Abstract
Locked Nucleic Acids (LNA) considerably enhance the thermodynamic stability of DNA and RNA duplexes. We report the thermodynamic stabilities of LNA-2'-O-methyl RNA/RNA duplexes designed to provide insight into the contributions of stacking and hydrogen bonding interactions to the enhanced stability. The results show that hydrogen bonding of LNA nucleotides is similar to that of 2'-O-methyl RNA nucleotides, whereas the 3'-stacking interactions are on average about 0.7 kcal/mol more favorable at 37 °C than for 2'-O-methyl or RNA nucleotides. Moreover, NMR spectra suggest helical pre-organization of the single stranded tetramer, CLAMALUM, probably due to restriction of some torsion angles. Thus enhanced stacking interactions and helical pre-organization of single stranded oligonucleotides contribute to the extraordinary stabilization of duplexes by LNA nucleotides.
Locked Nucleic Acids (LNA) are analogues of nucleic acids that provide large enhancements of thermodynamic stability for DNA and RNA duplexes (1–6). The methylene bridge between C4' and 2'-hydroxyl (Figure 1) results in restriction of ribose pseudorotation and exclusively C3'-endo conformers (2, 7). In consequence, the duplex adopts A-RNA structure. The unique thermodynamic properties of LNA led to applications in antisense, ribozyme, and microRNA regulation of the biological function and structure of RNA (8–13). Oligonucleotides containing LNA are also used as probes in isoenergetic 2'-O-methyl RNA (2'-OMe RNA) microarrays to study the structure and interactions of RNA (14, 15). Many thermodynamic properties of LNA/DNA and LNA/RNA duplexes have been reported (1, 5, 6, 16–19). The nearest neighbor thermodynamic parameters of chimeric LNA-DNA/DNA and LNA-2'-O-methyl RNA/RNA duplexes allow prediction of the thermodynamic stability of LNA modified duplexes (4, 6), (www.ibch.poznan.pl/kierzek).
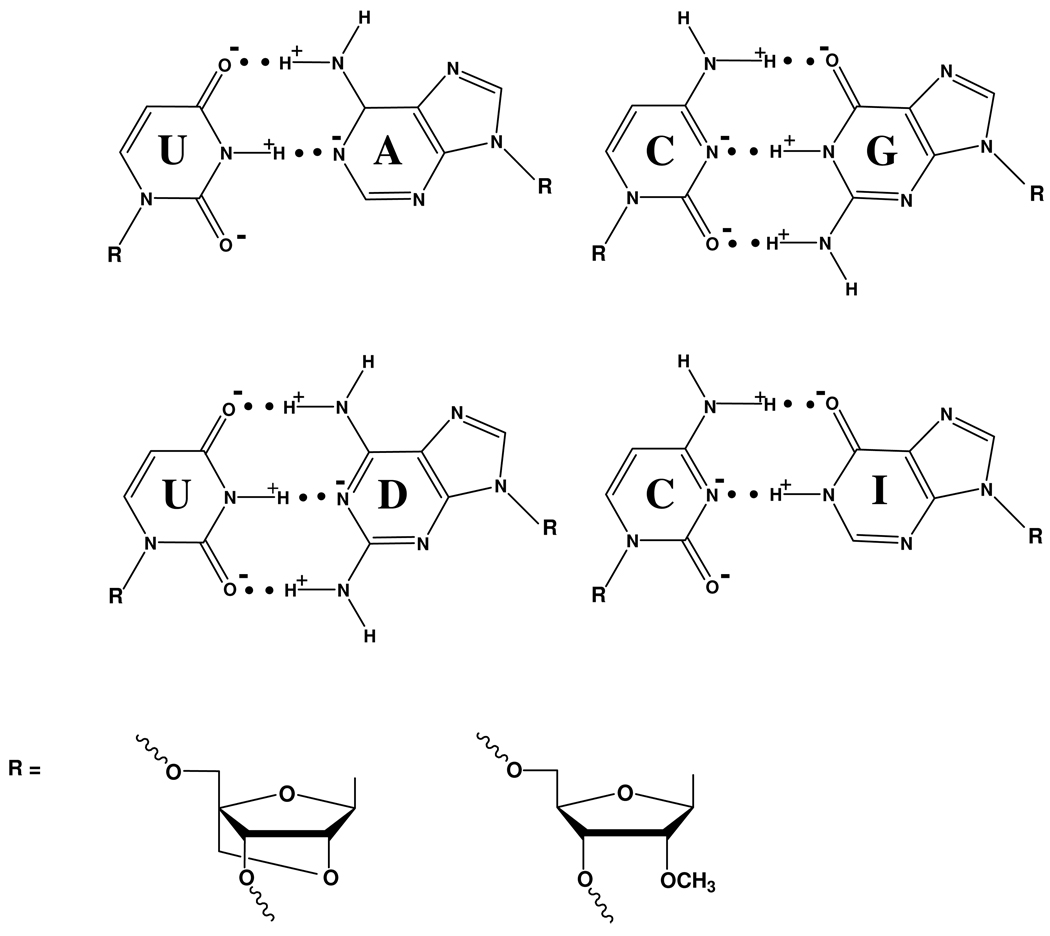
Figure 1.
The hydrogen bonding within A-U, G-C, D-U and I-C base pairs. The signs of partial charges are indicated to illustrate the presence of cross-strand secondary electrostatic interactions (72–77). The magnitudes of the partial charges on all atoms of the bases are given in Supporting Information
Several NMR structures of LNA/DNA and LNA/RNA duplexes have been published (20–22). They show that LNA nucleotides also cause the 3'-adjacent nucleotide to adopt C3'-endo conformation. In consequence, duplexes with RNA or DNA adopt A-RNA or A/B-type structures. A crystal structure reveals an extra water bridged hydrogen bond between the LNA-type ribose and the 3'-adjacent nucleotide (23). Differential Scanning Calorimetry (DSC) revealed that LNA/DNA duplexes have lower uptake of water in comparison to unmodified duplexes, which indirectly suggests that LNA modified duplexes are less hydrated (17).
The LNA properties described above could result in pre-organization of single stranded oligonucleotides containing LNA. Such pre-organization would result in a more favorable initiation free energy for duplex formation (6, 24). Such pre-organization has been detected in NMR studies of single stranded oligodeoxynucleotides modified with LNA or with 5-propynyldeoxyuridine and 5-propynyldeoxycytidine and is thought to enhance thermodynamic stability of 5-propynylated DNA duplexes (25, 26).
Results presented here demonstrate that hydrogen bonding in LNA/RNA and RNA/RNA base pairs is similar, but some stacking interactions are enhanced by LNA. Moreover, the presence of the O2',C4'-methylene bridge limits flexibility of LNA nucleotides and reduces degrees of the freedom of some torsion angles relative to ribose. That restriction of ribose flexibility should affect the entropy of duplex formation and in consequence could explain a significant part of the enhancement of stability of duplexes by LNA (27). Additionally, preliminary NMR studies of CAAU as single stranded RNA, 2'-O-Me RNA, and chimera LNA-2'-O-Me RNA tetramers demonstrated that the structure of the single stranded oligonucleotide, CLAMALUM, containing two LNA nucleotides is most rigid and likely adopting a pre-A-RNA structure.
EXPERIMENTAL PROCEDURES
General methods
The 3'-O-phosphoramidites of LNA nucleotides were synthesized according to published procedures with some minor modifications (2, 6, 28). Mass spectra of nucleosides and oligonucleotides were obtained on an LC MS Hewlett Packard series 1100 MSD with API-ES detector or an MALDI TOF MS, model Autoflex (Bruker).
Synthesis and purification of oligonucleotides
Oligonucleotides were synthesized on an Applied Biosystems DNA/RNA synthesizer, using β-cyanoethyl phosphoramidite chemistry (29, 30). Commercially available A, C, G, U, and I phosphoramidites with 2'-O-tertbutyldimethylsilyl or 2'-O-methyl groups were used for synthesis of RNA and 2'-O-methyl RNA, respectively (Glen Research, Azco, Proligo). LNA phosphoramidites were prepared according to described procedures (28). Thin-layer chromatography (TLC) purification of the oligonucleotides was carried out on Merck 60 F254 TLC plates with the mixture 1-propanol/aqueous ammonia/water = 55:35:10 (v/v/v). The details of deprotection and purification of oligoribonucleotides were described previously (24).
UV melting
Oligonucleotide duplexes, in 10−3 to 10−6 M concentration range, were melted in a buffer containing 100 mM NaCl, 20 mM sodium cacodylate, 0.5 mM Na2EDTA, pH 7.0. The relatively low NaCl concentration kept melting temperatures in the reasonable range and allowed comparison to previous experiments (1, 4–6). Oligonucleotide single strand concentrations were calculated from absorbance above 80 °C with single strand extinction coefficients approximated by a nearest-neighbor model (31, 32). Absorbance vs. temperature melting curves were measured at 260 nm with a heating rate of 1 °C/min from 0 to 90 °C on a Beckman DU 640 spectrophotometer with a thermoprogrammer. Melting curves were analyzed and thermodynamic parameters were calculated from a two-state model with the program MeltWin 3.5 (24, 33, 34). For most duplexes, the ΔH° derived from TM−1. ln (CT/4) plots is within 15% of that derived from averaging the fits to individual melting curves (see Supporting Information), as expected if the two-state model is reasonable.
NMR spectroscopy
CLAMALUM, CMAMAMUM and CAAU at ca. 1 mM concentration were dissolved in buffer containing 100 mM NaCl, 10 mM sodium phosphate (pH 6.8), 0.1 mM EDTA and placed in Shigemi tubes. NMR spectra were collected on a Bruker Avance II 400 MHz spectrometer. The residual signal from HOD was suppressed using low-power presaturation. NOESY spectra in D2O were recorded with mixing times of 400 ms, 2048 complex points in t2 and 512 FIDs in t1 were collected with spectral width of 3800 Hz and recycle delay of 3 s. The chemical shifts are referenced to 2,2-dimethyl-2-silapentane-5-sulfonic acid (DSS).
Methods of computations
Partial charge calculations for inosine (I) and 2,6-diaminopurine riboside (D)
In order to calculate the partial charges for inosine and 2,6-diaminopurine riboside, the C2′-endo and C3′-endo conformations for each residue were created. The RESP protocol (35–38) with multiconformational fitting was applied to these residues. During the RESP protocol, the charges of the sugar atoms, except C1′ and H1′, were fixed to the amber99 charges because only C1′ and H1′ charges may be significantly affected by the substitutions in the base (39). For completeness, a residue library for inosine and 2,6-diaminopurine riboside was created, which includes the nucleoside (XN), 3′-end side (X3), and 5′-end side (X5) (where X means I or D) versions of the residues, as well as the inter-residue version (X), which was the only form used for the calculations reported here. The missing force field parameters for inosine were taken from Sponer data (40). The missing force field parameters for 2,6-diaminopurine were taken from the amber 99 parameter set by analogy (See Supporting Information).
