PKA, other protein kinases have also been shown to phosphorylate HSL and regulate enzyme activity. The list includes extracellular signal-regulated kinase, glycogen synthase kinase-4 [139], Ca2+/calmodulin-dependent kinase II, and AMP-activated kinase [140]. Second, phosphorylated HSL interacts with the LD protein perilipin-1, which itself is a target of PKA phosphorylation.
The translocation of the phosphorylated enzyme to the LD in WAT is mediated by perilipin-1. In the basal, non-hormonally stimulated state, perilipin-1 is not phosphorylated and prevents the binding of HSL to LDs. In perilipin-1-deficient mice this barrier function is not present, basal lipolysis in WAT is increased, and the animals are leaner than wild-type mice [141–143]. In response to β-adrenergic stimulation, perilipin is phosphorylated on six consensus serine residues by PKA [144]. Specifically the phosphorylation of serines 81, 222, and 276 induce the binding of HSL to perilipin-1 and access to the LD [145]. The interaction domain of perilipin with HSL involves the N-terminal region of perilipin-1 although the exact region is controversial [145,146]. Consistent with this pro-lipolytic activity of phosphorylated perilipin-1, perilipin-1-deficient mice are unable to sufficiently recruit HSL to the LD and incapable to induce lipolysis in WAT upon hormone stimulation [142,143]. After full hormonal stimulation, HSL phosphorylation and the perilipin-1 mediated translocation of the enzyme to the LD causes a ∼100-fold induction of HSL activity in WAT [79].
In non-adipose cells lacking perilipin-1 the role of HSL is less well characterized. Generally, HSL expression in these cells and tissues is quite low. However, in some of them a physiological function of HSL has been described, e.g. in muscle and macrophages [135,147–153]. The interaction of HSL with LDs in tissues lacking perilipin-1 may involve perilipin-2 and perilipin-5 [145].
3.3. Regulation of MGL
To date, no evidence exists that cellular MGL mRNA concentrations or enzyme activities are regulated by either hormones or the energy state of the cell. MGL is highly expressed in many tissues. High MAG hydrolase activity levels are constitutively present in adipocytes, hepatocytes, and muscle cells suggesting that this activity is not subject to extensive regulation. MGL is necessary for the complete breakdown of TAG in in vitro experiments [154]. Yet, other enzymes such as HSL and α/β hydrolase domain containing protein 6 (ABHD6) also exhibit MAG hydrolase activity and it is therefore not clear whether in vivo MGL is the only relevant MAG hydrolase. The generation of an MGL-deficient mouse model has recently been published [23]. In this study the role of MGL in the breakdown of the endocannabinoid 2-arachidonoyl glycerol (2-AG) has been confirmed. Yet, whether or not a block of 2-AG catabolism in these mice affects appetite regulation and energy homeostasis and whether MGL is rate-limiting in the breakdown of MAG in adipose tissues has not been addressed.
4. Lipolysis: physiology and pathophysiology
4.1. ATGL is essential for TAG catabolism to provide NEFAs as energy fuel
Since ATGL is highly expressed in WAT and BAT it was thought to be of particular importance in these tissues [10]. However, the phenotype of ATGL-deficient mice [16] broadened this view and provided solid evidence for a crucial role of the enzyme in non-adipose tissues. ATGL-deficiency in mice is associated with reduced lipolysis resulting in excessive fat deposition in virtually all tissues including fat, liver, muscle, kidney, spleen, and lung. This implicates that the ATGL mediated catabolism of TAG is required in essentially all cell types of the body. Massive TAG accumulation in the heart of ATGL-deficient mice caused cardiac myopathy and premature death starting with the age of ∼12 weeks. The animals are moderately obese when kept on a low fat chow diet. The size of BAT, in contrast, increases dramatically (∼20-fold) due to the massive accumulation of lipid. Interestingly, ATGL-deficient mice are similarly active as wild-type mice when fed ad libitum. However, the situation changes dramatically under conditions of increased energy demand such as fasting, cold exposure, or physical exercise. Under these conditions, ATGL is essential for the provision of NEFAs as metabolic fuel. Accordingly, ATGL-deficient mice are unable to increase thermogenesis in response to cold exposure. Fasting or exercise normally induces lipolysis resulting in increased plasma NEFA levels. This increase does not occur in ATGL-deficient animals [155–157]. With increasing fasting time, ATGL-deficient animals develop hypoglycemia and show a gradual decrease in oxygen consumption starting ∼8 h after food deprivation [155]. After overnight fasting, oxygen consumption is reduced by 70–80% and mice exhibit hypothermia indicating energy starvation. The respiratory quotient of starved ATGL-deficient mice is elevated in comparison to wild-type mice indicative for an increased usage of carbohydrates for energy conversion. This change in energy substrate utilization can also be observed in response to physical exercise [155]. Moderately exercised animals rapidly deplete their liver glycogen stores and develop hypoglycemia under conditions which barely affect glycogen stores of wild-type animals [155,157]. Together, these observations demonstrate that ATGL-deficient mice are not capable of mobilizing sufficient energy in form of NEFAs to maintain normal energy metabolism during fasting or exercise. Conversely, tissue-specific overexpression of the enzyme in adipose tissue of transgenic mice results in increased lipolytic rates and decreased body weight [158].
It has long been known that NEFA metabolism strongly affects glucose utilization in skeletal muscle, the most efficient energy dissipating organ in the body. Randle et al. [159] proposed a “glucose–fatty acid cycle” in which the utilization of glucose is directly inhibited by the presence of NEFAs and vice versa. This mechanism adapts substrate utilization to substrate availability in coordination with hormones controlling energy metabolism [160]. Obesity and type II diabetes are commonly associated with high circulating NEFA levels. The chronic overexposure of non-adipose tissues to NEFAs promotes the accumulation of TAGs and NEFA-derived metabolites in liver, muscle, pancreatic β-cells, and other tissues. Additionally, it compromises insulin signaling and cell function, a process which is often referred to as lipotoxicity [161]. However, ATGL-deficient mice disproved the hypothesis that TAG accumulation per se causes insulin resistance or lipotoxicity because these animals exhibit increased insulin sensitivity and increased glucose tolerance despite massive accumulation of TAG in muscle and liver [16]. Furthermore, ATGL-deficient animals support the concept of the glucose–fatty acid cycle, since in this mouse model reduced NEFA levels are associated with increased carbohydrate oxidation. ATGL affects glucose metabolism by different mechanisms and in a tissue-specific manner: (i) measures of insulin signaling are increased in skeletal muscle and WAT, but decreased in BAT and liver [162]. (ii) In pancreatic islets, ATGL deficiency causes TAG accumulation and impairs fuel and non-fuel-stimulated insulin secretion, which is consistent with the concept that ATGL is required for the provision of NEFAs as energy source for the ATP demanding process of insulin secretion. Thus, the lack of ATGL leads to hypoinsulinemia in ATGL-deficient mice [163]. (iii) Serum insulin levels in ATGL-deficient mice are drastically reduced during glucose tolerance tests despite improved glucose tolerance [16]. This suggests that enhanced glucose tolerance is at least in part mediated by non-insulin dependent mechanisms.
In addition to serum NEFAs, glucose, and insulin levels, retinol-binding protein 4 (RBP4) has been implicated in the development of insulin resistance [164,165]. Human association studies and the characterization of mutant mouse lines that lack or overexpress RBP4 established a direct link between elevated serum RBP4 levels and insulin resistance [165–172]. Interestingly, in ATGL-deficient mice insulin sensitivity is not only associated with reduced plasma NEFA levels but also with decreased serum RBP4 concentrations [162]. Despite these initial observations, the question as to how ATGL activity regulates insulin sensitivity, insulin secretion, or RBP4 levels requires further attention.
4.2. The role of ATGL in human physiology – identification of ATGL and CGI-58 as causative genes for the development of NLSD
Following the discovery of ATGL, the question whether the enzyme plays the same significant role in human adipose and non-adipose tissues as in mouse tissues was somewhat controversial [173]. Recently, however, Langin and coworkers re-addressed the issue [174], examining the roles of HSL and ATGL/CGI-58 in basal and forskolin-stimulated lipolysis in a human white adipocyte model (hMADS cells). In an elegant study, the authors concluded that ATGL/CGI-58 acts independently of HSL and precedes its action in the sequential hydrolysis of TAG [174]. These results are consistent with the observations in mice and argue for an essential role of ATGL in the initiation of lipolysis.
Further evidence for an essential role of ATGL in human lipid metabolism came from the characterization of patients with neutral lipid storage disease (NLSD). This lipid disorder is characterized by massive TAG accumulation in many tissues and lymphocytes [175]. Interestingly, some patients with NLSD also developed ichthyosis, a massive skin defect, while other patients did not. NLSD patients without ichthyosis instead suffered from more severe skeletal and cardiac myopathy [176]. The first attempts to delineate the molecular basis for excessive TAG accumulation goes back to studies conducted in the 1980s and 1990s by the labs of Douste-Blazy [177,178], Salvayre [179–181], and Coleman [176,177,182–186]. The groups of Douste-Blazy and Salvayre used fibroblasts obtained from a patient suffering from “multisystemic lipid storage myopathy” without signs of ichthyosis and concluded from their pulse-chase experiments that the TAG accumulation may be explained by defective catabolism of cytoplasmic TAG [177,180]. Coleman’s group, using fibroblasts from a patient suffering from NLSD with ichthyosis, concluded quite the opposite. They found that the massive TAG accumulation is more likely to be caused by defects in phospholipid metabolism rather than TAG catabolism [182,183]. In 2001, the group of Fischer finally identified mutations in the gene coding for CGI-58 as causative for Chanarin-Dorfman syndrome (CDS) [187], now referred to as NLSD with ichthyosis (NLSDI) [14]. Later, CGI-58 was shown to specifically activate TAG hydrolysis by ATGL and biochemical studies with CGI-58 mutants known to be associated with NLSDI revealed that mutated CGI-58 variants completely lost their capability to stimulate ATGL [8]. This study provided the biochemical function for CGI-58 in TAG catabolism and a rational mechanism for the lipid storage phenotype in patients with CGI-58 deficiency. The functional link between CGI-58 and ATGL also suggested that in a subgroup of patients NLSD might be caused by a mutation of ATGL. This prediction was verified in 2007 [14]. Patients with mutations in the ATGL gene develop a similar phenotype as ATGL-deficient mice: they suffer from massive TAG accumulation in multiple tissues and develop severe myopathy. Cardiac myopathy is often lethal and can only be prevented by heart transplantation. Importantly, however, and in contrast to patients with defects in CGI-58, patients with mutations in ATGL do not develop ichthyosis [14]. Because of these phenotypical differences and to better differentiate between two distinct monogenic disorders, NLSD caused by mutations in the ATGL gene is now referred to as NLSD with myopathy (NLSDM). So far, 10 different mutations causative for NLSDM have been described (reviewed in Schweiger et al. [15]). In most of the patients mutations affect the C-terminal half of the enzyme leading to truncated forms of ATGL. In these variants the patatin domain, containing the putative catalytic dyad (Ser47–Asp166), and the αβα sandwich domain are intact and the proteins may retain remnant enzyme activity in vivo [48]. In accordance with the NLSDM phenotype, however, the truncated enzymes exhibit defective LD binding, suggesting that reduced lipolysis is caused by mislocalization of ATGL [47]. In one patient a mutation occurred within the patatin domain of ATGL and this individual may entirely lack a functional enzyme [188].
