Abstract
GATA4, a transcription factor expressed in the proximal small intestine but not in the distal ileum, maintains proximal-distal distinctions by multiple processes involving gene repression, gene activation, and cell fate determination. Friend of GATA (FOG) is an evolutionarily conserved family of cofactors whose members physically associate with GATA factors and mediate GATA-regulated repression in multiple tissues. Using a novel, inducible, intestine-specific Gata4 knock-in model in mice, in which wild-type GATA4 is specifically inactivated in the small intestine, but a GATA4 mutant that does not bind FOG cofactors (GATA4ki) continues to be expressed, we found that ileal-specific genes were significantly induced in the proximal small intestine (P<0.01); in contrast, genes restricted to proximal small intestine and cell lineage markers were unaffected, indicating that GATA4-FOG interactions contribute specifically to the repression function of GATA4 within this organ. Fog1 mRNA displayed a proximal-distal pattern that parallels that of Gata4, and FOG1 protein was co-expressed with GATA4 in intestinal epithelial cells, implicating FOG1 as the likely mediator of GATA4 function in the small intestine. Our data are the first to indicate FOG function and expression in the mammalian small intestine.
Keywords: GATA4, Friend of GATA, FOG, intestinal differentiation, apical sodium-dependent bile acid transporter, ASBT
INTRODUCTION
The mammalian small intestine is lined by a highly specialized epithelium that displays a wide ranging, yet tightly regulated functional diversity along its cephalo-caudal axis (Gordon et al., 1992). This functional diversity is linked to a continuous renewal process in which stem cells located at or near the base of crypts produce transit amplifying cells that ultimately differentiate into four principal cell types. Absorptive enterocytes, which constitute the majority of intestinal epithelial cells, enteroendocrine cells, and goblet cells migrate up the villi and are shed into the intestinal lumen every three to five days, whereas Paneth cells reside at the base of crypts and turn over at a slower rate. Specific absorptive enterocyte genes that encode proteins that mediate absorption of bile salts are localized to the distal ileum, including the apical sodium-dependent bile acid transporter (ASBT) (Shneider, 2001) and ileal lipid binding protein (ILBP) (Crossman et al., 1994). Proteins responsible for the terminal digestion and absorption of most nutrients are localized to jejunum and proximal ileum, as exemplified by lactase-phlorizin hydrolase (LPH) (Krasinski et al., 1997) and liver fatty acid binding protein (FABP1) (Simon et al., 1993). Goblet cells are more numerous in distal small intestine (Specian and Oliver, 1991), and enteroendocrine subpopulations display a functional diversity characterized by the regional segregation of hormones that activate (e.g. cholecystokinin, CCK) or repress (e.g. peptide YY, PYY) gastrointestinal processes (Schonhoff et al., 2004). Maintenance of a dynamic diversity in gene expression and cell fate allocation along the cephalo-caudal axis is necessary for the normal functioning of the small intestine.
Recently, we found that GATA4 is a key regulator of regional gene expression and cell fate allocation in the adult mouse small intestine (Bosse et al., 2006a). GATA4 is a member of a conserved family of transcription factors that contain a pair of zinc fingers that mediate binding to their consensus DNA sequence, WGATAR, in the regulatory region of target genes (Molkentin, 2000). In the small intestine of adult rodents and humans, GATA4 is expressed at high levels in proximal regions, but is undetectable in distal ileum (Bosse et al., 2006a; van Wering et al., 2004). Conditional, inducible inactivation of Gata4 in the adult mouse jejunum results in a generalized transformation to an ileal-like phenotype that is characterized by an induction of ileal-specific genes, including Asbt and Ilbp, and attenuation of genes restricted to proximal small intestine, including Lph and Fabp1. Furthermore, secretory cell fate also becomes ileal-like as indicated by a significant increase in goblet cell number and the abundance of the mRNA for Math1, a secretory cell mediator (Yang et al., 2001), and a trend toward an increase in Pyy mRNA and a decrease in Cck mRNA (Bosse et al., 2006a). This novel finding establishes a fundamental plasticity in the adult mammalian small intestine not previously realized, and highlights multiple levels of regulation by GATA4 in this organ involving gene repression, gene activation, and cell fate determination. However, the precise mechanisms by which GATA4 regulates these diverse functions in the mature small intestine are currently unknown.
Friend of GATA (FOG) is an evolutionarily conserved multi-zinc finger cofactor family whose members physically associate with GATA factors, and mediate GATA function in a broad array of tissues and cell types (Cantor and Orkin, 2005). The GATA-FOG interaction is conserved in Drosophila where the FOG homolog, U-shaped, physically associates with pannier, a GATA homolog (Haenlin et al., 1997), indicating the fundamental importance of this interaction. Using a split two-hybrid screen, a GATA1 mutant (GATA1ki) with a valine-to-glycine substitution at position 205 in the N-terminal zinc finger was identified that has attenuated binding affinity for FOG cofactors, but normal DNA binding function (Crispino et al., 1999). Gata1ki/ki mice that express GATA1ki in place of GATA1 die during embryogenesis due to anemia caused by disrupted erythroid maturation and megakaryocyte abnormalities (Crispino et al., 1999). This phenotype is similar to that in Gata1−/− (Chang et al., 2002; Fujiwara et al., 1996; Pevny et al., 1991) or Fog1−/− (Tsang et al., 1998) mice, indicating that GATA1-FOG1 interaction is required for hematopoiesis. A FOG1 mutant that restores GATA1-FOG1 interaction rescues this phenotype, providing in vivo evidence that the Gata1 knock-in mutation specifically disrupts the ability of GATA1 to bind FOG cofactors, and that GATA1ki is otherwise functional (Cantor et al., 2002; Crispino et al., 1999). In addition, a Gata4 knock-in model was designed with the analogous valine-to-glycine substitution at position 217 in GATA4 (GATA4ki) that disrupts the interaction between GATA4 and FOG cofactors (Crispino et al., 2001; Tevosian et al., 2002). Gata4ki/ki mice show an embryonic lethal cardiac phenotype, very similar to that found in Gata4−/− (Kuo et al., 1997; Molkentin et al., 1997) and Fog2−/− (Svensson et al., 2000; Tevosian et al., 2000) mice, providing evidence that GATA4-FOG2 interaction is required for cardiogenesis. Furthermore, the Gata4ki model has revealed that GATA4-FOG interaction is important in gonadal differentiation (Manuylov et al., 2007; Tevosian et al., 2002) and gastric epithelial development (Jacobsen et al., 2005).
Using a novel, inducible, intestine-specific Gata4 knock-in model in mice, in which wild-type GATA4 is specifically inactivated in the small intestine, but a GATA4 mutant that does not bind FOG cofactors (GATA4ki) continues to be expressed, we found that ileal-specific genes were significantly induced in the proximal small intestine (P<0.01), but genes restricted to proximal small intestine and cell lineage markers were unaffected, indicating that GATA4-FOG interactions contribute specifically to the repression function of GATA4 within this organ. Fog1 mRNA displayed a proximal-distal pattern that parallels that of Gata4, and FOG1 protein was co-expressed with GATA4 in intestinal epithelial cells, implicating FOG1 as the likely mediator of GATA4 function in the small intestine. These data are the first to indicate FOG function and expression in the mammalian small intestine.
MATERIALS AND METHODS
Mice
Mice were housed under standard conditions in the Animal Research at Children’s Hospital (ARCH) facility and provided food and water ad libitum. Approval was obtained from the Institutional Animal Care and Use Committee. Four Gata4 alleles were utilized in this study (Fig. 1A). The Gata4flox allele, which contains loxP sites flanking the translational start site and the region encoding the activation domains of GATA4, expresses wild-type GATA4 (Pu et al., 2004). Exposure of Gata4flox to the recombinase CRE results in exon2 excision (Gata4Δex2), and the subsequent utilization of an alternative in-frame ATG in exon3 leading to the synthesis of a truncated, transcriptionally inactive form of GATA4 (GATA4Δex2) devoid of its activation domains (Bosse et al., 2006a). The Gata4ki allele encodes a mutant GATA4 (GATA4ki) that contains a single amino acid substitution (V217G) in the N-terminal zinc finger rendering it unable to bind specifically with FOG cofactors (Crispino et al., 2001). The wild-type allele (Gata4wt), was also utilized in the study. Genotyping to distinguish among each of the four Gata4 alleles was conducted by PCR on DNA extracted from tail biopsies as described (Bosse et al., 2006b) using primers specific for exon2 or exon5 of Gata4 (see supplemental Fig. 1 (Fig. S1)).
Fig. 1.