Explicit solvent simulations
The structures of the RNA/RNA duplexes: (i) 5'ACUDACA/3'UGAUUGU, (ii) 5'ACUAACA/3'UGAUUGU, (iii) 5'ACUCACA/3'UGAGUGU, and (iv) 5'ACUCACA/3'UGAIUGU were modeled with the nucgen module of AMBER ver. 9 (41). They were solvated with TIP3PBOX water molecules (42) in a truncated octahedral box. Systems with (i) and (ii) have 3332 water molecules, while systems with (iii) and (iv) have 3331 water molecules. Twelve sodium cations were used to neutralize each system. The parameter/topology files for each system were created with the x-leap module (41). Two types of molecular dynamics (MD) simulations, unrestrained and restrained, were done using the amber 99 force field.
Minimization
The structures were minimized in two steps. For each system, the same protocol was used: (1) with the RNA held fixed with a restraint force of 500 kcal/mol-Å2, steepest descent minimization of 2500 steps was followed by a conjugate gradient minimization of 2500 steps. Constant volume dynamics with a cutoff of 8 Å was chosen. (2) With all restraints removed, steepest descent minimization of 2500 steps was followed by a conjugate gradient minimization of 2500 steps. Constant volume dynamics with a cutoff of 8 Å was chosen again.
Pressure Regulation
After the minimization, two steps of pressure equilibration were done on each system: (1) RNA structures were held fixed with a restraint force of 10 kcal/molÅ2. Constant volume dynamics with a cutoff of 8 Å was used. SHAKE (43) was turned on for bonds involving hydrogen atoms. Temperature was raised from 0 to 300 K in 20 ps. Langevin dynamics with a collision frequency of 1 ps−1 was used. A total of 20 ps of MD were run with a 2 fs time step. (2) The same conditions as above were chosen, except that constant pressure dynamics with isotropic position scaling was turned on. Reference pressure was set to 1 atm with a pressure relaxation time of 2 ps. A total of 100 ps of MD were run with a 2 fs time step. Particle Mesh Ewald (PME) method was always on with the default values.
Molecular dynamics simulations
Initially, no restraints were used in the simulations, but this resulted in large conformational changes for the terminal base pairs within 10 ns. Because the region of interest is the middle of the duplexes, distance restraints were used on the terminal AU base pairs to keep them hydrogen bonded. In particular, the A1H61⋯U14O4, and A7N1⋯U8H3 distances were restrained to be between 1.34 and 2.34 Å, and 1.45 and 2.45 Å, respectively (See Supporting Information). For each simulation, the last structure of pressure regulation was taken as the initial structure. Constant pressure dynamics was chosen with a cutoff of 8 Å. A total of 30 ns of MD were run with a 1 fs time step. Energy information, restart, and trajectory files were written every 250, 5000, and 5000 steps, respectively. Another 15 ns of MD with a 1 fs time step was run for the structure of 5'ACUAACA/3'UGAUUGU to reach convergence, yielding a total of 45 ns of MD. Translational center-of-mass was removed after every 5000 steps. NRESPA was set to 1. Pressure relaxation time (TAUP) was set to 2 ps. Reference pressure (PRES0) was set to 1 atm.
Analysis of Computations
MMPBSA/GBSA analysis (44–50)
Only the results from restrained simulations were analyzed. The first 1120 ps of each simulation were omitted from the calculations to allow equilibration. The mm_pbsa.pl script (41) was used to create the structures and calculate the binding free energies. A total of 2900 structures were extracted from the trajectory files, except for the 5'ACUAACA/3'UGAUUGU simulation where 4400 structures were extracted. Molecular mechanics energies (MM=1), desolvation energies using GB and PB models (GB=1, PB=1), and nonpolar contributions to desolvation using molsurf (MS=1) were used in the calculations. The average binding energy with respect to time and the values at times t and t/2 indicate convergence (see Supporting Information). No normal-mode analysis was done for the systems.
RMSD Analysis
RMSD calculations were done with the ptraj module of AMBER 9 (41). First, water and sodium cations were stripped out of the trajectory files. All atoms of the RNA were included in the RMSD calculations. The trajectory file was RMSD fitted to the final structure from equilibration (See Supporting Information).
RESULTS
Stability increments from stacking of terminal unpaired nucleotides on LNA-2′-O-Me RNA/RNA helixes
One way to evaluate stacking is to measure stability increments of terminal unpaired nucleotides, so called “dangling ends” (51–53). Table 1 provides the thermodynamics of formation for duplexes with and without various dangling ends and Table 2 summarizes the free energy increments for 3'-dangling ends at 37 °C. Stability increments for 3'-LNA dangling ends are listed with a superscript b in Table 2 and are similar for DL, AL, and GL when they are stacked on the same type of base pair. For 3' GL dangling ends in duplexes, 5'CMCMGMUMCMYMGL/3'GGCAGZ, where YM is AM, UM, CM, or GM, and Z is the Watson-Crick complement of Y, thermodynamic stability at 37 °C is enhanced by 1.2, 1.8, 2.0, and 3.0 kcal/mol for terminal AM-U, UM-A, GM-C, and CM-G base pairs, respectively. For the same duplexes but with a 3'-dangling AL, stabilities are enhance by 1.1, 2.2, 2.0 and 2.5 kcal/mol, respectively, when next to terminal AM-U, UM-A, GM-C, and CM-G base pairs. Similar enhancements of 1.4, 2.5, 2.4 and 2.2 kcal/mol were observed for 3'-dangling DL next to terminal AM-U, UM-A, GM-C, and CM-G base pairs, respectively. The 3'-dangling pyrimidines have a smaller effect than purines on stability. (Compare values with superscript b in Table 2.) UL enhances stabilities by 0.7, 0.9, 1.0 and 1.4 kcal/mol and CL by 0.7, 1.0, 0.9 and 1.2 kcal/mol when next to AM-U, UM-A, GM-C, and CM-G base pairs, respectively.
Table 1.
Thermodynamic parameters of helix formation with RNA and 2'-O-Me oligoribonucleotides. The effect of LNA 3'- and 5'-dangling endsa.
| LNA-2′OMe RNA/RNA duplexes | TM−1 vs log CT plots | LNA-2′OMe RNA / RNA duplexes | TM−1 vs log CT plots | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LNA-2′OMe RNA (5′-3′) |
RNA (3′-5′) |
−ΔH° (kcal/mol) |
−ΔS° (eu) |
−ΔG°37 (kcal/mol) |
TMb (°C) |
ΔΔG°37 (kcal/ mol) |
LNA-2′OMe RNA (5′-3′) |
RNA (3′-5′) |
−ΔH° (kcal/mol) |
−ΔS° (eu) |
−ΔG°37 (kcal/mol) |
TMb (°C) |
ΔΔG°37 (kcal/ mol) |
|
A. Thermodynamic parameters of helix formation with various 3′-dangling ends at 2′-OMeRNA/RNA and 2′-OMeRNA/2′-OMeRNA helical cores. | |||||||||||||
| CMCMGMUMCMGM | GGCAGC | 51.6±1.7 | 138.9±5.4 | 8.49±0.05 | 49.2 | 0 | CMCMGMUMCMCM | GGCAGG | 57.9±3.3 | 157.0±9.9 | 9.21±0.15 | 52.1 | 0 |
| CMCMGMUMCMGMDM | GGCAGC | 60.8±1.9 | 165.1±6.1 | 9.64±0.08 | 53.7 | −1.15 | CMCMGMUMCMCMDM | GGCAGG | 61.4±3.3 | 163.5±10.0 | 10.70±0.19 | 59.5 | −1.49 |