Interestingly, and in contrast to mice lacking ATGL, patients affected with mutations in the ATGL gene are not obese [14]. Whether these results from remnant ATGL activity in WAT of most NLSDM patients or whether the activity of alternative lipases can compensate for the absence of ATGL (e.g. HSL) is currently unknown. It is also conceivable that either a compensatory down-regulation of lipid synthesis prevents excessive fat accumulation in WAT when lipolysis is defective or that behavioral changes lead to decreased food intake and, thus, prevent WAT accumulation. Interestingly and in good agreement with the observations in ATGL-deficient mice, NLSDM patients do not develop insulin resistance despite massive hepatic and ectopic lipid accumulation [14].
4.3. Physiological role of CGI-58: CGI-58 is essential for functional TAG catabolism and skin barrier development
The phenotypical difference in the skin between patients with NLSDI and NLSDM strongly argued for an ATGL-independent function of CGI-58 in the skin. To better define this function, CGI-58-deficient mice were generated and characterized with regard to systemic lipid metabolism and skin physiology. Unexpectedly and in contrast to humans, CGI-58 deficient mice die between 12 and 16 h after birth [189]. Consequently, biochemical and physiological studies of CGI-58 deficiency in mice were restrained to new-born pups and disclosed a dual function of CGI-58 in lipid metabolism. First, systemic TAG accumulation and severe hepatic steatosis in newborn CGI-58-deficient mice confirmed a rate-limiting role for CGI-58 in TAG hydrolysis. This function of CGI-58 in TAG catabolism is directly linked to its ability to activate ATGL [8]. Second, CGI-58-deficient mice suffer from a severe skin permeability barrier defect, which leads to rapid desiccation after birth causing postnatal death. These phenotypes strongly resemble those observed in human subjects with defective CGI-58. In contrast to mice, however, the skin defect in humans is not lethal. Possible reasons for this phenotypical difference could be that humans nurse their offsprings more intensely, they have increased volume to surface ratio, and the human skin might be less permeable since it lacks a protective fur.
Extensive characterization of the biochemical defect in the epidermis of CGI-58-deficient mice revealed that the skin barrier defect is linked to the impaired hydrolysis of epidermal TAG. This defect can be partially restored by addition of recombinant CGI-58. The hydrolytic defect in the CGI-58-deficient epidermis leads to the sequestration of NEFAs in LDs. NEFAs, however, are essential for the synthesis of skin lipids and normal barrier function. They are required for the activation of PPARs during keratinocyte differentiation [190–194], the formation of long-chain fatty acid ω-OH-ceramides [195,196], and the subsequent generation of acylceramides. The massive accumulation of ω-OH-ceramides, the complete lack of acylceramides, and drastically reduced levels of protein-bound ceramides in the corneocyte lipid envelope suggested that LD-derived NEFAs are an absolute requirement for the formation of a functional skin permeability barrier [189]. Importantly, the skin defect in NLSDI patients appears to arise from the same biochemical defect since samples of human epidermis of NLSDI patients also lack acylceramides [197]. Consistent with the defect in TAG catabolism [188,198,199], the epidermal layer of NLSDI patients accumulates TAG as reported in [200]. In a more detailed study, Akiyama et al. [199] reported that CGI-58 is packaged into lipid transport and secretion granules (lamellar granules) of keratinocytes during keratinization. CGI-58-deficiency leads to the malformation of lamellar granules and thus might play a role in remodeling of granular lipids before or even after secretion. Taken together, these findings are consistent with the concept that CGI-58 activates a currently unknown epidermal TAG hydrolase, which catabolizes TAG and provides NEFAs for the synthesis of acylceramides. The lack of acylceramides in CGI-58-deficient mice or humans impedes the assembly of a functional corneocyte lipid envelope leading to the observed severe skin barrier defect.
Beside the essential role of CGI-58 in TAG catabolism, it is also conceivable that the acyl-CoA-dependent LPAAT activity [97] of the protein (see above) contributes to the skin defect in CGI-58 deficiency [98]. This reaction may channel fatty acids from the breakdown of TAG into glycerophospholipids [93,201]. The observation of Coleman and colleagues that NLSD fibroblasts, which express a C-terminal truncated CGI-58 variant [8], are defective in phospholipid metabolism coheres with this mechanism. Yet, Ghosh et al. [97] reported that mutant CGI-58 variants, known to be associated with the development of NLSDI, are not compromised in LPAAT activity. Thus, the significance of CGI-58 as LPAAT in skin lipid metabolism and the development of ichthyosis remains to be elucidated.
5. Conclusions
Over the last 5 years important discoveries extended our knowledge and led to a revision of the lipolytic pathway catabolizing cellular fat stores. They included the discovery of ATGL as an essential TAG hydrolase and the identification of the coactivator CGI-58 and the corepressor G0S2. Additionally, a large number of protein factors have been characterized mediating the correct enzyme targeting (e.g. the ARF/COP system) to LDs, the access to the substrate (e.g. PAT proteins), and the regulation of enzyme activity (e.g. Fsp27).
Important questions, however, remain to be addressed: the hormonal regulation of ATGL and its coordinated interplay with the other lipolytic and lipogenic enzymes require further investigation. Particularly the role of phosphorylation for enzyme activity needs to be assessed in more detail. In more general terms, it will be important to elucidate the role of lipolysis for the generation of lipid intermediates required for cell signaling processes. In this context, it is conceivable that specific lipolysis-derived NEFAs, DAGs, or MAGs act as ligands or precursors for ligands of nuclear receptors. Similarly, it is possible that the controlled release of polyunsaturated NEFAs from LDs regulates the synthesis and biological action of eicosanoids and prostaglandins. Specific stereoisomers of DAGs regulate various isoforms of protein kinase-C and insulin action. It remains to be determined whether the lipolytic system can participate in the generation or degradation of these signaling lipids. Finally, the role of lipolysis in human disease is not sufficiently delineated. Further studies are needed to understand the role of the newly identified lipolytic players in the pathogenesis of insulin resistance, diabetes, and obesity. Similarly, the role of lipolysis in the uncontrolled loss of adipose tissue during cachexia requires clarification.
In summary, the increasing number of enzymes and interacting partners on LD indicates that lipolysis comprises a large network of factors coordinately acting within a “lipolytic machinery”. The current pace of new discoveries will augment the complexity of this machinery and disclose its involvement in the pathogenesis of lipid-associated disorders.
Acknowledgements
This work was supported by the Grant “GOLD: Genomic Of Lipid-associated Disorders”, which is part of the Austrian Genome Project “GEN-AU: Genome Research in Austria” funded by the Austrian Ministry of Science and Research and the FFG. This research was also supported by the Grants P21296, P22170, F30 SFB LIPOTOX, and the Wittgenstein Award Z136, which are funded by the Austrian Science Foundation. Additional funding was obtained from the European Commission Grant Agreements Nos. 201608 (TOBI) and 202272 (LipidomicNet). We thank Mag. Caroline Schober-Trummler and Mag. Claudia Lanschuetzer for their careful and critical reading of the manuscript.