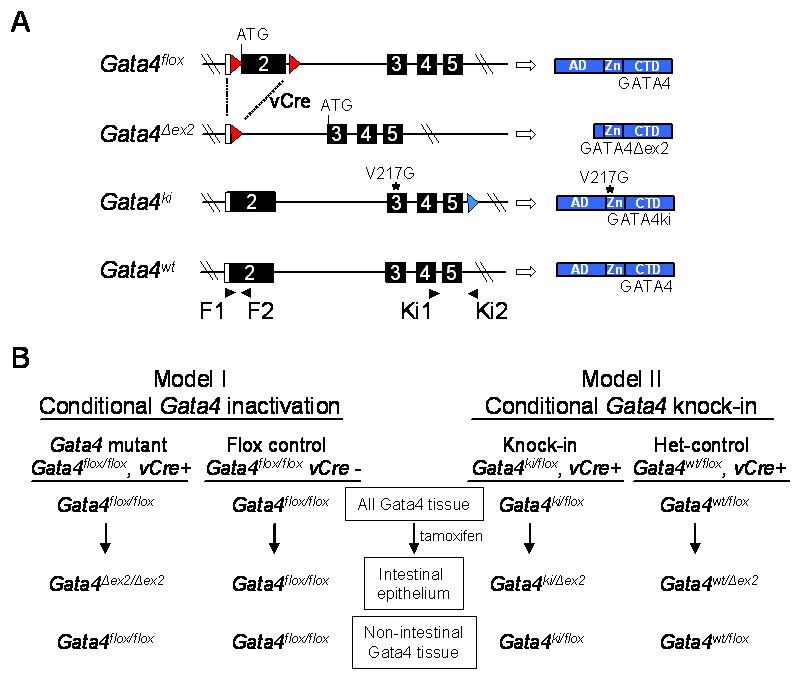
In vivo mouse models. (A) Schematic representation of the different Gata4 alleles used in this study, and their protein products. Open boxes indicate untranslated region. Filled boxes indicate translated region. Numbers indicate exons. Red arrowheads indicate loxP sites used for excision of the GATA4 activation domains in the Gata4flox allele. Blue arrowhead indicates a residual loxP site previously used to remove a neomycin cassette from the Gata4ki allele (Crispino et al., 2001). Blue boxes indicate protein product. AD, activation domain; Zn, zinc fingers; CTD, C-terminal domain. Filled arrowheads indicate PCR primers. (B) Models used in this study showing the Gata4 alleles in different tissue in test and control mice before and after tamoxifen treatment. vCre indicates the VillinCreERT2 transgene.
Two different conditional, inducible genetic mouse models were used for this study, including a previously validated conditional Gata4 inactivation model (Bosse et al., 2006a), and a novel conditional Gata4 knock-in model (Fig. 1B). Both models were established in a VillinCreERT2 transgenic background (el Marjou et al., 2004) in which CRE-mediated excision of floxed Gata4 DNA occurs specifically in intestinal and colonic epithelium after tamoxifen treatment (Bosse et al., 2006a). Adult mice (6–8 weeks) were treated with five single intraperitoneal injections of tamoxifen (100µl, 10mg/ml) per day for 5 consecutive days and sacrificed for tissue collection 14 days after the last injection (Bosse et al., 2006a), unless indicated otherwise. In the Gata4 inactivation model (Fig. 1B, Model I), Gata4flox/flox, VillinCreERT2-positive mice were treated with tamoxifen resulting in exon2 deletion (Gata4Δex2) and subsequent conditional Gata4 inactivation in the intestinal epithelium (Bosse et al., 2006a). Gata4flox/flox, VillinCreERT2-negative mice treated with tamoxifen were used as controls. In the Gata4 knock-in model (Fig. 1B, Model II), Gata4ki/flox knock-in mice and Gata4wt/flox controls (het-controls) were established in a VillinCreERT2 background. Before tamoxifen treatment, both knock-in mice and het-controls produce wild-type GATA4 from at least one allele in all GATA4 expressing tissue, including the small intestine. After tamoxifen treatment of knock-in mice, GATA4 and GATA4ki are expressed in all tissues that normally express GATA4, except in the intestinal epithelium, where GATA4 from the Gata4flox allele is specifically inactivated by CRE-mediated excision, but GATA4ki continues to be expressed from the Gata4ki allele. After tamoxifen treatment of het-controls, GATA4 is expressed from both alleles in all tissues that normally express GATA4, except in the intestinal epithelium, where Gata4flox is excised by CRE, but GATA4 continues to be expressed from Gata4wt allele. This approach creates a conditional, inducible Gata4 knock-in model.
RNA isolation
RNA was isolated from heart, stomach, and small intestine using the RNeasy kit (Qiagen) as described previously (Bosse et al., 2006a). Intestinal segments (0.5 to 1.0 cm) were obtained from the most proximal region adjacent to the pylorus (segment 1), the 25% mark (segment 2), the geometric center (segment 3), the 75% mark (segment 4), and the most distal region adjacent to the ileocecal junction (segment 5).
Sequencing
To confirm CRE-mediated excision and expression of Gata4ki, sequencing analyses was conducted. Complementary DNA (cDNA) was synthesized from jejunal RNA obtained from segment 3, and the region encompassing the knock-in mutation was amplified by PCR. The PCR product was separated on an agarose gel by electrophoresis, extracted using the QIAquick Gel Extraction Kit (Qiagen), re-amplified using the same primers, and purified using ExoSap-IT (USB Corporation). The purified PCR product was sequenced at the Molecular Genetics Core (Children’s Hospital Boston) using a nested primer.
RT-PCR
To quantify mRNA abundances, semi-quantitative and real-time RT-PCR were conducted as described previously (Bosse et al., 2006a). Primer pairs (Fig. S2) were designed using Beacon Design software (Biosoft International) and optimized. Real-time RT-PCR was carried out using an iCycler and iQ SYBR Green Supermix (Bio-Rad). Gapdh mRNA abundance was measured for each sample and used to normalize the data. All data were expressed relative to a calibrator as indicated in the figure legends.
Immunoblotting
Crude nuclear extracts were isolated from four quarters of intestine (proximal to distal: I – IV) and Western analysis was conducted as described previously (van Wering et al., 2004) using 100 µg of extract. The membranes were blocked for 1 h at room temperature in 5% nonfat dried milk in PBS and incubated with goat anti-FOG1 (1:1500, Santa Cruz) with or without FOG1 blocking peptide (1:1500, Santa Cruz) for 1 h. Membranes were stripped and re-probed using mouse anti-β-actin (1:4000, Santa Cruz). Horseradish peroxidase-linked secondary antibodies and chemiluminescence solution (Pierce West Femto Kit) were used to visualize FOG1 or β-actin signals.
In situ hybridization
RNA probes were prepared by in vitro transcription of a partial cDNA insert from the Fog1 library plasmid M10 subcloned into pBluescript KS (Stratagene) (Tsang et al., 1997) using digoxigenin-UTP (Roche Molecular Biochemicals), and T3 (antisense) or T7 (sense) polymerase as described (Katz et al., 2003). In situ hybridization assays were conducted as described previously (Tevosian et al., 2000).
Immunohistochemistry
Intestinal segments were fixed in ice-cold 4% paraformaldehyde in PBS for 4 h, dehydrated overnight as described (Bosse et al., 2006a), embedded in paraffin, and sectioned (5 µm) in the Department of Pathology at Children’s Hospital Boston. After deparaffinization and antigen retrieval (Bosse et al., 2006a; Bosse et al., 2006b), the sections were incubated with the primary antibody for 1 h at 37°C, rinsed, and then incubated with the secondary antibody for 1 h at 37°C. For immunofluorescence, sections were incubated in a solution containing 4’,6-diamino-2-phenylindol dihydrochloride (DAPI, 2 µg/ml, Molecular Probes) in PBS for 15 min at room temperature, washed in PBS, and mounted in Mowiol mounting medium (Calbiochem). For immunohistochemistry, biotinylated secondary antibodies were linked to avidin-horseradish peroxidase or avidin-alkaline phosphatase conjugates (Vector Labs), and visualized using 3,3’-diamino benzidine (DAB) for 2–5 min or 4-nitro blue tetrazolium chloride (NBT)/5-bromo-4-chloro-3indolyl-phosphate (BCIP) for 20–90 min, respectively. For selected sections, the tissue was lightly counterstained with methyl green.
The primary antibodies included rabbit anti-ASBT (1:500, kind gift of Dr. P.A. Dawson, Wake Forest School of Medicine), goat anti-FOG1 (1:200, Santa Cruz), mouse anti-GATA4 (1:400, Santa Cruz), goat anti-GATA4 (1:400, Santa Cruz), rabbit anti-chromogranin A (1:1000, Immunostar), rabbit anti-lysozyme (1:200, Zymed), and rabbit anti-Ki67 (1:100, Zymed). The secondary antibodies included Alexa fluor 488 anti-rabbit IgG (1:500, Invitrogen), biotinylated anti-goat IgG (1:500, Vector Labs), and biotinylated anti-rabbit IgG (1:500, Vector Labs).
Statistical analyses
A total of 16 knock-in and 10 het-control mice were analyzed. Due to unequal variances for certain data sets, the median and individual data points are presented, and statistically significant differences, indicated by a P-value of less than 0.05, were determined by the nonparametric Mann-Whitney U-test. For data sets in which statistical analysis was not performed, the mean and standard deviation are indicated.