| CMCMGMUMCMGMDM | GGCAGC | 65.5±1.9 | 176.1±5.9 | 10.88±0.11 | 59.1 | −2.39 | CMCMGMUMCMCMDL | GGCAGG | 62.7±0.9 | 165.4±2.8 | 11.39±0.06 | 63.1 | −2.18 |
| CMCMGMUMCMGMAM | GGCAGC | 55.9±1.4 | 149.6±4.2 | 9.54±0.05 | 54.6 | −0.96 | CMCMGMUMCMCMAM | GGCAGG | 63.3±2.9 | 169.3±9.0 | 10.76±0.17 | 59.2 | −1.55 |
| CMCMGMUMCMGMAL | GGCAGC | 60.4±2.4 | 161.2±7.4 | 10.43±0.12 | 58.4 | −1.94 | CMCMGMUMCMCMAL | GGCAGG | 66.2±1.8 | 175.8±5.4 | 11.69±0.12 | 63.2 | −2.48 |
| CMCMGMUMCMGMGM | GGCAGC | 55.8±2.0 | 149.8±6.1 | 9.32±0.07 | 53.3 | −0.83 | CMCMGMUMCMCMGM | GGCAGG | 62.4±1.7 | 167.1±5.1 | 10.60±0.09 | 58.6 | −1.39 |
| CMCMGMUMCMGMGL | GGCAGC | 60.7±1.2 | 161.9±3.5 | 10.49±0.05 | 59.1 | −2.00 | CMCMGMUMCMCMGL | GGCAGG | 70.6±1.3 | 188.1±3.8 | 12.24±0.10 | 64.3 | −3.03 |
| CMCMGMUMCMGMUM | GGCAGC | 54.7±1.8 | 146.7±5.5 | 9.15±0.07 | 52.6 | −0.66 | CMCMGMUMCMCMUM | GGCAGG | 60.9±2.1 | 163.1±6.4 | 10.35±0.11 | 57.7 | −1.14 |
| CMCMGMUMCMGMUL | GGCAGC | 57.7±0.7 | 155.4±2.1 | 9.49±0.03 | 53.8 | −1.00 | CMCMGMUMCMCMUL | GGCAGG | 59.8±1.5 | 158.3±4.7 | 10.64±0.09 | 59.9 | −1.43 |
| CMCMGMUMCMGMCM | GGCAGC | 54.2±4.1 | 145.7±12.8 | 9.01±0.15 | 51.9 | −0.52 | CMCMGMUMCMCMCM | GGCAGG | 60.2±3.7 | 162.5±11.6 | 9.78±0.15 | 54.7 | −0.57 |
| CMCMGMUMCMGMCL | GGCAGC | 60.7±4.3 | 165.2±13.4 | 9.41±0.17 | 52.5 | −0.92 | CMCMGMUMCMCMCL | GGCAGG | 61.0±1.6 | 162.9±4.9 | 10.45±0.09 | 58.3 | −1.24 |
| CMCMGMUMCMGM | GMGMCMAMGMCM | 47.6±1.2 | 125.9±3.9 | 8.57±0.04 | 50.9 | 0 | CMCMGMUMCMCM | GMGMCMAMGMGM | 54.6±1.0 | 145.8±3.0 | 9.40±0.05 | 54.2 | 0 |
| CMCMGMUMCMGMGM | GMGMCMAMGMCM | 53.3±2.4 | 141.6±7.3 | 9.34±0.11 | 54.3 | −0.77 | CMCMGMUMCMCMGM | GMGMCMAMGMGM | 60.4±1.0 | 160.0±3.1 | 10.78±0.06 | 60.5 | −1.38 |
| CMCMGMUMCMGMGL | GMGMCMAMCM | 55.6±1.6 | 145.5±4.7 | 10.46±0.09 | 60.6 | −1.89 | CMCMGMUMCMCMGL | GMGMCMAMGMGM | 66.5±1.1 | 174.8±+3.3 | 12.32±0.09 | 66.6 | −2.92 |
| CMCMGMUMCMUM | GGCAGA | 43.2±1.2 | 114.7±3.9 | 7.63±0.02 | 45.1 | 0 | CMCMGMUMCMAM | GGCAGU | 47.5±1.6 | 127.8±5.1 | 7.83±0.04 | 45.7 | 0 |
| CMCMGMUMCMUMDM | GGCAGA | 54.9±2.1 | 148.8±6.5 | 8.72±0.06 | 49.9 | −1.09 | CMCMGMUMCMAMDM | GGCAGU | 51.1±0.6 | 137.9±1.8 | 8.34±0.01 | 48.4 | −0.61 |
| CMCMCMUMCMUMDL | GGCAGA | 62.5±1.8 | 169.0±5.7 | 10.09±0.09 | 55.7 | −2.46 | CMCMGMUMCMAMDL | GGCAGU | 58.1±1.2 | 157.7±+3.8 | 9.24±0.04 | 52.2 | −1.41 |
| CMCMGMUMCMUMAM | GGCAGA | 56.5±1.2 | 154.3±3.8 | 8.67±0.03 | 49.2 | −1.04 | CMCMGMUMCMAMAM | GGCAGU | 51.8±0.8 | 140.2±2.4 | 8.35±0.02 | 48.3 | −0.52 |
| CMCMGMUMCMUMAL | GGCAGA | 62.5±1.3 | 169.7±3.9 | 9.87±0.05 | 54.5 | −2.24 | CMCMGMUMCMAMAL | GGCAGU | 54.5±1.2 | 146.9±3.7 | 8.95±0.05 | 51.4 | −1.12 |
| CMCMGMUMCMUMGM | GGCAGA | 52.4±0.9 | 141.6±2.8 | 8.48±0.02 | 49.0 | −0.85 | CMCMGMUMCMAMGM | GGCAGU | 51.3±3.1 | 138.1±9.8 | 8.50±0.09 | 49.4 | −0.67 |
| CMCMGMUMCMUMGL | GGCAGA | 58.3±1.0 | 157.6±3.2 | 9.44±0.04 | 53.3 | −1.81 | CMCMGMUMCMAMGL | GGCAGU | 56.1±1.1 | 151.7±3.5 | 9.00±0.04 | 51.3 | −1.17 |
| CMCMGMUMCMUMUM | GGCAGA | 62.3±5.7 | 172.6±5.7 | 8.81±0.10 | 48.8 | −1.18 | CMCMGMUMCMAMUM | GGCAGU | 47.2±1.2 | 125.3±3.9 | 8.31±0.03 | 49.2 | −0.48 |
| CMCMGMUMCMUMUL | GGCAGA | 54.0±2.2 | 146.6±6.9 | 8.53±0.07 | 48.9 | −0.90 | CMCMGMUMCMAMUL | GGCAGU | 50.5±1.9 | 135.4±6.0 | 8.50±0.08 | 49.6 | −0.67 |
| CMCMGMUMCMUMCM | GGCAGA | 50.5±2.7 | 136.4±8.4 | 8.17±0.05 | 47.4 | −0.54 | CMCMGMUMCMAMCM | GGCAGU | 49.8±1.0 | 133.5±3.3 | 8.44±0.03 | 49.4 | −0.61 |
| CMCMGMUMCMUMCL | GGCAGA | 51.5±1.8 | 138.2±5.6 | 8.64±0.06 | 50.3 | −1.01 | CMCMGMUMCMAMCL | GGCAGU | 50.4±2.1 | 135.1±6.6 | 8.54±0.05 | 49.9 | −0.71 |
| CMCMGMUMCMUM | CMGMCMAMGMAM | 48.0±1.7 | 129.3±5.5 | 7.94±0.03 | 46.4 | 0 | CMCMGMUMCMAM | GMGMCMAMGMUM | 45.1±1.0 | 119.9±3.1 | 7.94±0.02 | 47.0 | 0 |
| CMCMGMUMCMUMGM | GMGMCMAMGMAM | 51.8±0.8 | 139.5±2.7 | 8.55±0.02 | 49.6 | −0.61 | CMCMGMUMCMAMGM | GMGMCMAMGMUM | 51.1±0.5 | 137.3±1.6 | 8.51±0.01 | 49.5 | −0.57 |
| CMCMGMUMCMUMGL | GMGMCMAMGMAM | 57.6±0.9 | 154.5±2.7 | 9.72±0.04 | 55.2 | −1.78 | CMCMGMUMCMAMGL | GMGMCMAMGMUM | 57.2±1.5 | 154.1±4.5 | 9.42±0.06 | 53.5 | −1.48 |
| B. Thermodynamic parameters of helix formation with various 5’-dangling ends at 2’-OMeRNA/RNA and 2’-OMeRNA/2’-OMeRNA helical cores. | C. Thermodynamic parameters of helix formation with GL 3’-dangling ends at RNA/RNA helical core. | ||||||||||||
| CMCMGMUMCMGM | GGCAGC | 51.6±1.7 | 138.9±5.4 | 8.49±0.05 | 49.2 | 0 | CCGUCA | GGCAGU | 51.2±2.5 | 142.3±8.1 | 7.09±0.05 | 40.4 | 0 |
| GMCMCMGMUMCMGM | GGCAGC | 51.1±1.3 | 137.1±4.0 | 8.57±0.04 | 49.9 | −0.08 | CCGUCAGL | GGCAGU | 58.7±1.1 | 163.0±3.6 | 8.17±0.02 | 45.9 | −1.08 |
| GLCMCMGMUMCMGM | GGCAGC | 50.0±1.3 | 133.4±3.9 | 8.61±0.04 | 50.4 | −0.12 | CCGUCU | GGCAGA | 46.0±1.0 | 126.3±3.3 | 6.88±0.01 | 39.4 | 0 |
| CMCMGMUMCMGM | GMGMCMAMGMCM | 47.6±1.2 | 125.9±3.9 | 8.57±0.04 | 50.9 | 0 | CCGUCUGL | GGCAGA | 61.8±0.8 | 172.2±2.6 | 8.39±0.02 | 46.6 | −1.51 |
| GMCMCMGMUMCMGM | GMGMCMAMGMCM | 49.0±0.8 | 130.1±2.4 | 8.69±0.03 | 51.3 | −0.12 | |||||||
| GLCMCMGMUMCMGM | GMGMCMAMGMCM | 47.3±1.8 | 124.6±5.6 | 8.67±0.06 | 51.7 | −0.10 | |||||||
– solutions are 100 mM NaCl, 20 mM sodium cacodylate and 0.5 mM Na2EDTA, pH 7. Listed errors are standard deviations from reported measurements assuming no correlation of errors in slope and intercept and are therefore underestimates. Estimated errors from all sources are ±10%, ±10%, ±2%, and 1 °C for ΔH°, ΔS°, ΔG°, and Tm, respectively, b – calculated for 10−4M oligomer concentration.
Table 2.
Free energy increments (ΔΔG037 in kcal/mol) for unpaired LNA and 2'-O-methyl dangling nucleotides in 0.1 M NaCl.
| X = DM/L | X = AM/L | X = GM/L | X = UM/L | X = CM/L | ||
|---|---|---|---|---|---|---|
| ---AMX | −0.6a | −0.5a | −0.7a [−0.6]d | −0.5a | −0.6a | |
| −0.8c | −0.8c | −0.6c | −0.5c | |||
| ---U | ||||||
| −1.4b | −1.1b | −1.2b (−1.5)e | −0.7b | −0.7b | ||
| ---UMX | −1.1a | −1.0a | −0.9a [−0.6]d | −1.2a | −0.5a | |
| −0.7c | −0.8c | −0.3c | −0.1c | |||
| ---A | ||||||
| −2.5b | −2.2b | −1.8b (−1.8)e | −0.9b | −1.0b | ||
| ---GMX | −1.2a | −1.0a | −0.8a [−0.8]d | −0.7a | −0.5a | |
| −1.1c | −1.3c | −0.6c | −0.4c | |||
| ---C | ||||||
| −2.4b | −2.0b | −2.0b (−1.9)e | −1.0b | −0.9b | ||
| ---CMX | −1.5a | −1.6a | −1.4a [−1.4]d | −1.1a | −0.6a | |
| −1.7c | −1.7c | −1.1c | −0.8c | |||
| ---G | ||||||
| −2.2b | −2.5b | −3.0b (−2.9)e | −1.4b | −1.2b | ||
Thermodynamic parameters of 3’-dangling ends for the following duplexes: a - 2’-O-methylRNA/RNA duplexes (top line),
chimera LNA-2’-O-methylRNA/RNA duplexes with X as LNA (bottom line),
– RNA/RNA duplexes in 1 M NaCl (references (71, 72, 79),
2’-O-methylRNA/2’-O-methylRNA duplexes (square brackets),
chimera LNA-2’-O-methylRNA/2’-O-methylRNA duplexes with X as LNA (parenthesis).