References
- 1.Unger R.H., Clark G.O., Scherer P.E., Orci L. Lipid homeostasis, lipotoxicity and the metabolic syndrome. Biochim Biophys Acta. 2010;1801:209–214. doi: 10.1016/j.bbalip.2009.10.006. [DOI] [PubMed] [Google Scholar]
- 2.Slawik M., Vidal-Puig A.J. Lipotoxicity, overnutrition and energy metabolism in aging. Ageing Res Rev. 2006;5:144–164. doi: 10.1016/j.arr.2006.03.004. [DOI] [PubMed] [Google Scholar]
- 3.DeFronzo R.A. Dysfunctional fat cells, lipotoxicity and type 2 diabetes. Int J Clin Pract Suppl. 2004:9–21. doi: 10.1111/j.1368-504x.2004.00389.x. [DOI] [PubMed] [Google Scholar]
- 4.Lelliott C., Vidal-Puig A.J. Lipotoxicity, an imbalance between lipogenesis de novo and fatty acid oxidation. Int J Obes Relat Metab Disord. 2004;28(Suppl. 4):S22–S28. doi: 10.1038/sj.ijo.0802854. [DOI] [PubMed] [Google Scholar]
- 5.Duncan R.E., Ahmadian M., Jaworski K., Sarkadi-Nagy E., Sul H.S. Regulation of lipolysis in adipocytes. Annu Rev Nutr. 2007;27:79–101. doi: 10.1146/annurev.nutr.27.061406.093734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Langin D. Control of fatty acid and glycerol release in adipose tissue lipolysis. CR Biol. 2006;329:598–607. doi: 10.1016/j.crvi.2005.10.008. [discussion 653–655] [DOI] [PubMed] [Google Scholar]
- 7.Zechner R., Strauss J.G., Haemmerle G., Lass A., Zimmermann R. Lipolysis: pathway under construction. Curr Opin Lipidol. 2005;16:333–340. doi: 10.1097/01.mol.0000169354.20395.1c. [DOI] [PubMed] [Google Scholar]
- 8.Lass A., Zimmermann R., Haemmerle G., Riederer M., Schoiswohl G., Schweiger M. Adipose triglyceride lipase-mediated lipolysis of cellular fat stores is activated by CGI-58 and defective in Chanarin-Dorfman syndrome. Cell Metab. 2006;3:309–319. doi: 10.1016/j.cmet.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 9.Holm C. Molecular mechanisms regulating hormone-sensitive lipase and lipolysis. Biochem Soc Trans. 2003;31:1120–1124. doi: 10.1042/bst0311120. [DOI] [PubMed] [Google Scholar]
- 10.Zimmermann R., Strauss J.G., Haemmerle G., Schoiswohl G., Birner-Gruenberger R., Riederer M. Fat mobilization in adipose tissue is promoted by adipose triglyceride lipase. Science. 2004;306:1383–1386. doi: 10.1126/science.1100747. [DOI] [PubMed] [Google Scholar]
- 11.Haemmerle G., Zimmermann R., Hayn M., Theussl C., Waeg G., Wagner E. Hormone-sensitive lipase deficiency in mice causes diglyceride accumulation in adipose tissue, muscle, and testis. J Biol Chem. 2002;277:4806–4815. doi: 10.1074/jbc.M110355200. [DOI] [PubMed] [Google Scholar]
- 12.Osuga J., Ishibashi S., Oka T., Yagyu H., Tozawa R., Fujimoto A. Targeted disruption of hormone-sensitive lipase results in male sterility and adipocyte hypertrophy, but not in obesity. Proc Natl Acad Sci USA. 2000;97:787–792. doi: 10.1073/pnas.97.2.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karlsson M., Contreras J.A., Hellman U., Tornqvist H., Holm C. CDNA cloning, tissue distribution, and identification of the catalytic triad of monoglyceride lipase. Evolutionary relationship to esterases, lysophospholipases, and haloperoxidases. J Biol Chem. 1997;272:27218–27223. doi: 10.1074/jbc.272.43.27218. [DOI] [PubMed] [Google Scholar]
- 14.Fischer J., Lefevre C., Morava E., Mussini J.M., Laforet P., Negre-Salvayre A. The gene encoding adipose triglyceride lipase (PNPLA2) is mutated in neutral lipid storage disease with myopathy. Nat Genet. 2007;39:28–30. doi: 10.1038/ng1951. [DOI] [PubMed] [Google Scholar]
- 15.Schweiger M., Lass A., Zimmermann R., Eichmann T.O., Zechner R. Neutral lipid storage disease: genetic disorders caused by mutations in adipose triglyceride lipase/PNPLA2 or CGI-58/ABHD5. Am J Physiol Endocrinol Metab. 2009;297:E289–E296. doi: 10.1152/ajpendo.00099.2009. [DOI] [PubMed] [Google Scholar]
- 16.Haemmerle G., Lass A., Zimmermann R., Gorkiewicz G., Meyer C., Rozman J. Defective lipolysis and altered energy metabolism in mice lacking adipose triglyceride lipase. Science. 2006;312:734–737. doi: 10.1126/science.1123965. [DOI] [PubMed] [Google Scholar]
- 17.Yang X., Lu X., Lombes M., Rha G.B., Chi Y.I., Guerin T.M. The G(0)/G(1) switch gene 2 regulates adipose lipolysis through association with adipose triglyceride lipase. Cell Metab. 2010;11:194–205. doi: 10.1016/j.cmet.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu X., Yang X., Liu J. Differential control of ATGL-mediated lipid droplet degradation by CGI-58 and G0S2. Cell Cycle. 2010;9:2719–2725. doi: 10.4161/cc.9.14.12181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jenkins C.M., Mancuso D.J., Yan W., Sims H.F., Gibson B., Gross R.W. Identification, cloning, expression, and purification of three novel human calcium-independent phospholipase A2 family members possessing triacylglycerol lipase and acylglycerol transacylase activities. J Biol Chem. 2004;279:48968–48975. doi: 10.1074/jbc.M407841200. [DOI] [PubMed] [Google Scholar]
- 20.Wilson P.A., Gardner S.D., Lambie N.M., Commans S.A., Crowther D.J. Characterization of the human patatin-like phospholipase family. J Lipid Res. 2006;47:1940–1949. doi: 10.1194/jlr.M600185-JLR200. [DOI] [PubMed] [Google Scholar]
- 21.Okazaki H., Osuga J., Tamura Y., Yahagi N., Tomita S., Shionoiri F. Lipolysis in the absence of hormone-sensitive lipase: evidence for a common mechanism regulating distinct lipases. Diabetes. 2002;51:3368–3375. doi: 10.2337/diabetes.51.12.3368. [DOI] [PubMed] [Google Scholar]
- 22.Kinsey SG, Long JZ, Cravatt BF, Lichtman AH. Fatty acid amide hydrolase and monoacylglycerol lipase inhibitors produce anti-allodynic effects in mice through distinct cannabinoid receptor mechanisms. J Pain 2010; in press. [DOI] [PMC free article] [PubMed]
- 23.Schlosburg J.E., Blankman J.L., Long J.Z., Nomura D.K., Pan B., Kinsey S.G. Chronic monoacylglycerol lipase blockade causes functional antagonism of the endocannabinoid system. Nat Neurosci. 2010;13:1113–1119. doi: 10.1038/nn.2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Long J.Z., Nomura D.K., Vann R.E., Walentiny D.M., Booker L., Jin X. Dual blockade of FAAH and MAGL identifies behavioral processes regulated by endocannabinoid crosstalk in vivo. Proc Natl Acad Sci USA. 2009;106:20270–20275. doi: 10.1073/pnas.0909411106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pan B., Wang W., Long J.Z., Sun D., Hillard C.J., Cravatt B.F. Blockade of 2-arachidonoylglycerol hydrolysis by selective monoacylglycerol lipase inhibitor 4-nitrophenyl 4-(dibenzo[d][1,3]dioxol-5-yl(hydroxy)methyl)piperidine-1-carboxylate (JZL184) enhances retrograde endocannabinoid signaling. J Pharmacol Exp Ther. 2009;331:591–597. doi: 10.1124/jpet.109.158162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Long J.Z., Nomura D.K., Cravatt B.F. Characterization of monoacylglycerol lipase inhibition reveals differences in central and peripheral endocannabinoid metabolism. Chem Biol. 2009;16:744–753. doi: 10.1016/j.chembiol.2009.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kinsey S.G., Long J.Z., O’Neal S.T., Abdullah R.A., Poklis J.L., Boger D.L. Blockade of endocannabinoid-degrading enzymes attenuates neuropathic pain. J Pharmacol Exp Ther. 2009;330:902–910. doi: 10.1124/jpet.109.155465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schlosburg J.E., Carlson B.L., Ramesh D., Abdullah R.A., Long J.Z., Cravatt B.F. Inhibitors of endocannabinoid-metabolizing enzymes reduce precipitated withdrawal responses in THC-dependent mice. AAPS J. 2009;11:342–352. doi: 10.1208/s12248-009-9110-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Long J.Z., Li W., Booker L., Burston J.J., Kinsey S.G., Schlosburg J.E. Selective blockade of 2-arachidonoylglycerol hydrolysis produces cannabinoid behavioral effects. Nat Chem Biol. 2009;5:37–44. doi: 10.1038/nchembio.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Blankman J.L., Simon G.M., Cravatt B.F. A comprehensive profile of brain enzymes that hydrolyze the endocannabinoid 2-arachidonoylglycerol. Chem Biol. 2007;14:1347–1356. doi: 10.1016/j.chembiol.2007.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Villena J.A., Roy S., Sarkadi-Nagy E., Kim K.H., Sul H.S. Desnutrin, an adipocyte gene encoding a novel patatin domain-containing protein, is induced by fasting and glucocorticoids: ectopic expression of desnutrin increases triglyceride hydrolysis. J Biol Chem. 2004;279:47066–47075. doi: 10.1074/jbc.M403855200. [DOI] [PubMed] [Google Scholar]
- 32.Rogalska E., Cudrey C., Ferrato F., Verger R. Stereoselective hydrolysis of triglycerides by animal and microbial lipases. Chirality. 1993;5:24–30. doi: 10.1002/chir.530050106. [DOI] [PubMed] [Google Scholar]
- 33.Shewry P.R. Tuber storage proteins. Ann Bot (Lond) 2003;91:755–769. doi: 10.1093/aob/mcg084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rydel T.J., Williams J.M., Krieger E., Moshiri F., Stallings W.C., Brown S.M. The crystal structure, mutagenesis, and activity studies reveal that patatin is a lipid acyl hydrolase with a Ser–Asp catalytic dyad. Biochemistry. 2003;42:6696–6708. doi: 10.1021/bi027156r. [DOI] [PubMed] [Google Scholar]