RESULTS
GATA4-mediated repression of Asbt gene expression occurs in differentiated absorptive enterocytes
As previously shown (Bosse et al., 2006a), conditional, inducible Gata4 inactivation by excision of the activation domains of GATA4 (synthesis of GATA4Δex2), results in an induction in jejunum of absorptive enterocyte genes normally restricted to distal ileum, including Asbt and Ilbp. Since GATA4 is expressed in both crypt and villus epithelial cells (Bosse et al., 2006a; Divine et al., 2004; Dusing and Wiginton, 2005; van Wering et al., 2004), it is uncertain whether GATA4 represses ileal-specific genes in proximal small intestine by a process that occurs in differentiated absorptive enterocytes on villi, and/or is determined early in the differentiation program in crypt progenitor cells. To localize the site of action for GATA4-mediated repression of intestinal genes, the induction of Asbt expression during the initial phases of Gata4 inactivation was characterized. Gata4flox/flox mice positive for the VillinCreERT2 transgene were treated for one day or two days with tamoxifen, and sacrificed 24 h after the last injection. In mid-jejunum, CRE-mediated excision of Gata4 was complete after only one injection of tamoxifen (Fig. 2A), and Asbt mRNA was induced (Fig. 2B). Because of the 3-day crypt-to-villus tip cell migration time in mice (Gordon et al., 1992), we hypothesized that if Asbt was induced directly in differentiated absorptive enterocytes, then ASBT would first be detected in a random, patchy pattern throughout the villi. Conversely, if Asbt was induced by a process that originated in crypts, then it would first appear in lower villi and migrate up the villi over time. ASBT, not normally present in mid-jejunum (Fig. 2C) (Dawson et al., 2003; Shneider et al., 1995), was first detected in the microvillus membrane of single cells scattered throughout the villi after one day of Gata4 inactivation (Fig. 2D–F), and was increasingly expressed in a patchy pattern along the length of the villi extending to the villus tip after two days of Gata4 inactivation (Fig. 2G–I). ASBT induction in the mid-jejunum of Gata4 mutant mice is shown as a control (Fig. 2J). Although the design and outcome of this experiment cannot rule out the possibility that a component of GATA4-mediated Asbt repression in the jejunum is directed by GATA4 in crypt cells, these data are consistent with the hypothesis that GATA4 mediates Asbt repression in differentiated absorptive enterocytes on villi.
Fig. 2.
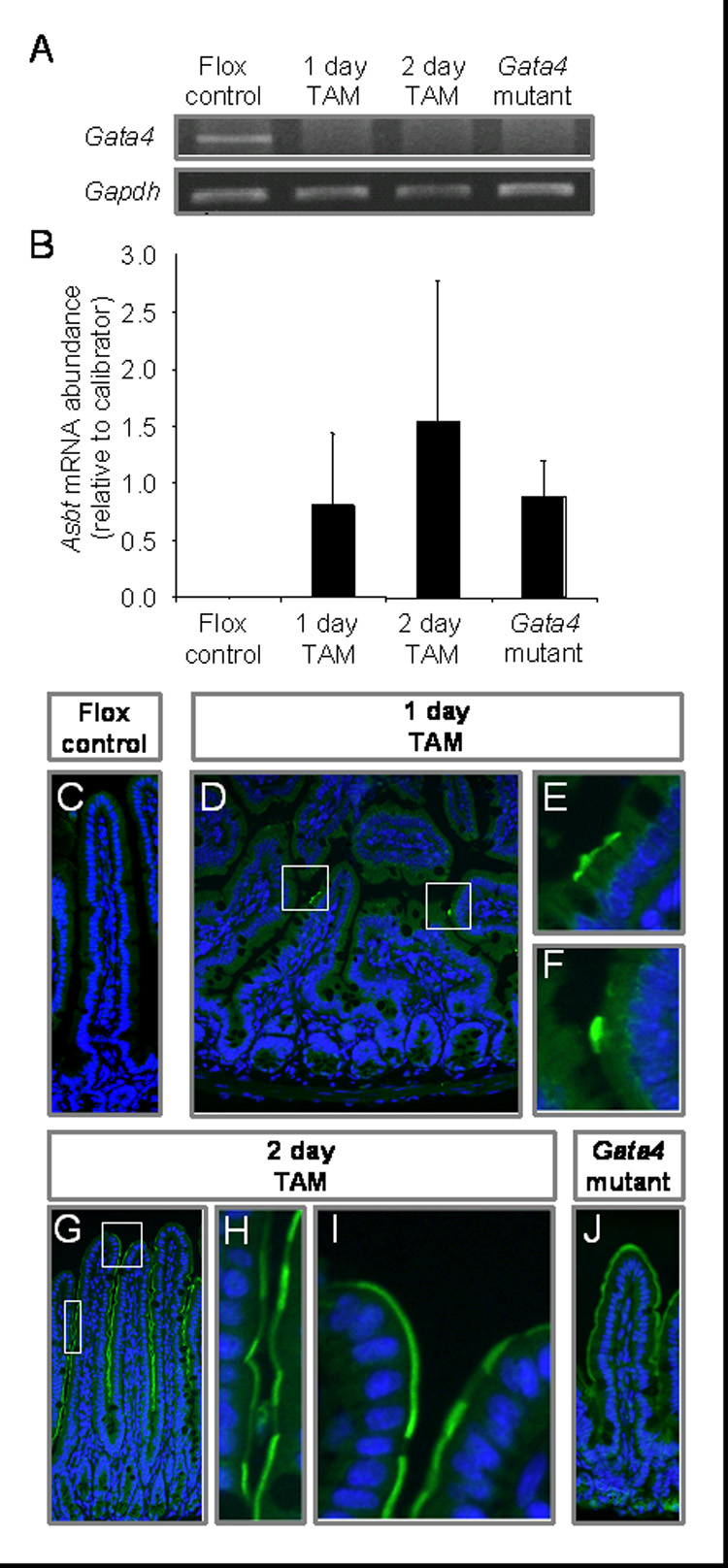
GATA4-mediated repression of Asbt expression occurs in differentiated absorptive enterocytes. Gata4flox/flox, VillinCreERT2-positive mice were treated with tamoxifen for 1 day (1 day TAM) or 2 days (2 day TAM) and sacrificed 24 h after the last injection. Gata4flox/flox mice, negative (Flox control) or positive (Gata4 mutant) for the VillinCreERT2 transgene were treated with tamoxifen for 5 days and sacrificed 2 wk later as described (Bosse et al., 2006a). All samples were collected from the geometric center of the small intestine (mid-jejunum). (A) Gata4 mRNA abundance, determined by semi-quantitative RT-PCR using primers specific for exon2, reveals complete CRE-mediated excision after only one treatment of tamoxifen. Gapdh was used as a positive control. This finding was replicated on 3 different sets of mice. (B) Asbt mRNA is induced within 1 day of a single dose of tamoxifen as determined by real-time RT-PCR (n=3 in each group). RNA from jejunum of a Gata4 mutant mouse was used as a calibrator. (C–J) ASBT is induced in the microvillus membrane of enterocytes on villi 1 and 2 days after tamoxifen treatment as determined by immunofluorescence (green). Nuclei were counterstained with DAPI (blue).
Inducible VillinCreERT2-mediated recombination of the Gata4flox allele in Gata4ki/flox mice results in an intestine-specific Gata4 knock-in model
FOG cofactors mediate GATA-regulated repression of specific genes in multiple non-intestinal systems (Crispino et al., 1999; Grass et al., 2003; Hong et al., 2005; Letting et al., 2004; Lu et al., 1999; Roche et al., 2008; Svensson et al., 1999; Tsang et al., 1997), but their function and expression in the small intestine is unknown. To test the hypothesis that FOG cofactors mediate the GATA4-regulated repression of ileal-specific absorptive enterocyte genes in the proximal small intestine, we established Gata4ki/flox, VillinCreERT2-positive (knock-in) mice, and Gata4wt/flox, VillinCreERT2-positive heterozygous controls (het-controls) (Fig. 1). All genotypes were confirmed by PCR using primer pairs that distinguish the Gata4ki from the Gata4flox or Gata4wt alleles, as well as the Gata4flox from the Gata4ki or Gata4wt alleles (Fig. S3A).
To validate intestine-specific CRE-mediated excision of the Gata4flox alleles, and specific allelic expression from the Gata4ki allele in the knock-in mice, and from the Gata4wt allele in the het-controls, RT-PCR and cDNA sequencing was conducted on RNA samples from mid-jejunum. The Gata4flox allele, common to both knock-in and het-control mice, was excised in jejunum but not heart after tamoxifen treatment (Fig. S3B), verifying intestine-specific CRE-mediated excision. To distinguish expression from the Gata4ki vs. Gata4wt alleles, the region encompassing the V217G knock-in mutation was amplified by RT-PCR using the Ki3 and Ki4 primers (Fig. S3C). Because Ki3 hybridizes to exon2 sequence, and because this exon is excised from the Gata4flox allele after tamoxifen treatment producing the recombined Gata4Δex2 allele, the Gata4Δex2 cDNA is not amplified. Using a nested primer (Ki5), the knock-in or wild-type cDNA was then confirmed by sequencing (Fig. S3C). For all mice in this study, there was no evidence of band ambiguity at the knock-in site for Gata4 knock-in mice, verifying that CRE-mediated excision was complete.
The expression of Gata4, Fog1, and Fog2 in the knock-in mice and het-controls was determined (Fig. 3). Because the Gata4ki, Gata4flox, and Gata4wt alleles are all under the control of the endogenous Gata4 promoter, we quantified total Gata4 mRNA abundance as the most sensitive indicator of a possible knock-in effect on Gata4 expression. Total Gata4 and Fog1 mRNA abundances were not significantly different between the two groups, but Fog2 mRNA was significantly lower (60%, P<0.05) in the knock-in mice as compared to the het-controls. Noteworthy, the cycle threshold (CT) for Fog1 was ~9 cycles lower as compared to Fog2 indicating that Fog1 mRNA abundance is 1000-fold higher than that of Fog2. These data suggest that although Fog2 mRNA is expressed at low levels, its abundance is dependent on GATA4-FOG interactions.
Fig. 3.