The above results can be compared with values measured for the same duplexes but with 2'-O-methyl 3'-dangling ends (Tables 1 and values with superscript a in Table 2). For GM 3'-dangling ends, thermodynamic stability at 37 °C is enhanced by 0.7, 0.9, 0.8, and 1.4 kcal/mol, respectively, when YM-Z are AM-U, UM-A, GM-C, and CM-G base pairs. A 3'-dangling AM enhances stability by 0.5, 1.0, 1.0, and 1.5 kcal/mol, respectively, for the same series. Similar enhancements of 0.6, 1.1, 1.1, and 1.5 kcal/mol, respectively, were observed for 3'-dangling DM. For the same duplexes, but with a UM 3'-dangling end, stabilities at 37 °C are enhanced by 0.5, 1.2, 0.7, and 1.1 kcal/mol for duplexes with terminal AM-U, UM-A, GM-C, and CM-G base pairs, respectively, while a 3'-dangling CM increases duplex stability by 0.6, 0.5, 0.5, and 0.6 kcal/mol, respectively. Evidently, the sequence dependence of 2'-O-methyl 3'-dangling ends is considerably smaller than for 3'-dangling end LNAs. While 3'-dangling CL, CM, UL and UM have stability increments that differ on average by only 0.3 kcal/mol, GL, AL, and DL add on average 1.0 kcal/mol more to stability than GM, AM, and DM.
The effect of changes in the backbone on stability increments for dangling ends was determined for several cases (Table 1) (1, 4, 16, 24, 54, 55). When the RNA backbone was changed to 2'-O-methyl RNA to give duplexes, 5'CMCMGMUMCMYMGL/M/3'GMGMCMAMGMZM with YM-ZM either AM-UM or UM-AM, the free energy increment for the 3'-dangling GL or GM was essentially the same as when bound to the RNA hexamers. Essentially unchanged free energy increments are also observed for a 3'-dangling GL when both strands are RNA in the duplexes, 5'CCGUCAGL/3'GGCAGU and 5'CCGUCUGL/3'GGCAGA. All these duplexes are expected to form A-like helixes so it is not surprising that these backbone variations have little effect on 3'-dangling end stability.
A 5'-dangling end negligibly enhances stability of RNA duplexes (56, 57) but can stabilize DNA duplexes (58). The influence of GL and GM as 5'-dangling ends was measured in 5'GL/MCMCMGMUMCMGM/3'GGCAGC and 5'GL/MCMCMGMUMCMGM/3'GMGMCMAMGMCM duplexes (Table 1). There was negligible enhancement of about 0.1 kcal/mol in all cases, consistent with results for RNA/RNA duplexes.
Stability increments for hydrogen bonds within LNA-2′-O-Me RNA/RNA duplexes
The stability increments for dangling end stacking can be compared with stability increments for adding a terminal or internal hydrogen bond to a duplex (59–62). A previous study has reported the stability enhancement when 2,6-diaminopurine riboside (D) is substituted for A in an A-U base pair at a terminal or internal position (6) and those results are summarized in Table 3 and in Supporting Information. A D-U pair has three hydrogen bonds whereas an A-U pair has two (Figure 1).
Table 3.
The differences in hydrogen bonding revealed by substituting 2,6-diaminopurine riboside (D) for A or inosine (I) for G.
| Context (R = A or D) | ΔΔGo 37, HB C R = (DM-AM) or R = (DL-AL) | Context (R = G or I) | ΔΔGo 37,HB R = (G–I) |
|---|---|---|---|
| 5'R C | 0.5a | 5'CLCM | 1.4 |
| 3'U G | 0.4b | 3'R G | |
| 5'C R | 0.4a | 5'CMCL | 1.7 |
| 3'G U | 0.1b | 3'G R | |
| 5'UMCLAM | 2.4 | ||
| 3'A R U | |||
| 5'UM CL UM | 2.4 | ||
| 3'A R A | |||
| Context (R = A or D) | ΔΔG° 37, HB C | Context (R = A or D) | ΔΔG° 37,HB C |
| (DM-AM) or (DL-AL) | (DM-AM) or (DL-AL) | ||
| 5'A R C | 0.8a | 5'A R A | 1.2a |
| 3'U U G | 1.0b | 3'U U U | 1.3b |
| 5'G R C | 0.9a | 5'G R A | 0.7a |
| 3'C U G | 0.9b | 3'C U U | 1.0b |
| 5'C R C | 0.7a | 5'C R A | 0.8a |
| 3'G U G | 1.5b | 3'G U U | 0.8b |
| 5'U R C | 1.0a | 5'U R A | 1.0a |
| 3'A U G | 1.1b | 3'A U U | 1.1b |
| 5'U R G | 1.0a | 5'U R U | 1.1a |
| 3'A U C | 1.0b | 3'A U A | 1.2b |
the difference in free energy of hydrogen bonding increments (ΔΔG°37, HB) when R is a 2'-O-methylated nucleotide
the difference in free energy of hydrogen bonding increments (ΔΔG°37, HB) when R is an LNA nucleotide
reference (6).
In the duplex, 5'XCMUMAMCMCMAM/3'UGAUGGU where X is AM, DM, AL, or DL, substitution of AM or AL with DM or DL in the terminal AU pair enhances duplex stability at 37 °C by 0.5 and 0.4 kcal/mol, respectively (Table 3). The same substitutions in the duplex, 5'AMCMUMAMCMCMX/3'UGAUGGU, enhance stability by 0.4 and 0.1 kcal/mol, respectively. Evidently, the stability increment for the extra hydrogen bond in a terminal DL-U base pair is essentially the same as for a terminal DM-U base pair. The average is 0.4 kcal/mol.
Table 3 also lists stability increments for the additional hydrogen bond in internal D-U pairs as a function of flanking base pairs. These values are obtained from measurements of duplex formation for 5'AMCMZMXYMCMAM/3'UGYUZGU, where X is AM, DM, AL, or DL paired with an internal U, and ZM, YM are 2'-O-methyl nucleotides that form A-U or G-C base pairs with the RNA strand (6). Substitution of AM by DM enhances thermodynamic stability between 0.7 and 1.2 kcal/mol at 37 °C, with an average of 0.9 kcal/mol. Substitution of AL by DL enhances stability between 0.9 and 1.5 kcal/mol with an average of 1.1 kcal/mol. Evidently, the neighboring base pair has a modest effect on the stability increment for the additional hydrogen bond in a D-U pair, but the difference between DM and DL is small.
The stability increment from the G amino to C carbonyl hydrogen bond (60, 69) in a G-CL pair was measured by substituting inosine (I) for G at either a terminal or internal position in the RNA strand (Table 4). The inosine substitutions in 5'UGGUAGI and 5'IGGUAGU destabilized the duplex with the complementary 2'-O-methyl/LNA strand by 1.4 and 1.7 kcal/mol, respectively, at 37 °C (Table 3 and Table 4). Thus, in terminal pairs, the G amino to C carbonyl hydrogen bond enhances stability by about 1 kcal/mol more than the D 2-amino to U 2-carbonyl hydrogen bond. The inosine substitutions in 5'UGUIAGU and 5'UGAIAGU destabilized each duplex by 2.4 kcal/mol, which is equivalent to a 50-fold decrease in equilibrium constant. This change in stabilities is 1.0 and 1.6 kcal/mol larger than measured by substitution of DL-U with AL-U in the middle of otherwise identical LNA-2'-O-Me RNA/RNA duplexes. Evidently in both terminal and internal positions, the G amino to C carbonyl hydrogen bond enhances stability by roughly 1 kcal/mol more than the D 2-amino to U 2-carbonyl hydrogen bond.
Table 4.
Thermodynamic parameters of helix formation with RNA and 2'-OMe oligoribonucleotides. The effect of inosine-LNA-cytidine base pair formation a.
| LNA-2'OMeRNA/RNA duplexes | TM−1 vs log CT plots | |||||
|---|---|---|---|---|---|---|
| LNA-2'OMe RNA | RNA | −ΔH° | −ΔS° | −ΔG37 | TMa | ΔΔG°37 |
| (5'-3') | (3'-5') | (kcal/mol) | (eu) | (kcal/mol) | (°C) | (kcal/mol) |
| CLCMUMAMCMCMAM, C | GGAUGGU | 60.9±5.6 | 165.9±17.2 | 9.47±0.28 | 52.7 | 0 |
| CLCMUMAMCMCMAM | IGAUGGU | 50.1±0.7 | 135.6±2.1 | 8.03±0.01 | 46.6 | 1.44 |
| AMCMUMAMCMCMCL,C | UGAUGGG | 76.6±7.0 | 216.8±21.9 | 9.34±0.27 | 48.8 | 0 |
| AMCMUMAMCMCMCL | UGAUGGI | 50.5±2.0 | 138.4±6.4 | 7.60±0.03 | 43.7 | 1.74 |
| AMCMUMCLAMCMAM, C | UGAGUGU | 56.3±2.3 | 153.8±7.4 | 8.65±0.04 | 49.1 | 0 |
| AMCMUMCLAMCMAM | UGAIUGU | 45.2±1.9 | 125.5±6.4 | 6.25±0.05 | 35.1 | 2.40 |
| AMCMUMCLUMCMAM, C | UGAGAGU | 58.1±1.7 | 158.0±5.3 | 9.14±0.04 | 51.6 | 0 |
| AMCMUMCLUMCMAM | UGAIAGU | 46.2±1.3 | 126.9±4.1 | 6.79±0.02 | 38.8 | 2.35 |
solutions are 100 mM NaCl, 20 mM sodium cacodylate and 0.5 mM Na2EDTA, pH 7
calculated for 10−4 M oligomer concentration,
reference (4)
Comparison to predictions from MMPBSA and MMGBSA calculations
The experimental data indicate that stability increments from an extra amino to carbonyl hydrogen bond depend on whether the extra hydrogen bond is in a D-U or G-C pair (Table 3). To test if this difference is expected on the basis of computational predictions, MMPBSA and MMGBSA calculations were done to predict the thermodynamics of duplex formation for RNA/RNA duplexes 5'ACUCACA/3'UGAGUGU, 5'ACUCACA/3'UGAIUGU, 5'ACUDACA/3'UGAUUGU, and 5'ACUAACA/3'UGAUUGU (Table 5). The calculations do not mimic exactly the experiments because they assume that the separated single strands retain the same A-form structure as assumed for the duplex. Moreover, the calculations employ the force field that has been developed for RNA whereas the experiments are on LNA-2'-O-methyl RNA/RNA duplexes. Because the strength of a hydrogen bond is presumably manifested as enthalpy and MMPBSA and MMGBSA are best at predicting ΔH°, only ΔH° was computed. The ΔΔH° measured experimentally assumes that ΔH° is independent of temperature, which is not true (59, 63). Despite the approximations, predictions of ΔH° follow the same trends observed experimentally. Adding a hydrogen bond stabilizes the duplex, and the effect is predicted to be larger for G-C than for D-U base pairs (Table 5).
Table 5.