- 35.Dessen A., Tang J., Schmidt H., Stahl M., Clark J.D., Seehra J. Crystal structure of human cytosolic phospholipase A2 reveals a novel topology and catalytic mechanism. Cell. 1999;97:349–360. doi: 10.1016/s0092-8674(00)80744-8. [DOI] [PubMed] [Google Scholar]
- 36.Notari L., Baladron V., Aroca-Aguilar J.D., Balko N., Heredia R., Meyer C. Identification of a lipase-linked cell membrane receptor for pigment epithelium-derived factor. J Biol Chem. 2006;281:38022–38037. doi: 10.1074/jbc.M600353200. [DOI] [PubMed] [Google Scholar]
- 37.Chen J.F., Dai L.H., Xu N.Y., Xiong Y.Z., Jiang S.W. Assignment of the patatin-like phospholipase domain containing 2 gene (PNPLA2) to porcine chromosome 2p17 with radiation hybrids. Cytogenet Genome Res. 2006;112:342G. doi: 10.1159/000089897. [DOI] [PubMed] [Google Scholar]
- 38.Saarela J., Jung G., Hermann M., Nimpf J., Schneider W.J. The patatin-like lipase family in Gallus gallus. BMC Genomics. 2008;9:281. doi: 10.1186/1471-2164-9-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gronke S., Mildner A., Fellert S., Tennagels N., Petry S., Muller G. Brummer lipase is an evolutionary conserved fat storage regulator in Drosophila. Cell Metab. 2005;1:323–330. doi: 10.1016/j.cmet.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 40.Eastmond P.J. Sugar-dependent1 encodes a patatin domain triacylglycerol lipase that initiates storage oil breakdown in germinating Arabidopsis seeds. Plant Cell. 2006;18:665–675. doi: 10.1105/tpc.105.040543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kurat C.F., Natter K., Petschnigg J., Wolinski H., Scheuringer K., Scholz H. Obese yeast: triglyceride lipolysis is functionally conserved from mammals to yeast. J Biol Chem. 2006;281:491–500. doi: 10.1074/jbc.M508414200. [DOI] [PubMed] [Google Scholar]
- 42.Athenstaedt K., Daum G. Tgl4p and Tgl5p, two triacylglycerol lipases of the yeast Saccharomyces cerevisiae are localized to lipid particles. J Biol Chem. 2005;280:37301–37309. doi: 10.1074/jbc.M507261200. [DOI] [PubMed] [Google Scholar]
- 43.Kim J.Y., Tillison K., Lee J.H., Rearick D.A., Smas C.M. The adipose tissue triglyceride lipase ATGL/PNPLA2 is downregulated by insulin and TNF-alpha in 3T3–L1 adipocytes and is a target for transactivation by PPARgamma. Am J Physiol Endocrinol Metab. 2006;291:E115–E127. doi: 10.1152/ajpendo.00317.2005. [DOI] [PubMed] [Google Scholar]
- 44.Lake A.C., Sun Y., Li J.L., Kim J.E., Johnson J.W., Li D. Expression, regulation, and triglyceride hydrolase activity of adiponutrin family members. J Lipid Res. 2005;46:2477–2487. doi: 10.1194/jlr.M500290-JLR200. [DOI] [PubMed] [Google Scholar]
- 45.Kershaw E.E., Hamm J.K., Verhagen L.A., Peroni O., Katic M., Flier J.S. Adipose triglyceride lipase: function, regulation by insulin, and comparison with adiponutrin. Diabetes. 2006;55:148–157. [PMC free article] [PubMed] [Google Scholar]
- 46.Duncan R.E., Wang Y., Ahmadian M., Lu J., Sarkadi-Nagy E., Sul H.S. Characterization of desnutrin functional domains: critical residues for triacylglycerol hydrolysis in cultured cells. J Lipid Res. 2010;51:309–317. doi: 10.1194/jlr.M000729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schweiger M., Schoiswohl G., Lass A., Radner F.P., Haemmerle G., Malli R. The C-terminal region of human adipose triglyceride lipase affects enzyme activity and lipid droplet binding. J Biol Chem. 2008;283:17211–17220. doi: 10.1074/jbc.M710566200. [DOI] [PubMed] [Google Scholar]
- 48.Kobayashi K., Inoguchi T., Maeda Y., Nakashima N., Kuwano A., Eto E. The lack of the C-terminal domain of adipose triglyceride lipase causes neutral lipid storage disease through impaired interactions with lipid droplets. J Clin Endocrinol Metab. 2008;93:2877–2884. doi: 10.1210/jc.2007-2247. [DOI] [PubMed] [Google Scholar]
- 49.Vaughan M., Berger J.E., Steinberg D. Hormone-sensitive lipase and monoglyceride lipase activities in adipose tissue. J Biol Chem. 1964;239:401–409. [PubMed] [Google Scholar]
- 50.Bjorntorp P., Furman R.H. Lipolytic activity in rat epididymal fat pads. Am J Physiol. 1962;203:316–322. doi: 10.1152/ajplegacy.1962.203.2.316. [DOI] [PubMed] [Google Scholar]
- 51.Hollenberg C.H., Raben M.S., Astwood E.B. The lipolytic response to corticotropin. Endocrinology. 1961;68:589–598. doi: 10.1210/endo-68-4-589. [DOI] [PubMed] [Google Scholar]
- 52.MA R.I.Z.A.C.K. Activation of an epinephrine-sensitive lipolytic activity from adipose tissue by adenosine 3′,5′-phosphate. J Biol Chem. 1964;239:392–395. [PubMed] [Google Scholar]
- 53.Yeaman S.J., Smith G.M., Jepson C.A., Wood S.L., Emmison N. The multifunctional role of hormone-sensitive lipase in lipid metabolism. Adv Enzyme Regul. 1994;34:355–370. doi: 10.1016/0065-2571(94)90022-1. [DOI] [PubMed] [Google Scholar]
- 54.Yeaman S.J. Hormone-sensitive lipase – a multipurpose enzyme in lipid metabolism. Biochim Biophys Acta. 1990;1052:128–132. doi: 10.1016/0167-4889(90)90067-n. [DOI] [PubMed] [Google Scholar]
- 55.Kraemer F.B., Patel S., Saedi M.S., Sztalryd C. Detection of hormone-sensitive lipase in various tissues. I. Expression of an HSL/bacterial fusion protein and generation of anti-HSL antibodies. J Lipid Res. 1993;34:663–671. [PubMed] [Google Scholar]
- 56.Kraemer F.B., Patel S., Singh-Bist A., Gholami S.S., Saedi M.S., Sztalryd C. Detection of hormone-sensitive lipase in various tissues. II. Regulation in the rat testis by human chorionic gonadotropin. J Lipid Res. 1993;34:609–616. [PubMed] [Google Scholar]
- 57.Holm C., Belfrage P., Fredrikson G. Human adipose tissue hormone-sensitive lipase: identification and comparison with other species. Biochim Biophys Acta. 1989;1006:193–197. doi: 10.1016/0005-2760(89)90195-1. [DOI] [PubMed] [Google Scholar]
- 58.Tsujita T., Ninomiya H., Okuda H. P-Nitrophenyl butyrate hydrolyzing activity of hormone-sensitive lipase from bovine adipose tissue. J Lipid Res. 1989;30:997–1004. [PubMed] [Google Scholar]
- 59.Anthonsen M.W., Degerman E., Holm C. Partial purification and identification of hormone-sensitive lipase from chicken adipose tissue. Biochem Biophys Res Commun. 1997;236:94–99. doi: 10.1006/bbrc.1997.6923. [DOI] [PubMed] [Google Scholar]
- 60.Khoo J.C., Reue K., Steinberg D., Schotz M.C. Expression of hormone-sensitive lipase mRNA in macrophages. J Lipid Res. 1993;34:1969–1974. [PubMed] [Google Scholar]
- 61.Sztalryd C., Kraemer F.B. Regulation of hormone-sensitive lipase during fasting. Am J Physiol. 1994;266:E179–E185. doi: 10.1152/ajpendo.1994.266.2.E179. [DOI] [PubMed] [Google Scholar]
- 62.Langin D., Laurell H., Holst L.S., Belfrage P., Holm C. Gene organization and primary structure of human hormone-sensitive lipase: possible significance of a sequence homology with a lipase of Moraxella TA144, an antarctic bacterium. Proc Natl Acad Sci USA. 1993;90:4897–4901. doi: 10.1073/pnas.90.11.4897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Laurin N.N., Wang S.P., Mitchell G.A. The hormone-sensitive lipase gene is transcribed from at least five alternative first exons in mouse adipose tissue. Mamm Genome. 2000;11:972–978. doi: 10.1007/s003350010185. [DOI] [PubMed] [Google Scholar]
- 64.Osterlund T., Danielsson B., Degerman E., Contreras J.A., Edgren G., Davis R.C. Domain-structure analysis of recombinant rat hormone-sensitive lipase. Biochem J. 1996;319(Pt 2):411–420. doi: 10.1042/bj3190411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shen W.J., Sridhar K., Bernlohr D.A., Kraemer F.B. Interaction of rat hormone-sensitive lipase with adipocyte lipid-binding protein. Proc Natl Acad Sci USA. 1999;96:5528–5532. doi: 10.1073/pnas.96.10.5528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fu Y., Luo L., Luo N., Garvey W.T. Lipid metabolism mediated by adipocyte lipid binding protein (ALBP/aP2) gene expression in human THP-1 macrophages. Atherosclerosis. 2006;188:102–111. doi: 10.1016/j.atherosclerosis.2005.10.041. [DOI] [PubMed] [Google Scholar]
- 67.Shen W.J., Patel S., Hong R., Kraemer F.B. Hormone-sensitive lipase functions as an oligomer. Biochemistry. 2000;39:2392–2398. doi: 10.1021/bi992283h. [DOI] [PubMed] [Google Scholar]
- 68.Shen W.J., Liang Y., Hong R., Patel S., Natu V., Sridhar K. Characterization of the functional interaction of adipocyte lipid-binding protein with hormone-sensitive lipase. J Biol Chem. 2001;276:49443–49448. doi: 10.1074/jbc.M104095200. [DOI] [PubMed] [Google Scholar]
- 69.Contreras J.A., Karlsson M., Osterlund T., Laurell H., Svensson A., Holm C. Hormone-sensitive lipase is structurally related to acetylcholinesterase, bile salt-stimulated lipase, and several fungal lipases. Building of a three-dimensional model for the catalytic domain of hormone-sensitive lipase. J Biol Chem. 1996;271:31426–31430. doi: 10.1074/jbc.271.49.31426. [DOI] [PubMed] [Google Scholar]
- 70.Osterlund T., Contreras J.A., Holm C. Identification of essential aspartic acid and histidine residues of hormone-sensitive lipase: apparent residues of the catalytic triad. FEBS Lett. 1997;403:259–262. doi: 10.1016/s0014-5793(97)00063-x. [DOI] [PubMed] [Google Scholar]