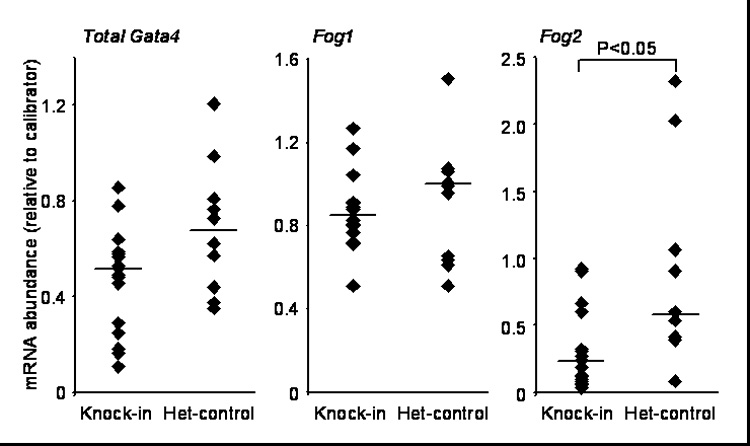
Fog2 mRNA abundance is lower in Gata4 knock-in mice as compared to het-controls. Gata4, Fog1, and Fog2 mRNA abundances were determined by real-time RT-PCR of RNA isolated from the mid-jejunum of Gata4 knock-in mice and het-controls. Filled diamonds represent single data points from individual mice. Bars indicate medians. RNA from jejunum of a het-control mouse was used as a calibrator for Gata4 and Fog1, whereas pooled RNA from ileum of 3 wild-type mice was used as a calibrator for Fog2.
Asbt and Ilbp expression is induced in Gata4 knock-in mice
To test the hypothesis that FOG cofactors specifically mediate GATA4-regulated repression of ileal-specific absorptive enterocyte genes in proximal small intestine, the jejunal expression of absorptive enterocyte target genes and lineage markers were compared between Gata4 knock-in mice and het-controls. As shown in Fig. 4, the mRNA abundances of Asbt and Ilbp (Fig. 4A) in jejunum of the knock-in mice were induced 4-fold (P<0.01) and 14-fold (P<0.001), respectively, compared to the het-controls, consistent with a requirement of GATA4-FOG interactions for repression of ileal-specific genes in the jejunum. Noteworthy, Asbt and Ilbp mRNAs, which are known to be undetectable in wild-type mid-jejunum (Bosse et al., 2006a; Dawson et al., 2003; Sacchettini et al., 1990; Shneider et al., 1995), were induced in the jejunum of the het-control mice, indicating either a heterozygous effect, or a dominant negative effect of GATA4Δex2 on wild-type GATA4, suggesting that repression of Asbt and Ilbp by GATA4 is dose-dependent in vivo. ASBT was highly induced on the microvillus membrane of villus enterocytes in the Gata4 knock-in mice (Fig. 4B), and was detected in isolated cells in the het-controls (Fig. 4C), generally consistent with Asbt mRNA abundances in the two groups. Undetectable expression in wild-type jejunum (Fig. 4D) and endogenous expression in wild-type ileum (Fig. 4E) are shown for reference.
Fig. 4.
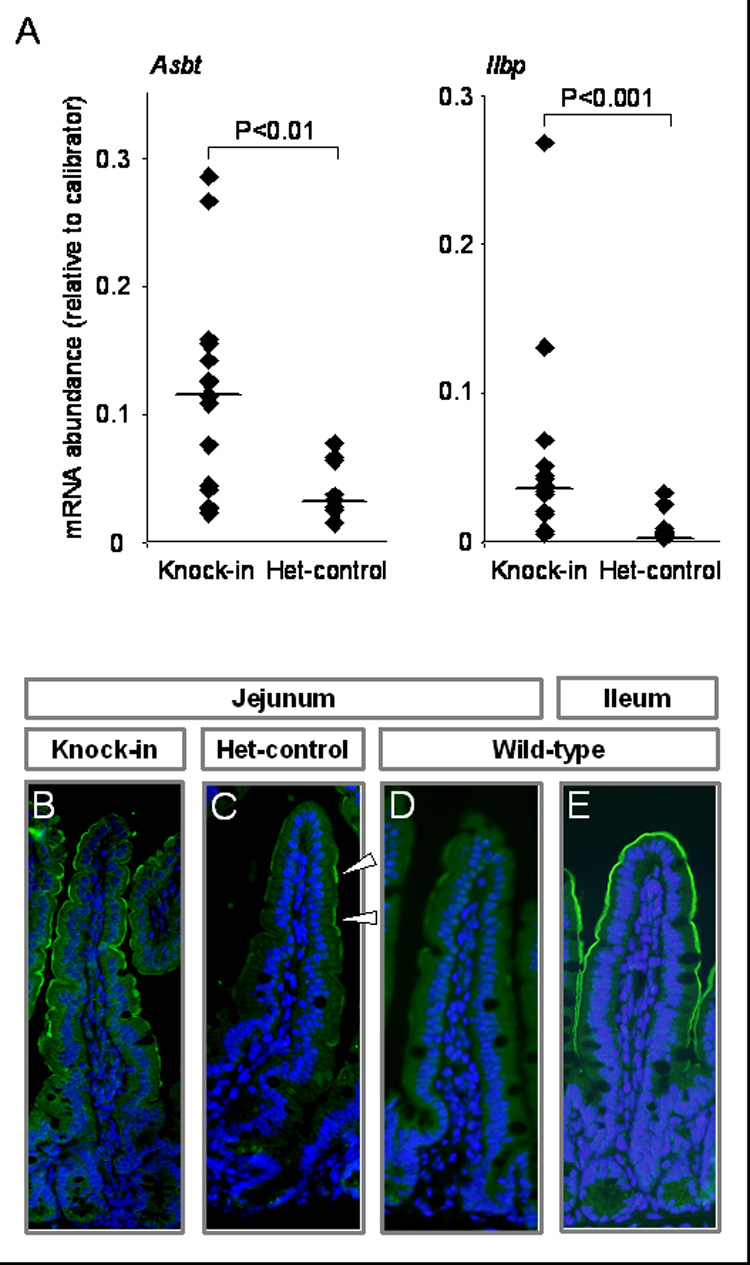
Asbt and Ilbp expression is induced in the Gata4 knock-in mice as compared to het-controls. (A) Asbt and Ilbp mRNA abundances were determined by real-time RT-PCR on RNA isolated from the mid-jejunum of Gata4 knock-in mice and het-controls. Data are represented as indicated in the legend for Fig. 3. Pooled RNA from ileum of 3 wild-type mice was used as a calibrator. (B–E) ASBT was identified by immunofluorescence (green) for representative samples of knock-in jejunum (B), het-control jejunum (C), wild-type jejunum (D) and wild-type ileum (E). Nuclei were counterstained with DAPI (blue). White arrowhead indicates positive ASBT immunofluorescence (C).
The mRNA abundances for the GATA4-mediated activation pathway in absorptive enterocytes (Lph and Fabp1), and for cell lineage markers (Math1, Muc2, Cck and Pyy) did not reveal any statistically significant differences (Fig. 5), suggesting that GATA4-FOG interaction is not required for GATA4-mediated activation of absorptive enterocyte genes, or maintenance of jejunal cell lineage distribution patterns.
Fig. 5.
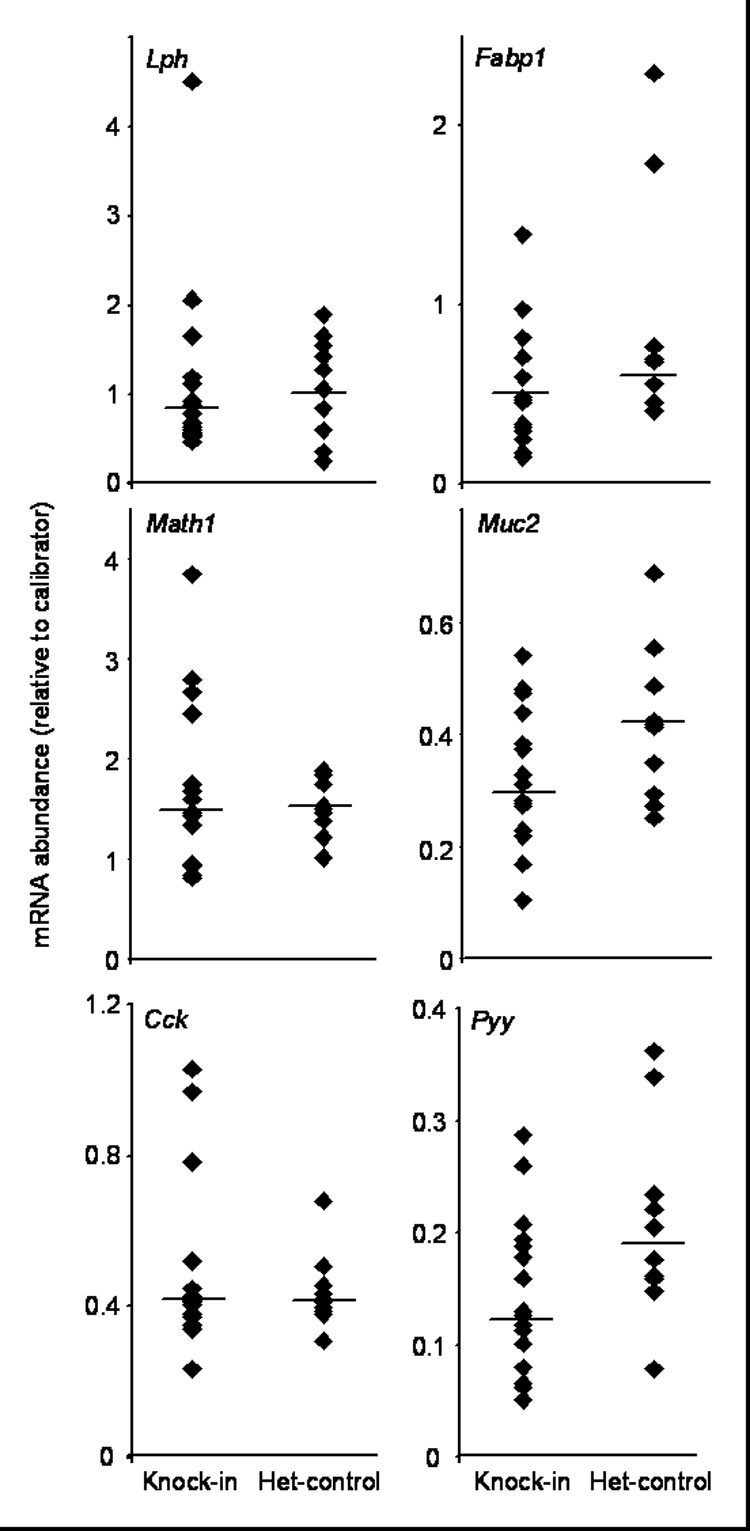
GATA4-mediated activation pathway in absorptive enterocytes and cell lineage markers were not significantly different between Gata4 knock-in mice and het-controls. The mRNAs for the GATA4-regulated activation pathway in absorptive enterocytes (Lph and Fabp1) and for cell lineage (Math1, Muc2, Cck, Pyy) were determined by real-time RT-PCR on RNA isolated from the mid-jejunum of Gata4 knock-in mice and het-controls. Data are represented as indicated in the legend for Fig. 3. Pooled RNA from jejunum of 3 wild-type mice was used as a calibrator for Lph, Fabp1, Math1 and Cck, whereas pooled RNA from ileum of 3 wild-type mice was used as a calibrator for Muc2 and Pyy.