Predicted increments for hydrogen bonds in RNA/RNA duplexes calculated by MMPBSA and MMGBSA (in parentheses) methodsa.
| Duplex | Measuredb | Predictedc | Measuredb | Predictedc |
|---|---|---|---|---|
| ΔH° (kcal/mol) | ΔH° (kcal/mol) | ΔΔH° (kcal/mol) | ΔΔH° (kcal/mol) | |
| 5'ACUCACA | −56.3±2.3 | −56.7 | 0 | 0 |
| 3'UGAGUGU | (−48.8) | (0) | ||
| 5'ACUCACA | −45.2±1.9 | −51.8 | −11.1±3.0 | −4.9 |
| 3'UGAIUGU | (−44.0) | (−4.8) | ||
| 5'ACUDACA | −51.0±0.5 | −57.0 | 0 | 0 |
| 3'UGAUUGU | (−47.8) | (0) | ||
| 5'ACUAACA | −49.5±1.8 | −52.9 | −1.5±1.9 | −4.1 |
| 3'UGAUUGU | (−44.5) | (−3.3) | ||
MMPBSA is Molecular Mechanics Poisson Boltzman Surface Area and MMGBSA is Molecular Mechanics Generalized Born Surface Area.
Measurements are for 2'-O-methyl RNA/RNA duplexes (Table 4 and Supporting Information).
Predictions are for RNA/RNA duplexes. Standard error of mean is 0.07 kcal/mol.
NMR spectra of CAAU, CMAMAMUM, and CLAMALUM
Helical preorganization of single stranded oligonucleotides containing LNA could enhance duplex stability by reducing the unfavorable entropy of duplex formation (17, 25, 26). To gain insight into the structural changes induced by introduction of 2'-O-methyl groups and LNA residues into short single strands, one-dimensional NMR spectra of CAAU, CMAMAMUM, and CLAMALUM tetramers were measured from 2 to 50 °C. The sequence was designed to minimize the possibility of self-association. Aromatic and anomeric resonances were assigned on the basis on 2D spectra (see Supporting Information).
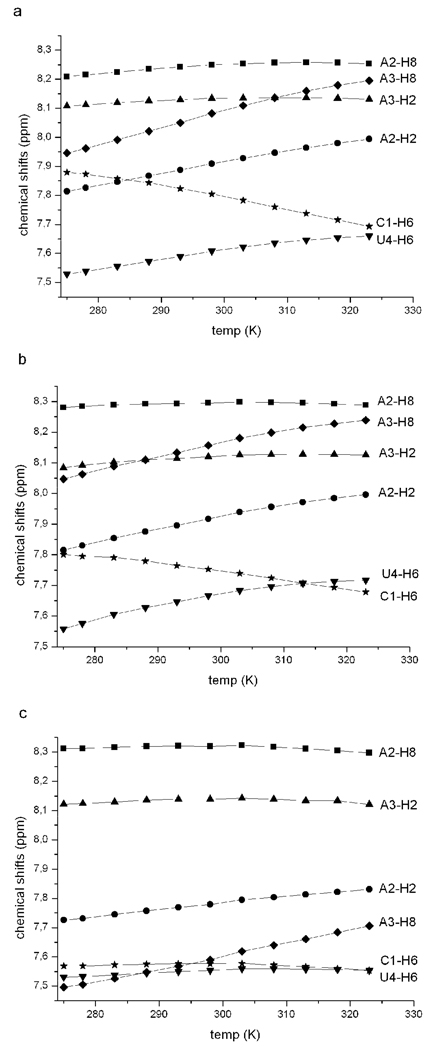
Figure 2 shows the profile and magnitude of chemical shift changes of the base protons as a function of temperature and Table 6 summarizes the results. Most base protons exhibit decreased shielding upon increased temperature, as expected (64). Such shielding changes are usually attributed to less stacking at higher temperatures. The temperature profiles and chemical shifts are similar for CAAU and CMAMAMUM, but somewhat different for CLAMALUM (Figure 2 and Table 6). For CLAMALUM, only A2H2, A3H8, and U4H5 have meaningful changes of chemical shift with increasing temperature. C1H5, C1H6, A2H2, U4H5, and U4H6 change less than their counterparts in CAAU and CMAMAMUM. Moreover, the chemical shifts for A3H8 in CLAMALUM at 5 and 50 °C are about 0.5 ppm up field of those in CAAU and CMAMAMUM, suggesting a different stacking pattern for the A3 LNA base.
Figure 2.
Temperature dependence of the chemical shift changes of base protons of (a) CAAU, (b) CMAMAMUM and (c) CLAMALUM.
Table 6.
Chemical shifts (ppm) at 5 and 50 °C and changes in chemical shifts between 5 and 50 °C
| 5 °C | 50 °C | |||||
|---|---|---|---|---|---|---|
| CLAMALUM | CMAMAMUM | CAAU | CLAMALUM | CMAMAMUM | CAAU | |
| C1H5 | 5.52 (0.05) | 5.52 (0.17) | 5.60 (0.17) | 5.57 | 5.69 | 5.77 |
| C1H6 | 7.57 (−0.02) | 7.80 (−0.12) | 7.87 (−0.18) | 7.55 | 7.68 | 7.69 |
| A2H2 | 7.73 (0.10) | 7.81 (0.18) | 7.81 (0.18) | 7.83 | 7.99 | 7.99 |
| A2H8 | 8.31 (−0.02) | 8.28 (0.01) | 8.21 (0.04) | 8.29 | 8.29 | 8.25 |
| A3H2 | 8.12 (0.01) | 8.09 (0.04) | 8.11 (0.02) | 8.13 | 8.13 | 8.13 |
| A3H8 | 7.50 (0.21) | 8.06 (0.19) | 7.96 (0.23) | 7.71 | 8.24 | 8.19 |
| U4H5 | 5.18 (0.14) | 5.46 (0.22) | 5.31 (0.29) | 5.32 | 5.68 | 5.60 |
| U4H6 | 7.53 (0.02) | 7.55 (0.16) | 7.53 (0.13) | 7.55 | 7.71 | 7.66 |
Data in parentheses reflect changes in chemical shifts between 5 and 50 °C
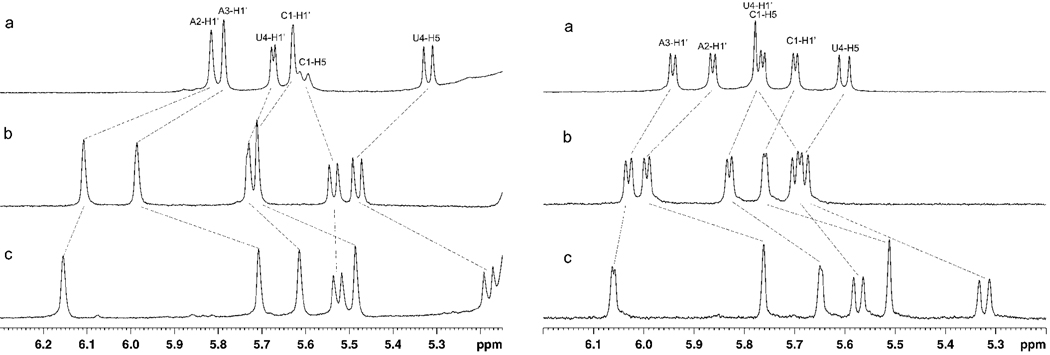
The anomeric region of the one-dimensional spectra (Figure 3) provides information about sugar puckers. At 5 °C, all H1' resonances except the terminal U4H1' of CAAU are singlets, indicating predominantly C3'-endo sugar puckers (65, 66). At 50 °C, the anomeric resonances of CAAU and CMAMAMUM split into doublets with average 3JH1'-H2' of about 4 Hz, indicating dynamical C3'-endo/C2'-endo character of sugars. For CLAMALUM at 50 °C, however, all the H1' resonances are only slightly broadened by splitting with H2' protons. This indicates greater pre-organization of CLAMALUM at 50 °C. It is consistent with previous NMR studies on DNA-LNA chimeras showing a preference for sugar rings to adopt C3'-endo conformation when they flank an LNA nucleotide (26, 67, 68).
Figure 3.
Comparison of anomeric regions of 1H NMR spectra of (a) CAAU, (b) CMAMAMUM and (c) CLAMALUM at the temperatures 5 °C (left) and 50 °C (right).
NOE spectra with 400 ms mixing time were recorded at 5 °C to probe further the differences between the tetramers (Supporting Information). More NOEs are detected for CMAMAMUM and CLAMALUM than for CAAU. Moreover, CLAMALUM exhibits the most cross-peaks with intensity typical for an A-form helix. This pattern suggests that CLAMALUM is the most pre-organized and CAAU is the least ordered.
DISCUSSION
LNA modified DNA and RNA duplexes are thermodynamically particularly stable (1–6). Exceptional thermodynamic stability of LNA duplexes, coupled with simplicity of chemical synthesis and purification of LNA oligonucleotides, along with chemical and nuclease stability, results in the use of LNA oligonucleotides for many applications (69). Thermodynamic stabilities of nucleic acid helixes depend on hydrogen bonding, intra- and interstrand stacking, and electrostatic interactions of phosphodiesters (27). Insight into the origins of enhanced thermodynamic stability of oligonucleotides containing LNA will be useful for guiding design of thermodynamically stable nucleic acids for many applications.
The effect of LNA on dangling end stacking is sequence and context dependent
When a dangling nucleotide is added to the 5'-end of a duplex, there is almost no enhancement of duplex stability, independent of whether the 5'-nucleotide is LNA, 2'-O-Me, or RNA (Table 2). This is expected for an A-form helix because there is essentially no overlap of a 5'-dangling end with the opposite strand (56, 57) and femtosecond spectroscopy experiments support this interpretation (70). Larger and more sequence dependent enhancements are seen for 5'-dangling ends in DNA duplexes, which have B-form conformations (58).
In contrast to 5'-dangling ends, the stability increments of 3'-dangling ends depend on sequence as expected from cross-strand overlap in A-form (56, 57) and from femtosecond spectroscopy experiments (70), but also depend on type of sugar (Table 1 and Table 2). Stacking propensity of unpaired nucleotides in three dimensional structures of RNA correlate with dangling end stacking increments (71). In particular, 83% of sequences with dangling end stacking increments more favorable than −0.7 kcal/mol are stacked in three-dimensional structures compared with only 34% when the increment is less favorable than −0.4 kcal/mol. As shown in Table 2, 2'-O-methyl modification enhances the free energy increment for the 3'-dangling end U of 5'UU/3'A by 0.9 kcal/mol relative to that in RNA (51, 72), which suggests that this modification can have a drastic effect on the three dimensional structures of natural and designed RNAs (73). This may be one reason that certain nucleotides are modified to be 2'-O-methyl in natural RNAs.