- 71.Anthonsen M.W., Ronnstrand L., Wernstedt C., Degerman E., Holm C. Identification of novel phosphorylation sites in hormone-sensitive lipase that are phosphorylated in response to isoproterenol and govern activation properties in vitro. J Biol Chem. 1998;273:215–221. doi: 10.1074/jbc.273.1.215. [DOI] [PubMed] [Google Scholar]
- 72.Greenberg A.S., Shen W.J., Muliro K., Patel S., Souza S.C., Roth R.A. Stimulation of lipolysis and hormone-sensitive lipase via the extracellular signal-regulated kinase pathway. J Biol Chem. 2001;276:45456–45461. doi: 10.1074/jbc.M104436200. [DOI] [PubMed] [Google Scholar]
- 73.Shen W.J., Patel S., Natu V., Kraemer F.B. Mutational analysis of structural features of rat hormone-sensitive lipase. Biochemistry. 1998;37:8973–8979. doi: 10.1021/bi980545u. [DOI] [PubMed] [Google Scholar]
- 74.Hemila H., Koivula T.T., Palva I. Hormone-sensitive lipase is closely related to several bacterial proteins, and distantly related to acetylcholinesterase and lipoprotein lipase: identification of a superfamily of esterases and lipases. Biochim Biophys Acta. 1994;1210:249–253. doi: 10.1016/0005-2760(94)90129-5. [DOI] [PubMed] [Google Scholar]
- 75.Tornqvist H., Belfrage P. Purification and some properties of a monoacylglycerol-hydrolyzing enzyme of rat adipose tissue. J Biol Chem. 1976;251:813–819. [PubMed] [Google Scholar]
- 76.Bertrand T., Auge F., Houtmann J., Rak A., Vallee F., Mikol V. Structural basis for human monoglyceride lipase inhibition. J Mol Biol. 2010;396:663–673. doi: 10.1016/j.jmb.2009.11.060. [DOI] [PubMed] [Google Scholar]
- 77.Labar G., Bauvois C., Borel F., Ferrer J.L., Wouters J., Lambert D.M. Crystal structure of the human monoacylglycerol lipase, a key actor in endocannabinoid signaling. Chembiochem. 2010;11:218–227. doi: 10.1002/cbic.200900621. [DOI] [PubMed] [Google Scholar]
- 78.Dinh T.P., Carpenter D., Leslie F.M., Freund T.F., Katona I., Sensi S.L. Brain monoglyceride lipase participating in endocannabinoid inactivation. Proc Natl Acad Sci USA. 2002;99:10819–10824. doi: 10.1073/pnas.152334899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Schweiger M., Schreiber R., Haemmerle G., Lass A., Fledelius C., Jacobsen P. Adipose triglyceride lipase and hormone-sensitive lipase are the major enzymes in adipose tissue triacylglycerol catabolism. J Biol Chem. 2006;281:40236–40241. doi: 10.1074/jbc.M608048200. [DOI] [PubMed] [Google Scholar]
- 80.Kienesberger P.C., Oberer M., Lass A., Zechner R. Mammalian patatin domain containing proteins: a family with diverse lipolytic activities involved in multiple biological functions. J Lipid Res. 2009;50(Suppl.):S63–S68. doi: 10.1194/jlr.R800082-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Romeo S., Huang-Doran I., Baroni M.G., Kotronen A. Unravelling the pathogenesis of fatty liver disease: patatin-like phospholipase domain-containing 3 protein. Curr Opin Lipidol. 2010;21:247–252. doi: 10.1097/mol.0b013e328338ca61. [DOI] [PubMed] [Google Scholar]
- 82.Hoekstra M., Li Z., Kruijt J.K., Van Eck M., Van Berkel T.J., Kuiper J. The expression level of non-alcoholic fatty liver disease-related gene PNPLA3 in hepatocytes is highly influenced by hepatic lipid status. J Hepatol. 2010;52:244–251. doi: 10.1016/j.jhep.2009.11.004. [DOI] [PubMed] [Google Scholar]
- 83.He S., McPhaul C., Li J.Z., Garuti R., Kinch L., Grishin N.V. A sequence variation (I148M) in PNPLA3 associated with nonalcoholic fatty liver disease disrupts triglyceride hydrolysis. J Biol Chem. 2010;285:6706–6715. doi: 10.1074/jbc.M109.064501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Huang Y., He S., Li J.Z., Seo Y.K., Osborne T.F., Cohen J.C. A feed-forward loop amplifies nutritional regulation of PNPLA3. Proc Natl Acad Sci USA. 2010;107:7892–7897. doi: 10.1073/pnas.1003585107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chen W., Chang B., Li L., Chan L. Patatin-like phospholipase domain-containing 3/adiponutrin deficiency in mice is not associated with fatty liver disease. Hepatology. 2010;52:1134–1142. doi: 10.1002/hep.23812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gao J.G., Simon M. A comparative study of human GS2, its paralogues, and its rat orthologue. Biochem Biophys Res Commun. 2007;360:501–506. doi: 10.1016/j.bbrc.2007.06.089. [DOI] [PubMed] [Google Scholar]
- 87.Gao J., Simon M. Identification of a novel keratinocyte retinyl ester hydrolase as a transacylase and lipase. J Invest Dermatol. 2005;124:1259–1266. doi: 10.1111/j.0022-202X.2005.23761.x. [DOI] [PubMed] [Google Scholar]
- 88.Lee W.C., Salido E., Yen P.H. Isolation of a new gene GS2 (DXS1283E) from a CpG island between STS and KAL1 on Xp22.3. Genomics. 1994;22:372–376. doi: 10.1006/geno.1994.1397. [DOI] [PubMed] [Google Scholar]
- 89.Shapiro L.J., Yen P., Pomerantz D., Martin E., Rolewic L., Mohandas T. Molecular studies of deletions at the human steroid sulfatase locus. Proc Natl Acad Sci USA. 1989;86:8477–8481. doi: 10.1073/pnas.86.21.8477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Okazaki H., Igarashi M., Nishi M., Tajima M., Sekiya M., Okazaki S. Identification of a novel member of the carboxylesterase family that hydrolyzes triacylglycerol: a potential role in adipocyte lipolysis. Diabetes. 2006;55:2091–2097. doi: 10.2337/db05-0585. [DOI] [PubMed] [Google Scholar]
- 91.Gilham D., Alam M., Gao W., Vance D.E., Lehner R. Triacylglycerol hydrolase is localized to the endoplasmic reticulum by an unusual retrieval sequence where it participates in VLDL assembly without utilizing VLDL lipids as substrates. Mol Biol Cell. 2005;16:984–996. doi: 10.1091/mbc.E04-03-0224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gruber A., Cornaciu I., Lass A., Schweiger M., Poeschl M., Eder C. The N-terminal region of comparative gene identification-58 (CGI-58) is important for lipid droplet binding and activation of adipose triglyceride lipase. J Biol Chem. 2010;285:12289–12298. doi: 10.1074/jbc.M109.064469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Granneman J.G., Moore H.P., Krishnamoorthy R., Rathod M. Perilipin controls lipolysis by regulating the interactions of AB-hydrolase containing 5 (Abhd5) and adipose triglyceride lipase (Atgl) J Biol Chem. 2009;284:34538–34544. doi: 10.1074/jbc.M109.068478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Yang X., Lu X., Liu J. Identification of a novel splicing isoform of murine CGI-58. FEBS Lett. 2010;584:903–910. doi: 10.1016/j.febslet.2009.12.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ollis D.L., Cheah E., Cygler M., Dijkstra B., Frolow F., Franken S.M. The alpha/beta hydrolase fold. Protein Eng. 1992;5:197–211. doi: 10.1093/protein/5.3.197. [DOI] [PubMed] [Google Scholar]
- 96.Granneman J.G., Moore H.P., Granneman R.L., Greenberg A.S., Obin M.S., Zhu Z. Analysis of lipolytic protein trafficking and interactions in adipocytes. J Biol Chem. 2007;282:5726–5735. doi: 10.1074/jbc.M610580200. [DOI] [PubMed] [Google Scholar]
- 97.Ghosh A.K., Ramakrishnan G., Chandramohan C., Rajasekharan R. CGI-58, the causative gene for Chanarin-Dorfman syndrome, mediates acylation of lysophosphatidic acid. J Biol Chem. 2008;283:24525–24533. doi: 10.1074/jbc.M801783200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Montero-Moran G., Caviglia J.M., McMahon D., Rothenberg A., Subramanian V., Xu Z. CGI-58/ABHD5 is a coenzyme A-dependent lysophosphatidic acid acyltransferase. J Lipid Res. 2010;51:709–719. doi: 10.1194/jlr.M001917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Russell L., Forsdyke D.R. A human putative lymphocyte G0/G1 switch gene containing a CpG-rich island encodes a small basic protein with the potential to be phosphorylated. DNA Cell Biol. 1991;10:581–591. doi: 10.1089/dna.1991.10.581. [DOI] [PubMed] [Google Scholar]
- 100.Zandbergen F., Mandard S., Escher P., Tan N.S., Patsouris D., Jatkoe T. The G0/G1 switch gene 2 is a novel PPAR target gene. Biochem J. 2005;392:313–324. doi: 10.1042/BJ20050636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Welch C., Santra M.K., El-Assaad W., Zhu X., Huber W.E., Keys R.A. Identification of a protein, G0S2, that lacks Bcl-2 homology domains and interacts with and antagonizes Bcl-2. Cancer Res. 2009;69:6782–6789. doi: 10.1158/0008-5472.CAN-09-0128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kurat C.F., Wolinski H., Petschnigg J., Kaluarachchi S., Andrews B., Natter K. Cdk1/Cdc28-dependent activation of the major triacylglycerol lipase Tgl4 in yeast links lipolysis to cell-cycle progression. Mol Cell. 2009;33:53–63. doi: 10.1016/j.molcel.2008.12.019. [DOI] [PubMed] [Google Scholar]
- 103.Haemmerle G., Zimmermann R., Strauss J.G., Kratky D., Riederer M., Knipping G. Hormone-sensitive lipase deficiency in mice changes the plasma lipid profile by affecting the tissue-specific expression pattern of lipoprotein lipase in adipose tissue and muscle. J Biol Chem. 2002;277:12946–12952. doi: 10.1074/jbc.M108640200. [DOI] [PubMed] [Google Scholar]
- 104.Mulder H., Sorhede-Winzell M., Contreras J.A., Fex M., Strom K., Ploug T. Hormone-sensitive lipase null mice exhibit signs of impaired insulin sensitivity whereas insulin secretion is intact. J Biol Chem. 2003;278:36380–36388. doi: 10.1074/jbc.M213032200. [DOI] [PubMed] [Google Scholar]