FOG1 demonstrates a decreasing proximal-to-distal expression pattern in adult mouse small intestine
To begin to elucidate which FOG cofactor may be responsible for mediating GATA4 function, the expression patterns of Fog1 and Fog2 were determined in the adult mouse small intestine. Fog1 mRNA abundance demonstrated a decreasing proximal-to-distal pattern (Fig. 6A, upper) similar to that of Gata4 (Bosse et al., 2006a; van Wering et al., 2004). The abundance of Fog1 mRNA in proximal duodenum (segment 1) was similar to that in stomach, but in distal ileum (segment 5) was ~10% of that in stomach, and similar to the low level of expression in heart. Fog2 mRNA was low proximally and increased distally (Fig. 6A, lower), a pattern reciprocal to that of Gata4 and Fog1. Intestinal Fog2 mRNA abundance was similar or less than that in stomach, and ranged from 7–20% of that in heart.
Fig. 6.
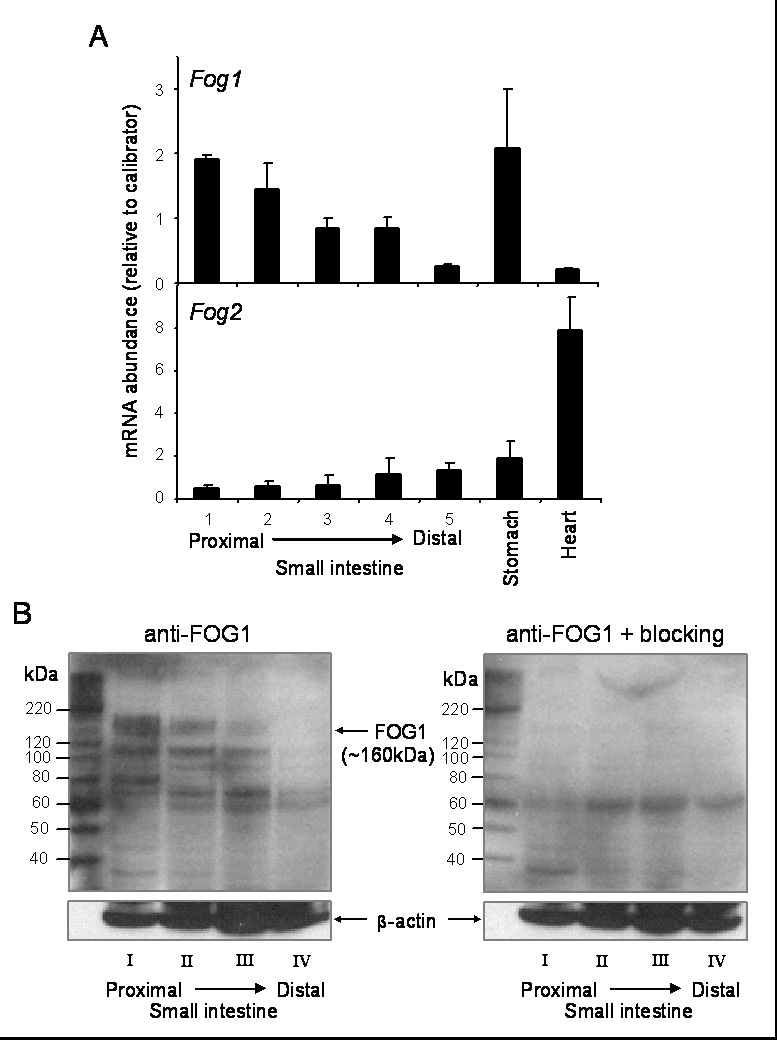
FOG1 demonstrates a decreasing proximal-to-distal expression pattern in adult mouse small intestine. (A) Fog1 (upper) and Fog2 (lower) mRNAs were determined by real-time RT-PCR on RNA obtained from segments 1–5 from wild-type mouse small intestine, stomach, and heart (n=3 in each group). RNA from jejunum of a wild-type mouse was used as a calibrator. (B) FOG1 was identified by Western blot analysis on crude nuclear extracts isolated from quarters I–IV (proximal-to-distal) of wild-type mouse small intestine using a goat anti-FOG1 antibody (Santa Cruz) and an anti-goat IgG secondary antibody (Santa Cruz), without (left) or with (right) an epitope-specific blocking peptide. Blots were re-probed with an anti-β-actin antibody.
To determine whether FOG proteins mimic their mRNA distributions, Western blot analyses were carried out. Since Fog2 mRNA abundance was low in the small intestine (~1000-fold less than Fog1), and FOG2 protein could not be detected by immunohistochemistry (data not shown), only protein patterns for FOG1 were analyzed. As shown in Fig. 6B, a 160 kDa band corresponding to FOG1 (Cantor et al., 2002) demonstrated a declining proximal-to-distal pattern similar to that of its mRNA profile. The 160 kDa band, as well as bands with faster mobilities, were specifically blocked by an epitope-specific polypeptide, verifying specific antigen detection. Taken together, these data demonstrate that FOG1 has a declining proximal-to-distal expression pattern in the adult small intestine that is similar to that of GATA4.
To define Fog1 mRNA expression at the cellular level, in situ hybridization assays were carried out on stomach and small intestine of wild-type mice (Fig. 7). Fog1 mRNA was highly expressed in the stomach with the strongest signal localized to the base of the gastric gland (Fig. 7A). Fog1 mRNA was detected throughout the small intestine, but revealed a clear proximal-to-distal decrease in signal intensity and cellular localization (Fig. 7B–D), corroborating the quantitative cephalo-caudal decrease in Fog1 mRNA and protein. Fog1 mRNA was detected in crypts and villi in duodenum, crypts and lower villi in jejunum, and only in crypts in ileum. A sense control was used to indicate background (Fig. 7E–H).
Fig. 7.
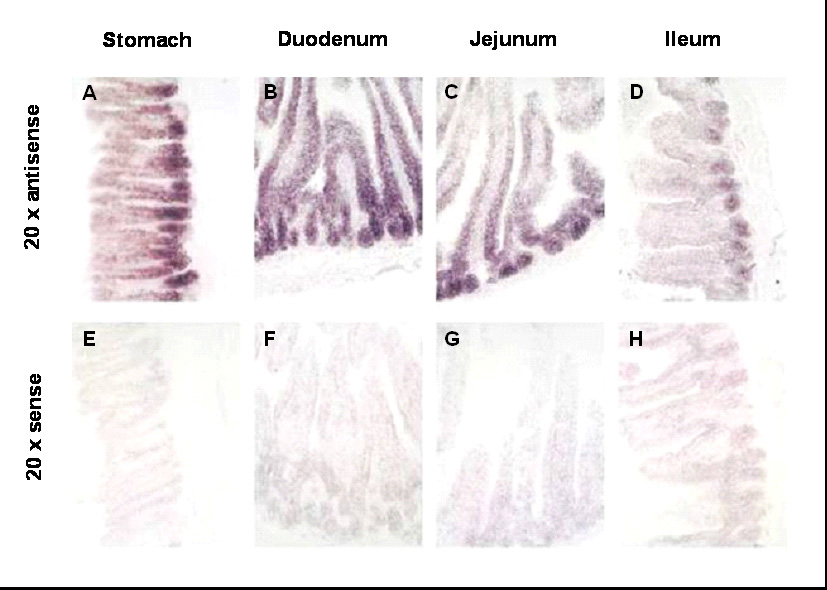
Fog1 mRNA is expressed in a distinct crypt-villus and proximal-distal pattern in the adult mouse small intestine. In situ hybridization assays were conducted using antisense (A–D) and sense (E–H) Fog1 probes, and images at 20x magnification are shown. Analysis of stomach (A,E), duodenum (B,F), jejunum (C,G), and ileum (D,H) reveal a generally decreasing proximal-to-distal signal intensity with the greatest intensity localized to the base of the gastric gland (stomach) and in crypts (small intestine).