Table 7 lists the differences between LNA and 2'-O-methyl 3'-dangling end stability increments. On average, an LNA 3'-dangling end U or C is only 0.3 kcal/mol more stable than a 2'-O-methyl U or C, which is smaller than the average stability enhancement of 1.0 kcal/mol for 3'-dangling end LNA purines. The 0.3 kcal/mol LNA increment for a 3'-dangling pyrimidine is similar to the expected value of –RT ln 2 = −0.4 kcal/mol at 37 °C if LNA preorganizes the sugar to a C3'-endo conformation instead of allowing equal populations of C3'-endo and C2'-endo in the single strands.
Table 7.
Free energy enhancements (ΔΔG°37 kcal/mol) when LNA is substituted for an 2'-O-methyl 3'-dangling end.
| X | |||||
|---|---|---|---|---|---|
| X = DM/L | X = AM/L | X = GM/L | X = UM/L | X = CM/L | |
| ---AMX | 0.8 | 0.6 | 0.5 | 0.2 | 0.1 |
| ---U | |||||
| ---UMX | 1.4 | 1.2 | 1.0 | 0.3 | 0.5 |
| ---A | |||||
| ---GMX | 1.2 | 0.9 | 1.2 | 0.3 | 0.4 |
| ---C | |||||
| ---CMX | 0.7 | 0.9 | 1.6 | 0.3 | 0.7 |
| ---G | |||||
The largest dependence of stability increments on sequence and sugar type is observed for 3'-dangling end purines, D, A, and G (Table 1, Table 2, and Table 7). For a given sugar and adjacent base pair, stability increments for D, A, and G 3'-dangling ends are similar, but a 3'-dangling end LNA purine adds an additional 1 kcal/mol on average at 37 °C for both 2'-O-Me RNA/RNA and 2'-O-Me RNA/2'-O-Me RNA duplexes. The average LNA enhancement of 1 kcal/mol is larger than the 0.4 kcal/mol expected if the sugar conformation is restricted to C3'-endo, but close to that expected if an LNA also preorganizes an adjacent sugar. It is also close to the average LNA enhancements of −1.2 kcal/mol for 3'-terminal AL-U, CL-G, GL-C, and DL-U base pairs and −1.4 kcal/mol measured for internal Watson-Crick like base pairs (4, 6). Evidently, LNA facilitation of stacking is largely responsible for the LNA enhanced stability of Watson-Crick like base pairs. The large favorable free energy for 3'-dangling end LNA purines suggests they may be useful for ensuring stacking in designed structures. The above results can be compared to preliminary measurements by Wengel et al. (74) on the change in melting temperature due to adding dT or LNA T as dangling end at the 5'- or 3'-end of 5'-d(GTGATATGC) hybridized to either 3'-d(CACTATACG) or 3'-r(CACUAUACG). The 5'-dangling ends gave little or no change in Tm. The 3'-dangling ends increased Tm by 2–4 °C with LNA T having a larger effect than dT with DNA target, but not with the RNA target. This suggest that the relative effects of LNA dangling ends depend on the backbone context.
NMR spectra reveal preorganization of single stranded CLAMALUM
An LNA nucleotide is locked into a C3'-endo conformation and in LNA-DNA chimera/DNA duplexes and single stranded chimeric LNA-DNA oligonucleotides also favors adoption of C3'-endo conformation for the 3'-adjacent nucleotide (26, 68). The NMR results for CLAMALUM extend this expectation to single stranded LNA-2'-O-Me oligonucleotides (Figure 3). At 50 °C, which is close to the melting temperature of most of the duplexes studied, all the sugars are almost entirely in a C3'-endo conformation. Thus, the single strand is preorganized to form an Aform duplex. Each preorganized sugar is expected to enhance duplex formation by roughly 0.4 kcal/mol relative to CAAU and CMAMAMUM which have roughly equal populations of C3'-endo and C2'-endo sugars at 50 °C (Figure 3).
Hydrogen bonding between the 2-amino and 2-carbonyl of a D-U pair is relatively unaffected by substitution of LNA for a 2′-O-methyl ribose
As shown in Table 3, the stability increment associated with replacing an AL-U pair with a DL-U pair is essentially the same as for replacing an AM-U pair with a DM-U pair (6). This is not surprising because both are expected to be in an A-form helix. This comparison is consistent with the suggestion that the stability enhancement of base pairs due to LNA is largely due to stacking, which contains a contribution from preorganization of the sugar residue.
The hydrogen bond increment for the G amino to C carbonyl group in a G-CL pair is larger than that for the 2-amino to 2-carbonyl in a DL-U pair
Differences in duplex stabilities measured when a hydrogen bonding group is added as in D-U vs. A-U, or deleted, as in G-C vs. I-C, provide free energy increments for hydrogen bonds (27, 60, 62). As shown in Table 3 and Table 4, replacing G with I in the contexts, 5'UMCLAM/3'ARU or 5'UMCLUM/3'ARA, where R is a purine, makes duplex formation less favorable by 2.4 kcal/mol at 37 °C. In contrast, replacing D with A in the contexts 5'UMRLAM/3'AUU or 5'UMRLUM/3'AUA makes duplex formation less favorable by 1.1 and 1.2 kcal/mol, respectively (6) (Table 3 and Supporting Information). Similarly, replacing a terminal G with I in the terminal contexts 5'CLCM/3'RG or 5'CMCL/3'GR makes duplex formation less favorable by 1.4 and 1.7 kcal/mol, respectively, while replacing D with A in the contexts 5'RLCM/3'UG or 5'CMRL/3'GU makes duplex formation less favorable by only 0.4 and 0.1 kcal/mol, respectively (Table 3 and Table 4). Thus, it appears that the G amino to C carbonyl hydrogen bond in a G-C pair is stronger than the 2-amino to 2-carbonyl hydrogen bond in a D-U pair. This can be at least partially attributed to secondary electrostatic interactions as suggested by Jorgensen and Pranata (75), quantified for base pairs by Sponer and colleagues (76, 77), captured by MMPBSA and MMGBSA calculations (Table 5), and illustrated in Figure 1.
Increments for adding hydrogen bonds in nucleic acids are expected to be dependent on sequence and context and fall roughly within the range of 0 to 2 kcal/mol at 37 °C (60, 78). The Individual Nearest Neighbor – Hydrogen Bond (INN-HB) model for predicting RNA/RNA duplex stabilities assumes that hydrogen bonding is similar in internal and terminal base pairs and assigns a value of 0.9 kcal/mol to the difference in hydrogen bonding between G-C and A-U pairs (24). The results presented here suggest that internal hydrogen bonds are more favorable than terminal hydrogen bonds. Results here and elsewhere (56, 60), however, suggest that the value of 0.9 kcal/mol is a reasonable approximation for the difference between hydrogen bonding in G-C and A-U pairs.
Supplementary Material
Tables and figures with complete data. This material is available free of charge via the internet at http://pubs.acs.org.
ABBREVIATIONS
- LNA
Locked Nucleic Acids
- NMR
nuclear magnetic resonance
- NOE
Nuclear Overhauser Effect
- TLC
thin layer chromatography
- MMPBSA
molecular mechanics Poisson Boltzman surface area
- MMGBSA
molecular mechanics generalized Born surface area
- PME
particle mesh ewald
- RMSD
root mean square deviation.
Footnotes
This work was supported by Ministry of Science and Higher Education, grant NN 3013383 33 to E.K. and grant PBZ-MNiSW-07/I/2007 to R.K and by National Institute of Health grant GM 22939 to D.H.T.