- 105.Bickel P.E., Tansey J.T., Welte M.A. PAT proteins, an ancient family of lipid droplet proteins that regulate cellular lipid stores. Biochim Biophys Acta. 2009;1791:419–440. doi: 10.1016/j.bbalip.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Yamaguchi T., Matsushita S., Motojima K., Hirose F., Osumi T. MLDP, a novel PAT family protein localized to lipid droplets and enriched in the heart, is regulated by peroxisome proliferator-activated receptor alpha. J Biol Chem. 2006;281:14232–14240. doi: 10.1074/jbc.M601682200. [DOI] [PubMed] [Google Scholar]
- 107.Wolins N.E., Quaynor B.K., Skinner J.R., Tzekov A., Croce M.A., Gropler M.C. OXPAT/PAT-1 is a PPAR-induced lipid droplet protein that promotes fatty acid utilization. Diabetes. 2006;55:3418–3428. doi: 10.2337/db06-0399. [DOI] [PubMed] [Google Scholar]
- 108.Dalen K.T., Dahl T., Holter E., Arntsen B., Londos C., Sztalryd C. LSDP5 is a PAT protein specifically expressed in fatty acid oxidizing tissues. Biochim Biophys Acta. 2007;1771:210–227. doi: 10.1016/j.bbalip.2006.11.011. [DOI] [PubMed] [Google Scholar]
- 109.Subramanian V., Rothenberg A., Gomez C., Cohen A.W., Garcia A., Bhattacharyya S. Perilipin A mediates the reversible binding of CGI-58 to lipid droplets in 3T3-L1 adipocytes. J Biol Chem. 2004;279:42062–42071. doi: 10.1074/jbc.M407462200. [DOI] [PubMed] [Google Scholar]
- 110.Listenberger L.L., Ostermeyer-Fay A.G., Goldberg E.B., Brown W.J., Brown D.A. Adipocyte differentiation-related protein reduces the lipid droplet association of adipose triglyceride lipase and slows triacylglycerol turnover. J Lipid Res. 2007;48:2751–2761. doi: 10.1194/jlr.M700359-JLR200. [DOI] [PubMed] [Google Scholar]
- 111.Bell M., Wang H., Chen H., McLenithan J.C., Gong D.W., Yang R.Z. Consequences of lipid droplet coat protein downregulation in liver cells: abnormal lipid droplet metabolism and induction of insulin resistance. Diabetes. 2008;57:2037–2045. doi: 10.2337/db07-1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Tansey J.T., Sztalryd C., Gruia-Gray J., Roush D.L., Zee J.V., Gavrilova O. Perilipin ablation results in a lean mouse with aberrant adipocyte lipolysis, enhanced leptin production, and resistance to diet-induced obesity. Proc Natl Acad Sci USA. 2001;98:6494–6499. doi: 10.1073/pnas.101042998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wolins N.E., Skinner J.R., Schoenfish M.J., Tzekov A., Bensch K.G., Bickel P.E. Adipocyte protein S3-12 coats nascent lipid droplets. J Biol Chem. 2003;278:37713–37721. doi: 10.1074/jbc.M304025200. [DOI] [PubMed] [Google Scholar]
- 114.Wolins N.E., Quaynor B.K., Skinner J.R., Schoenfish M.J., Tzekov A., Bickel P.E. S3-12, adipophilin, and TIP47 package lipid in adipocytes. J Biol Chem. 2005;280:19146–19155. doi: 10.1074/jbc.M500978200. [DOI] [PubMed] [Google Scholar]
- 115.Granneman J.G., Moore H.P., Mottillo E.P., Zhu Z. Functional interactions between Mldp (LSDP5) and Abhd5 in the control of intracellular lipid accumulation. J Biol Chem. 2009;284:3049–3057. doi: 10.1074/jbc.M808251200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Puri V., Konda S., Ranjit S., Aouadi M., Chawla A., Chouinard M. Fat-specific protein 27, a novel lipid droplet protein that enhances triglyceride storage. J Biol Chem. 2007;282:34213–34218. doi: 10.1074/jbc.M707404200. [DOI] [PubMed] [Google Scholar]
- 117.Keller P., Petrie J.T., De Rose P., Gerin I., Wright W.S., Chiang S.H. Fat-specific protein 27 regulates storage of triacylglycerol. J Biol Chem. 2008;283:14355–14365. doi: 10.1074/jbc.M708323200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Guo Y., Walther T.C., Rao M., Stuurman N., Goshima G., Terayama K. Functional genomic screen reveals genes involved in lipid-droplet formation and utilization. Nature. 2008;453:657–661. doi: 10.1038/nature06928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Beller M., Sztalryd C., Southall N., Bell M., Jackle H., Auld D.S. COPI complex is a regulator of lipid homeostasis. PLoS Biol. 2008;6:e292. doi: 10.1371/journal.pbio.0060292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Soni K.G., Mardones G.A., Sougrat R., Smirnova E., Jackson C.L., Bonifacino J.S. Coatomer-dependent protein delivery to lipid droplets. J Cell Sci. 2009;122:1834–1841. doi: 10.1242/jcs.045849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Bartz R., Zehmer J.K., Zhu M., Chen Y., Serrero G., Zhao Y. Dynamic activity of lipid droplets: protein phosphorylation and GTP-mediated protein translocation. J Proteome Res. 2007;6:3256–3265. doi: 10.1021/pr070158j. [DOI] [PubMed] [Google Scholar]
- 122.Narbonne P., Roy R. Caenorhabditis elegans dauers need LKB1/AMPK to ration lipid reserves and ensure long-term survival. Nature. 2009;457:210–214. doi: 10.1038/nature07536. [DOI] [PubMed] [Google Scholar]
- 123.Kershaw E.E., Schupp M., Guan H.P., Gardner N.P., Lazar M.A., Flier J.S. PPARgamma regulates adipose triglyceride lipase in adipocytes in vitro and in vivo. Am J Physiol Endocrinol Metab. 2007;293:E1736–E1745. doi: 10.1152/ajpendo.00122.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Festuccia W.T., Laplante M., Berthiaume M., Gelinas Y., Deshaies Y. PPARgamma agonism increases rat adipose tissue lipolysis, expression of glyceride lipases, and the response of lipolysis to hormonal control. Diabetologia. 2006;49:2427–2436. doi: 10.1007/s00125-006-0336-y. [DOI] [PubMed] [Google Scholar]
- 125.Chakrabarti P., Kandror K.V. FoxO1 controls insulin-dependent adipose triglyceride lipase (ATGL) expression and lipolysis in adipocytes. J Biol Chem. 2009;284:13296–13300. doi: 10.1074/jbc.C800241200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Liu L.F., Purushotham A., Wendel A.A., Koba K., Deiuliis J., Lee K. Regulation of adipose triglyceride lipase by rosiglitazone. Diabetes Obes Metab. 2009;11:131–142. doi: 10.1111/j.1463-1326.2008.00916.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Jocken J.W., Langin D., Smit E., Saris W.H., Valle C., Hul G.B. Adipose triglyceride lipase and hormone-sensitive lipase protein expression is decreased in the obese insulin-resistant state. J Clin Endocrinol Metab. 2007;92:2292–2299. doi: 10.1210/jc.2006-1318. [DOI] [PubMed] [Google Scholar]
- 128.Kralisch S., Klein J., Lossner U., Bluher M., Paschke R., Stumvoll M. Isoproterenol, TNFalpha, and insulin downregulate adipose triglyceride lipase in 3T3-L1 adipocytes. Mol Cell Endocrinol. 2005;240:43–49. doi: 10.1016/j.mce.2005.06.002. [DOI] [PubMed] [Google Scholar]
- 129.Chakrabarti P., English T., Shi J., Smas C.M., Kandror K.V. The mTOR complex 1 suppresses lipolysis, stimulates lipogenesis and promotes fat storage. Diabetes. 2010;59:775–781. doi: 10.2337/db09-1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Reidy S.P., Weber J. Leptin: an essential regulator of lipid metabolism. Comp Biochem Physiol A Mol Integr Physiol. 2000;125:285–298. doi: 10.1016/s1095-6433(00)00159-8. [DOI] [PubMed] [Google Scholar]
- 131.Shen J., Tanida M., Niijima A., Nagai K. In vivo effects of leptin on autonomic nerve activity and lipolysis in rats. Neurosci Lett. 2007;416:193–197. doi: 10.1016/j.neulet.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 132.Krempler F., Breban D., Oberkofler H., Esterbauer H., Hell E., Paulweber B. Leptin, peroxisome proliferator-activated receptor-gamma, and CCAAT/enhancer binding protein-alpha mRNA expression in adipose tissue of humans and their relation to cardiovascular risk factors. Arterioscler Thromb Vasc Biol. 2000;20:443–449. doi: 10.1161/01.atv.20.2.443. [DOI] [PubMed] [Google Scholar]
- 133.Li Y.C., Zheng X.L., Liu B.T., Yang G.S. Regulation of ATGL expression mediated by leptin in vitro in porcine adipocyte lipolysis. Mol Cell Biochem. 2010;333:121–128. doi: 10.1007/s11010-009-0212-4. [DOI] [PubMed] [Google Scholar]
- 134.Kraemer F.B., Shen W.J. Hormone-sensitive lipase: control of intracellular tri-(di-)acylglycerol and cholesteryl ester hydrolysis. J Lipid Res. 2002;43:1585–1594. doi: 10.1194/jlr.r200009-jlr200. [DOI] [PubMed] [Google Scholar]
- 135.Watt M.J., Spriet L.L. Regulation and role of hormone-sensitive lipase activity in human skeletal muscle. Proc Nutr Soc. 2004;63:315–322. doi: 10.1079/PNS2004360. [DOI] [PubMed] [Google Scholar]
- 136.Donsmark M., Langfort J., Holm C., Ploug T., Galbo H. Regulation and role of hormone-sensitive lipase in rat skeletal muscle. Proc Nutr Soc. 2004;63:309–314. doi: 10.1079/PNS2004359. [DOI] [PubMed] [Google Scholar]
- 137.Langfort J., Donsmark M., Ploug T., Holm C., Galbo H. Hormone-sensitive lipase in skeletal muscle: regulatory mechanisms. Acta Physiol Scand. 2003;178:397–403. doi: 10.1046/j.1365-201X.2003.01155.x. [DOI] [PubMed] [Google Scholar]
- 138.Krintel C., Osmark P., Larsen M.R., Resjo S., Logan D.T., Holm C. Ser649 and Ser650 are the major determinants of protein kinase A-mediated activation of human hormone-sensitive lipase against lipid substrates. PLoS One. 2008;3:e3756. doi: 10.1371/journal.pone.0003756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Olsson H., Stralfors P., Belfrage P. Phosphorylation of the basal site of hormone-sensitive lipase by glycogen synthase kinase-4. FEBS Lett. 1986;209:175–180. doi: 10.1016/0014-5793(86)81106-1. [DOI] [PubMed] [Google Scholar]