FOG1 is co-expressed with GATA4 in absorptive enterocytes on villi, and in lysozyme-positive and proliferating cells in the crypt of the adult mouse small intestine
To delineate the cellular expression patterns of FOG1 in the small intestine, and its co-localization with GATA4, immunostaining for FOG1 and GATA4, along with lineage-specific markers, was conducted on proximal jejunum of adult mouse small intestine. We had previously demonstrated that GATA4 is expressed in absorptive enterocytes on villi, and in proliferating and lysozyme-positive crypt cells, but is not expressed in goblet or enteroendocrine cells (Bosse et al., 2006a). Using an alkaline phosphatase substrate detection approach, FOG1 was identified in the nuclei of epithelial cells throughout the crypts and villi (Fig. 8A), and serial section immunostaining revealed co-localization with GATA4 (Fig. 8B) in both compartments. FOG1 immunostaining could not be detected in the nuclei of goblet cells (Fig. 8C–E), as with GATA4 (Bosse et al., 2006a), but FOG1 immunostaining was detected in the nuclei of chromograninA-positive enteroendocrine cells (Fig. 8F–H), contrasting with the absence of GATA4 immunostaining in this lineage (Fig. 8I–K) (Bosse et al., 2006a). FOG1 immunostaining was also detected in lysozyme-positive (Fig. 8L,M) and proliferating (Fig. 8N,O) crypt cells, and is thus co-localized with GATA4 in this compartment (Bosse et al., 2006a). Taken together, these data show that FOG1 and GATA4 are co-localized in absorptive enterocytes on villi, and in lysozyme-positive and proliferating crypt cells of the adult mouse small intestine, demonstrating a topographic basis for possible interactions. These data also reveal divergent expression in chromagraninA-positive enteroendocrine cells, suggesting that FOG1 has functions in this lineage that are independent of GATA4.
Fig. 8.
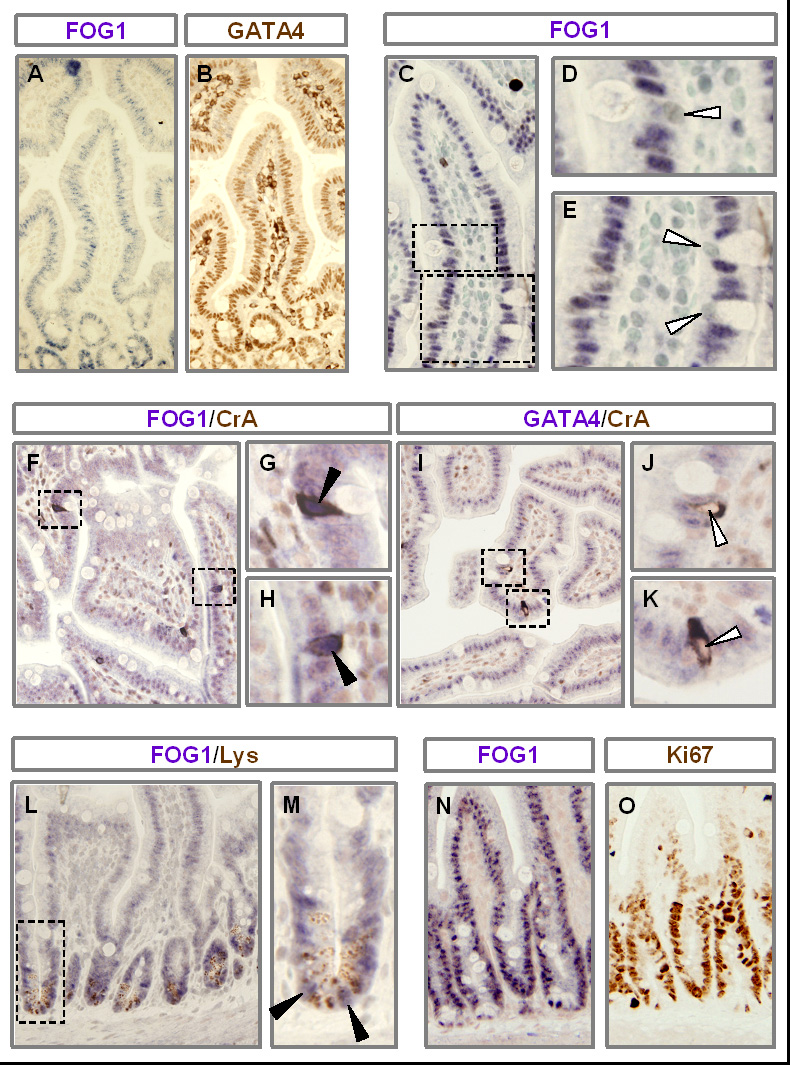
FOG1 is co-expressed with GATA4 in absorptive enterocytes on villi and in lysozyme-positive and proliferating crypt cells. Jejunal sections obtained from adult wild-type mice were used for all immunostaining. Immunostaining for FOG1 (A) or GATA4 (B) on serial sections demonstrate that FOG1 and GATA4 are co-expressed. Nuclei (counterstained with methyl green) of goblet cells do not stain for FOG1 (C–E). Immunostaining with antibodies for FOG1 (F–H) or GATA4 (I–K), co-stained with chromogranin-A (F–K), show that FOG1 is expressed in the nuclei of chromogranin-A positive enteroendocrine cells, but GATA4 is not. Immunostaining with antibodies for FOG1 and lysozyme (L,M) demonstrate that FOG1 is expressed in lysozyme-positive cells. Serial sections stained for FOG1 (N) or Ki67 (O) show that FOG1 is expressed in Ki67-positive proliferating cells in crypts. Open arrowheads indicate an absence of FOG1 or GATA4 staining; filled arrowheads indicate FOG1-positive immunostaining.
DISCUSSION
GATA4, a transcription factor expressed in the proximal small intestine but not in the distal ileum, maintains proximal-distal distinctions in the mature small intestine by multiple processes involving gene repression, gene activation, and cell fate determination (Bosse et al., 2006a). Since interactions between GATA factors and FOG cofactors result in the repression of GATA-mediated transcriptional activation of hematopoietic and cardiac target promoters (Crispino et al., 1999; Grass et al., 2003; Hong et al., 2005; Letting et al., 2004; Lu et al., 1999; Roche et al., 2008; Svensson et al., 1999; Tsang et al., 1997), we tested the hypothesis that FOG cofactors are necessary for GATA4-mediated repression of ileal-specific absorptive enterocyte genes in the small intestine in vivo. We engineered a novel, inducible, intestine-specific Gata4 knock-in model (Fig. 1), in which wild-type GATA4 is specifically inactivated in the intestine, but a GATA4 mutant that does not bind FOG cofactors (GATA4ki) continues to be expressed. We found that ileal-specific genes are induced in the proximal small intestine (Fig. 4), whereas genes restricted to proximal small intestine and cell lineage markers are unaffected (Fig. 5), indicating that GATA4-FOG interactions contribute specifically to the repression function of GATA4 within this organ. Furthermore, co-expression of GATA4 and FOG1 in the nuclei of absorptive enterocytes on villi, and throughout the crypt epithelium (Fig. 8), suggests that FOG1 mediates GATA4-regulated repression of intestinal genes, although a role for FOG2 in this process cannot be discounted. These findings provide the first indication of FOG function and expression in the mammalian small intestine.
Repression of specific genes by GATA factors is well documented (Crispino et al., 1999; Grass et al., 2003; Letting et al., 2004; Lu et al., 1999; Svensson et al., 1999; Tsang et al., 1997), and generally occurs by recruitment of FOG cofactors to target gene promoters, which, in turn, mediates recruitment of the nucleosome remodelling and histone deacetylase (NuRD) complex leading to the deacetylation of local histones and gene silencing (Hong et al., 2005; Roche et al., 2008). Our data show that, GATA4 and FOG cofactors, specifically FOG1, are co-expressed in differentiated absorptive enterocytes on villi (Fig. 8), and are consistent with the hypothesis that GATA4 mediates Asbt repression in differentiated absorptive enterocytes on villi rather than by an upstream process dictated earlier in the differentiation process in crypt progenitor cells (Fig. 2). These findings have led us to hypothesize that GATA4 mediates Asbt repression by binding directly to the Asbt promoter. While it is possible that an indirect pathway within absorptive enterocytes, such as repression of another activator, or activation of a repressor, could mediate Asbt repression, we have previously shown that the mRNAs for known activators of Asbt, including hepatocyte nuclear factor-alpha (HNF1α) (Shih et al., 2001), liver receptor homolog-1 (LRH1) (Chen et al., 2003), and c-FOS (Chen et al., 2001), are not decreased in our conditional Gata4 inactivation model (Bosse et al., 2006a). Thus, we hypothesize that GATA4-regulated repression of specific intestinal genes is mediated by promoter-dependent recruitment of GATA4-FOG complexes that may, in turn, promote histone deacetylation and gene silencing by recruitment of the NuRD complex. This model assumes that GATA4-FOG repression in the proximal small intestine hierarchically overrides activation by HNF1α, LRH1, c-FOS, or any other as yet unknown activator of Asbt gene expression.