REFERENCES
- 1.Kierzek E, Mathews DH, Ciesielska A, Turner DH, Kierzek R. Nearest neighbor parameters for Watson-Crick complementary heteroduplexes formed between 2'-O-methyl RNA and RNA oligonucleotides. Nucleic Acids Res. 2006;34:3609–3614. doi: 10.1093/nar/gkl232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Koshkin AA, Singh SK, Nielsen P, Rajwanshi VK, Kumar R, Meldgaard M, Olsen CE, Wengel J. LNA (Locked Nucleic Acids): Synthesis of the adenine, cytosine, guanine, 5-methylcytosine, thymine and uracil bicyclonucleoside monomers, oligomerisation, and unprecedented nucleic acid recognition. Tetrahedron. 1998;54:3607–3630. [Google Scholar]
- 3.Vester B, Wengel J. LNA (Locked Nucleic Acid): High-affinity targeting of complementary RNA and DNA. Biochemistry. 2004;43:13233–13241. doi: 10.1021/bi0485732. [DOI] [PubMed] [Google Scholar]
- 4.Kierzek E, Ciesielska A, Pasternak K, Mathews DH, Turner DH, Kierzek R. The influence of locked nucleic acid residues on the thermodynamic properties of 2'-O-methyl RNA/RNA heteroduplexes. Nucleic Acids Res. 2005;33:5082–5093. doi: 10.1093/nar/gki789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pasternak A, Kierzek E, Pasternak K, Fratczak A, Turner DH, Kierzek R. The thermodynamics of 3'-terminal pyrene and guanosine for the design of isoenergetic 2'-O-methyl-RNA-LNA chimeric oligonucleotide probes of RNA structure. Biochemistry. 2008;47:1249–1258. doi: 10.1021/bi701758z. [DOI] [PubMed] [Google Scholar]
- 6.Pasternak A, Kierzek E, Pasternak K, Turner DH, Kierzek R. A chemical synthesis of LNA-2,6-diaminopurine riboside, and the influence of 2'-O-methyl-2,6-diaminopurine and LNA-2,6-diaminopurine ribosides on the thermodynamic properties of 2'-O-methyl RNA/RNA heteroduplexes. Nucleic Acids Res. 2007;35:4055–4063. doi: 10.1093/nar/gkm421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Obika S, Nanbu D, Hari Y, Morio K, In Y, Ishida T, Imanishi T. Synthesis of 2'-O,4'-C-methyleneuridine and -cytidine. Novel bicyclic nucleosides having a fixed C-3'-endo sugar puckering. Tetrahedron Lett. 1997;38:8735–8738. [Google Scholar]
- 8.Kurreck J, Wyszko E, Gillen C, Erdmann VA. Design of antisense oligonucleotides stabilized by locked nucleic acids. Nucleic Acids Res. 2002;30:1911–1918. doi: 10.1093/nar/30.9.1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vester B, Lundberg LB, Sorensen MD, Babu RB, Douthwaite S, Wengel J. LNAzymes: incorporation of LNA-type monomers into DNAzymes markedly increases RNA cleavage. J. Am. Chem. Soc. 2002;124 doi: 10.1021/ja0276220. 13682-12683. [DOI] [PubMed] [Google Scholar]
- 10.Frieden M, Orum H. Locked nucleic acid holds promise in the treatment of cancer. Cur. Pharm. Design. 2008;14:1138–1142. doi: 10.2174/138161208784246234. [DOI] [PubMed] [Google Scholar]
- 11.Fabani MM, Gait MJ. miR-122 targeting with LNA/2'-O-methyl oligonucleotide mixmers, peptide nucleic acids (PNA), and PNA-peptide conjugates. RNA. 2008;14:336–346. doi: 10.1261/rna.844108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fluiter K, Frieden M, Vreijling J, Rosenbohm C, De Wissel MB, Christensen SM, Koch T, Orum H, Baas F. On the in vitro and in vivo properties of four locked nucleic acid nucleotides incorporated into an anti-H-Ras antisense oligonucleotide. Chembiochem. 2005;6:1104–1109. doi: 10.1002/cbic.200400419. [DOI] [PubMed] [Google Scholar]
- 13.Orom UA, Kauppinen S, Lund AH. LNA-modified oligonucleotides mediate specific inhibition of microRNA function. Gene. 2006;372:137–141. doi: 10.1016/j.gene.2005.12.031. [DOI] [PubMed] [Google Scholar]
- 14.Kierzek E, Kierzek R, Turner DH, Catrina IE. Facilitating RNA structure prediction with microarrays. Biochemistry. 2006;45:581–593. doi: 10.1021/bi051409+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kierzek E, Kierzek R, Moss WN, Christensen SM, Eickbush TH, Turner DH. Isoenergetic penta- and hexanucleotide microarray probing and chemical mapping provide a secondary structure model for an RNA element orchestrating R2 retrotransposon protein function. Nucleic Acids Res. 2008;36:1770–1782. doi: 10.1093/nar/gkm1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McTigue PM, Peterson RJ, Kahn JD. Sequence-dependent thermodynamic parameters; for locked nucleic acid (LNA)-DNA duplex formation. Biochemistry. 2004;43:5388–5405. doi: 10.1021/bi035976d. [DOI] [PubMed] [Google Scholar]
- 17.Kaur H, Arora A, Wengel J, Maiti S. Thermodynamic, counterion, and hydration effects for the incorporation of locked nucleic acid nucleotides into DNA duplexes. Biochemistry. 2006;45:7347–7355. doi: 10.1021/bi060307w. [DOI] [PubMed] [Google Scholar]
- 18.Kaur H, Wengel J, Maiti S. LNA-modified oligonucleotides effectively drive intramolecular stable hairpin to intermolecular-duplex state. Biochem. Biophys. Res. Commun. 2007;352:118–122. doi: 10.1016/j.bbrc.2006.10.155. [DOI] [PubMed] [Google Scholar]
- 19.Kaur H, Wengel J, Maiti S. Thermodynamics of DNA-RNA heteroduplex formation: Effects of locked nucleic acid nucleotides incorporated into the DNA strand. Biochemistry. 2008;47:1218–1227. doi: 10.1021/bi700996z. [DOI] [PubMed] [Google Scholar]
- 20.Nielsen CB, Singh SK, Wengel J, Jacobsen JP. The solution structure of a locked nucleic acid (LNA) hybridized to DNA. J. Biomol. Struct. Dyn. 1999;17:175–191. doi: 10.1080/07391102.1999.10508352. [DOI] [PubMed] [Google Scholar]
- 21.Nielsen KE, Rasmussen J, Kumar R, Wengel J, Jacobsen JP, Petersen M. NMR studies of fully modified locked nucleic acid (LNA) hybrids: Solution structure of an LNA : RNA hybrid and characterization of an LNA : DNA hybrid. Bioconjugate Chem. 2004;15:449–457. doi: 10.1021/bc034145h. [DOI] [PubMed] [Google Scholar]
- 22.Petersen M, Bondensgaard K, Wengel J, Jacobsen JP. Locked nucleic acid (LNA) recognition of RNA : NMR solution structures of LNA : RNA hybrids. J. Am. Chem. Soc. 2002;124:5974–5982. doi: 10.1021/ja012288d. [DOI] [PubMed] [Google Scholar]
- 23.Egli M, Minasov G, Teplova M, Kumar R, Wengel J. X-ray crystal structure of a locked nucleic acid (LNA) duplex composed of a palindromic 10-mer DNA strand containing one LNA thymine monomer. Chem. Commun. 2001:651–652. [Google Scholar]
- 24.Xia TB, SantaLucia J, Burkard ME, Kierzek R, Schroeder SJ, Jiao XQ, Cox C, Turner DH. Thermodynamic parameters for an expanded nearest-neighbor model for formation of RNA duplexes with Watson-Crick base pairs. Biochemistry. 1998;37:14719–14735. doi: 10.1021/bi9809425. [DOI] [PubMed] [Google Scholar]
- 25.Znosko BM, Barnes TW, Krugh TR, Turner DH. NMR studies of DNA single strands and DNA : RNA hybrids with and without 1-propynylation at C5 of oligopyrimidines. J. Am. Chem. Soc. 2003;125:6090–6097. doi: 10.1021/ja021285d. [DOI] [PubMed] [Google Scholar]
- 26.Petersen M, Nielsen CB, Nielsen KE, Jensen GA, Bondensgaard K, Singh SK, Rajwanshi VK, Koshkin AA, Dahl BM, Wengel J, Jacobsen JP. The conformations of locked nucleic acids (LNA) J. Mol. Recognit. 2000;13:44–53. doi: 10.1002/(SICI)1099-1352(200001/02)13:1<44::AID-JMR486>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 27.Turner DH. Conformational Changes. In: Bloomfield VA, Crothers DM, Tinoco I, editors. Nucleic Acids - Structures, Properties and Functions. California: University Science Books, Sausalito; 2000. pp. 259–334. [Google Scholar]
- 28.Pedersen DS, Rosenbohm C, Koch T. Preparation of LNA phosphoramidites. Synthesis. 2002:802–808. [Google Scholar]
- 29.McBride LJ, Kierzek R, Beaucage SL, Caruthers MH. Amidine protecting groups for oligonucleotide synthesis. J. Am. Chem. Soc. 1986;108:2040–2048. [Google Scholar]
- 30.Kierzek R, Caruthers MH, Longfellow CE, Swinton D, Turner DH, Freier SM. Polymer-supported RNA synthesis and its application to test the nearest-neighbor model for duplex stability. Biochemistry. 1986;25:7840–7846. doi: 10.1021/bi00372a009. [DOI] [PubMed] [Google Scholar]
- 31.Borer PN. Optical properties of nucleic acids, absorption and circular dichroism spectra. In: Fasman GD, editor. CRC Handbook of Biochemistry and Molecular Biology: Nucleic Acids. 3rd edn. CRC Press: Cleveland, OH; 1975. pp. 589–595. [Google Scholar]
- 32.Richards EG. Use of tables in calculations of absorption, optical rotatory dispersion and circular dichroism of polyribonucleotides. In: Fasman GD, editor. CRC Handbook of Biochemistry and Molecular Biology: Nucleic Acids. 3rd edn. CRC Press: Cleveland, OH; 1975. pp. 596–603. [Google Scholar]
- 33.McDowell JA, Turner DH. Investigation of the structural basis for thermodynamic stabilities of tandem GU mismatches: Solution structure of (rGAGGUCUC)2 by two-dimensional NMR and simulated annealing. Biochemistry. 1996;35:14077–14089. doi: 10.1021/bi9615710. [DOI] [PubMed] [Google Scholar]
- 34.SantaLucia J, Turner DH. Measuring the thermodynamics of RNA secondary structure formation. Biopolymers. 1997;44:309–319. doi: 10.1002/(SICI)1097-0282(1997)44:3<309::AID-BIP8>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 35.Cieplak P, Cornell WD, Bayly C, Kollman PA. Application of the multimolecule and multiconformational RESP methodology to biopolymers - charge derivation for DNA, RNA, and proteins. J. Comput. Chem. 1995;16:1357–1377. [Google Scholar]
- 36.Cornell WD, Cieplak P, Bayly CI, Kollman PA. Application of RESP charges to calculate conformational energies, hydrogen-bond energies, and freeenergies of solvation. J. Am. Chem. Soc. 1993;115:9620–9631. [Google Scholar]
- 37.Bayly CI, Cieplak P, Cornell WD, Kollman PA. A well-behaved electrostatic potential based method using charge restraints for deriving atomic charges - the RESP model. J. Phys. Chem. 1993;97:10269–10280. [Google Scholar]
- 38.Cornell WD, Cieplak P, Bayly CI, Gould IR, Merz KM, Ferguson DM, Spellmeyer DC, Fox T, Caldwell JW, Kollman PA. A second generation force field for the simulation of proteins, nucleic acids, and organic molecules. J. Am. Chem. Soc. 1995;117:5179–5197. [Google Scholar]
- 39.Wang JM, Cieplak P, Kollman PA. How well does a restrained electrostatic potential (RESP) model perform in calculating conformational energies of organic and biological molecules ? J Comput. Chem. 2000;21:1049–1074. [Google Scholar]