- 140.Garton A.J., Campbell D.G., Carling D., Hardie D.G., Colbran R.J., Yeaman S.J. Phosphorylation of bovine hormone-sensitive lipase by the AMP-activated protein kinase. A possible antilipolytic mechanism. Eur J Biochem. 1989;179:249–254. doi: 10.1111/j.1432-1033.1989.tb14548.x. [DOI] [PubMed] [Google Scholar]
- 141.Martinez-Botas J., Anderson J.B., Tessier D., Lapillonne A., Chang B.H., Quast M.J. Absence of perilipin results in leanness and reverses obesity in Lepr(db/db) mice. Nat Genet. 2000;26:474–479. doi: 10.1038/82630. [DOI] [PubMed] [Google Scholar]
- 142.Tansey J.T., Huml A.M., Vogt R., Davis K.E., Jones J.M., Fraser K.A. Functional studies on native and mutated forms of perilipins. A role in protein kinase A-mediated lipolysis of triacylglycerols. J Biol Chem. 2003;278:8401–8406. doi: 10.1074/jbc.M211005200. [DOI] [PubMed] [Google Scholar]
- 143.Sztalryd C., Xu G., Dorward H., Tansey J.T., Contreras J.A., Kimmel A.R. Perilipin A is essential for the translocation of hormone-sensitive lipase during lipolytic activation. J Cell Biol. 2003;161:1093–1103. doi: 10.1083/jcb.200210169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Miyoshi H., Perfield J.W., 2nd, Souza S.C., Shen W.J., Zhang H.H., Stancheva Z.S. Control of adipose triglyceride lipase action by serine 517 of perilipin A globally regulates protein kinase A-stimulated lipolysis in adipocytes. J Biol Chem. 2007;282:996–1002. doi: 10.1074/jbc.M605770200. [DOI] [PubMed] [Google Scholar]
- 145.Wang H., Hu L., Dalen K., Dorward H., Marcinkiewicz A., Russell D. Activation of hormone-sensitive lipase requires two steps, protein phosphorylation and binding to the PAT-1 domain of lipid droplet coat proteins. J Biol Chem. 2009;284:32116–32125. doi: 10.1074/jbc.M109.006726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Shen W.J., Patel S., Miyoshi H., Greenberg A.S., Kraemer F.B. Functional interaction of hormone-sensitive lipase and perilipin in lipolysis. J Lipid Res. 2009;50:2306–2313. doi: 10.1194/jlr.M900176-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Kiens B. Skeletal muscle lipid metabolism in exercise and insulin resistance. Physiol Rev. 2006;86:205–243. doi: 10.1152/physrev.00023.2004. [DOI] [PubMed] [Google Scholar]
- 148.Donsmark M., Langfort J., Holm C., Ploug T., Galbo H. Hormone-sensitive lipase as mediator of lipolysis in contracting skeletal muscle. Exerc Sport Sci Rev. 2005;33:127–133. doi: 10.1097/00003677-200507000-00005. [DOI] [PubMed] [Google Scholar]
- 149.Buchebner M., Pfeifer T., Rathke N., Chandak P.G., Lass A., Schreiber R. Cholesteryl ester hydrolase activity is abolished in HSL−/− macrophages but unchanged in macrophages lacking KIAA1363. J Lipid Res. 2010;51:2896–2908. doi: 10.1194/jlr.M004259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Hansson O., Strom K., Guner N., Wierup N., Sundler F., Hoglund P. Inflammatory response in white adipose tissue in the non-obese hormone-sensitive lipase null mouse model. J Proteome Res. 2006;5:1701–1710. doi: 10.1021/pr060101h. [DOI] [PubMed] [Google Scholar]
- 151.Kraemer F.B., Shen W.J. Hormone-sensitive lipase knockouts. Nutr Metab (Lond) 2006;3:12. doi: 10.1186/1743-7075-3-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Yeaman S.J. Hormone-sensitive lipase – new roles for an old enzyme. Biochem J. 2004;379:11–22. doi: 10.1042/BJ20031811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Johnson W.J., Jang S.Y., Bernard D.W. Hormone sensitive lipase mRNA in both monocyte and macrophage forms of the human THP-1 cell line. Comp Biochem Physiol B Biochem Mol Biol. 2000;126:543–552. doi: 10.1016/s0305-0491(00)00220-0. [DOI] [PubMed] [Google Scholar]
- 154.Fredrikson G., Tornqvist H., Belfrage P. Hormone-sensitive lipase and monoacylglycerol lipase are both required for complete degradation of adipocyte triacylglycerol. Biochim Biophys Acta. 1986;876:288–293. doi: 10.1016/0005-2760(86)90286-9. [DOI] [PubMed] [Google Scholar]
- 155.Schoiswohl G., Schweiger M., Schreiber R., Gorkiewicz G., Preiss-Landl K., Taschler U. Adipose triglyceride lipase plays a key role in the supply of the working muscle with fatty acids. J Lipid Res. 2010;51:490–499. doi: 10.1194/jlr.M001073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Olivecrona G. The crucial role of ATGL for energy supply of muscles. J Lipid Res. 2010;51:449–450. doi: 10.1194/jlr.E004770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Huijsman E., van de Par C., Economou C., van der Poel C., Lynch G.S., Schoiswohl G. Adipose triacylglycerol lipase deletion alters whole body energy metabolism and impairs exercise performance in mice. Am J Physiol Endocrinol Metab. 2009;297:E505–E513. doi: 10.1152/ajpendo.00190.2009. [DOI] [PubMed] [Google Scholar]
- 158.Ahmadian M., Duncan R.E., Varady K.A., Frasson D., Hellerstein M.K., Birkenfeld A.L. Adipose overexpression of desnutrin promotes fatty acid use and attenuates diet-induced obesity. Diabetes. 2009;58:855–866. doi: 10.2337/db08-1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Randle P.J., Garland P.B., Hales C.N., Newsholme E.A. The glucose fatty-acid cycle. Its role in insulin sensitivity and the metabolic disturbances of diabetes mellitus. Lancet. 1963;1:785–789. doi: 10.1016/s0140-6736(63)91500-9. [DOI] [PubMed] [Google Scholar]
- 160.Hue L., Taegtmeyer H. The Randle cycle revisited: a new head for an old hat. Am J Physiol Endocrinol Metab. 2009;297:E578–E591. doi: 10.1152/ajpendo.00093.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Schaffer J.E. Lipotoxicity: when tissues overeat. Curr Opin Lipidol. 2003;14:281–287. doi: 10.1097/00041433-200306000-00008. [DOI] [PubMed] [Google Scholar]
- 162.Kienesberger P.C., Lee D., Pulinilkunnil T., Brenner D.S., Cai L., Magnes C. Adipose triglyceride lipase deficiency causes tissue-specific changes in insulin signaling. J Biol Chem. 2009;284:30218–30229. doi: 10.1074/jbc.M109.047787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Peyot M.L., Guay C., Latour M.G., Lamontagne J., Lussier R., Pineda M. Adipose triglyceride lipase is implicated in fuel- and non-fuel-stimulated insulin secretion. J Biol Chem. 2009;284:16848–16859. doi: 10.1074/jbc.M109.006650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Tang J., Kriz R.W., Wolfman N., Shaffer M., Seehra J., Jones S.S. A novel cytosolic calcium-independent phospholipase A2 contains eight ankyrin motifs. J Biol Chem. 1997;272:8567–8575. doi: 10.1074/jbc.272.13.8567. [DOI] [PubMed] [Google Scholar]
- 165.Yang Q., Graham T.E., Mody N., Preitner F., Peroni O.D., Zabolotny J.M. Serum retinol binding protein 4 contributes to insulin resistance in obesity and type 2 diabetes. Nature. 2005;436:356–362. doi: 10.1038/nature03711. [DOI] [PubMed] [Google Scholar]
- 166.Jourdan M., Jaleel A., Karakelides H., Ford G.C., Kahn B.B., Nair K.S. Impact of type 1 diabetes and insulin treatment on plasma levels and fractional synthesis rate of retinol-binding protein 4. J Clin Endocrinol Metab. 2009;94:5125–5130. doi: 10.1210/jc.2009-1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Goodman E., Graham T.E., Dolan L.M., Daniels S.R., Goodman E.R., Kahn B.B. The relationship of retinol binding protein 4 to changes in insulin resistance and cardiometabolic risk in overweight black adolescents. J Pediatr. 2009;154:67–73. doi: 10.1016/j.jpeds.2008.07.018. [e1] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Krzyzanowska K., Zemany L., Krugluger W., Schernthaner G.H., Mittermayer F., Schnack C. Serum concentrations of retinol-binding protein 4 in women with and without gestational diabetes. Diabetologia. 2008;51:1115–1122. doi: 10.1007/s00125-008-1009-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Hammarstedt A., Pihlajamaki J., Graham T.E., Kainulainen S., Kahn B.B., Laakso M. High circulating levels of RBP4 and mRNA levels of aP2, PGC-1alpha and UCP-2 predict improvement in insulin sensitivity following pioglitazone treatment of drug-naive type 2 diabetic subjects. J Intern Med. 2008;263:440–449. doi: 10.1111/j.1365-2796.2007.01914.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Kovacs P., Geyer M., Berndt J., Kloting N., Graham T.E., Bottcher Y. Effects of genetic variation in the human retinol binding protein-4 gene (RBP4) on insulin resistance and fat depot-specific mRNA expression. Diabetes. 2007;56:3095–3100. doi: 10.2337/db06-1647. [DOI] [PubMed] [Google Scholar]
- 171.Balagopal P., Graham T.E., Kahn B.B., Altomare A., Funanage V., George D. Reduction of elevated serum retinol binding protein in obese children by lifestyle intervention: association with subclinical inflammation. J Clin Endocrinol Metab. 2007;92:1971–1974. doi: 10.1210/jc.2006-2712. [DOI] [PubMed] [Google Scholar]
- 172.Graham T.E., Yang Q., Bluher M., Hammarstedt A., Ciaraldi T.P., Henry R.R. Retinol-binding protein 4 and insulin resistance in lean, obese, and diabetic subjects. N Engl J Med. 2006;354:2552–2563. doi: 10.1056/NEJMoa054862. [DOI] [PubMed] [Google Scholar]