Our data show that while the gene repression pathway is dependent on GATA4-FOG interactions (Fig. 4), the gene activation pathway is independent of this interaction (Fig. 5). We and others have previously shown using in vitro and cell culture models that GATA4 physically associates with HNF1α and synergistically activates the Lph and Fabp1 promoters (Divine et al., 2004; van Wering et al., 2004) through an evolutionarily conserved mechanism (van Wering et al., 2002). Furthermore, both genes are strongly attenuated in intestine of mice in which Gata4 is conditionally inactivated (Bosse et al., 2007; Bosse et al., 2006a), or Hnf1α is knocked out (Bosse et al., 2007; Bosse et al., 2006b). These findings support our original hypothesis that the overlapping expression of GATA4 and HNF1α in the intestinal epithelium, combined with specific promoter signatures in target genes, results in the activation of a specific subset of genes in proximal small intestine (Krasinski et al., 2001; van Wering et al., 2004; van Wering et al., 2002). It is intriguing to speculate that the promoter configurations in genes activated by GATA4-HNF1α cooperativity specifically exclude the recruitment of GATA4-FOG complexes.
We had previously shown that conditional inactivation of Gata4 results in a significant increase in goblet cell number in the jejunum, and an increase in the expression of Math1 (Bosse et al., 2006a), a mediator of secretory lineages in the intestine (Yang et al., 2001), consistent with a jejunum-to-ileum transformation in cell fate allocation. We also previously found in the jejunum a trend toward an increase in the mRNA abundance of Muc2, a goblet cell marker (Specian and Oliver, 1991), and a trend toward a decease in the mRNA for Cck and increase in that for Pyy, both of which are enteroendocrine cell markers that normally show an increasing and decreasing proximal-distal gradient (Schonhoff et al., 2004), respectively. In the present study, we found no evidence that FOG cofactors are required for GATA4-regulated cell fate allocation in the intestine (Fig. 5). However, it should be noted that jejuno-ileal differences in cell lineage composition are subtle, and that these differences may be obscured by a gene dosage effect due to the heterozygous nature of the controls, and thus a role for GATA4-FOG interactions in these processes cannot be ruled out.
Both Fog1 and Fog2 mRNAs are expressed in the adult mouse small intestine, but Fog1 is more abundant (~1000-fold), and demonstrates a quantitative pattern that is similar to that of Gata4 (Fig. 6). In humans, Fog1 mRNA is expressed in the proximal small intestine (Freson et al., 2003), whereas Fog2 mRNA can not be detected (Holmes et al., 1999), supporting the relative differential abundances of Fog1 and Fog2 mRNAs in the mouse small intestine. FOG1 and GATA4 are co-expressed in absorptive enterocytes on villi, and throughout the proliferating and lysozyme-positive Paneth cells in crypts (Fig. 8), suggesting that FOG1 mediates GATA4 function. While FOG1 is generally associated with GATA1, and FOG2 with GATA4, FOG1 was shown to be necessary for heart development (Katz et al., 2003) presumably reflecting cooperation with the GATA4/5/6 subfamily. FOG1 is also expressed in the chromagraninA-positive enteroendocrine cells while GATA4 is not expressed in these cells (Bosse et al., 2006a), suggesting that FOG1 has functions in these cells that are independent of GATA4, possibly through interaction with GATA6, which is expressed in this lineage (Dusing and Wiginton, 2005). Fog2 mRNA is detectible in the adult mouse small intestine (Fig. 6A), and is decreased in the Gata4 knock-in mice as compared to the het-controls (Fig. 3), suggesting that its levels are regulated by GATA4-FOG interactions. Although our data support a role for FOG1 in mediating GATA4 function, we cannot rule out the possibility that FOG2, or both FOG1 and FOG2, are required for GATA4 function in the intestine. Correlation of our knock-in results with those of conditional Fog1 and Fog2 single and double knockout will allow a precise determination of FOG1 and FOG2 requirements for GATA4 function in this organ.
The demonstration of a GATA4-FOG requirement for gene expression in the small intestine adds to the growing list of tissues and cell types that require GATA-FOG interactions for normal function. In addition to the well documented role of GATA1-FOG1 interactions in hematopoiesis (Cantor and Orkin, 2005), GATA4-FOG interaction is required for coronary vasculature initiation, cardiac morphogenesis and valve formation during cardiogenesis (Crispino et al., 2001), differentiation of precursors into XY-specific Sertoli cells during gonad development (Tevosian et al., 2002), and epithelial-mesenchymal signaling during stomach development (Jacobsen et al., 2005). While these studies all highlight the critical roles of GATA4-FOG interactions for specific aspects of embryonic development, our study, due to the inducible nature of our model, is the first to demonstrate a role for GATA4-FOG interactions in adult intestine. Taken together, these studies underscore the diverse nature of GATA4-FOG functions, including morphogenesis, cell fate determination, and maintenance of terminal differentiation.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Drs. W. T. Pu (Children’s Hospital Boston, Boston, MA) for the floxed Gata4 mouse line, S. Robine (Institut Curie, Paris, France) for the VillinCreERT2 transgenic mouse line, and P.A. Dawson (Wake Forest School of Medicine, Winson-Salem, NC) for the ASBT antibody. We also thank Lena Liu (Department of Pathology, Children’s Hospital Boston, Boston, MA) for technical support.
This work was supported by National Institute of Diabetes and Digestive and Kidney Diseases Grants R01-DK-061382 (SDK) and R37-DK-32658 (RJG), the Harvard Digestive Disease Center (5P30-DK-34854), and the Nutricia Research Foundation (EB and TB), the Foundation De Drie Lichten (EB) and the Foundation Doctor Catharine van Tussenbroek (EB) in The Netherlands.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Bosse T, et al. Gata4 and Hnf1alpha are partially required for the expression of specific intestinal genes during development. Am J Physiol Gastrointest Liver Physiol. 2007;292:G1302–G1314. doi: 10.1152/ajpgi.00418.2006. [DOI] [PubMed] [Google Scholar]
- Bosse T, et al. Gata4 is essential for the maintenance of jejunal-ileal identities in the adult mouse small intestine. Mol Cell Biol. 2006a;26:9060–9070. doi: 10.1128/MCB.00124-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosse T, et al. Hepatocyte nuclear factor-1alpha is required for expression but dispensable for histone acetylation of the lactase-phlorizin hydrolase gene in vivo. Am J Physiol Gastrointest Liver Physiol. 2006b;290:G1016–G1024. doi: 10.1152/ajpgi.00359.2005. [DOI] [PubMed] [Google Scholar]
- Cantor AB, et al. Distinct domains of the GATA-1 cofactor FOG-1 differentially influence erythroid versus megakaryocytic maturation. Mol Cell Biol. 2002;22:4268–4279. doi: 10.1128/MCB.22.12.4268-4279.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantor AB, Orkin SH. Coregulation of GATA factors by the Friend of GATA (FOG) family of multitype zinc finger proteins. Semin Cell Dev Biol. 2005;16:117–128. doi: 10.1016/j.semcdb.2004.10.006. [DOI] [PubMed] [Google Scholar]
- Chang AN, et al. GATA-factor dependence of the multitype zinc-finger protein FOG-1 for its essential role in megakaryopoiesis. Proc Natl Acad Sci U S A. 2002;99:9237–9242. doi: 10.1073/pnas.142302099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, et al. The role of AP-1 in the transcriptional regulation of the rat apical sodium-dependent bile acid transporter. J Biol Chem. 2001;276:38703–38714. doi: 10.1074/jbc.M104511200. [DOI] [PubMed] [Google Scholar]
- Chen F, et al. Liver receptor homologue-1 mediates species- and cell line-specific bile acid-dependent negative feedback regulation of the apical sodium-dependent bile acid transporter. J Biol Chem. 2003;278:19909–19916. doi: 10.1074/jbc.M207903200. [DOI] [PubMed] [Google Scholar]
- Crispino JD, et al. Use of altered specificity mutants to probe a specific protein-protein interaction in differentiation: the GATA-1:FOG complex. Mol Cell. 1999;3:219–228. doi: 10.1016/s1097-2765(00)80312-3. [DOI] [PubMed] [Google Scholar]
- Crispino JD, et al. Proper coronary vascular development and heart morphogenesis depend on interaction of GATA-4 with FOG cofactors. Genes Dev. 2001;15:839–844. doi: 10.1101/gad.875201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crossman MW, et al. The mouse ileal lipid-binding protein gene: a model for studying axial patterning during gut morphogenesis. J Cell Biol. 1994;126:1547–1564. doi: 10.1083/jcb.126.6.1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson PA, et al. Targeted deletion of the ileal bile acid transporter eliminates enterohepatic cycling of bile acids in mice. J Biol Chem. 2003;278:33920–33927. doi: 10.1074/jbc.M306370200. [DOI] [PubMed] [Google Scholar]