- 40.Stefl R, Spackova N, Berger I, Koca J, Sponer J. Molecular dynamics of DNA quadruplex molecules containing inosine, 6-thioguanine and 6-thiopurine. Biophys. J. 2001;80:455–468. doi: 10.1016/S0006-3495(01)76028-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Case DA, Darden TA, Cheatham TEI, Simmerling CL, Wang J, Duke RE, Luo R, Merz KM, Pearlman DA, Crowley M, Walker RC, Zhang W, Wang B, Hayik S, Roitberg A, Seabra G, Wong KF, Paesani F, Wu X, Brozell S, Tsui V, Gohlke H, Yang L, Tan C, Mongan J, Hornak V, Cui G, Beroza P, Mathews DH, Schafmeister C, Ross WS, Kollman PA. San Francisco: University of California; 2006. [Google Scholar]
- 42.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- 43.Ryckaert JP, Ciccotti G, Berendsen HJC. Numerical integration of cartesian equations of motion of a system with constraints - molecular dynamics of Nalkanes. J. Comput. Phys. 1977;23:327–341. [Google Scholar]
- 44.Srinivasan J, Cheatham TE, Cieplak P, Kollman PA, Case DA. Continuum solvent studies of the stability of DNA, RNA, and phosphoramidate -DNA helices. J. Am. Chem. Soc. 1998;120:9401–9409. [Google Scholar]
- 45.Huo S, Massova I, Kollman PA. Computational alanine scanning of the 1 : 1 human growth hormone-receptor complex. J. Comput. Chem. 2002;23:15–27. doi: 10.1002/jcc.1153. [DOI] [PubMed] [Google Scholar]
- 46.Kollman PA, Massova I, Reyes C, Kuhn B, Huo SH, Chong L, Lee M, Lee T, Duan Y, Wang W, Donini O, Cieplak P, Srinivasan J, Case DA, Cheatham TE. Calculating structures and free energies of complex molecules: Combining molecular mechanics and continuum models. Accounts Chem. Res. 2000;33:889–897. doi: 10.1021/ar000033j. [DOI] [PubMed] [Google Scholar]
- 47.Lee MR, Duan Y, Kollman PA. Use of MM-PB/SA in estimating the free energies of proteins: Application to native, intermediates, and unfolded villin headpiece. Proteins. 2000;39:309–316. [PubMed] [Google Scholar]
- 48.Reyes CM, Kollman PA. Structure and thermodynamics of RNAprotein binding: Using molecular dynamics and free energy analyses to calculate the free energies of binding and conformational change. J. Mol. Biol. 2000;297:1145–1158. doi: 10.1006/jmbi.2000.3629. [DOI] [PubMed] [Google Scholar]
- 49.Wang JM, Morin P, Wang W, Kollman PA. Use of MM-PBSA in reproducing the binding free energies to HIV-1 RT of TIBO derivatives and predicting the binding mode to HIV-1 RT of efavirenz by docking and MM-PBSA. J. Am. Chem. Soc. 2001;123:5221–5230. doi: 10.1021/ja003834q. [DOI] [PubMed] [Google Scholar]
- 50.Wang W, Kollman PA. Free energy calculations on dimer stability of the HIV protease using molecular dynamics and a continuum solvent model. J. Mol. Biol. 2000;303:567–582. doi: 10.1006/jmbi.2000.4057. [DOI] [PubMed] [Google Scholar]
- 51.Sugimoto N, Kierzek R, Turner DH. Sequence dependence for the energetics of dangling ends and terminal base-pairs in ribonucleic acid. Biochemistry. 1987;26:4554–4558. doi: 10.1021/bi00388a058. [DOI] [PubMed] [Google Scholar]
- 52.Ziomek K, Kierzek E, Biala E, Kierzek R. The influence of various modified nucleotides placed as 3'-dangling end on thermal stability of RNA duplexes. Biophys. Chem. 2002;97:243–249. doi: 10.1016/s0301-4622(02)00073-x. [DOI] [PubMed] [Google Scholar]
- 53.Freier SM, Burger BJ, Alkema D, Neilson T, Turner DH. Effects of 3'-dangling end stacking on the stability of GGCC and CCGG double helices. Biochemistry. 1983;22:6198–6206. [Google Scholar]
- 54.SantaLucia J, Jr., Allawi HT, Seneviratne A. Improved nearestneighbor parameters for predicting DNA duplex stability. Biochemistry. 1996;35:3555–3562. doi: 10.1021/bi951907q. [DOI] [PubMed] [Google Scholar]
- 55.Sugimoto N, Nakano S, Katoh M, Matsumura A, Nakamuta H, Ohmichi T, Yoneyama M, Sasaki M. Thermodynamic parameters to predict stability of RNA/DNA hybrid duplexes. Biochemistry. 1995;34:11211–11216. doi: 10.1021/bi00035a029. [DOI] [PubMed] [Google Scholar]
- 56.Freier SM, Sugimoto N, Sinclair A, Alkema D, Neilson T, Kierzek R, Caruthers MH, Turner DH. Stability of XGCGCp, GCGCYp, and XGCGCYp helixes - an empirical estimate of the energetics of hydrogen-bonds in nucleic acids. Biochemistry. 1986;25:3214–3219. doi: 10.1021/bi00359a020. [DOI] [PubMed] [Google Scholar]
- 57.Freier SM, Alkema D, Sinclair A, Neilson T, Turner DH. Contributions of dangling end stacking and terminal base-pair formation to the stabilities of XGGCCp, XCCGGp, XGGCCYp, and XCCGGYp helixes. Biochemistry. 1985;24:4533–4539. doi: 10.1021/bi00338a008. [DOI] [PubMed] [Google Scholar]
- 58.Bommarito S, Peyret N, SantaLucia J., Jr. Thermodynamic parameters for DNA sequences with dangling ends. Nucleic Acids Res. 2000;28:1929–1934. doi: 10.1093/nar/28.9.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Petersheim M, Turner DH. Base-stacking and base-pairing contributions to helix stability - thermodynamics of double-helix formation with CCGG, CCGGp, CCGGAp, ACCGGp, CCGGUp, and ACCGGUp. Biochemistry. 1983;22:256–263. doi: 10.1021/bi00271a004. [DOI] [PubMed] [Google Scholar]
- 60.Turner DH, Sugimoto N, Kierzek R, Dreiker SD. Free energy increments for hydrogen-bonds in nucleic acid base pairs. J. Am. Chem. Soc. 1987;109:3783–3785. [Google Scholar]
- 61.Aboulela F, Koh D, Tinoco I, Martin FH. Base-base mismatches -thermodynamics of double helix formation for DCA3XA3g + DCT3YT3G (X, Y = A,C,G,T) Nucleic Acids Res. 1985;13:4811–4824. doi: 10.1093/nar/13.13.4811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Martin FH, Castro MM, Aboulela F, Tinoco I. Base-pairing involving deoxyinosine - implications for probe design. Nucleic Acids Res. 1985;13:8927–8938. doi: 10.1093/nar/13.24.8927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mikulecky PJ, Feig AL. Heat capacity changes associated with nucleic acid folding. Biopolymers. 2006;82:38–58. doi: 10.1002/bip.20457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wuthrich K. NMR of Proteins and Nucleic Acids. John Wiley & Sons: 1986. [Google Scholar]
- 65.Varani L, Hasegawa M, Spillantini MG, Smith MJ, Murrell JR, Ghetti B, Klug A, Goedert M, Varani G. Structure of tau exon 10 splicing regulatory element RNA and destabilization by mutations of frontotemporal dementia and parkinsonism linked to chromosome 17. Proc. Natl. Acad. Sci. U. S. A. 1999;96:8229–8234. doi: 10.1073/pnas.96.14.8229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wijmenga SS, van Buuren BNM. The use of NMR methods for conformational studies of nucleic acids. Prog. Nucl. Magn. Res. Spec. 1998;32:287–387. [Google Scholar]
- 67.Nielsen KE, Singh SK, Wengel J, Jacobsen JP. Solution structure of an LNA hybridized to DNA : NMR study of the d(CTLGCTLTLCTLGC) : d(GCAGAAGCAG) duplex containing four locked nucleotides. Bioconjugate Chem. 2000;11:228–238. doi: 10.1021/bc990121s. [DOI] [PubMed] [Google Scholar]
- 68.Jensen GA, Singh SK, Kumar R, Wengel J, Jacobsen JP. A comparison of the solution structures of an LNA : DNA duplex and the unmodified DNA : DNA duplex. J. Chem. Soc.-Perkin Trans. 2001;2:1224–1232. [Google Scholar]
- 69.Wengel J, Petersen M, Frieden M, Koch T. Chemistry of locked nucleic acids (LNA): Design, synthesis, and biophysical properties. Lett. Peptide Sci. 2003;10:237–253. [Google Scholar]
- 70.Liu JD, Zhao L, Xia TB. The dynamic structural basis of differential enhancement of conformational stability by 5'- and 3'-dangling ends in RNA. Biochemistry. 2008;47:5962–5975. doi: 10.1021/bi800210t. [DOI] [PubMed] [Google Scholar]
- 71.Burkard ME, Kierzek R, Turner DH. Thermodynamics of unpaired terminal nucleotides on short RNA helixes correlates with stacking at helix termini in larger RNAs. J. Mol. Biol. 1999;290:967–982. doi: 10.1006/jmbi.1999.2906. [DOI] [PubMed] [Google Scholar]
- 72.O'Toole AS, Miller S, Haines N, Zink MC, Serra MJ. Comprehensive thermodynamic analysis of 3' double nucleotide overhangs neighboring Watson-Crick terminal base pairs. Nucleic Acids Res. 2006;34:3338–3344. doi: 10.1093/nar/gkl428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kawai G, Yamamoto Y, Kamimura T, Masegi T, Sekine M, Hata T, Iimori T, Watanabe T, Miyazawa T, Yokoyama S. Conformational rigidity of specific pyrimidine residues in tRNA arises from posttranscriptional modifications that enhance steric interaction between the base and the 2'-hydroxyl group. Biochemistry. 1992;31:1040–1046. doi: 10.1021/bi00119a012. [DOI] [PubMed] [Google Scholar]
- 74.Wengel J, Petersen M, Nielsen KE, Jensen GA, Hakansson AE, Kumar R, Sorensen MD, Rajwanshi VK, Bryld T, Jacobsen JP. LNA (locked nucleic acid) and the diastereoisomeric alpha-L-LNA: Conformational tuning and high-affinity recognition of DNA/RNA targets. Nucleosides Nucleotides Nucleic Acids. 2001;20:389–396. doi: 10.1081/NCN-100002312. [DOI] [PubMed] [Google Scholar]
- 75.Jorgensen WL, Pranata J. Importance of secondary interactions in triply hydrogen-bonded complexes - guanine-cytosine vs uracil-2,6-diaminopyridine. J. Am. Chem. Soc. 1990;112:2008–2010. [Google Scholar]
- 76.Sponer J, Leszczynski J, Hobza P. Structures and energies of hydrogenbonded DNA base pairs. A nonempirical study with inclusion of electron correlation. J. Phys. Chem. 1996;100:1965–1974. [Google Scholar]
- 77.Sponer J, Jurecka P, Hobza P. Accurate interaction energies of hydrogen-bonded nucleic acid base pairs. J. Am. Chem. Soc. 2004;126:10142–10151. doi: 10.1021/ja048436s. [DOI] [PubMed] [Google Scholar]
- 78.Santalucia J, Kierzek R, Turner DH. Context dependence of hydrogen-bond free-energy revealed by substitutions in an RNA hairpin. Science. 1992;256:217–219. doi: 10.1126/science.1373521. [DOI] [PubMed] [Google Scholar]
- 79.Turner DH, Sugimoto N, Freier SM. RNA structure prediction. Ann. Rev. Biophys. Biophys. Chem. 1988;17:167–192. doi: 10.1146/annurev.bb.17.060188.001123. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables and figures with complete data. This material is available free of charge via the internet at http://pubs.acs.org.