- 173.Langin D., Dicker A., Tavernier G., Hoffstedt J., Mairal A., Ryden M. Adipocyte lipases and defect of lipolysis in human obesity. Diabetes. 2005;54:3190–3197. doi: 10.2337/diabetes.54.11.3190. [DOI] [PubMed] [Google Scholar]
- 174.Bezaire V., Mairal A., Ribet C., Lefort C., Girousse A., Jocken J. Contribution of adipose triglyceride lipase and hormone-sensitive lipase to lipolysis in hMADS adipocytes. J Biol Chem. 2009;284:18282–18291. doi: 10.1074/jbc.M109.008631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Chanarin I., Patel A., Slavin G., Wills E.J., Andrews T.M., Stewart G. Neutral-lipid storage disease: a new disorder of lipid metabolism. Br Med J. 1975;1:553–555. doi: 10.1136/bmj.1.5957.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Igal R.A., Rhoads J.M., Coleman R.A. Neutral lipid storage disease with fatty liver and cholestasis. J Pediatr Gastroenterol Nutr. 1997;25:541–547. doi: 10.1097/00005176-199711000-00011. [DOI] [PubMed] [Google Scholar]
- 177.Radom J., Salvayre R., Negre A., Maret A., Douste-Blazy L. Metabolism of neutral lipids in cultured fibroblasts from multisystemic (or type 3) lipid storage myopathy. Eur J Biochem. 1987;164:703–708. doi: 10.1111/j.1432-1033.1987.tb11183.x. [DOI] [PubMed] [Google Scholar]
- 178.Radom J., Salvayre R., Levade T., Douste-Blazy L. Influence of chain length of pyrene fatty acids on their uptake and metabolism by Epstein-Barr-virus-transformed lymphoid cell lines from a patient with multisystemic lipid storage myopathy and from control subjects. Biochem J. 1990;269:107–113. doi: 10.1042/bj2690107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Hilaire N., Salvayre R., Thiers J.C., Bonnafe M.J., Negre-Salvayre A. The turnover of cytoplasmic triacylglycerols in human fibroblasts involves two separate acyl chain length-dependent degradation pathways. J Biol Chem. 1995;270:27027–27034. doi: 10.1074/jbc.270.45.27027. [DOI] [PubMed] [Google Scholar]
- 180.Hilaire N., Negre-Salvayre A., Salvayre R. Cellular uptake and catabolism of high-density-lipoprotein triacylglycerols in human cultured fibroblasts: degradation block in neutral lipid storage disease. Biochem J. 1994;297(Pt 3):467–473. doi: 10.1042/bj2970467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Hilaire N., Negre-Salvayre A., Salvayre R. Cytoplasmic triacylglycerols and cholesteryl esters are degraded in two separate catabolic pools in cultured human fibroblasts. FEBS Lett. 1993;328:230–234. doi: 10.1016/0014-5793(93)80933-l. [DOI] [PubMed] [Google Scholar]
- 182.Igal R.A., Coleman R.A. Acylglycerol recycling from triacylglycerol to phospholipid, not lipase activity, is defective in neutral lipid storage disease fibroblasts. J Biol Chem. 1996;271:16644–16651. doi: 10.1074/jbc.271.28.16644. [DOI] [PubMed] [Google Scholar]
- 183.Igal R.A., Coleman R.A. Neutral lipid storage disease: a genetic disorder with abnormalities in the regulation of phospholipid metabolism. J Lipid Res. 1998;39:31–43. [PubMed] [Google Scholar]
- 184.Williams M.L., Monger D.J., Rutherford S.L., Hincenbergs M., Rehfeld S.J., Grunfeld C. Neutral lipid storage disease with ichthyosis: lipid content and metabolism of fibroblasts. J Inherit Metab Dis. 1988;11:131–143. doi: 10.1007/BF01799862. [DOI] [PubMed] [Google Scholar]
- 185.Williams M.L., Coleman R.A., Placezk D., Grunfeld C. Neutral lipid storage disease: a possible functional defect in phospholipid-linked triacylglycerol metabolism. Biochim Biophys Acta. 1991;1096:162–169. doi: 10.1016/0925-4439(91)90055-e. [DOI] [PubMed] [Google Scholar]
- 186.Bergman R., Aviram M., Shemer A., Oiknine Y., Vardi D.A., Friedman-Birnbaum R. Enhanced low-density lipoprotein degradation and cholesterol synthesis in monocyte-derived macrophages of patients with adult xanthogranulomatosis. J Invest Dermatol. 1993;101:880–882. doi: 10.1111/1523-1747.ep12371711. [DOI] [PubMed] [Google Scholar]
- 187.Lefevre C., Jobard F., Caux F., Bouadjar B., Karaduman A., Heilig R. Mutations in CGI-58, the gene encoding a new protein of the esterase/lipase/thioesterase subfamily, in Chanarin-Dorfman syndrome. Am J Hum Genet. 2001;69:1002–1012. doi: 10.1086/324121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Akiyama M., Sakai K., Ogawa M., McMillan J.R., Sawamura D., Shimizu H. Novel duplication mutation in the patatin domain of adipose triglyceride lipase (PNPLA2) in neutral lipid storage disease with severe myopathy. Muscle Nerve. 2007;36:856–859. doi: 10.1002/mus.20869. [DOI] [PubMed] [Google Scholar]
- 189.Radner F.P., Streith I.E., Schoiswohl G., Schweiger M., Kumari M., Eichmann T.O. Growth retardation, impaired triacylglycerol catabolism, hepatic steatosis, and lethal skin barrier defect in mice lacking comparative gene identification-58 (CGI-58) J Biol Chem. 2010;285:7300–7311. doi: 10.1074/jbc.M109.081877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Jiang Y.J., Barish G., Lu B., Evans R.M., Crumrine D., Schmuth M. PPARdelta activation promotes stratum corneum formation and epidermal permeability barrier development during late gestation. J Invest Dermatol. 2010;130:511–519. doi: 10.1038/jid.2009.245. [DOI] [PubMed] [Google Scholar]
- 191.Jiang Y.J., Uchida Y., Lu B., Kim P., Mao C., Akiyama M. Ceramide stimulates ABCA12 expression via peroxisome proliferator-activated receptor delta in human keratinocytes. J Biol Chem. 2009;284:18942–18952. doi: 10.1074/jbc.M109.006973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Demerjian M., Choi E.H., Man M.Q., Chang S., Elias P.M., Feingold K.R. Activators of PPARs and LXR decrease the adverse effects of exogenous glucocorticoids on the epidermis. Exp Dermatol. 2009;18:643–649. doi: 10.1111/j.1600-0625.2009.00841.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Schmuth M., Jiang Y.J., Dubrac S., Elias P.M., Feingold K.R. Thematic review series: skin lipids. Peroxisome proliferator-activated receptors and liver X receptors in epidermal biology. J Lipid Res. 2008;49:499–509. doi: 10.1194/jlr.R800001-JLR200. [DOI] [PubMed] [Google Scholar]
- 194.Man M.Q., Barish G.D., Schmuth M., Crumrine D., Barak Y., Chang S. Deficiency of PPARbeta/delta in the epidermis results in defective cutaneous permeability barrier homeostasis and increased inflammation. J Invest Dermatol. 2008;128:370–377. doi: 10.1038/sj.jid.5701026. [DOI] [PubMed] [Google Scholar]
- 195.Coderch L., Lopez O., de la Maza A., Parra J.L. Ceramides and skin function. Am J Clin Dermatol. 2003;4:107–129. doi: 10.2165/00128071-200304020-00004. [DOI] [PubMed] [Google Scholar]
- 196.Mizutani Y., Mitsutake S., Tsuji K., Kihara A., Igarashi Y. Ceramide biosynthesis in keratinocyte and its role in skin function. Biochimie. 2009;91:784–790. doi: 10.1016/j.biochi.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 197.Elias P.M., Williams M.L., Holleran W.M., Jiang Y.J., Schmuth M. Pathogenesis of permeability barrier abnormalities in the ichthyoses: inherited disorders of lipid metabolism. J Lipid Res. 2008;49:697–714. doi: 10.1194/jlr.R800002-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Demerjian M., Crumrine D.A., Milstone L.M., Williams M.L., Elias P.M. Barrier dysfunction and pathogenesis of neutral lipid storage disease with ichthyosis (Chanarin-Dorfman syndrome) J Invest Dermatol. 2006;126:2032–2038. doi: 10.1038/sj.jid.5700332. [DOI] [PubMed] [Google Scholar]
- 199.Akiyama M., Sawamura D., Nomura Y., Sugawara M., Shimizu H. Truncation of CGI-58 protein causes malformation of lamellar granules resulting in ichthyosis in Dorfman-Chanarin syndrome. J Invest Dermatol. 2003;121:1029–1034. doi: 10.1046/j.1523-1747.2003.12520.x. [DOI] [PubMed] [Google Scholar]
- 200.Elias P.M., Williams M.L. Neutral lipid storage disease with ichthyosis. Defective lamellar body contents and intracellular dispersion. Arch Dermatol. 1985;121:1000–1008. [PubMed] [Google Scholar]
- 201.Yamaguchi T., Omatsu N., Morimoto E., Nakashima H., Ueno K., Tanaka T. CGI-58 facilitates lipolysis on lipid droplets but is not involved in the vesiculation of lipid droplets caused by hormonal stimulation. J Lipid Res. 2007;48:1078–1089. doi: 10.1194/jlr.M600493-JLR200. [DOI] [PubMed] [Google Scholar]
- 202.Notredame C., Higgins D.G., Heringa J. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J Mol Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- 203.Clamp M., Cuff J., Searle S.M., Barton G.J. The Jalview Java alignment editor. Bioinformatics. 2004;20:426–427. doi: 10.1093/bioinformatics/btg430. [DOI] [PubMed] [Google Scholar]
- 204.Kiefer F., Arnold K., Kunzli M., Bordoli L., Schwede T. The SWISS-MODEL repository and associated resources. Nucleic Acids Res. 2009;37:D387–D392. doi: 10.1093/nar/gkn750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Zou J., Hallberg B.M., Bergfors T., Oesch F., Arand M., Mowbray S.L. Structure of Aspergillus niger epoxide hydrolase at 1.8 A resolution: implications for the structure and function of the mammalian microsomal class of epoxide hydrolases. Structure. 2000;8:111–122. doi: 10.1016/s0969-2126(00)00087-3. [DOI] [PubMed] [Google Scholar]