- Divine JK, et al. GATA-4, GATA-5, and GATA-6 activate the rat liver fatty acid binding protein gene in concert with HNF-1alpha. Am J Physiol Gastrointest Liver Physiol. 2004;287:G1086–G1099. doi: 10.1152/ajpgi.00421.2003. [DOI] [PubMed] [Google Scholar]
- Dusing MR, Wiginton DA. Epithelial lineages of the small intestine have unique patterns of GATA expression. J Mol Histol. 2005;36:15–24. doi: 10.1007/s10735-004-2908-9. [DOI] [PubMed] [Google Scholar]
- el Marjou F, et al. Tissue-specific and inducible Cre-mediated recombination in the gut epithelium. Genesis. 2004;39:186–193. doi: 10.1002/gene.20042. [DOI] [PubMed] [Google Scholar]
- Freson K, et al. Molecular cloning and characterization of the GATA1 cofactor human FOG1 and assessment of its binding to GATA1 proteins carrying D218 substitutions. Hum Genet. 2003;112:42–49. doi: 10.1007/s00439-002-0832-1. [DOI] [PubMed] [Google Scholar]
- Fujiwara Y, et al. Arrested development of embryonic red cell precursors in mouse embryos lacking transcription factor GATA-1. Proc Natl Acad Sci U S A. 1996;93:12355–12358. doi: 10.1073/pnas.93.22.12355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon JI, et al. Studies of intestinal stem cells using normal, chimeric, and transgenic mice. Faseb J. 1992;6:3039–3050. doi: 10.1096/fasebj.6.12.1521737. [DOI] [PubMed] [Google Scholar]
- Grass JA, et al. GATA-1-dependent transcriptional repression of GATA-2 via disruption of positive autoregulation and domain-wide chromatin remodeling. Proc Natl Acad Sci U S A. 2003;100:8811–8816. doi: 10.1073/pnas.1432147100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haenlin M, et al. Transcriptional activity of pannier is regulated negatively by heterodimerization of the GATA DNA-binding domain with a cofactor encoded by the u-shaped gene of Drosophila. Genes Dev. 1997;11:3096–3108. doi: 10.1101/gad.11.22.3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes M, et al. hFOG-2, a novel zinc finger protein, binds the co-repressor mCtBP2 and modulates GATA-mediated activation. J Biol Chem. 1999;274:23491–23498. doi: 10.1074/jbc.274.33.23491. [DOI] [PubMed] [Google Scholar]
- Hong W, et al. FOG-1 recruits the NuRD repressor complex to mediate transcriptional repression by GATA-1. Embo J. 2005;24:2367–2378. doi: 10.1038/sj.emboj.7600703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen CM, et al. GATA-4:FOG interactions regulate gastric epithelial development in the mouse. Dev Dyn. 2005;234:355–362. doi: 10.1002/dvdy.20552. [DOI] [PubMed] [Google Scholar]
- Katz SG, et al. Endothelial lineage-mediated loss of the GATA cofactor Friend of GATA 1 impairs cardiac development. Proc Natl Acad Sci U S A. 2003;100:14030–14035. doi: 10.1073/pnas.1936250100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasinski SD, et al. Rat lactase-phlorizin hydrolase/human growth hormone transgene is expressed on small intestinal villi in transgenic mice. Gastroenterology. 1997;113:844–855. doi: 10.1016/s0016-5085(97)70179-3. [DOI] [PubMed] [Google Scholar]
- Krasinski SD, et al. Differential activation of intestinal gene promoters: functional interactions between GATA-5 and HNF-1 alpha. Am J Physiol Gastrointest Liver Physiol. 2001;281:G69–G84. doi: 10.1152/ajpgi.2001.281.1.G69. [DOI] [PubMed] [Google Scholar]
- Kuo CT, et al. GATA4 transcription factor is required for ventral morphogenesis and heart tube formation. Genes Dev. 1997;11:1048–1060. doi: 10.1101/gad.11.8.1048. [DOI] [PubMed] [Google Scholar]
- Letting DL, et al. Context-dependent regulation of GATA-1 by friend of GATA-1. Proc Natl Acad Sci U S A. 2004;101:476–481. doi: 10.1073/pnas.0306315101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, et al. FXR-activating ligands inhibit rabbit ASBT expression via FXR-SHP-FTF cascade. Am J Physiol Gastrointest Liver Physiol. 2005;288:G60–G66. doi: 10.1152/ajpgi.00170.2004. [DOI] [PubMed] [Google Scholar]
- Lu JR, et al. FOG-2, a heart- and brain-enriched cofactor for GATA transcription factors. Mol Cell Biol. 1999;19:4495–4502. doi: 10.1128/mcb.19.6.4495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manuylov NL, et al. The regulation of Sox9 gene expression by the GATA4/FOG2 transcriptional complex in dominant XX sex reversal mouse models. Dev Biol. 2007;307:356–367. doi: 10.1016/j.ydbio.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molkentin JD. The zinc finger-containing transcription factors GATA-4, -5, and -6. Ubiquitously expressed regulators of tissue-specific gene expression. J Biol Chem. 2000;275:38949–38952. doi: 10.1074/jbc.R000029200. [DOI] [PubMed] [Google Scholar]
- Molkentin JD, et al. Requirement of the transcription factor GATA4 for heart tube formation and ventral morphogenesis. Genes Dev. 1997;11:1061–1072. doi: 10.1101/gad.11.8.1061. [DOI] [PubMed] [Google Scholar]
- Pevny L, et al. Erythroid differentiation in chimaeric mice blocked by a targeted mutation in the gene for transcription factor GATA-1. Nature. 1991;349:257–260. doi: 10.1038/349257a0. [DOI] [PubMed] [Google Scholar]
- Pu WT, et al. GATA4 is a dosage-sensitive regulator of cardiac morphogenesis. Dev Biol. 2004;275:235–244. doi: 10.1016/j.ydbio.2004.08.008. [DOI] [PubMed] [Google Scholar]
- Roche AE, et al. The zinc finger and C-terminal domains of MTA proteins are required for FOG-2-mediated transcriptional repression via the NuRD complex. J Mol Cell Cardiol. 2008;44:352–360. doi: 10.1016/j.yjmcc.2007.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacchettini JC, et al. Developmental and structural studies of an intracellular lipid binding protein expressed in the ileal epithelium. J Biol Chem. 1990;265:19199–19207. [PubMed] [Google Scholar]
- Schonhoff SE, et al. Minireview: Development and differentiation of gut endocrine cells. Endocrinology. 2004;145:2639–2644. doi: 10.1210/en.2004-0051. [DOI] [PubMed] [Google Scholar]
- Shih DQ, et al. Hepatocyte nuclear factor-1alpha is an essential regulator of bile acid and plasma cholesterol metabolism. Nat Genet. 2001;27:375–382. doi: 10.1038/86871. [DOI] [PubMed] [Google Scholar]
- Shneider BL. Intestinal bile acid transport: biology, physiology, and pathophysiology. J Pediatr Gastroenterol Nutr. 2001;32:407–417. doi: 10.1097/00005176-200104000-00002. [DOI] [PubMed] [Google Scholar]
- Shneider BL, et al. Cloning and molecular characterization of the ontogeny of a rat ileal sodium-dependent bile acid transporter. J Clin Invest. 1995;95:745–754. doi: 10.1172/JCI117722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon TC, et al. Use of transgenic mice to map cis-acting elements in the liver fatty acid-binding protein gene (Fabpl) that regulate its cell lineage-specific, differentiation-dependent, and spatial patterns of expression in the gut epithelium and in the liver acinus. J Biol Chem. 1993;268:18345–18358. [PubMed] [Google Scholar]
- Specian RD, Oliver MG. Functional biology of intestinal goblet cells. Am J Physiol. 1991;260:C183–C193. doi: 10.1152/ajpcell.1991.260.2.C183. [DOI] [PubMed] [Google Scholar]
- Svensson EC, et al. A syndrome of tricuspid atresia in mice with a targeted mutation of the gene encoding Fog-2. Nat Genet. 2000;25:353–356. doi: 10.1038/77146. [DOI] [PubMed] [Google Scholar]
- Svensson EC, et al. Molecular cloning of FOG-2: a modulator of transcription factor GATA-4 in cardiomyocytes. Proc Natl Acad Sci U S A. 1999;96:956–961. doi: 10.1073/pnas.96.3.956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tevosian SG, et al. Gonadal differentiation, sex determination and normal Sry expression in mice require direct interaction between transcription partners GATA4 and FOG2. Development. 2002;129:4627–4634. doi: 10.1242/dev.129.19.4627. [DOI] [PubMed] [Google Scholar]
- Tevosian SG, et al. FOG-2, a cofactor for GATA transcription factors, is essential for heart morphogenesis and development of coronary vessels from epicardium. Cell. 2000;101:729–739. doi: 10.1016/s0092-8674(00)80885-5. [DOI] [PubMed] [Google Scholar]
- Tsang AP, et al. Failure of megakaryopoiesis and arrested erythropoiesis in mice lacking the GATA-1 transcriptional cofactor FOG. Genes Dev. 1998;12:1176–1188. doi: 10.1101/gad.12.8.1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang AP, et al. FOG, a multitype zinc finger protein, acts as a cofactor for transcription factor GATA-1 in erythroid and megakaryocytic differentiation. Cell. 1997;90:109–119. doi: 10.1016/s0092-8674(00)80318-9. [DOI] [PubMed] [Google Scholar]
- van Wering HM, et al. Complex regulation of the lactase-phlorizin hydrolase promoter by GATA-4. Am J Physiol Gastrointest Liver Physiol. 2004;287:G899–G909. doi: 10.1152/ajpgi.00150.2004. [DOI] [PubMed] [Google Scholar]
- van Wering HM, et al. Physical interaction between GATA-5 and hepatocyte nuclear factor-1alpha results in synergistic activation of the human lactase-phlorizin hydrolase promoter. J Biol Chem. 2002;277:27659–27667. doi: 10.1074/jbc.M203645200. [DOI] [PubMed] [Google Scholar]
- Yang Q, et al. Requirement of Math1 for secretory cell lineage commitment in the mouse intestine. Science. 2001;294:2155–2158. doi: 10.1126/science.1065718. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


